Molecular-crowding effects on single-molecule RNA folding/unfolding thermodynamics and kinetics (original) (raw)
Significance
The cell cytoplasm is a multicomponent solution that is much more concentrated than typically sampled in conventional in vitro studies. The presence of cosolutes can strongly influence biomolecular folding due to “molecular-crowding” effects, which are poorly understood and yet crucial to understand. In this study, the thermodynamics/kinetics of “crowding” effects are probed at the single-molecule level for an isolated RNA tertiary interaction. We observe dramatic stabilization of the folded state and, for the first time (to our knowledge), establish the kinetic origin of molecular crowding to be dominated by entropic acceleration of folding.
Keywords: scaled particle theory, fluorescence, PEG
Abstract
The effects of “molecular crowding” on elementary biochemical processes due to high solute concentrations are poorly understood and yet clearly essential to the folding of nucleic acids and proteins into correct, native structures. The present work presents, to our knowledge, first results on the single-molecule kinetics of solute molecular crowding, specifically focusing on GAAA tetraloop–receptor folding to isolate a single RNA tertiary interaction using time-correlated single-photon counting and confocal single-molecule FRET microscopy. The impact of crowding by high–molecular-weight polyethylene glycol on the RNA folding thermodynamics is dramatic, with up to ΔΔ_G_° ∼ −2.5 kcal/mol changes in free energy and thus >60-fold increase in the folding equilibrium constant (_K_eq) for excluded volume fractions of 15%. Most importantly, time-correlated single-molecule methods permit crowding effects on the kinetics of RNA folding/unfolding to be explored for the first time (to our knowledge), which reveal that this large jump in _K_eq is dominated by a 35-fold increase in tetraloop–receptor folding rate, with only a modest decrease in the corresponding unfolding rate. This is further explored with temperature-dependent single-molecule RNA folding measurements, which identify that crowding effects are dominated by entropic rather than enthalpic contributions to the overall free energy change. Finally, a simple “hard-sphere” treatment of the solute excluded volume is invoked to model the observed kinetic trends, and which predict ΔΔ_G_° ∼ −5 kcal/mol free-energy stabilization at excluded volume fractions of 30%.
In recent years, the number of functional roles for noncoding RNAs has grown rapidly, forever changing the perception of RNA in biology (1). New activities attributed to RNA include gene regulation through self-splicing, ligand sensing, and silencing via RNA interference (2). Due to the well-known relationship between structure formation and function, many studies have focused on the kinetics and thermodynamics that govern both secondary and tertiary structure formation in RNAs (3). However, the intimate coupling of polyanionic RNA to the surrounding medium makes descriptions of the “RNA-plus-solution” system unusually complicated, requiring intense, ongoing efforts to understand quantitatively how complex solution mixtures perturb the dynamics and kinetics of RNA structure formation.
In the cell, many RNAs function in the cytoplasm, a heterogeneous solution of biopolymers (proteins and nucleic acids), inorganics (K+, Na+, Mg2+, Cl−, HCO3−, etc.), and other small cosolutes (amino acids, NTPs) (4). This multicomponent mixture differs substantially from the dilute, buffered, aqueous conditions in which biomolecular reactions are often studied in the laboratory. The presence of these cosolutes, however, is known to significantly affect the equilibrium properties and kinetics of RNA structure. In particular, cations interact strongly with the polyanionic RNA backbone and thus ion–nucleic acid interactions have been studied intensively (5–7). However, even in the absence of any strong intermolecular chemical interaction potential, other cosolutes can also influence the equilibrium and kinetics of macromolecules. The most relevant example in molecular biology can be seen in the cytoplasm, where proteins and nucleic acids exist in high concentrations (>350 mg/mL). By occupying a large fraction of the 3D solution, these cosolutes can significantly limit the space that RNAs have for conformational sampling, resulting in “molecular-crowding” effects (8). Due to the extreme differences in cosolute concentration between in vitro biochemical experiments and the crowded cytosol environment in vivo, a fundamental “first-principles” understanding of these effects is obviously crucial.
Previous studies have documented the stabilizing effects of molecular crowding on the equilibrium thermodynamics of biomolecular folding; however, the majority of the work has been focused on protein systems (9–11). In a more limited scope, excluded volume ideas have also been applied to the thermodynamics of nucleic acid systems, including DNA duplex/hairpin motifs, RNA ribozymes, and telomerase pseudoknot RNA (12–15). The observation common to all of these studies is that high–molecular-weight PEGs increase the thermodynamic stability of the folded, more compact nucleic acid structures, as observed via increase in _T_m or decrease in cation-titration transition midpoints (13, 14). Interestingly, despite considerable progress made by such efforts, molecular-crowding effects on nucleic acid folding have been limited to thermodynamic perspectives, with no clear picture emerging for the underlying kinetic origins of this stabilization, which reflects a major goal of the present study.
