DNA Enrichment by Allele-Specific Hybridization (DEASH): A Novel Method for Haplotyping and for Detecting Low-Frequency Base Substitutional Variants and Recombinant DNA Molecules (original) (raw)
Abstract
Detecting rare sequence variants in genomic DNA is central to the analysis of de novo mutation and recombination events and the detection of rare pathological mutations in mixed cell populations. Current PCR techniques suffer from noise that limits detection to variants present at a frequency of at least 10-4-10-5 per cell. We nowdescribe an alternative approach that recovers genomic DNA molecules containing a known single-nucleotide variant by hybridization selection using a biotinylated allele-specific oligonucleotide, followed by hybrid capture on streptavidin-coated paramagnetic beads and subsequent analysis by PCR. This technique of DNA enrichment by allele-specific hybridization (DEASH) is fast, effective for all tested single-nucleotide polymorphisms (SNPs), and can recover large (>10 kb) single-stranded molecules. A single round of DEASH is effective in separating haplotypes from genomic DNA and can not only readily detect and validate DNA molecules containing a single base change at a frequency of 10-5 per cell, but can also place these changes within the context of an extended haplotype. This technique offers a newapproach to the analysis of mutation and recombination, and has the potential to detect very rare de novo base substitutions.
The detection of sequence variants present at very low frequency in genomic DNA remains a major challenge in molecular genetics, of direct relevance to the detection of germline and somatic de novo mutation events and other forms of spontaneous genomic rearrangement (Tiemann-Boege et al. 2002). It is also central to the detection and characterization of rare recombinant molecules in bulk genomic DNA, essential for high-resolution analysis of meiotic crossover and gene conversion (Jeffreys et al. 1998a, 2001). Rare variant detection is also important for the early detection of pathological mutations, particularly in cancer (Sidransky et al. 1991, 1992; Tada et al. 1993; Boyle et al. 1994; Mao et al. 1994; Mills et al. 1995; Smith-Ravin et al. 1995), and for the identification of tumor cells in patients in relapse. It is also relevant to prenatal diagnosis using fetal cells present in maternal blood (Adinolfi et al. 1989; Lo et al. 1994; Hengstschlager et al. 1997; Zhang et al. 2000; Reed et al. 2002), and to studies of the accumulation of somatic mutations in aging individuals (see Crow 1999; Knight 2000; Vijg 2000; Berdanier and Everts 2001).
Many methods for detecting low-frequency variants in genomic DNA use the polymerase chain reaction (PCR) to amplify mutant plus wild-type target, followed by interrogating these products by sequencing, oligonucleotide ligation, restriction digestion, mass spectrometry, or hybridization with allele-specific oligonucleotides to identify the variant against the background of wild-type DNA (see, e.g., Saiki et al. 1986; Haliassos et al. 1989; Laken et al. 1998; Gerry et al. 1999; Vogelstein and Kinzler 1999). Other methods use allele-specific PCR to amplify products selectively from the low-frequency variant, with or without additional selection, for example by digesting PCR products with a restriction enzyme that specifically cleaves and renders unamplifiable the wild-type product (Gibbs et al. 1989; Newton et al. 1989; Cha et al. 1992; Fox et al. 1998; Haque et al. 1998; Tiemann-Boege et al. 2002). Both approaches have inherent limitations due to base misincorporation during PCR which creates false mutant products from the wild-type allele, and to the lack of total specificity of allele-specific primers. As a result, all current approaches have limited sensitivity, with the most sensitive assays being capable of detecting at best one mutant molecule against a background of 104-105 wild-type molecules. Although this is adequate for many applications, increased sensitivity is essential for detecting very rare spontaneous mutation events, particularly in the human germline, where the only currently feasible approach is the direct detection of mutations in gametic (sperm) DNA (Tiemann-Boege et al. 2002), and it would also offer potential for the very early detection of pathological mutations.
Most mutation detection systems yield an assay signal that is difficult to validate in terms of the number of mutant molecules detected. This can be overcome in part by analyzing multiple samples, each containing limited DNA (typically 50 genome equivalents), yielding direct information on numbers of mutant molecules (digital PCR; Vogelstein and Kinzler 1999). However, the large number of PCR reactions required, together with background noise arising from PCR misincorporation, is likely to limit this approach to detection levels of about 10-3. Another limitation of many mutation detection procedures is that they replace the mutant site with a PCR primer sequence and yield short amplicons containing little if any information other than the presence of a putative mutant allele.
The unifying problem behind all of these PCR approaches for detecting rare variants is replication infidelity during amplification. The solution therefore is to collect mutant DNA molecules from genomic DNA prior to analyzing them by PCR. We now describe a rapid method that achieves this goal and which should be applicable to any base-substitutional variant. This method is a modification of traditional techniques for enriching nucleic acids by hybridization with biotinylated DNA probes (see, e.g., Abe 1992), and uses allele-specific oligonucleotides to fractionate DNA molecules differing by a single base substitution. This method of DNA enrichment by allele-specific hybridization (DEASH) is capable of substantial purification of DNA molecules carrying any given base substitution, and further can recover large DNA molecules containing additional marker information relevant to studies, for example, of recombination and of the parent-of-origin of a de novo mutation.
RESULTS
The Principles of DEASH
The DEASH procedure is outlined in Figure 1. The input is double-stranded DNA containing a mixture of molecules that differ in the chosen single-base substitution; this mixture could be due to SNP heterozygosity in a single individual, or could represent a mixed DNA sample, or be due to de novo mutation. This DNA is mixed with a biotinylated allele-specific oligonucleotide (bio-ASO) directed to the chosen variant, plus a competitor ASO complementary to the other allele to increase the specificity of hybridization. The mixture is thermally denatured and hybridized, and bio-ASO/target hybrids are captured on streptavidin-coated paramagnetic Dynabeads. The captured hybrid is then thermally eluted from the beads in low-salt buffer, yielding single-stranded target enriched for the chosen variant and free from bio-ASO. Additional target can be recovered by adding bio-ASO to the unbound DNA and subjecting it to further cycles of re-extraction using DEASH. In addition, the enriched target can be purified further simply by adding buffer, bio-ASO, and competitor ASO and proceeding with a second round of DEASH (reenrichment; Fig. 1).
