Reconstruction of the Mouse Otocyst and Early Neuroblast Lineage at Single Cell Resolution (original) (raw)
. Author manuscript; available in PMC: 2015 May 8.
Summary
The otocyst harbors progenitors for most cell types of the mature inner ear. Developmental lineage analysis and gene expression studies suggest that distinct progenitor populations are compartmentalized to discrete axial domains in the early otocyst. Here, we conducted highly parallel quantitative RT-PCR reactions on 382 individual cells from the developing otocyst and neuroblast lineages to assay 96 genes representing established otic markers, signaling pathway associated transcripts, and novel otic-specific genes. By applying multivariate cluster, principal component and network analyses to the data matrix, we were able to readily distinguish the delaminating neuroblasts, and to describe progressive states of gene expression in this population at single cell resolution. It further established a three-dimensional model of the otocyst where each individual cell can be precisely mapped into spatial expression domains. Our bioinformatic modeling revealed spatial dynamics of different signaling pathways active during early neuroblast development and prosensory domain specification.
Introduction
In this study, we use the otocyst, the precursor of the vertebrate inner ear, as a model system to explore quantitative single cell transcriptional characterization for 96 genes at the spatial, temporal, and functional level. The otocyst is a three-dimensional structure that arises from the otic placode, adjacent to the developing hindbrain (Fritzsch et al., 2002; Morsli et al., 1998). It harbors the vast majority of cells that give rise to the inner ear as well as the vestibular and cochlear neurons (Corwin and Cotanche, 1989; Groves and Fekete, 2012; Swanson et al., 1990).
Despite the wealth of knowledge accumulated by studies of individual gene expression patterns (Alsina et al., 2009; Radde-Gallwitz et al., 2004), it is not clear whether the specific cell populations located at distinct positions in the otocyst such as dorsal or ventral are homogenous or whether they can be further subdivided into smaller and spatially defined groups of cells. Likewise, it has been hypothesized that the developing sensory organs and neuroblasts that arise from the otocyst are the product of regional synergistic relationships between cells or groups of cells, effects of surrounding tissues, as well as cell fate restrictions (Brigande et al., 2000; Fekete and Wu, 2002; Groves and Fekete, 2012; Wu and Kelley, 2012). Population-based approaches do not recognize rare cell types nor do they reveal spatial correlations of genes that define cell identities with active signaling pathways. In contrast, single cell analysis technologies provide a powerful method to study global cell heterogeneity and to describe mechanisms on a local level (Tischler and Surani, 2013). Our aim was to use the mouse otocyst as an example of a simple but highly organized system of cells, and to apply single cell quantitative gene expression analysis in order to gain insight into regional cell identities, dynamic processes, and areas of active signaling. We analyzed 382 individual mouse otocyst and neuroblast cells by performing 36,672 individual quantitative RT-PCR reactions conducted on microfluidic arrays. Using three complementary analyses of correlation, principal components and network topology, we defined the dynamic architecture of neuroblast development inherited in cell-specific transcription motifs. We further applied bioinformatic methods in the context of well-established spatial gene expression patterns to computationally reconstruct an otocyst organ model that provides in-depth biological insight at single cell resolution. Our analyses describe temporal and spatial components of otic development. This allowed us to organize high-dimensional data into simple models that contribute to a better understanding of the cellular heterogeneity.
Results
Transcriptional Profiling of Individual Otocyst and Neuroblast Cells
During mammalian inner ear development, expression of the transcription factor Pax2 is first detectable in the otic placode and continues to be expressed in the otocyst as development progresses (Hidalgo-Sanchez et al., 2000). In Pax2Cre+/−;Gt(ROSA)26SormtdTomato,mEGFP reporter mice (Muzumdar et al., 2007; Ohyama and Groves, 2004), the progeny of the otic placode including all otocyst cells as well as delaminating neuroblasts express membrane-EGFP, whereas the surrounding non-otic cells continue to express membrane-tdTomato fluorescent protein (Figure 1A,A′). Using fluorescence-activated cell sorting (FACS), we collected 384 individual membrane-EGFP(+)/membrane-tdTomato(−) cells from the otocyst and the immediate neighboring tissue of embryonic day 10.5 (E10.5) embryos (Figures 1B and S1). We quantitatively measured expression of 96 different transcripts utilizing a microfluidic quantitative PCR platform. Included were transcripts with known expression in the mouse otocyst, potentially novel otocyst-enriched transcripts identified in an independent microarray study, as well as genes associated with five major signaling pathways implicated in inner ear development (Notch, Shh, Fgf, Tgfβ, canonical Wnt) (Table S1). The performance of each primer pair was validated for technical reproducibility and specific signal generation (Figure S2 and Supplementary Methods: Primer Validation). Single cell cDNA was analyzed in 36,864 individual qPCR reactions. 382 cells passed a number of stringent quality control assessments that ensured high quality single cell data and were included for subsequent analyses (Figure S3 and Supplementary Methods: Quality Control and Initial Data Processing).
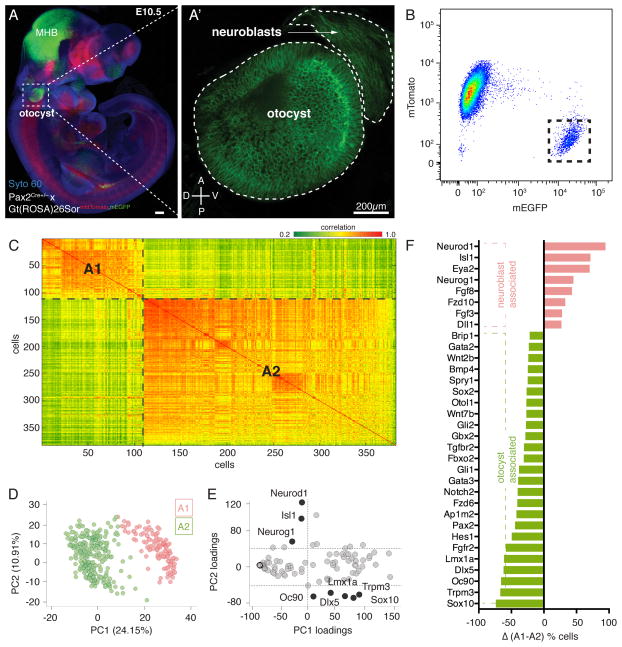
Figure 1. Sorted Single Cells can be Grouped into Corresponding Cell Identities using Multivariate Analyses.
(A) Representative image of E10.5-old Pax2Cre+/−;Gt(ROSA)26SormtdTomato,mEGFP embryo. Green fluorescence (mEGFP) indicates Cre-mediated recombination labeling the otic lineage from placode to otocyst. The midbrain-hindbrain boundary (MHB) area is also notably labeled. (A′) Otocyst and delaminated neuroblast cells on the ventro-anterior region are mEGFP positive. (B) FACS plot shows two main cell populations: mTomato+/mEGFP− and mTomato−/mEGFP+, which were gated for single cell sorting. (C) Pearson Correlation of 382 single cells from otocysts and neuroblasts; 2 cells of the originally collected population of 384 were excluded from the analysis. Red indicates high positive correlation. Green represents high negative correlation. (D) PCA of 382 cells projected onto the first two components. (E) Genes projected onto first two principal component loadings. Thresholds of 0 (PC1), and −40 (PC2) were applied to determine transcripts along first PC loading. Thresholds values of +40 (PC1) and 0 (PC2) were used to determine transcripts along second PC loading. (F) Binary analysis of different genes whose expression was on or off in each group (A1/A2). Shown are the proportion differences of cells between cluster A1 and A2 in descending order from top to bottom. Red indicates genes that are overrepresented in cells of the A1 cluster. Green represents genes that are expressed in relatively more cells in cluster A2. See also Figures S1–S4 and Tables S1 and S2E.
Single Cell Transcriptional Profiling Distinguishes Between Otocyst and Neuroblast Cells
Neuroblast specification is one of the earliest cell fate decisions in inner ear development. The otocyst harbors precursor cells that delaminate from the ventro-anterior region and migrate ventro-medially to accumulate, proliferate, and differentiate into the neurons that innervate the cochlea and vestibular organs of the inner ear (Rubel and Fritzsch, 2002). The process of delamination and migration begins around E9.5 and persists for at least 1.5 days until E11 (Kim et al., 2001; Ma et al., 1998). Neuroblasts express a number of hallmark genes such as Neurog1 (Ma et al., 1998), Neurod1 (Liu et al., 2000) and Isl1 (Li et al., 2004). At E10.5, we expected to isolate both delaminating and migrating neuroblast cells and we anticipated them to be distinguishable from the otocyst cells based on their distinct gene expression patterns.