In this work, the coupling of an isolated GAAA tetraloop–receptor (Fig. 1_A_) to the surrounding solution is studied in mixtures of high molecular weight polyethylene glycols (PEGs) [average molecular weight () = 8,000 amu], which have often been used as a proxy for effects of a crowded intracellular environment (8, 13, 14). Specifically, PEGs are known to increase the bulk solution viscosity, while at the same time provide strong equilibrium stabilization for folded conformations of proteins and nucleic acids (ΔΔ_G_° < 0) (13, 14, 16, 17). One powerful advantage of the present studies is that, in addition to such shifts in equilibrium behavior, single-molecule FRET (smFRET) measurements also permit direct observation of the kinetic effects of PEG on conformational transitions for an isolated GAAA tetraloop–receptor tertiary interaction. Furthermore, the shifts in smFRET equilibrium and kinetic behavior have been explored as a function of temperature, which allow the crowding free-energy changes to be cleanly isolated in terms of their enthalpic and entropic contributions. Of particular interest, the present experiments identify the predominant thermodynamic origin of such crowding based equilibrium and kinetic behavior as a reduction in the unfolded RNA-plus-solution conformational entropy (8, 18, 19).
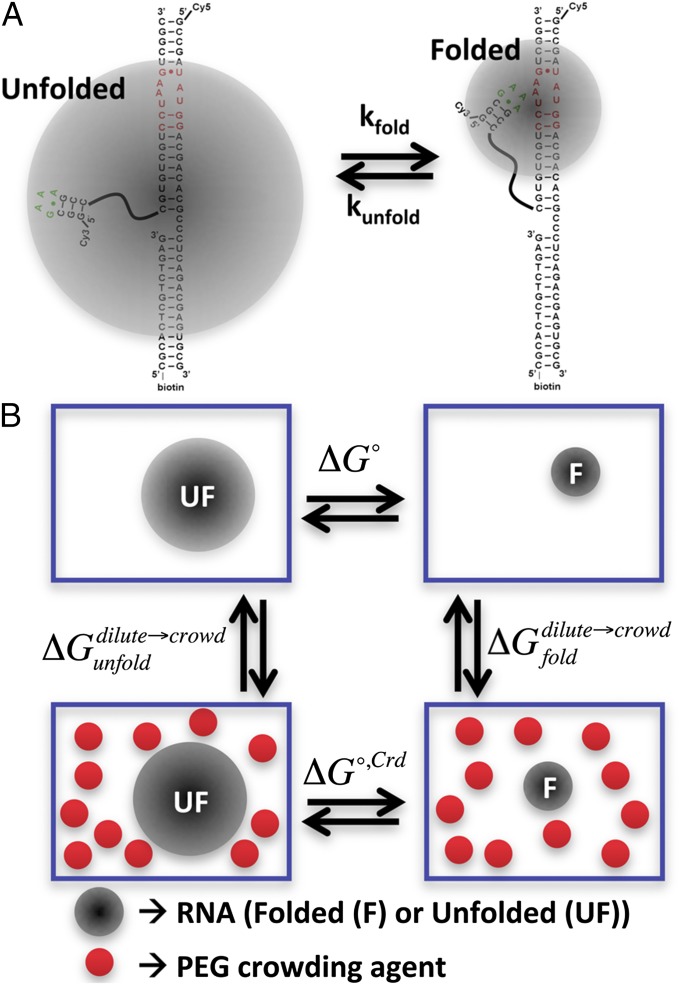
Fig. 1.
(A) The single-molecule GAAA tetraloop–receptor construct is shown. The shaded circles represent the approximate effective volumes occupied by the folded and unfolded states of the tetraloop and receptor. The GAAA tetraloop is connected to the receptor helix via a PEG6 linker. (B) A thermodynamic cycle describes the differential change in folding free energy between dilute and crowded solution conditions.
As a theoretical framework for analyzing and extracting insight from the experimental data, it is instructive to consider the simplest physical model for the RNA–PEG cosolute interactions. In particular, we exploit a “hard-sphere” interaction model (i.e., scaled particle theory) to approximate the limited conformational space that RNA samples when surrounded by PEG cosolutes (Fig. 1_B_) (8, 18, 19). This model was originally adapted by Minton to describe the biophysical effects of crowding conditions on multiple protein systems (8, 20). By virtue of limiting the space available, and restricting the RNA to occupy the cavities between the cosolutes, there is an implicit free-energy increase (i.e., “cost”) between the RNA molecule in dilute solution conditions and a solution crowded with large cosolutes.