Figure 1.
The principles of DEASH. (A) DNA containing a mixture of molecules differing by the chosen base substitution (circle) is mixed with a biotinylated ASO (gray, with biotin shown as a square) and the corresponding competitor ASO (black) directed to the alternative variant, then denatured and hybridized. (B) Streptavidin-coated paramagnetic beads (Dynal) are added to bind the bio-ASO/target hybrid and are then separated with a magnet (C). Bound hybrid is thermally eluted (D), releasing the enriched single-stranded target and leaving the bio-ASO still bound to the beads. Further target can be recovered by adding bio-ASO to the unbound DNA, denaturing, and processing as above (re-extraction). Enriched target can be purified further by adding buffer, bio-ASO, and competitor ASO, followed by DEASH (re-enrichment).
To minimize damage to target DNA, the DEASH protocol uses a modified high-pH buffer based on a standard PCR buffer (Jeffreys et al. 1988). Pilot experiments indicated that PCR-amplifiable single-stranded DNA is thermally stable in this buffer; human genomic DNA subjected to 2 min at 96°C followed by 15 min at 70°C showed no detectable loss of an amplifiable 4-kb target compared with untreated genomic DNA (data not shown). Recovered single-stranded target DNA is stable and can be stored for at least 6 months at -20°C with no loss of amplifiable DNA. The DEASH buffer has the added advantage of being fully compatible with PCR analysis.
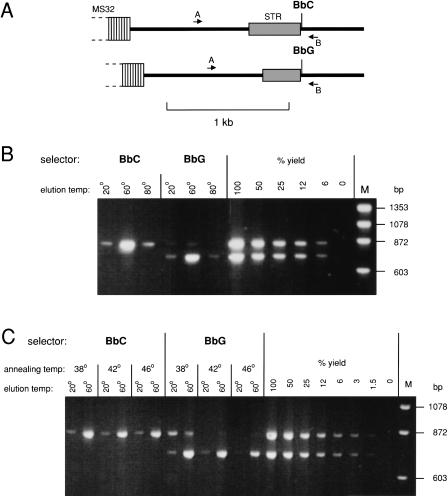
Separation of Haplotypes From PCR Products
DEASH was optimized using PCR products from a region adjacent to human minisatellite MS32(Fig. 2A; Jeffreys et al. 1998b). A person was chosen who was heterozygous for a SNP (BbC/G) adjacent to a length-heterozygous short tandem repeat (STR), the latter providing a simple assay for haplotype separation using bio-ASOs directed to BbC or BbG. Various parameters of the DEASH protocol were optimized on PCR products from this person (concentrations of bio-ASO and competitor ASO, annealing temperature, time of annealing and subsequent binding to streptavidin magnetic beads, bead washing conditions, and the elution temperature and time required to release single-stranded target DNA). Yields of each haplotype were estimated by PCR amplification of input and recovered DNA, limiting PCR to the exponential phase of amplification and comparing PCR product yields by agarose gel electrophoresis. Yield estimation was adjusted for the fact that unfractionated double-stranded DNA yields twice as much PCR product as from an equal number of single-stranded molecules. The optimized protocol (see Methods) was rapid, allowing fractionation in 20 min and giving excellent yields (>50%) of DNA eluted at 60°C, with little target being released in the 20°C pre-elution or 80°C post-elution washes (Fig. 2B). Substantial separation of haplotypes was achieved (>100-fold enrichment using BbC and 50-fold enrichment with BbG). Enrichment was robust over a broad range of annealing temperatures (Fig. 2C), with improved haplotype separation but reduced yield of BbG-selected DNA at higher annealing temperatures.
Figure 2.
Use of DEASH to separate haplotypes from PCR products. (A) The target region adjacent to human minisatellite MS32 (Jeffreys et al. 1998b) in the person analyzed, including a heterozygous SNP (BbC/G) used for DEASH selection and a length-heterozygous STR locus. This region was PCR-amplified from blood DNA using primers A and B. (B) Haplotype separation using BbC/G. Ten pg PCR products were fractionated by DEASH using biotinylated BbC or BbG ASOs, with annealing at 42°C. Hybrids retained on Dynabeads were eluted sequentially at the indicated temperatures. Aliquots of eluted single-stranded DNA (4% of each eluate) were amplified using primers A and B, along with decreasing inputs of unfractionated PCR products (200-12 fg DNA) to estimate yield of recovered DNA, and analyzed by agarose gel electrophoresis followed by staining with ethidium bromide. M, marker DNA. (C) Effects of varying the annealing temperature on yield and purification.
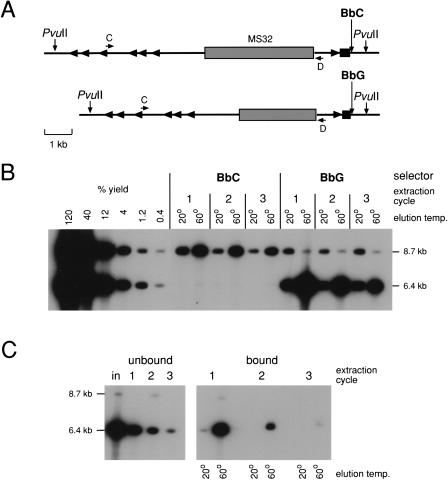
Separating Haplotypes From Genomic DNA
To test whether DEASH could also be applied to genomic DNA to recover much larger single-stranded DNA fragments, sperm DNA was fractionated from a person heterozygous for BbC/G and carrying two different length minisatellite MS32 alleles (Fig. 3A). BbC/G fractionation gave very low (<1%) yields of enriched DNA, as assayed by amplifying the minisatellite itself (data not shown). However, yields improved substantially using DNA digested with _Pvu_II, which cleaves upstream of the minisatellite and 582bp downstream of BbC/G (Fig. 3B). A single round of DEASH using biotinylated BbG gave 30% recovery of the 10.2-kb target, with 100-fold enrichment of the selected allele. Yields of the larger (12.5-kb) target using BbC were lower (∼8%) but the purification was excellent (200-fold). As predicted, more target could be recovered from the unbound DNA using additional cycles of DEASH (Fig. 3B); three cycles of re-extraction increased overall yields for BbG and BbC to about 60% and 20% respectively, with little if any decrease in the level of purification achieved at each round of fractionation.