To discriminate between neuroblasts and otocyst cells, we compared transcriptional profiles of all 382 cells across 96 genes. We used the Pearson correlation coefficients (Pearson, 1896) as measures of similarity among expression profiles of all individual cells, and produced a heat map reflecting correlation of each individual cell to all the others. Correlations revealed two distinct clusters of cells which we termed A1 (consisting of 110 cells) and A2 (consisting of 272 cells) (Figure 1C). In a parallel and independent mathematical analysis, we used principal component analysis (PCA (Jolliffe, 2002; Yeung and Ruzzo, 2001)) to distinguish between distinct cell groups. PCA is a multivariate technique that reduces the high-dimensionality of the data (here 92 genes (96 minus 4 control genes) corresponding to 92 dimensions) by determining new coordinates in a re-transformed multi-dimensional space, and selecting a smaller set of coordinates that still capture the variations in the original high-dimensional data. PCA allows for patterns to be recognized in a lower-dimensional space. We found that the first two principal components retained 35.12% of the original biological variability of the data, which is sufficient to partition two groups corresponding with A1 and A2 of the Pearson analysis (Figures S4A and 1D).
Each principal component consists of weighted contributions from all 92 genes. In PCA, the correlation of a variable with a given component is referred to as a loading, which represents the proportion of contribution of a given gene to the distribution of cells along the component. When we projected the first two principal component loadings for all 92 transcripts, we were able to categorize two distinguishing cohorts of genes based on high differential loadings between PC1 and PC2 (Figure 1E). Genes identified with this strategy included the neuroblast markers Neurog1, Neurod1, and Isl1 within one group, whereas the other group contained presumptive otocyst genes. Furthermore, comparison of the relative proportion of cells expressing individual genes reveal that neuroblast-associated transcripts are linked with cells belonging to A1, clearly distinguishing these cells from the A2 cluster (Figure 1F).
Bi-clustering Analysis Further Subdivides Otocyst and Neuroblast Populations
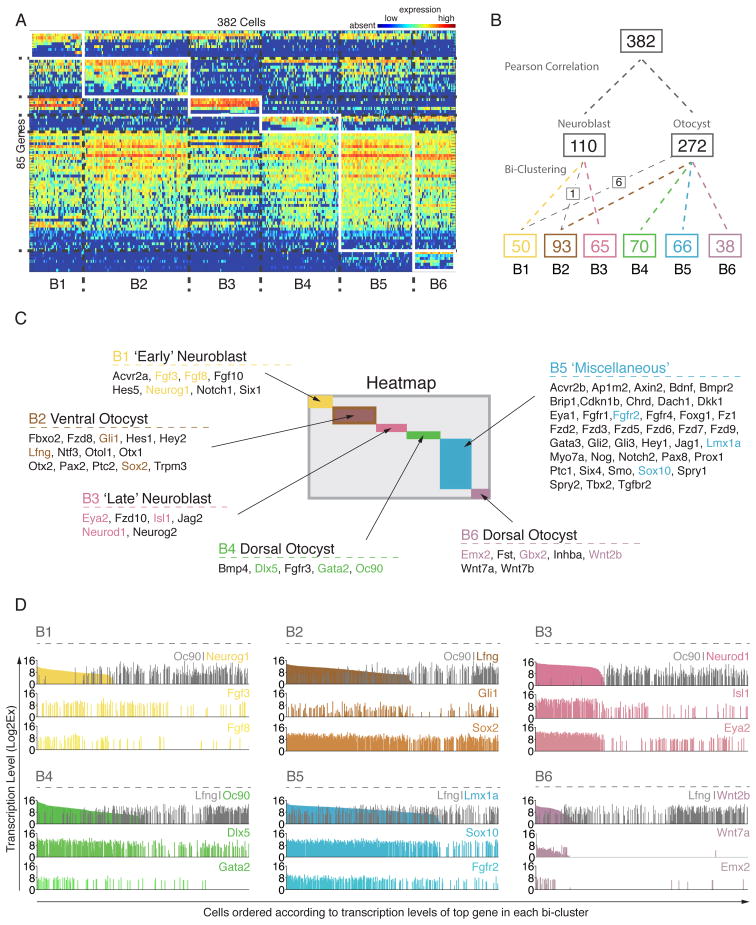
Visual inspection of the heat map generated by Pearson correlation suggested that the two major cell populations can be further subdivided. We therefore employed bi-clustering, an unbiased partitioning approach that allows simultaneous clustering of genes and cells. It resolves local rather than global gene association patterns and thus can identify subsets of genes with similar expression motifs across subsets of cells (Cheng and Church, 2000). Figure 2A shows the heat map generated by the bi-clustering algorithm, which assembled the data into six clusters of cells, designated B1–B6. B1 and B3 consisted of 50 and 65 cells, respectively, and contained all of the 110 A1 cells previously identified with Pearson correlation and PCA. 267 of the 272 A2 associated cells were found in B2 and B4–B6 (Figure 2B). The overall organization of the sub-cluster structure reinforces the idea that the global cellular heterogeneity of the otocyst and neuroblast cells can be computationally organized into transcriptionally and/or functionally related cell groups. Each of the six cell clusters is determined by differential expression of distinct subsets of genes expressed in a correlated manner. Clusters B1 and B3 show complete absence of dorsal otocyst markers and, as already suggested by Pearson correlation and PCA, are defined primarily by presence of neuroblast markers. Bi-clustering revealed distinguishing details between the B1 and B3 clusters. Interestingly, markers expressed during the early phase of neuroblast delamination such as Neurog1 and Fgf3 (Hatch et al., 2007; Ma et al., 1998) were largely expressed in B1 cells, whereas genes found in migrating and post-delaminated neuroblasts such as Isl1, Neurod1, and Eya1 (Li et al., 2004; Liu et al., 2000; Radde-Gallwitz et al., 2004) were transcripts delineating the B3 cluster of cells. Additionally, cluster B1 showed less expression of a group of genes that specifically described cluster B2 (Figure 2A, boxes in B1–3). Based on these observations, we hypothesized that B1 consists of early neuroblasts whereas B3 resembles cells of late neuroblast identity (Figure 2C).
Figure 2. Organizing the Cellular Heterogeneity of Otocyst Cells and Neuroblasts into Bi-Clusters with Distinct Transcriptional Profiles.
(A) Heatmap of all 382 cells and 85 genes after bi-cluster analysis. Two reference and 9 other genes were excluded from the analysis because they were expressed by fewer than 5 cells. Six distinguishable cell groups are clustered according to transcriptional gene signatures. (B) Cluster tree showing the overlapping partitions of cells generated by two independent and unbiased grouping algorithms. Pearson Correlation distinguishes two main groups. Bi-clustering resulted in six clusters, which almost perfectly represent sub-clusters of the two main groups generated by Pearson Correlation, except for 1 neuroblast and 6 otocyst cells that were reassigned as indicated. Numbers indicate cell number per group. (C) Genes that delineate each bi-cluster are listed. Expression of colored markers is shown in (D) at single cell resolution. (D) Six representative examples for 4 genes each show expression distribution (y-axis) across all 382 cells (X-axis). Cells from each cluster are ordered according to expression levels of the first gene (high to low expression) so that each cell is located at the same location on the X-axis. In grey are shown two hallmark genes of dorsal (Oc90) and ventral (Lfng) character for comparison. See also Figures S5 and S6, and Table S2E.
Cluster B2 consists of 93 cells that express ventral-associated markers including the prosensory genes Lfng, Sox2, and Pax2, as well as the sonic hedgehog (Shh) signaling effector Gli1 (Dabdoub et al., 2008; Morsli et al., 1998; Riccomagno et al., 2002). This suggests a ventro-medial otocyst origin, which is supported by absent or low expression of dorsal-associated genes such as Oc90 and Dlx5 (Depew et al., 1999; Verpy et al., 1999) (Figure 2C).
Cells grouped in B4 were categorized as dorsal otocyst and feature absence of ventrally associated and neuroblast markers, whereas genes with reported dorsal expression domains are strongly expressed, such as Bmp4, Dlx5, Gata2 and Oc90 (Fekete and Wu, 2002; Lillevali et al., 2004; Zheng et al., 2003).
Cluster B5 is characterized by a large number of genes, several of which are mainly associated with the otocyst and not specifically with delaminating neuroblasts. Many of these genes are expressed more widely, which made it difficult to readily assign specific domain identities.
Lastly, cluster B6 represents a distinct subpopulation of cells that uniquely express members of the Wnt/b-catenin family (Wnt2b, Wnt7a, Wnt7b). Wnt2b is described as specifically labeling the endolymphatic duct area (Hatch et al., 2007; Koo et al., 2009; Lin et al., 2005; Riccomagno et al., 2002). Cells in B6 also express dorsal genes Oc90 and Dlx5, as well as the medial marker Gbx2 (Hidalgo-Sanchez et al., 2000) suggesting that they might derive from these regions.