Figure 3.
Separating haplotypes from human genomic DNA. (A) The target region, including the selector site (BbC/G) and minisatellite MS32 used to test for haplotype separation. Alu repeats are marked with arrowheads. (B) Haplotype separation. Four μg sperm DNA from a man with the two haplotypes shown in (A) was digested with _Pvu_II and fractionated by DEASH using either bio-BbC or bio-BbG. Unbound DNA was subjected to two further cycles of extraction after adding bio-BbC or bio-BbG. Aliquots of eluted fractions (2% of each eluate) were assayed by long PCR amplification with primers C and D, followed by agarose gel electrophoresis and Southern blot hybridization with a 32P-labeled MS32 repeat probe. Yields were estimated by comparison with decreasing inputs of unfractionated _Pvu_II-digested sperm DNA (48-0.16ng). The autoradiograph was overexposed to show traces of contaminating haplotype. (C) Assaying depletion of target from unbound DNA during DEASH. BbG-fractionated single-stranded DNA from B (60°C eluates pooled from all three extraction cycles) was subjected to three cycles of extraction/re-extraction using bio-BbG. Unbound (left) and bound (right) DNA recovered after each cycle was analyzed by PCR as in B and compared to input DNA (in). Note that primers C and D are located away from the selector site, allowing the unbound DNA to be analyzed by PCR even though it contains competitor BbC ASO. The faint residual 8.7-kb signal in the re-extracted fraction 1 is that expected for a single molecule of DNA, and given repeat instability and recombinational activity at MS32 (Jeffreys et al. 1998a,b), may be a mutant/recombinant molecule containing BbG rather than a contaminating BbC allele. For this reason, the MS32 system cannot be used to estimate levels of additional purification achieved by re-extraction.
DEASH recovers only one of the two complementary strands of a target, with the other strand remaining in the unbound genomic DNA fraction (Fig. 1). Because both strands serve as templates for the PCR assay, it was not possible to assay target depletion from the unbound DNA during purification. To circumvent this problem and directly test levels of depletion, we recovered single-stranded DNA by selection with bio-BbG and used this as target in an additional round of enrichment with re-extractions using bio-BbG. Assaying both unbound and bound DNA showed that most of the target was recovered by a single cycle of enrichment; after two further cycles of re-extraction, about 50% of starting target was recovered in a PCR-amplifiable form, with less than 5% of target DNA remaining in the unbound DNA fraction (Fig. 3C). Proportionately more target was recovered in the first cycle of extraction compared to genomic DNA (Fig. 3, cf. B and C). This suggests that low yields seen particularly with unrestricted genomic DNA may be due to inter-molecular hybridization between repeat DNA that could sterically hinder binding of the bio-ASO/target hybrid to the streptavidin beads. Such hybridization would also create high-molecular-weight aggregates that might promote shearing of the bound target, again reducing yields of intact target DNA.
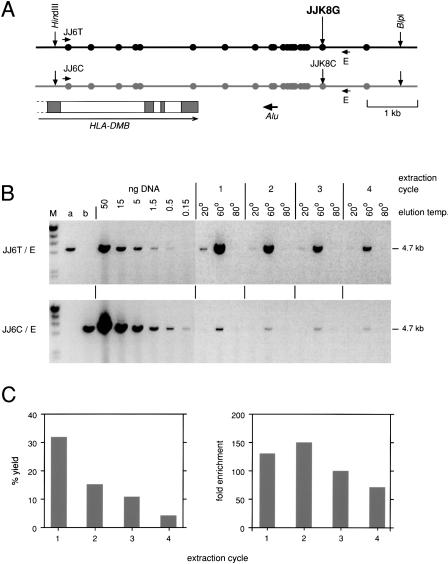
Bulk Fractionation of Genomic DNA
DEASH can be readily scaled to process large amounts of genomic DNA, as shown for SNP-mediated haplotype separation of a target within the class II region of the major histocompatibility complex (MHC) where, in contrast to the MS32 target, there was no length difference between haplotypes and where the selector site was located more internally within the target restriction fragment (Fig. 4A). Excellent haplotype separation was achieved using a bio-ASO directed to SNP JJK8. About 60% of target was recovered after four cycles of enrichment on 16 μg genomic DNA, as estimated both by allele-specific PCR analysis of the recovered target using the distal heterozygous SNP JJ6T/C (Fig. 4B) and by Poisson analysis of the number of amplifiable molecules in limiting dilutions of the purified DNA (data not shown; Jeffreys et al. 1998a). The correct haplotype was enriched 120-fold. Yields declined substantially in later cycles of re-extraction, and were accompanied by a modest decrease in the level of purification (Fig. 4C).
Figure 4.
Bulk separation of haplotypes within the human MHC. (A) The target region containing the end of the HLA-DMB gene and multiple heterozygous SNPs (circles) in the individual tested (Jeffreys et al. 2001). The region was released on a 6.6-kb fragment by digesting sperm DNA with _Hin_dIII plus _Blp_I. The black haplotype was fractionated from 16μg DNA using DEASH directed to allele JJK8G, using four cycles of extraction. (B) Haplotype separation. Recovery of the black haplotype was assayed by 24 cycles of long PCR using the allele-specific primer JJ6T plus primer E and comparing PCR product yields from aliquots of the eluates (3% of each eluate tested) with yields from decreasing inputs of unfractionated digested DNA. Contamination with the gray haplotype was similarly assayed using primer JJ6C (27 cycles). Genomic DNA from individuals homozygous for JJ6T (a) or JJ6C (b) (5 ng per PCR) provided a control for allele-specificity; these primers were chosen for their high specificity, with each showing a >1000 fold discrimination between alleles (data not shown). M, λ DNA × _Hin_dIII markers. (C) Yield and degree of enrichment estimated for each cycle of extraction/re-extraction.
This MHC target region contained multiple additional SNP heterozygosities in the individual tested (Fig. 4A). Linkage phase of these SNPs relative to the JJK8 selector site could be easily determined by ASO hybridization. Analysis of each SNP on dot blots of PCR products from the enriched target allowed complete haplotypes to be rapidly established (data not shown).
Properties of Bio-ASOs Used in DEASH
Twelve different bio-ASOs directed to various human SNPs near minisatellite MS32 (Jeffreys et al. 1998b), within the MHC (Jeffreys et al. 2001), and in the pseudoautosomal SHOX gene (May et al. 2002) were tested for their efficiency in separating haplotypes from restriction enzyme-digested genomic DNA (Table 1). All gave good yields of the enriched haplotype and, in most cases, excellent purification, in three instances so great (>200-300-fold) that the true level of enrichment could not be determined. Fractionation was successful for all classes of polymorphism tested (transitions, transversions, and an insertion/deletion variant), and could be successfully applied to SNPs within an Alu repeat (AB12/5 and AB13) and in simple-sequence DNA (AB2). Yields and levels of purification did not correlate with the GC content of the bio-ASO, and even very AT-rich ASOs could be used to fractionate DNA. Only two ASOs gave relatively low levels of enrichment (JJK10, JJK12; Table 1). The first is within a GT repeat sequence, and the second is in an extensive polypurine region which contains additional targets fairly well matched to the bio-ASO sequence (data not shown). Both SNPs were therefore tested for re-enrichment by subjecting enriched target DNA to a second round of DEASH (Fig. 1). The levels of purification increased in a multiplicative fashion, from sevenfold to 50-fold for JJK12, and from 13-fold to 140-fold for JJK10 (data not shown). Yields similarly showed a proportionate drop, from 60% to about 25%, indicating a fairly constant loss of amplifiable target per round of DEASH.
Table 1.
Efficiencies of Bio-ASOs Directed to Different Human SNPs
| SNP allele | Typea | Genomic region | Location | Bio-ASO sequence | Competitor ASO sequence | % GCb | Annealing temp.c | Target, kbd | % Yield | Purification |
|---|---|---|---|---|---|---|---|---|---|---|
| BbC | Tv | Minisatellite MS32 | Single Copy | Bio-AATAGGGAGACGATAAAG | AATAGGGAGAGGATAAAG | 39 | 42° | 12.5 | 20 | 200 × |
| BbG | Tv | Minisatellite MS32 | Single copy | Bio-AATAGGGAGAGGATAAAG | AATAGGGAGACGATAAAG | 39 | 42° | 10.2 | 60 | 100 × |
| AB2T | Tv | MHC HLA-DOA region | Single copy | Bio-GTTTGTTGTTTTTTGTTT | GTTTGTTGTTTGTTGTTT | 22 | 35° | 7.0 | 45 | >300 × |
| AB12/5AC | Tv+Ti | MHC HLA-DOA region | In Alu | Bio-AACCCCGTATCCACTGAA | AACCCCGTCTCTACTGAA | 50 | 40° | 7.0 | 50 | >300 × |
| AB13A | Tv | MHC HLA-DOA region | In Alu | Bio-TGAAAACACAAAAAAAAT | TGAAAACACACAAAAAAT | 17 | 35° | 7.0 | 65 | 40 × |
| AB14C | Ti | MHC HLA-DOA region | Single copy | Bio-ACTAATGATTCACTAACA | ACTAATGATTTACTAACA | 28 | 40° | 7.0 | 45 | 300 × |
| JJK6A | Ti | MHC HLA-DMB region | Single copy | Bio-GGTGAGAGGAAAGAAGTG | GGTGAGAGGAGAGAAGTG | 50 | 45° | 6.6 | 65 | 120 × |
| JJK8G | Tv | MHC HLA-DMB region | Single copy | Bio-AACGCGTTGAGCCCACAT | AACGCGTTGACCCCACAT | 56 | 50° | 6.6 | 60 | 120 × |
| JJK10C | Ti | MHC HLA-DMB region | Single copy | Bio-AAAAGGAGTGCGTGTGTG | AAAAGGAGTGTGTGTGTG | 50 | 50° | 6.6 | 60 | 13 × |
| JJK12- | 3 bp indel | MHC HLA-DMB region | Single | Bio-GAAAAAAAGAGGAGTGGA | AAAAAAAGAGAAGGAGTG | 39 | 49° | 6.6 | 60 | 7 × |
| 5053C | Tv | SHOX gene region | Single copy | Bio-CGGCTTTTGCCTTATGGA | CGGCTTTTGCGTTATGGA | 44 | 35° | 9.4 | 30 | 20 × |
| 5543G | Tv | SHOX gene region | Single copy | Bio-GGACGGGCTGGGGTTCCT | GGACGGGCTGCGGTTCCT | 72 | 55° | 9.4 | 30 | >200 × |
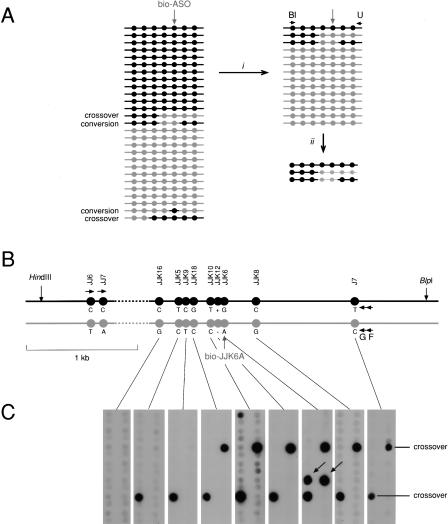
Use of DEASH to Detect Low-Frequency Changes in Human DNA