The above bi-cluster analysis allowed identification of genes that correlate with individual cell clusters. We next sought to directly compare expression of select groups of these definitive genes across all 382 individual cells. Figure 2D shows six examples of direct comparison for three cell cluster associated genes (color coded as in 2C) as well as one contrasting gene (grey) that is generally absent or expressed at low levels in the respective cluster (see Figure S5 for comprehensive list). In B1 for example, cells that express the early neuroblast gene Neurog1 (cells sorted descending according to its expression level) are generally negative for the dorsal marker Oc90. Additionally, Fgf3 and Fgf8 expression is partially correlated with Neurog1+ cells.
The B2-characterizing marker lunatic fringe is expressed in about 59% of cells in B2, the majority of which are Oc90 negative or low. Expression distributions of prosensory marker Sox2, ventro-medial marker Pax2, and sonic hedgehog associated marker Gli1 are consistent with a non-neural, ventral otocyst identity for cluster B2 (Figures 2D and S5B). Distributions of B3 marker genes Neurod1, Isl1 and Eya2 reveal that Neurod1 is expressed in a distinct subset of cells within B3 with an unimodal expression behavior. With respect to Oc90, Neurod1 expression is virtually mutually exclusive with only a few cells in cluster B3 positive for both markers. Distributions of Isl1 and Eya2 (cells ordered according to Neurod1 transcriptional levels) display a bimodal expression behavior (Figure 2D, see also Figure S6). Whereas the majority of cells positive for both genes also express Neurod1, a small sub-population of Isl1+/Neurod1− and Eya2+/Neurod1− exists; these cells generally express Isl1 and Eya2 at lower levels than the Neurod1+ population (Figures 2D and S5C).
Analysis of B4 associated genes and comparison of their expression across all cells together with ventral marker Lfng highlights the correlation between Oc90, Dlx5, and Gata2. A specific subpopulation of Oc90+ cells that is mainly Lfng negative also expresses dorsal marker Dlx5 (although a small Oc90−/Dlx5+ population also exists). The majority of cells that express Gata2 at a high level also express Oc90, and expression of Gata2 was mutually exclusive with Neurod1 (Figures 2D and S5D). In contrast, the bulk of genes that characterize cells in B5 are more broadly expressed in a gradient-like manner. Cells sorted according to expression levels of Lmx1a revealed a transcriptional correlation with Sox10 and Fgfr2. Additional juxtapositions between other marker genes identified even more detailed descriptive expression patterns (Figure S5E). Finally, B6 cells represent a distinct sub-population of Wnt active cells that exclusively co-express the endolymphatic duct marker Wnt2b and newly identified Wnt7a, but not Wnt7b, and are also positive for Emx2 which previously was not detectable in the otocyst by in situ hybridization (Holley et al., 2010) (Figures 2D and S5F).
In summary, bi-clustering analysis revealed different subpopulations within the otocyst and neuroblast cells. Gene-by-gene and cell-by-cell analyses divulge quite interesting and in-depth correlations, but it does not provide distinct relationships in the context of otic development. This prompted an analysis strategy, where we considered known gene expression information to establish models in which individual cells can be assigned to specific developmental stages and/or to their original spatial context in the otocyst.
Temporal Dynamics of Neuroblast Maturation Revealed by Network Analysis and One-Dimensional PCA
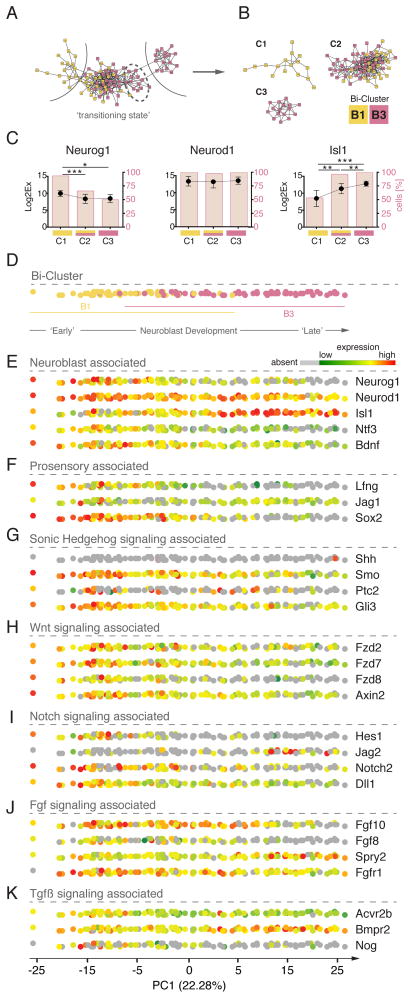
Neuroblasts originate in the ventro-anterior region of the otocyst where they delaminate and migrate to form the cochleo-vestibular ganglion (Fritzsch et al., 2002; Kim et al., 2001; Ma et al., 1998). Based on the bi-clustering results, we hypothesized that the two subsets of neural associated cells represent two temporally distinct neuroblast cell pools, an early neural progenitor population (B1) and a developmentally further advanced population (B3). Neuroblast differentiation is a dynamic process during which cells delaminate, migrate, proliferate, and differentiate. To visualize the developmental progression of B1 and B3 associated cells, we assumed that cells in similar differentiation stages show comparable gene expression patterns, recognizable by high co-expression scores determined with Pearson correlation. Connecting cells with high co-expression scores, we constructed a differentiation phase-similarity network comprised of all 115 B1 and B3 cells (Figure 3A, Supplementary Methods: Network Generation and Topology Analysis). Topological analysis of the phase-similarity network (Girvan and Newman, 2002) revealed three distinct community groups, (designated C1–C3) that correlated with previously established bi-clusters B1 and B3 (Figure 3B). The C1 and C3 groups comprised cells exclusively associated with clusters B1 and B3, respectively, consistent with early and later stages of neuroblast development. The C2 group of cells was topographically between C1 and C3, and consisted of B1 and B3 cells arranged asymmetrically. Interestingly, a smaller interconnected group of cells within C2 (dotted oval in Figure 3A) were all previously partitioned within the ‘late neuroblast’ bi-cluster B3, suggesting their temporal character is disparate from the presumably “younger” B1 cells found on the left side of C2. This network topology and correlation with bi-clustering is consistent with C2 representing a transitional state of otic neurogenesis.
Figure 3. Otic Neurogenesis can be Resolved Based on Gene Expression Changes at Single Cell Resolution.
(A) Co-expression gene network representation. Colors represent bi-cluster association. (B) Network topology analysis revealed three sub-networks. (C) Quantitation of expression level and fraction of cells per sub-network of three hallmark genes of neuroblast development. Black dots indicate transcriptional levels. Red bars represent cell proportions. Shown are means and standard deviations. P values are adjusted for multiple comparisons: * indicates p < 0.05, ** p < 0.01, *** p < 0.001. (D) PCA of 115 neuroblast cells. Cells are projected onto the first principal component and color-coded based on bi-cluster affiliation (B1 and B3). (E) Visualization of cellular transcriptional levels of neuroblast associated genes from high (red) to low (green) or absent (grey). (F) Expression of selected pro-sensory markers is visualized onto 1D PC projection. (G) – (K) Visualization of transcriptional levels of signaling pathway linked genes: Shh, Wnt, Notch, Fgf, Tgfβ. See also Figure S7 and Table S2E.
To validate the network topology analysis, we studied expression levels of individual marker genes associated with distinct phases of neuroblast maturation. The bHLH transcription factor Neurog1 is an early neuroblast marker necessary for neuroblast specification (Ma et al., 1998). Neurod1 is required for completion of neurogenesis and survival of neuroblasts (Liu et al., 2000). The LIM-Homeodomain transcription factor Isl1 is expressed in neuroblasts during delamination as well as in mature auditory and vestibular neurons (Li et al., 2004; Radde-Gallwitz et al., 2004). Cells in C1 expressed Neurog1 at a statistically significant 3-fold higher level over cells in the other two clusters (Figure 3C). Moreover, 93.3% of cells in C1 were positive for Neurog1, unlike cells grouped in C2 and C3 (65.9% and 50.0%, respectively). Consistent with its more protracted role in otic neurogenesis, Neurod1 was expressed in seemingly every cell in all three clusters at a high transcriptional level with no significant difference between groups. Isl1 expression, on the other hand, was increased 20-fold in the ‘late neuroblast’ associated cluster C3 compared to cells in the ‘early neuroblast’ cohort C1, and significantly increased in C2 compared with C1. These findings support the conclusion that the 115 cells employed for network analysis represent the inner ear neuroblast lineage in dynamic transition between early, intermediate, and late stages.
The ability to classify neuroblasts into presumptive stages of development prompted us to confine PCA to one single dimension, which we hypothesized would mainly represent the temporal axis and reveal dynamic expression profiles in a more continuous fashion. The first dimension (PC1) was highly informative, retaining a more substantial proportion of the original variability of the data when compared with analyses of other cell populations (Figure S4B). The sequence of cells along the PC1 axis highly correlated with the earlier bi-cluster analysis, where cells that previously were bi-clustered into B1 were positioned on the left side, which we marked ‘early’; whereas the previously B3 associated cells scattered along the right section of the vector, which we termed ‘late’ (Figure 3D).