To illustrate how DEASH can be used to detect rare changes in human genomic DNA, we chose the HLA-DMB region (Fig. 4), which we previously showed contains a strong meiotic crossover hot spot termed _DMB_2 detectable by analysis of sperm DNA (Jeffreys et al. 2001). We used bio-ASO fractionation as a novel method to investigate crossover and gene conversion events in this region (Fig. 5). The principle was to enrich sperm DNA for one SNP allele (JJK6A) within the hot spot, then use nested allele-specific PCR against two distal upstream markers from the opposite haplotype to amplify recombinant molecules that had undergone recombination between this marker pair and the marker used for bio-ASO enrichment. Dot-blot analysis of these PCR products by hybridization with labeled ASOs revealed crossover molecules at a frequency of 8×10-5 per sperm, as seen in previous studies using repulsion-phase allele-specific PCR to recover crossover molecules from sperm DNA (Jeffreys et al. 2001). In addition, molecules were detected that had undergone transfer of the selected SNP allele alone from one haplotype to the other. More extensive analyses of DNA enriched using one of five different bio-ASOs showed that these single-base switches (39 detected in total, in 560 PCR reactions) were only seen at the selected SNP, and were never observed at other, nonselected SNPs (data not shown). This provides strong evidence that these switches cannot be artifacts arising during PCR amplification of enriched DNA and must instead represent genuine single-base changes present in sperm DNA. Their frequency (4×10-5 per sperm for JJK6) is far higher than the estimated rate of de novo base substitution in the human germline (2×10-8 per nucleotide site; Nachman and Crowell 2000), and, together with their association with a recombination hot spot, strongly suggests that they have arisen by highly localized gene conversion events within the crossover hot spot.
Figure 5.
Use of DEASH to analyze recombination in human sperm. (A) The principle of detection. Sperm DNA molecules from a region with multiple SNP heterozygosities (circles) will contain a low frequency of recombinant molecules (crossovers and gene conversions). Sperm DNA fractionation (i) using a bio-ASO specific for the gray haplotype will recover gray haplotype molecules, plus any recombinant molecules that include the gray selector site, and will deplete though not totally eliminate molecules from the black haplotype. PCR using an allele-specific primer from the black haplotype (Bl) plus a universal (not allele-specific) primer (U) (ii) will generate PCR products only from recombinant molecules and from any remaining molecules of the black haplotype. (B) The HLA-DMB target region, as in Fig. 4, in the man tested. Sperm DNA (12 μg) was digested with _Hin_dIII plus _Blp_I and enriched using bio-JJK6A directed to the gray haplotype. Assays as in Fig. 4, as well as Poisson analyses of extreme dilutions of the DNA, showed that the enriched DNA contained 440,000 amplifiable molecules of the gray haplotype and was 130-fold enriched. Multiple aliquots of this DNA, each containing 4000 molecules of the gray haplotype plus 30 remaining molecules of the black haplotype, were amplified using the allele-specific primer JJ6C and the universal primer F, then re-amplified with primer JJ7C plus the universal primer G. (C) These re-amplified PCR products were dot-blot hybridized with 32P-labeled ASOs specific for each of the gray alleles. Four recombinants were detected, with hybridization signals consistent with them being present at a level of 3% of the PCR products from the remaining black haplotype. Two were crossovers with exchange points mapping to the intervals JJK9-JJK18 and JJK16-JJK5, respectively. The other two were conversions (arrowed) affecting only the selector site JJK6. These conversions were detected among 28 PCRs containing 112,000 molecules of the gray haplotype, giving a conversion rate of 2/112,000 = 1.8 × 10-5 per sperm.
DISCUSSION
DEASH is a powerful, flexible method for fractionating not only PCR products but also genomic DNA for known base substitutional variants. It can be applied equally efficiently to small amounts (100 ng or less) of genomic DNA and to bulk fractionation at the 10 μg level (data not shown), and could be readily scaled to process even larger amounts of DNA. DEASH provides a simple, rapid method for haplotype separation at the 10-kb scale, requiring only the presence of a known SNP heterozygosity and providing an approach to haplotype determination that complements traditional methods using pedigrees, allele-specific PCR, or chromosome separation in somatic cell hybrids (Papadopoulos et al. 1995) to resolve haplotypes. Further work will reveal whether larger genomic targets can be efficiently fractionated and haplotyped by DEASH.
Analysis of sperm recombination events has shown that just one round of DEASH enrichment giving 100-fold purification is sufficient to detect single base changes (arising by gene conversion) at a frequency of 10-5 per cell (Fig. 5; additional data not shown). Without fractionation, the conversion events detected in Figure 5 would have required the analysis of 2000 PCR reactions, each containing 50 molecules (using ASO hybridization, this is the pool size limit for reliably detecting a mutant molecule against the background of wild-type molecules). Furthermore, such large numbers of reactions would generate the occasional false “mutant” signal due to PCR misincorporation during the earliest rounds of PCR amplification, generating a background noise that would mask any low-frequency events. DEASH circumvents these problems by greatly reducing the numbers of molecules screened for mutation, and further validates the mutants by showing that a given mutant is only detected if DNA has been enriched using the relevant bio-ASO.
The sensitivity of DEASH in detecting rare variants reaches or surpasses the detection limits of all other mutation detection systems based on direct analysis of DNA. It is already sufficiently great to enable in principle the direct detection of the de novo germline mutations in sperm DNA that cause achondroplasia and Apert syndrome, both of which show remarkably high spontaneous mutation rates, of 3×10-5 and 1×10-5-5×10-5, respectively (Cohen et al. 1992; Czeizel et al. 1993; Bellus et al. 1995; Tiemann-Boege et al. 2002). One remarkable feature of DEASH is its potential for indefinite purification of any chosen target. The degree of enrichment at each cycle of DEASH is determined by the relative likelihoods that a correct and incorrect target molecule will be bound. In theory, these likelihoods will be constant at each cycle of DEASH (they are analogous to partition coefficients between the unbound and bound “phases”), allowing in principle the indefinite purification of any chosen target. This concept of multiplicative enrichment has been confirmed for two bio-ASOs giving relatively low levels of purification. If more generally true, then variant detection at very low levels should be readily achievable. For example, two rounds of enrichment at 100-fold purification per round should allow the detection of mutation events as rare as 10-7 per cell, and there is in theory no reason why additional rounds of purification should not increase sensitivity even further. The main limitation in detecting extremely rare mutation events will be the sheer mass of genomic DNA that would need to be analyzed, compounded by progressive decrease in yields due to incomplete binding of target and any damage caused to DNA during the DEASH procedure.
Quantitation of the rate of gene conversion events detected in sperm DNA was straightforward, given that the enrichment separated the two progenitor haplotypes and that the number of progenitor haplotype molecules in the purified DNA assayed for recombination was known (Fig. 5). In contrast, the enrichment and quantitation of de novo mutant molecules, or low-frequency variant molecules present in a mixed DNA sample, will involve the removal of almost all wild-type DNA and will not give information on the efficiency of recovery of mutant molecules. It will therefore be necessary to develop reporter constructs carrying the mutation that can be added to the starting genomic DNA, providing an internal control from which yields and thus mutation rates can be determined.
DEASH enrichment is flexible and could be readily extended to sequential enrichment using two or more different bio-ASOs. For instance, it should be possible to recover recombinant molecules without using PCR. Thus, if recombinants are required between haplotypes A-B and a-b, then bio-ASO fractionation with marker A followed by the repulsion-phase marker b would enrich for A-b recombinants. Another feature of DEASH is that it should be readily amenable to automation, allowing the parallel processing of multiple samples in, for example, the detection of low-frequency mutations in patient samples. Finally, DEASH is currently the only technology with the potential to detect directly very-low-frequency de novo base substitutional mutations in the human germline, an area of major importance in understanding the dynamics of heri-table mutation and the origin of pathological mutations.
METHODS
Oligonucleotides
All bio-ASOs and competitor ASOs were 18-nt-long, with the allele-specific base 11 nt from the 5′ end. 5′ biotinylated ASOs were HPLC-purified before use. Single- and triple-biotinylated ASOs (Oswel) gave very similar yields in DEASH, and most fractionations were performed with single-biotinylated ASOs.
MS32 PCR primers used were: A 5′-GTC AAG GCA GAT TTG AAG GGA G-3′; B 5′-GGT AGC CAC ATC TCT GCA TGA TCT C-3′; C 5′-CCA TGC CCA GCC TTC ATT CTT TTC-3′; D 5′-CTT CCT CGT TCT CCT CAG CCC TAG-3′. HLA-DMB primers were: E 5′-AGG AGC CCC CCA TCT GGG AGT AGT-3′; F 5′-GTC TGC CTC AGG TGC TGA CC-3′; G 5′-ATG GCT CCC ATG CTC CCT GG-3′; JJ6C 5′-CTG CTC TGG TGG TGT GGC-3′; JJ6T 5′-GCT CTG GTG GTG TGG T-3′; JJ7C 5′-CCC CCT TGC TTT GAA ATG AGG C-3′.
The DEASH Procedure
Genomic DNA was digested with restriction endonucleases according to the manufacturer's conditions, ethanol precipitated, and dissolved in 5 mM Tris-HCl at pH 7.5. DEASH buffer (1 × DEB) was 45 mM Tris-HCl at pH 8.8, 11 mM ammonium sulphate, 4.5 mM MgCl2, 6.7 mM 2-mercaptoethanol, 4.4 μM EDTA, plus 2μg/mL single-stranded high-molecular-weight herring sperm DNA as carrier. Target DNA (PCR products or up to 4 μg restricted genomic DNA) was added to a final volume of 20 μL 1 × DEB plus 0.38 μM bio-ASO and 1.5 μM competitor ASO in a 0.2-mL PCR tube (ABgene Thermo-Tube). Larger quantities of genomic DNA were processed in proportionately larger volumes. DNA was denatured at 96°C for 75 sec on a GeneAmp PCR System 9700 thermal cycler (PE Applied Biosystems), then step-down annealed from (A+9)°C to (A+1)°C over 3 min (where A°C is the optimal annealing temperature determined empirically), and held at A°C for 2 min. DNA was then transferred to a siliconized Eppendorf tube (Treff Lab polypropylene tube siliconized with dimethyldichlorosilane solution, BDH; siliconization is essential to prevent nonspecific recovery of single-stranded DNA) and held at A°C. Hybrids were captured by adding 0.18 mg (1.3 μL) Dynabeads M-280 Streptavidin (Dynal Biotech; pre-washed twice with 1 × DEB) and allowing binding to proceed for 10 min at A°C, with occasional gentle mixing to keep the beads suspended. Beads were immediately separated at room temperature (<30 sec) using a Dynal MPC-E magnetic particle concentrator, and unbound DNA transferred to a 0.2-mL PCR tube. Beads were washed at room temperature with 50 μL 1 × DEB containing 10 μg/mL BSA (Ultra Pure, Ambion), then with 50 μL DEB/BSA for 1 min at A°C. Washed beads were resuspended in 30 μL elution buffer (0.14 × DEB containing 5 μg/mL single-stranded herring DNA) and transferred to a fresh siliconized Eppendorf tube at 20°C. Beads were separated and the 20°C washings retained. Target DNA was released by incubating the beads in 30 μL elution buffer for 2 min at 60°C (70°C for bio-ASOs with >50% GC content), and a final wash was performed at 80°C. Eluants at 20°C, 60°C, and 80°C were stored in 0.2-mL PCR tubes at -20°C. Re-extractions were performed by adding bio-ASO to 0.38 μM to unbound DNA and proceeding as above, with an initial 1-min denaturation. Re-enrichments were similarly performed, after adding DEB, bio-ASO, and competitor ASO to recovered target DNA, as above with a 1-min denaturation.