We next examined the biological validity of our uni-dimensional model of the neuroblast population by examining expression levels of select developmental genes along the PC1 axis (Figures 3E–K).
The distribution of associated markers corroborated the previous finding for Neurog1, Neurod1, and Isl1, revealing that Neurog1 and Isl1 were expressed generally in opposing fashion. Cells with highest expression of early neuroblast markers distributed towards the left side whereas cells with high expression for Isl1 congregated on the right side of the first component vector (Figure 3E). Because the position of each cell along the axis is the composite result of contributions of all 92 genes used for the analysis, we were able to use the unidirectional presentation to inquire how other genes were expressed along the neuroblast lineage progression model (see Figure S7A for complete list). The neurotrophins Ntf3 and Bdnf for example are essential at later stages for inner ear neuron survival (Fritzsch et al., 2005), but their expression and role in the otocyst and delaminating neuroblasts has not been well characterized. These two factors declined from robust expression in virtually every early neuroblast to low levels or absence at later stages of otic neurogenesis.
Consistent with delamination from the otocyst and transitioning into the neuroblast lineage, the prosensory markers Lfng, Jag1, and Sox2 (Brooker et al., 2006; Hartman et al., 2010; Kiernan et al., 2005) became downregulated as developing neuroblasts advance along the temporal axis (Figure 3F). Lfng expression was rapidly lost in early neuroblasts, whereas downregulation of Jag1 and Sox2 occurred at a somewhat slower rate along the axis.
Analysis of assay genes representing major signaling pathways allowed us to correlate existing literature data and to delineate possible functional regulatory mechanisms. As expected (Bok et al., 2007), Shh expression was principally absent in otocyst or neuroblast cells analyzed in our assay (Figure 3G). Shh receptor genes Smo and Ptc2, and its effector Gli3, however, were expressed and became downregulated in cells of late neuroblast identity. This indicates that once neuroblasts adopt their lineage identity, they lose competence to respond to Shh.
Whereas virtually none of the 115 cells expressed any of the Wnt genes examined, gene expression mapping of Frizzled genes as well as the effector Axin2 support the hypothesis that neuroblast cells become unresponsive for Wnt signaling as they progress in development (Figure 3H). This is consistent with reports that increased responsiveness to Wnt signaling in Shh−/− mice leads to suppression of Neurog1 expression and failure of neuroblast development (Brown and Epstein, 2011); i.e. loss of Wnt responsiveness appears to be permissive for neuroblast development and our single cell analysis revealed that loss of frizzled expression and reduced expression of Wnt effector genes is a contributing mechanism ensuring proper neuroblast lineage development.
Notch signaling plays roles in regulating early regionalization and development of neuronal precursors in the inner ear (Abello et al., 2007; Daudet et al., 2007). We found that the neuroblast population displayed dynamic expression of several Notch related genes consistent with a temporal gradient of neuronal differentiation across PC1. Early neuroblast cells in particular were enriched for ligands Dll1 and Jag1, as well as Notch2 and the effectors Hes1, Hes5, and Hey2 (Figure 3I and S7A). The expression of these Notch genes declined along the temporal PC1 axis, with Notch2 and the effectors declining more sharply than the ligands (Dll1 and Jag1). Interestingly, Notch1 was detected in only a few early neuroblast cells, which appeared to be exclusive for Notch2, suggesting receptors may be differentially regulated (Figure S7A). In contrast to the above Notch genes, the ligand Jag2 and effector Hey1 did not display a decline along PC1, but rather appeared to increase slightly, which suggests these particular Notch genes may function in later stages of auditory and vestibular ganglion development.
The occurrence of potential subpopulations of cells towards later stages of neuroblast development revealed an important limitation of our interpretation that the single principle component axis represents a time line. Although successive development is very likely an important contributor to the order of cells along the axis, we cannot exclude that the neuroblast lineage diverges into multiple cell types. In fact, one would anticipate such a split, which has been reported as early as E12 for vestibular and auditory ganglia, the earliest time point investigated so far (Lu et al., 2011). Our data revealed that several genes including Foxg1 and Jag2 were strongly expressed in a group of cells positioned on the principal component 1 axis corresponding to later stages of neuroblast development (Figure S7A). This prompted us to focus only on the population of late neuroblast cells (B3). Two-dimensional PCA recognized two distinct cell populations within B3, characterized by asymmetric expression of Foxg1 and Jag2 (Figure S7B,C). Although speculative, these two populations could be indicative of an early separation of vestibular and spiral ganglion neurons. Profiled gene expression analyses between spiral and vestibular ganglia detected differences in expression of Foxg1 and Jag2 as early as E12 (Lu et al., 2011).
Regarding Fgf signaling, we noted that expression of Fgf8 and Fgf10 was higher in cells that reside in the early and intermediate phase of the presumed developmental time line (Figure 3J). Fgf receptor 1 (Fgfr1) and antagonist Spry2 show an opposing distribution in which Fgfr1 expression was high in cells of early neuroblast identity and declined over time, whereas Spry2 expression increased and was more abundant in cells with a late neuroblast state. Other Fgf signaling members that we investigated did not display such pronounced differences in distribution (Figure S7A). The distinct changes in expression of key otic Fgfs, the main receptor, and a prominent antagonist are in agreement with findings in chicken otic neuroblasts where Fgf signaling is essential for early events during neuroblast determination (Alsina et al., 2004). Loss of Fgf ligands and receptors as well as upregulation of Spry2 at the late neuroblast stage indicate that Fgf signaling ceases at this stage of development.
The most pronounced changes in expression of Tgfβ signaling genes were detectable for the inhibitor noggin, which declined from high expression during early stages of neuroblast development to absence in later stage neuroblasts (Figure 3K). Expression levels of activin receptor type-2B were reduced in late neuroblasts compared to younger stages, whereas Bmpr2 expression increased from moderate to high levels in later stage neuroblasts. The reversed expression pattern of Bmp4 antagonist noggin and Bmp receptor 2 suggest an active role of Tgfβ signaling in specifying a cellular neuroblast identity at later stages.
3D Reconstruction of the Otocyst
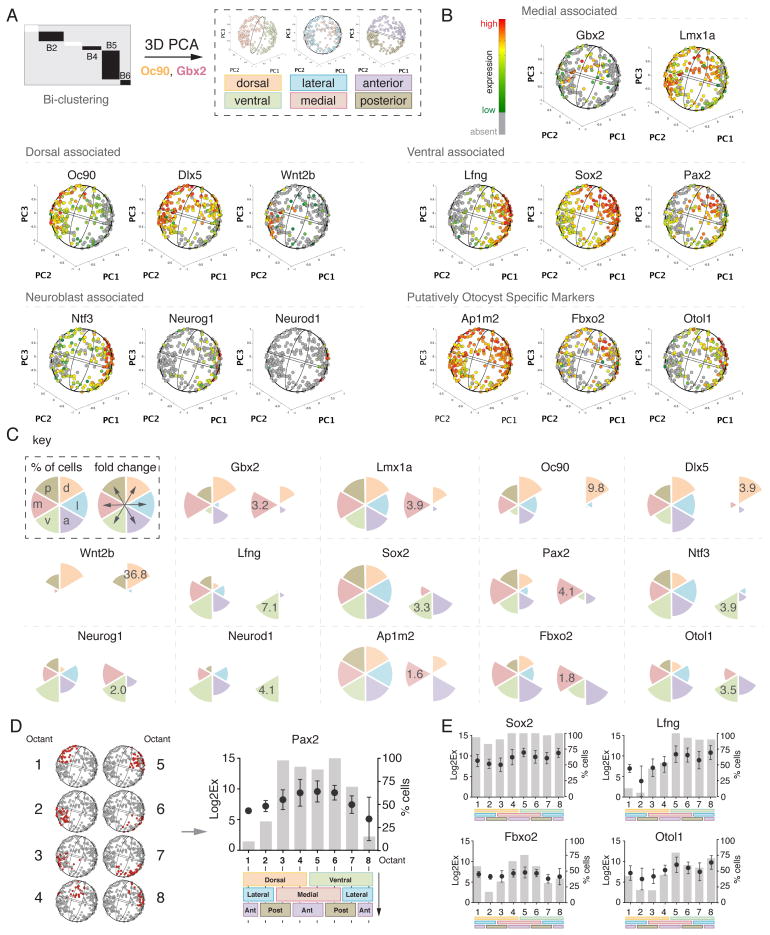
The morphology of the otocyst resembles a sphere and contains transcriptionally distinct expression domains commonly associated with at least one of the three major body axes, dorsal/ventral, medial/lateral, and posterior/anterior. To date, delineation of transcriptionally active regions occurs generally at low throughput in two-dimensional space. We sought to characterize expression domains of all assayed genes in three dimensions. To achieve this, we carried out PCA on 267 cells of the non-neuroblast clusters B2 and B4–B6 shown in Figure 2A,B. Next, we projected each of the 267 cells onto the surface of a sphere in a three dimensional coordinate system and approximated the three major axes with the help of single cell expression data for genes whose expression territories were confined to particular sides of the otocyst. Because several genes of our assay were selected to define known expression domains such as dorsal (Oc90) and medial (Gbx2), we were able to define the three major axes (Figure 4A; Supplementary Methods: Three-Dimensional Projection of PCA Data). To assess whether a 3D projection based on principal component coordinates could serve as a coarse reconstruction of the otocyst, we visualized the expression of marker genes quantitatively and analyzed the localization of marker-expressing cells in the sphere (Figure 4B and Movies S1–S4 for all).
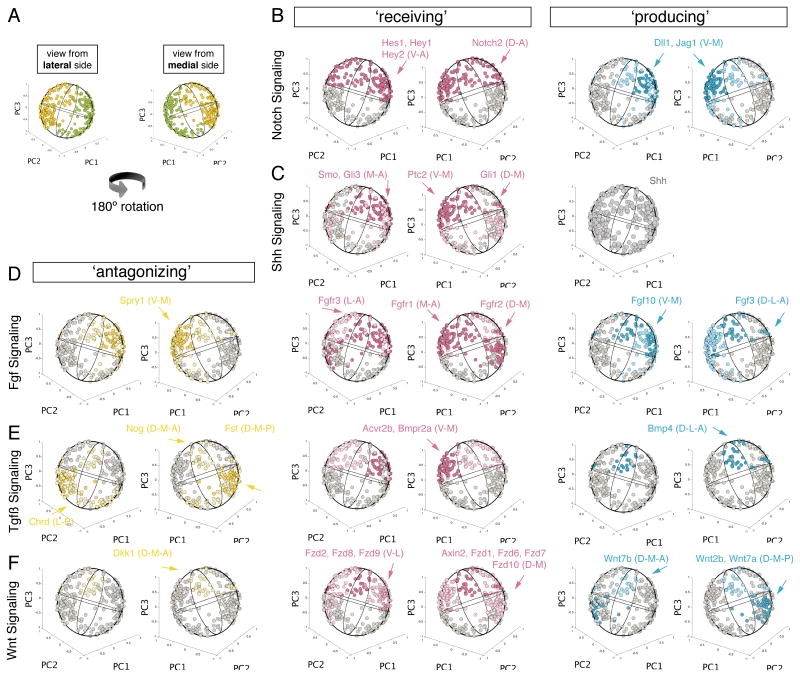
Figure 4. Three-Dimensional Reconstruction of the Mouse Otocyst Using PCA.
(A) Schematic overview of the bioinformatics algorithm to compute a three-dimensional model of the otocyst. (B) Transcriptional levels of selected marker genes projected onto 3D model representation. (C) Quantitation of % proportion of cells (left radial pie graph) and fold change between opposing body axes (right radial pie graph) for selected genes as shown in (B). (D) Representative illustration of octant analysis. Corresponding octant number is visualized in red in the 3D otocyst model. Quantitation of expression level and relative number of cells is octant-based. Color-code: d=dorsal (orange), v=ventral (green), l=lateral (blue), m=medial (red), p=posterior (brown), a=anterior (purple). (E) Octant analysis for two candidate genes from the pro-sensory (Sox2 and Lfng) and novel marker category, respectively. See also Table S2, and movies S1–S4.
Dorsal markers Oc90, Dlx5 and endolymphatic duct marker Wnt2b were found to be primarily projected to the dorsal half of the sphere model with significant differences in expression levels of 9.8-fold, 3.9-fold, and 36.8-fold, respectively, when compared with the opposite ventral half of the sphere (Figure 4C). Conversely, ventral markers such as Lfng and Sox2, were found to label cells on the ventral side. Lfng was expressed nearly exclusively in ventral otocyst cells, which accurately correlates with expression data from the literature (Hurd et al., 2010). Sox2 was detectable in virtually all cells, but expression was significantly higher by 3.3-fold in cells located in the ventral portion of the projection when compared to the dorsal half. Analysis of Pax2 expression revealed a 4.1-fold higher level on the medial side, which also corresponds with previous reports (Zheng et al., 2003).
Analysis of markers with reported medial expression such as Gbx2 and Lmx1a (Koo et al., 2009; Lin et al., 2005) revealed expression nuances previously not detectable with traditional methods. Whereas Gbx2 expression is restricted to cells located in the medial half of the otocyst, we also identified a tendency of this marker to be enriched in otocyst cells that express dorsal markers (Figure 4C). Lmx1a has been previously reported as a general marker for the majority of otocyst cells, except for the ventro-anterior domain (Koo et al., 2009). Our analysis confirmed that this gene is expressed in the majority of otocyst cells, but also revealed the highest expression of Lmx1a in the medial domain (3.9-fold higher when compared to the lateral half). Finally, cells that expressed neuroblast associated genes Ntf3, Neurog1, and Neurod1 were found in the ventral half of the projection, which agrees with published reports (Fritzsch, 2003). These cells are candidates for ventrally residing otocyst cells that have not yet fully transitioned into the neuroblast lineage.
After validating the spatial accuracy of the 3D model, we tested the ability of the model to define expression domains of genes previously not characterized in the mouse otocyst, such as Ap1m2, Fbxo2, and Otol1 (Figure 4B,C). Ap1m2 is expressed in virtually all otocyst cells with a small bias for higher expression levels at the medial-anterior region; the gene is downregulated as neuroblasts mature (Figure S7A). Fbxo2 and Otol1, on the other hand, display a spatially more confined expression with highest expression found in the medial and ventral domains, potentially indicating association with cells that ultimately will progress into prosensory domains as development continues.
Determination of the three major axes dividing the otocyst into six defined hemispheres (dorsal, ventral, medial, lateral, posterior, and anterior) presents an opportunity to further subdivide the otocyst into distinct octants (Figure 4D). This allowed us to display for each octant the number of cells expressing the gene as well as the mean expression value of a given gene. Pax2, for example, is significantly more highly expressed in octants representing the medial side of the model (p<0.0001, octants 3–6) (Figure 4D). Additionally, we determined expression domains of prosensory markers Lfng and Sox2, which both are predominantly expressed in all four octants corresponding to the ventral half of the otocyst. Expression was highest in octants 5 and 8, which correspond mainly to a ventro-anterior character (Figure 4D and see Table S2). Cells that expressed Fbxo2 and Otol1, two otic-specific candidate genes from our independent microarray study, predominantly reside in the medial-anterior and ventro-medio-anterior region, respectively, suggesting that this area of the otocyst that gives rise to the future prosensory domains is somewhat heterogeneous.
To further scrutinize the accuracy of the model, we measured gene expression profile distributions across all eight octants for each individual gene and compared the results to reported data in the literature. We calculated an ‘expression score’ which takes into account the relative proportion of cells positive for a marker and it’s associated transcriptional level. As a result, nearly all genes examined exhibit correct expression distributions as resolved by the 3D otocyst model (see Table S2).
Signaling Pathways can be Mapped to Spatially Defined Regions of the Otocyst
We used octant segmentation of the reconstructed otocyst to delineate signaling pathways by identifying cell groups that serve as originators or/and receivers of the following pathways: Notch, Shh, Fgf, Tgfβ, and Wnt.
Notch signaling is important in at least two major modalities during otic development (Murata et al., 2012), but has not been thoroughly analyzed at the otocyst stage. Our analysis mapped the Notch pathway to two distinct spatially confined territories (Figure 5A,B), one to the dorso-anterior side (Notch2) and one to the ventro-anterior side (Hes1, Hey1, Hey2). Several Notch signaling associated genes are expressed in neurogenic and prosensory progenitors, generally localized in the ventro-medial-anterior region. Thus, the prediction of the ventro-anterior spatial domain is consistent with available knowledge for Notch activity in this capacity. Notch2 is reportedly expressed in the early otocyst and mutation in this gene caused increased apoptosis in the developing ear, but the precise pattern of Notch2 expression has not been resolved (Hamada et al., 1999).
Figure 5. Regulatory Pathways Mapped onto Otocyst Reconstruction Model Confirms Distinct Regions of Signaling and Identifies New Spatially Defined Areas of Signaling.
(A) Schematic overview of 3D sphere rotational arrangement. The left sphere shows the view from the lateral (front) side in which the ventral domain (green) is positioned on the right side. (dorsal side shown in yellow on left side). The right sphere shows the view from the medial (back) side after a 180 degree rotation along PC3 axis. (B) Areas of Notch signaling are color-coded in an octant specific way. Red indicates areas of ‘receiving’ domains based on expression data of receptor or effector genes. Blue = domains of ‘producing’ fields based on expression data of ligand genes. Yellow = fields of ‘antagonizing’ domains based on expression data of signaling inhibiting genes. (C) – (F) Areas color-coded onto 3D model of active Shh, Fgf, Tgfβ, and Wnt signaling.