Analysis of Fractionated DNA
Long PCR analyses of enriched fractions using Taq plus Pfu polymerases were performed as described (Jeffreys et al. 1998b). The sperm recombination analysis (Fig. 5) involved a primary allele-specific PCR at 96°C for 20 sec followed by 22 cycles at 96°C for 20 sec and 65°C for 7.5 min. Products were diluted 20-fold with water, and 0.5 μL was used to seed a 15 μL secondary PCR which was cycled at 96°C for 1 min followed by 36 cycles at 96°C for 20 sec, 59°C for 30 sec, and 63°C for 8 min. Secondary PCR products were analyzed by dot-blot hybridization with ASOs as described (Jeffreys et al. 2000).
Acknowledgments
This work is the subject of a UK Patent Application. We thank J. Blower for providing semen samples, S. Mistry for oligonucleotide synthesis, and colleagues for helpful discussions. This work was supported by grants to A.J.J. from the Medical Research Council and Royal Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1214603.
References
- Abe, K. 1992. Rapid isolation of desired sequences from lone linker PCR amplified cDNA mixtures: Application to identification and recovery of expressed sequences in cloned genomic DNA. Mamm. Genome 2**:** 252-259. [DOI] [PubMed] [Google Scholar]
- Adinolfi, M., Camporese, C., and Carr, T. 1989. Gene amplification to detect fetal nucleated cells in pregnant women. Lancet 2**:** 328-329. [DOI] [PubMed] [Google Scholar]
- Bellus, G.A., Hefferon, T.W., Ortiz de Luna, R.I., Hecht, J.T., Horton, W.A., Machado, M., Kaitila, I., McIntosh, I., and Francomano, C.A. 1995. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am. J. Hum. Genet. 56**:** 368-373. [PMC free article] [PubMed] [Google Scholar]
- Berdanier, C.D. and Everts, H.B. 2001. Mitochondrial DNA in aging and degenerative disease. Mutat. Res. 475**:** 169-183. [DOI] [PubMed] [Google Scholar]
- Boyle, J.O., Mao, L., Brennan, J.A., Koch, W.M., Eisele, D.W., Saunders, J.R., and Sidransky, D. 1994. Gene mutations in saliva as molecular markers for head and neck squamous cell carcinomas. Am. J. Surg. 168**:** 429-432. [DOI] [PubMed] [Google Scholar]
- Cha, R.S., Zarbl, H., Keohavong, P., and Thilly, W.G. 1992. Mismatch amplification mutation assay (MAMA): Application to the c-H-ras gene. PCR Methods Appl. 2**:** 14-20. [DOI] [PubMed] [Google Scholar]
- Cohen, M.M., Kreiborg, S., Lammer, E.J., Cordero, J.F., Mastroiacovo, P., Erickson, J.D., Roeper, P., and Martinez-Frias, M.L. 1992. Birth prevalence study of the Apert syndrome. Am. J. Med. Genet. 42**:** 655-659. [DOI] [PubMed] [Google Scholar]
- Crow, J.F. 1999. Spontaneous mutation in man. Mutat. Res. 437**:** 5-9. [DOI] [PubMed] [Google Scholar]
- Czeizel, A.E., Elek, C., and Susanszky, E. 1993. Birth prevalence study of Apert syndrome. Am. J. Med. Genet. 45**:** 392-393. [DOI] [PubMed] [Google Scholar]
- Fox, J.C., England, J., White, P., Ellison, G., Callaghan, K., Charlesworth, N.R., Hehir, J., McCarthy, T.L., Smith-Ravin, J., Talbot, I.C., et al. 1998. The detection of K-ras mutations in colorectal cancer using the amplification-refractory mutation system. Br. J. Cancer 77**:** 1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerry, N.P., Witowski, N.E., Day, J., Hammer, R.P., Barany, G., and Barany, F. 1999. Universal DNA microarray method for multiplex detection of low abundance point mutations. J. Mol. Biol. 292**:** 251-262. [DOI] [PubMed] [Google Scholar]
- Gibbs, R.A., Nguyen, P.N., and Caskey, C.T. 1989. Detection of single DNA base differences by competitive oligonucleotide priming. Nucleic Acids Res. 17**:** 2437-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haliassos, A., Chomel, J.C., Grandjouan, S., Kruh, J., Kaplan, J.C., and Kitzis, A. 1989. Detection of minority point mutations by modified PCR technique: A new approach for a sensitive diagnosis of tumor-progression markers. Nucleic Acids Res. 17**:** 8093-8099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque, K., Hehir, J., Fox, J.C., Newton, C.R., and Little, S. 1998. Amplification refractory mutation system linear extension: A novel, gel-free, enzyme-linked immunoassay method for DNA genotyping. Diagn. Mol. Pathol. 7**:** 248-252. [DOI] [PubMed] [Google Scholar]
- Hengstschlager, M., Holzl, G., Ulm, B., and Bernaschek, G. 1997. Raising the sensitivity of fetal RhD typing and sex determination from maternal blood. J. Med. Genet. 34**:** 350-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys, A.J., Wilson, V., Neumann, R., and Keyte, J. 1988. Amplification of human minisatellites by the polymerase chain reaction: Towards DNA fingerprinting of single cells. Nucleic Acids Res. 16**:** 10953-10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys, A.J., Murray, J., and Neumann, R. 1998a. High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol. Cell 2**:** 267-273. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A.J., Neil, D.L., and Neumann, R. 1998b. Repeat instability at human minisatellites arising from meiotic recombination. EMBO J. 17**:** 4147-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys, A.J., Ritchie, A., and Neumann, R. 2000. High-resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum. Mol. Genet. 9**:** 725-733. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A.J., Kauppi, L., and Neumann, R. 2001. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 29**:** 217-222. [DOI] [PubMed] [Google Scholar]