None of the examined otocyst cells expressed Shh, which is in concordance with published data that reports the notochord as sole Shh source to date (Bok et al., 2007; Riccomagno et al., 2002). Shh receptor gene Ptc2 as well as the effector gene Gli3 were predominantly expressed in cells located in the ventral half of the otocyst model (Figure 5C). For Gli1 and Gli2, the region of highest expression extends dorsally and anteriorly, respectively.
Fgf signaling plays an important role in specifying neuroblast cells in the ventral domain of the otocyst (Alsina et al., 2004). Our analysis shows that Fgf3 and Fgf10 are being produced by a cohort of cells residing in the ventro-latero-anterior otocyst. Expression of the cell autonomous Fgf signaling antagonist Spry1 was confined to a more medial domain, whereas cells expressing Fgfr1, Fgfr2, and Fgfr3 were located medial-anterior, dorso-medial, and latero-anterior, respectively (Figure 5D).
The role of Tgfβ signaling in inner ear development has not been elucidated in detail and only selectively described in the context of Bmp signaling (Chang et al., 1999; Gerlach et al., 2000; Merlo et al., 2002). We confirmed and refined the previously reported expression pattern of Bmp4, which is reportedly confined to a dorso-lateral area of the mouse otocyst. Our analysis places Bmp4 expressing cells into the dorso-lateral-anterior domain (Figure 5E). Bmpr2 expression was predominantly detected in cells located in the medial-anterior domain, similar to Acvr2b. Follistatin, a secreted protein that modulates the activity of Tgfβ family members, is produced by a specific population of cells residing in the dorso-medial-posterior region of the otocyst. The Bmp antagonist chordin, however, is expressed by cells located in the latero-posterior domain of the otocyst. These findings show that Tgfβ signaling is potentially active in different regions of the otocyst.
Cells that preferentially reside at the dorso-medial-anterior area of the 3D model express an array of Wnt receptors (Fzd1, Fzd6, Fzd7, Fzd10) and the effector gene Axin2 at the highest levels suggesting their active state of receiving Wnt signaling (Figure 5F). Expression of Fzd2, Fzd8, and Fzd9 is highest in the ventro-anterior domain of the otocyst. The dorsally located Wnt-receiving cells might respond to Wnt1 and Wnt3a originating from the dorsal hindbrain (Riccomagno et al., 2005). A distinct group of cells that coexpress Wnt2b and Wnt7a specified another subset of cells that reside at the dorso-medial-posterior side. Given the endolymphatic duct character of Wnt2b, this suggests that this area of the 3D otocyst model symbolizes the future anlage of the vestibular system visible as an out-pocketing at this age.
Identification of a Distinct Ventral-Associated Subgroup of Potential Predecessors of Prosensory Cells
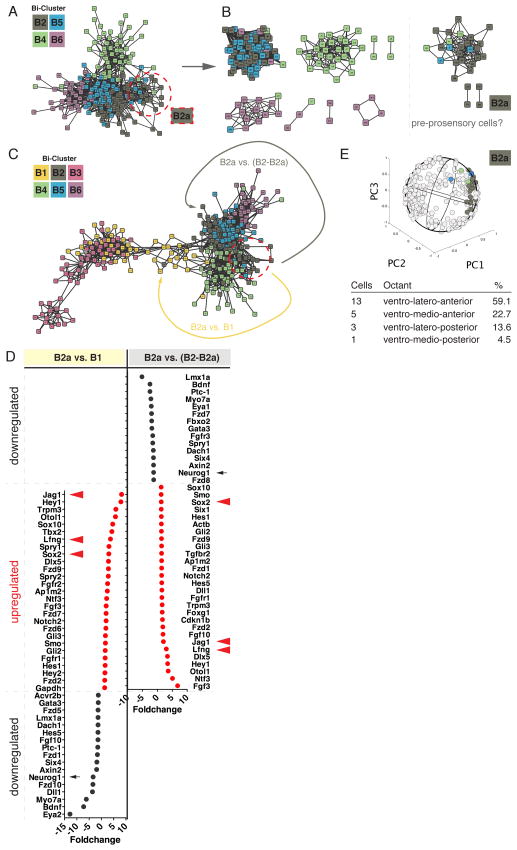
We sought to identify the pool of cells that harbors competency to give rise to the pro-sensory structures, which in turn will differentiate into vestibular and auditory hair cells and supporting cells. We focused on the ventral otocyst cells B2 (Figure 2C). The heat map generated with bi-clustering illustrates that this group represents a heterogeneous cell mixture (Figure 2A), and we hypothesized that only a portion of the 93 B2 associated cells are predecessors of prosensory domain cells. Next, we computed and visualized a coexpression network of all otocyst linked cells (Figure 6A). Topology analysis (Girvan and Newman, 2002) identified a group of 22 cells, which we named B2a, that was distinctively different from the bulk of cells in the center of the network, suggesting a potential different developmental commitment state (Figure 6B).
Figure 6. Characterization of an Otocyst Cell Population with Pre-Prosensory Cellular Identity.
(A) Co-expression network representation of 272 otocyst associated cells. Color code corresponds to bi-clusters shown in Figure 2. (B) Network topology analysis reveals a subnetwork primarily consisting of B2 associated cells (B2a). (C) Network architecture of all 382 otocyst/neuroblast cells. (D) Expression fold changes for two comparisons: B2a (‘pre-prosensory’) versus B1 (‘early neuroblast’), and B2a versus the remaining B2 cells (B2–B2a) (‘ventral otocyst’). Data in red indicates upregulation of genes in both juxtapositions. Pro-sensory markers are indicated by red arrowheads. Early neuroblast marker Neurog1 is indicated by a black arrow. (E) Visualization of B2a labeled cells onto 3D otocyst model reveals a ventral-anterior association at the medial/lateral boundary.
We investigated the two possible scenarios in which this cohort of cells either represents a very early population of pro-neural progenitor cells that take on a neuroblast developmental program or a different, non-neuronal population. The network architecture of all 382 cells implies the latter is true as the connecting arrangement between neuroblast associated (B1) and non-neuroblast associated cells is established by a group of cells other than the separating B2a cells (Figure 6C). We compared transcriptional levels of all 96 genes between B2a cells and the remaining B2 cells and between B2a cells and B1 cells. Figure 6D shows that the prosensory genes Jag1, Sox2, and Lfng are associated with cluster B2a in both comparisons. Furthermore, Neurog1 necessary for successful neuroblast commitment is downregulated in B2a cells compared to both related cohorts. B2a cells are mainly located in the ventro-anterior portion of our otocyst model (Figure 6E).
We noted that presumptive pre-prosensory B2a cells display Notch signaling activity reflected by expression of Hey1, Hey2, Hes1, Hes5, as well as Dll1, Jag1, and Notch2 (Figure 6D). Likewise, mediators of Shh hedgehog signaling Gli2, Gli3, and Smo are also associated with B2a cells. Finally, the increased expression of Fgf3 and Fgf10, as well as Spry1 and Spry2 suggests that B2a cells might serve as a source for Fgfs, but are likely antagonizing Fgf signaling in a cell autonomous fashion. These findings combined put forward a model in which Notch signaling is active during the initial phase of prosensory domain formation in combination with sustained response to Shh, and cell intrinsic inhibition of Fgf signaling, which is in agreement with the proposed role of Notch signaling in maintaining or expanding prosensory patches (Hartman et al., 2010; Murata et al., 2006; Neves et al., 2011) and with the observed downregulation of genes mediating response to Shh in neuroblasts (Brown and Epstein, 2011; Riccomagno et al., 2002)(Figure 4G). Whereas Notch signaling might not be responsible for induction of the prosensory domain (Basch et al., 2011; Kiernan et al., 2006), our data suggest that individual cells in the ventral otocyst integrate multiple signals at once, suggesting a complex balance of presumably permissive and partially inducing signals that lead to establishment of prosensory as well as neuroblast lineages in these regions.
Discussion
Single cell quantitative RT-PCR is a rapidly evolving method that bears promise to benefit many aspects of biology and medicine (Kalisky and Quake, 2011). Conventional approaches to study gene expression are generally limited to pooled cell populations or focus on tissues where expression of individual genes can be assessed at the transcript or protein level. Pooled cell populations have been successfully assayed for many genes using expression arrays and RNA deep sequencing techniques. These methods are restricted to the population and not individual cells. In situ hybridization and immunohistochemistry provide cellular information, yet are restricted to only a handful of genes. Integrating high-throughput with cellular resolution presents the ideal concept to study sparse cell derivatives with different biological identities. However, to date this requires dissociation of the tissues of interest, which in turn results in loss of critical spatial information. Our analysis strategies amalgamate existing spatial and temporal gene expression information with mathematical reconstruction models of developmental maturation (neuroblast) and three dimensional organ systems (otocyst). We used the mouse otocyst, the anlage of the inner ear because a simple map of broad expression domains that define major axes exists for multiple well-characterized genes exists. Knowledge about asymmetrically expressed genes such as markers for the delaminating neuroblast lineage, the precursors of the prosensory domains on the ventral side of the otocyst (Brooker et al., 2006; Kiernan et al., 2005), or the dorsally located endolymphatic duct (Hatch et al., 2007; Koo et al., 2009; Lin et al., 2005; Riccomagno et al., 2002) provided a framework for reconstruction of cell lineage relationships and location of individual otocyst cells in a 3D model.
Our analyses revealed that the cells we categorized as neuroblasts display a transitory character. The association of the first principal component with temporal changes allowed us to study this transition on a one-dimensional principal component vector. The accuracy of this novel informative analysis was consistent with existing gene expression data and revealed dynamics of many previously uncharacterized genes in a high-throughput context at single cell resolution. Ultimately, combination of the analysis techniques with assays and transgenic animals tailored to answer specific questions will without doubt have great impact on developmental biology and other fields.
Materials and Methods
Otocyst and Neuroblast Cell Isolation
Pax2Cre+/− males were mated with Gt(ROSA)26SormtdTomato,mEGFP females and checked for vaginal plugs daily. After positive plug confirmation (= E0.5), females were sacrificed 10 days thereafter and embryos were isolated (= E10.5). Embryos (n = 6), derived from one litter, were micro-dissected in ice cold Hank’s Balanced Salt Solution (HBSS). Successful Cre-mediated recombination was verified using a fluorescence dissection microscope. We ensured that all six embryos used for the study were very closely matched, based on overall size, completed otocyst closure, and clear manifestation of the endolymphatic duct. Otocysts (12 in total) and surrounding tissue were micro-dissected and incubated with thermolysine for 20min at 37°C to remove mesenchyme. Tissue was washed 1x with HBSS and treated with Accutase (Innovative Cell Technologies, Inc.) for 30min at 37°C. Cells were mechanically triturated and washed 2x with HBSS. Prior to cell sorting, the cell suspension was passed through a 35μm strainer (BD Biosciences) to remove residual cell clumps. Gating strategy was designed to maximize capture of one individual live cell per well (Figure S1).
Cell Sorting by FACS
Cells were stained with Propidium Iodide (Life Technologies) for dead cell exclusion and sorted with a FACSARIA II (BD Biosciences). After removal of debris and other none-cellular particles, doublets and multiplets were excluded on two consecutive gating steps (Forward-scatter height (FSC-H) versus forward-scatter area (FSC-A), side-scatter area (SSC-A) versus side-scatter width (SSC-W)). Dead cells were excluded based on Propidium Iodide uptake identified on the FSC-H versus PE-Cy5.5 profile, individual EGFP+/tdTomato− cells were sorted into different wells of 96-well PCR plates (USA Scientific) containing CellsDirect 2x Reaction mix (Invitrogen) supplemented with 0.05U of SUPERase-In RNase Inhibitor (Invitrogen). Flow rate was kept constant at 300 cells/sec (precision: single-cell, nozzle: 100 μm). 96-well plates were immediately sealed and stored at −80°C for subsequent RNA processing.
RNA Processing and qRT-PCR
RNA was reverse transcribed into cDNA with validated amplicon-specific DELTAgene Assays (Table S1) using SuperScript III RT Platinum Taq Mix plus 10X Primer Mix added to each sample. Reverse transcription and pre-amplification of target genes was performed in one step using 20 cycles. Samples were treated with Exonuclease I (NEB), diluted 5x for subsequent qPCR reaction and combined with sample pre-mix solution consisting of 2x SsoFast EvaGreen Supermix with Low ROX (Bio-Rad) and 20x DNA Binding Dye Sample Loading Reagent (Fluidigm). Assay Mix was formulated as follows: 2x Assay Loading Reagent (Fluidigm), 1x DNA Suspension Buffer, and pooled primer pairs (500 nM each). After priming the 96.96 dynamic array integrated fluidic circuit (IFC, Fluidigm) with control line fluid, the chip was loaded with assays and samples using an HX IFC controller (Fluidigm). The experiments were run on a Biomark HD (Fluidigm) for 30 cycles and subsequent melting curve generation. A master list of all cells and gene expression data is presented in Table S2E. See extended Materials and Methods in the Supplement for details.
Supplementary Material
01
02
03
04
05
06
4 Article Highlights.
- Early otic neuroblast lineage progression at single cell resolution
- Bioinformatic analyses describe time and space in development
- Refinement of expression domains of many known and novel otic genes
- Spatial dynamics of signaling pathways during development
Acknowledgments
We thank the staff at the Stanford Shared FACS Facility for their help and support. Patrick Raphael for his help with implementing movies. All members of the Heller laboratory for comments on the manuscript. This work was supported by NIH grants DC006167 and DC012250 to S.H., by P30 core support (DC010363), by the Stanford Initiative to Cure Hearing Loss, and in part by FP7-Health-2013-Innovation a cooperative grant by the European Commission. B.H.H. is supported by NIH F32 grant DC013210. J.W. is supported in part by a fellowship (WA3420/1) from the Deutsche Forschungsgemeinschaft.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abello G, Khatri S, Giraldez F, Alsina B. Early regionalization of the otic placode and its regulation by the Notch signaling pathway. Mech Dev. 2007;124:631–645. doi: 10.1016/j.mod.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Alsina B, Abello G, Ulloa E, Henrique D, Pujades C, Giraldez F. FGF signaling is required for determination of otic neuroblasts in the chick embryo. Dev Biol. 2004;267:119–134. doi: 10.1016/j.ydbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Alsina B, Giraldez F, Pujades C. Patterning and cell fate in ear development. Int J Dev Biol. 2009;53:1503–1513. doi: 10.1387/ijdb.072422ba. [DOI] [PubMed] [Google Scholar]
- Basch ML, Ohyama T, Segil N, Groves AK. Canonical Notch signaling is not necessary for prosensory induction in the mouse cochlea: insights from a conditional mutant of RBPjkappa. J Neurosci. 2011;31:8046–8058. doi: 10.1523/JNEUROSCI.6671-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok J, Dolson DK, Hill P, Ruther U, Epstein DJ, Wu DK. Opposing gradients of Gli repressor and activators mediate Shh signaling along the dorsoventral axis of the inner ear. Development. 2007;134:1713–1722. doi: 10.1242/dev.000760. [DOI] [PubMed] [Google Scholar]
- Brigande JV, Kiernan AE, Gao X, Iten LE, Fekete DM. Molecular genetics of pattern formation in the inner ear: do compartment boundaries play a role? Proc Natl Acad Sci U S A. 2000;97:11700–11706. doi: 10.1073/pnas.97.22.11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- Brown AS, Epstein DJ. Otic ablation of smoothened reveals direct and indirect requirements for Hedgehog signaling in inner ear development. Development. 2011;138:3967–3976. doi: 10.1242/dev.066126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Nunes FD, De Jesus-Escobar JM, Harland R, Wu DK. Ectopic noggin blocks sensory and nonsensory organ morphogenesis in the chicken inner ear. Dev Biol. 1999;216:369–381. doi: 10.1006/dbio.1999.9457. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Church GM. Biclustering of expression data. Proceedings/International Conference on Intelligent Systems for Molecular Biology; ISMB International Conference on Intelligent Systems for Molecular Biology. 2000;8:93–103. [PubMed] [Google Scholar]
- Corwin JT, Cotanche DA. Development of location-specific hair cell stereocilia in denervated embryonic ears. J Comp Neurol. 1989;288:529–537. doi: 10.1002/cne.902880402. [DOI] [PubMed] [Google Scholar]