- Knight, J.A. 2000. The biochemistry of aging. Adv. Clin. Chem. 35**:** 1-62. [DOI] [PubMed] [Google Scholar]
- Laken, S.J., Jackson, P.E., Kinzler, K.W., Vogelstein, B., Strickland, P.T., Groopman, J.D., and Friesen, M.D. 1998. Genotyping by mass spectrometric analysis of short DNA fragments. Nat. Biotechnol. 16**:** 1352-1356. [DOI] [PubMed] [Google Scholar]
- Lo, Y.M., Bowell, P.J., Selinger, M., Mackenzie, I.Z., Chamberlain, P., Gillmer, M.D., Elliott, P., Pratt, G., Littlewood, T.J., Fleming, K.A., et al. 1994. Prenatal determination of fetal rhesus D status by DNA amplification of peripheral blood of rhesus-negative mothers. Ann. NY Acad. Sci. 731**:** 229-236. [DOI] [PubMed] [Google Scholar]
- Mao, L., Hruban, R.H., Boyle, J.O., Tockman, M., and Sidransky, D. 1994. Detection of oncogene mutations in sputum precedes diagnosis of lung cancer. Cancer Res. 54**:** 1634-1637. [PubMed] [Google Scholar]
- May, C.A., Shone, A.C., Kalaydjieva, L., Sajantila, A., and Jeffreys, A.J. 2002. Crossover clustering and rapid decay of linkage disequilibrium in the Xp/Yp pseudoautosomal SHOX gene. Nat. Genet. 31**:** 272-275. [DOI] [PubMed] [Google Scholar]
- Mills, N.E., Fishman, C.L., Scholes, J., Anderson, S.E., Rom, W.N., and Jacobson, D.R. 1995. Detection of K-ras oncogene mutations in bronchoalveolar lavage fluid for lung cancer diagnosis. J. Natl. Cancer Inst. 87**:** 1056-1060. [DOI] [PubMed] [Google Scholar]
- Nachman, M.W., and Crowell, S.L. 2000. Estimate of the mutation rate per nucleotide in humans. Genetics 156**:** 297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, C.R., Graham, A., Heptinstall, L.E., Powell, S.J., Summers, C., Kalsheker, N., Smith, J.C., and Markham, A.F. 1989. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 17**:** 2503-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos, N., Leach, F.S., Kinzler, K.W., and Vogelstein, B. 1995. Monoallelic mutation analysis (MAMA) for identifying germline mutations. Nat. Genet. 11**:** 99-102. [DOI] [PubMed] [Google Scholar]
- Reed, W., Kong, D.Z., Lee, T.H., Cowan, M.J., Busch, M.P., and Baxter-Lowe, L.A. 2002. Noninvasive determination of the paternal HLA haplotype of a fetus using kinetic PCR to detect fetal microchimerism in maternal plasma. Bone Marrow Transplant 29**:** 527-529. [DOI] [PubMed] [Google Scholar]
- Saiki, R.K., Bugawan, T.L., Horn, G.T., Mullis, K.B., and Erlich, H.A. 1986. Analysis of enzymatically amplified β-globin and HLA-DQ α DNA with allele-specific oligonucleotide probes. Nature 324**:** 163-166. [DOI] [PubMed] [Google Scholar]
- Sidransky, D., Von Eschenbach, A., Tsai, Y.C., Jones, P., Summerhayes, I., Marshall, F., Paul, M., Green, P., Hamilton, S.R., Frost, P., et al. 1991. Identification of p53 gene mutations in bladder cancers and urine samples. Science 252**:** 706-709. [DOI] [PubMed] [Google Scholar]
- Sidransky, D., Tokino, T., Hamilton, S.R., Kinzler, K.W., Levin, B., Frost, P., and Vogelstein, B. 1992. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science 256**:** 102-105. [DOI] [PubMed] [Google Scholar]
- Smith-Ravin, J., England, J., Talbot, I.C., and Bodmer, W. 1995. Detection of c-Ki-ras mutations in faecal samples from sporadic colorectal cancer patients. Gut 36**:** 81-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada, M., Omata, M., Kawai, S., Saisho, H., Ohto, M., Saiki, R.K., and Sninsky, J.J. 1993. Detection of ras gene mutations in pancreatic juice and peripheral blood of patients with pancreatic adenocarcinoma. Cancer Res. 53**:** 2472-2474. [PubMed] [Google Scholar]
- Tiemann-Boege, I., Navidi, W., Grewal, R., Cohn, D., Eskenazi, B., Wyrobek, A.J., and Arnheim, N. 2002. The observed human sperm mutation frequency cannot explain the achondroplasia paternal age effect. Proc. Natl. Acad. Sci. 99**:** 14952-14957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg, J. 2000. Somatic mutations and aging: A re-evaluation. Mutat. Res. 447**:** 117-135. [DOI] [PubMed] [Google Scholar]
- Vogelstein, B. and Kinzler, K.W. 1999. Digital PCR. Proc. Natl. Acad. Sci. 96**:** 9236-9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Fidler, C., Murphy, M.F., Chamberlain, P.F., Sargent, I.L., Redman, C.W., Hjelm, N.M., Wainscoat, J.S., and Lo, Y.M. 2000. Determination of fetal RhD status by maternal plasma DNA analysis. Ann. NY Acad. Sci. 906**:** 153-155. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.le.ac.uk/genetics/ajj/HLA/index.html; MHC SNP data at the University of Leicester.
- http://www.le.ac.uk/ge/ajj/SHOX; SHOX gene SNP data at the University of Leicester.