- Dabdoub A, Puligilla C, Jones JM, Fritzsch B, Cheah KS, Pevny LH, Kelley MW. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci U S A. 2008;105:18396–18401. doi: 10.1073/pnas.0808175105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudet N, Ariza-McNaughton L, Lewis J. Notch signalling is needed to maintain, but not to initiate, the formation of prosensory patches in the chick inner ear. Development. 2007;134:2369–2378. doi: 10.1242/dev.001842. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Liu JK, Long JE, Presley R, Meneses JJ, Pedersen RA, Rubenstein JL. Dlx5 regulates regional development of the branchial arches and sensory capsules. Development. 1999;126:3831–3846. doi: 10.1242/dev.126.17.3831. [DOI] [PubMed] [Google Scholar]
- Fekete DM, Wu DK. Revisiting cell fate specification in the inner ear. Curr Opin Neurobiol. 2002;12:35–42. doi: 10.1016/s0959-4388(02)00287-8. [DOI] [PubMed] [Google Scholar]
- Fritzsch B. Development of inner ear afferent connections: forming primary neurons and connecting them to the developing sensory epithelia. Brain Res Bull. 2003;60:423–433. doi: 10.1016/s0361-9230(03)00048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW, Jones K, Farinas I, Maklad A, Lee J, Reichardt LF. Development and evolution of inner ear sensory epithelia and their innervation. J Neurobiol. 2002;53:143–156. doi: 10.1002/neu.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Pauley S, Matei V, Katz DM, Xiang M, Tessarollo L. Mutant mice reveal the molecular and cellular basis for specific sensory connections to inner ear epithelia and primary nuclei of the brain. Hear Res. 2005;206:52–63. doi: 10.1016/j.heares.2004.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach LM, Hutson MR, Germiller JA, Nguyen-Luu D, Victor JC, Barald KF. Addition of the BMP4 antagonist, noggin, disrupts avian inner ear development. Development. 2000;127:45–54. doi: 10.1242/dev.127.1.45. [DOI] [PubMed] [Google Scholar]
- Girvan M, Newman ME. Community structure in social and biological networks. Proc Natl Acad Sci U S A. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK, Fekete DM. Shaping sound in space: the regulation of inner ear patterning. Development. 2012;139:245–257. doi: 10.1242/dev.067074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada Y, Kadokawa Y, Okabe M, Ikawa M, Coleman JR, Tsujimoto Y. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development. 1999;126:3415–3424. doi: 10.1242/dev.126.15.3415. [DOI] [PubMed] [Google Scholar]
- Hartman BH, Reh TA, Bermingham-McDonogh O. Notch signaling specifies prosensory domains via lateral induction in the developing mammalian inner ear. Proc Natl Acad Sci U S A. 2010;107:15792–15797. doi: 10.1073/pnas.1002827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EP, Noyes CA, Wang X, Wright TJ, Mansour SL. Fgf3 is required for dorsal patterning and morphogenesis of the inner ear epithelium. Development. 2007;134:3615–3625. doi: 10.1242/dev.006627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo-Sanchez M, Alvarado-Mallart R, Alvarez IS. Pax2, Otx2, Gbx2 and Fgf8 expression in early otic vesicle development. Mech Dev. 2000;95:225–229. doi: 10.1016/s0925-4773(00)00332-4. [DOI] [PubMed] [Google Scholar]
- Holley M, Rhodes C, Kneebone A, Herde MK, Fleming M, Steel KP. Emx2 and early hair cell development in the mouse inner ear. Dev Biol. 2010;340:547–556. doi: 10.1016/j.ydbio.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd EA, Poucher HK, Cheng K, Raphael Y, Martin DM. The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development. 2010;137:3139–3150. doi: 10.1242/dev.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe IT. Principal component analysis. 2. New York: Springer; 2002. [Google Scholar]
- Kalisky T, Quake SR. Single-cell genomics. Nat Methods. 2011;8:311–314. doi: 10.1038/nmeth0411-311. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, Tease C, Lovell-Badge R, Steel KP, Cheah KS. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Xu J, Gridley T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2006;2:e4. doi: 10.1371/journal.pgen.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fritzsch B, Serls A, Bakel LA, Huang EJ, Reichardt LF, Barth DS, Lee JE. NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development. 2001;128:417–426. doi: 10.1242/dev.128.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo SK, Hill JK, Hwang CH, Lin ZS, Millen KJ, Wu DK. Lmx1a maintains proper neurogenic, sensory, and non-sensory domains in the mammalian inner ear. Dev Biol. 2009;333:14–25. doi: 10.1016/j.ydbio.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liu H, Sage C, Huang M, Chen ZY, Heller S. Islet-1 expression in the developing chicken inner ear. J Comp Neurol. 2004;477:1–10. doi: 10.1002/cne.20190. [DOI] [PubMed] [Google Scholar]
- Lillevali K, Matilainen T, Karis A, Salminen M. Partially overlapping expression of Gata2 and Gata3 during inner ear development. Dev Dyn. 2004;231:775–781. doi: 10.1002/dvdy.20185. [DOI] [PubMed] [Google Scholar]
- Lin Z, Cantos R, Patente M, Wu DK. Gbx2 is required for the morphogenesis of the mouse inner ear: a downstream candidate of hindbrain signaling. Development. 2005;132:2309–2318. doi: 10.1242/dev.01804. [DOI] [PubMed] [Google Scholar]
- Liu M, Pereira FA, Price SD, Chu MJ, Shope C, Himes D, Eatock RA, Brownell WE, Lysakowski A, Tsai MJ. Essential role of BETA2/NeuroD1 in development of the vestibular and auditory systems. Genes Dev. 2000;14:2839–2854. doi: 10.1101/gad.840500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CC, Appler JM, Houseman EA, Goodrich LV. Developmental profiling of spiral ganglion neurons reveals insights into auditory circuit assembly. J Neurosci. 2011;31:10903–10918. doi: 10.1523/JNEUROSCI.2358-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- Merlo GR, Paleari L, Mantero S, Zerega B, Adamska M, Rinkwitz S, Bober E, Levi G. The Dlx5 homeobox gene is essential for vestibular morphogenesis in the mouse embryo through a BMP4-mediated pathway. Dev Biol. 2002;248:157–169. doi: 10.1006/dbio.2002.0713. [DOI] [PubMed] [Google Scholar]
- Morsli H, Choo D, Ryan A, Johnson R, Wu DK. Development of the mouse inner ear and origin of its sensory organs. J Neurosci. 1998;18:3327–3335. doi: 10.1523/JNEUROSCI.18-09-03327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata J, Ikeda K, Okano H. Notch signaling and the developing inner ear. Adv Exp Med Biol. 2012;727:161–173. doi: 10.1007/978-1-4614-0899-4_12. [DOI] [PubMed] [Google Scholar]
- Murata J, Tokunaga A, Okano H, Kubo T. Mapping of notch activation during cochlear development in mice: implications for determination of prosensory domain and cell fate diversification. J Comp Neurol. 2006;497:502–518. doi: 10.1002/cne.20997. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Neves J, Parada C, Chamizo M, Giraldez F. Jagged 1 regulates the restriction of Sox2 expression in the developing chicken inner ear: a mechanism for sensory organ specification. Development. 2011;138:735–744. doi: 10.1242/dev.060657. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis. 2004;38:195–199. doi: 10.1002/gene.20017. [DOI] [PubMed] [Google Scholar]
- Pearson K. Mathematical Contributions to the Theory of Evolution. III. Regression, Heredity, and Panmixia. Philosophical Transactions of the Royal Society of London Series A, Containing Papers of a Mathematical or Physical Character. 1896;187:253–318. [Google Scholar]
- Radde-Gallwitz K, Pan L, Gan L, Lin X, Segil N, Chen P. Expression of Islet1 marks the sensory and neuronal lineages in the mammalian inner ear. J Comp Neurol. 2004;477:412–421. doi: 10.1002/cne.20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccomagno MM, Martinu L, Mulheisen M, Wu DK, Epstein DJ. Specification of the mammalian cochlea is dependent on Sonic hedgehog. Genes Dev. 2002;16:2365–2378. doi: 10.1101/gad.1013302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccomagno MM, Takada S, Epstein DJ. Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes Dev. 2005;19:1612–1623. doi: 10.1101/gad.1303905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubel EW, Fritzsch B. Auditory system development: primary auditory neurons and their targets. Annu Rev Neurosci. 2002;25:51–101. doi: 10.1146/annurev.neuro.25.112701.142849. [DOI] [PubMed] [Google Scholar]
- Swanson GJ, Howard M, Lewis J. Epithelial autonomy in the development of the inner ear of a bird embryo. Dev Biol. 1990;137:243–257. doi: 10.1016/0012-1606(90)90251-d. [DOI] [PubMed] [Google Scholar]
- Tischler J, Surani MA. Investigating transcriptional states at single-cell-resolution. Curr Opin Biotechnol. 2013;24:69–78. doi: 10.1016/j.copbio.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Verpy E, Leibovici M, Petit C. Characterization of otoconin-95, the major protein of murine otoconia, provides insights into the formation of these inner ear biominerals. Proc Natl Acad Sci U S A. 1999;96:529–534. doi: 10.1073/pnas.96.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DK, Kelley MW. Molecular mechanisms of inner ear development. Cold Spring Harbor perspectives in biology. 2012;4:a008409. doi: 10.1101/cshperspect.a008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung KY, Ruzzo WL. Principal component analysis for clustering gene expression data. Bioinformatics. 2001;17:763–774. doi: 10.1093/bioinformatics/17.9.763. [DOI] [PubMed] [Google Scholar]
- Zheng W, Huang L, Wei ZB, Silvius D, Tang B, Xu PX. The role of Six1 in mammalian auditory system development. Development. 2003;130:3989–4000. doi: 10.1242/dev.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05
06