At a more detailed level, where the RNA can occupy two distinct structures, as is the case with tertiary structure formation, the increase in free energy associated with putting the RNA system into a crowded solution results in a differential free energy change:
| ΔΔG=ΔGfolddilute→crowded−ΔGunfolddilute→crowded, | [1] |
|---|
which has the effect of shifting the folding equilibrium to the more compact state. Most importantly, we find that this differential free energy stabilization predominantly arises from entropic contributions and can be described by an analytical function of the relative “sizes,” modeled as hard-sphere radii, of the RNA states and cosolute. Although approximate, this simple model can be particularly useful when radius of gyration estimates of the various molecular species are known, thus enabling prediction of molecular-crowding effects for a variety of cosolute identities.
Results
Crowding Effects on the Tetraloop–Receptor Equilibrium.
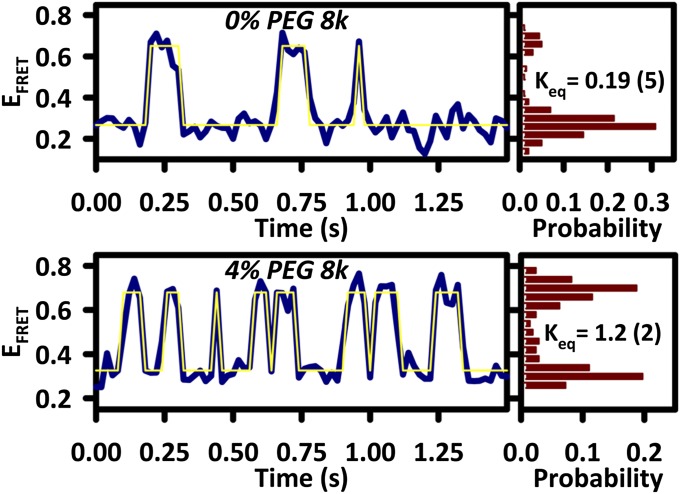
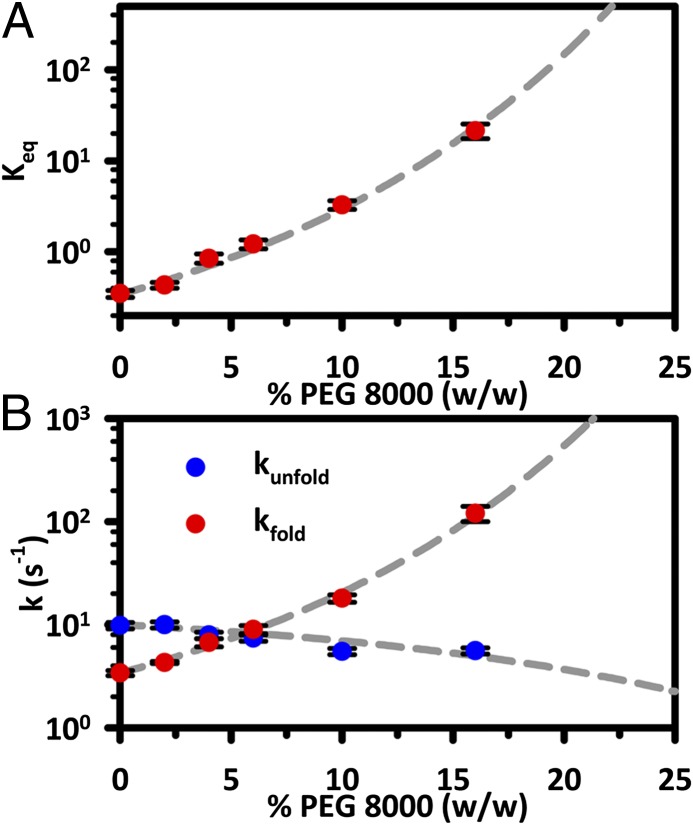
To develop and test a quantitative model of how large cosolutes “crowd” a single RNA tertiary interaction to modify its free-energy landscape, both (i) the equilibrium properties as well as (ii) the folding/unfolding kinetic rate constants of a doubly labeled GAAA tetraloop–receptor FRET construct (Fig. 1_A_) have been probed at the single-molecule level using photon color/polarization sorted time-correlated single-photon counting confocal microscopy (21–23). As illustrated in Fig. 2, _E_FRET time trajectories from these studies are shown for the tetraloop–receptor in buffered solution and with 4% (wt/wt) PEG 8000. The most obvious effect of adding PEG 8000 to the solution is a significant increase in the equilibrium constant, _K_eq = [folded]/[unfolded], from _K_eq = 0.19 (2) to 1.2 (2), as evidenced by histograms (right panels) for the individual _E_FRET trajectories. This experimental result is consistent with the many observations that high–molecular-weight PEGs preferentially stabilize the compact or folded states of biomolecules (9, 10, 13, 14). Even in a solution with relatively modest PEG 8000 content [4% (wt/wt)], this 6.3-fold change in _K_eq indicates a preferential stabilization of the folded tetraloop–receptor state by ΔΔ_G_° ∼ −1.1 kcal/mol (−1.8 RT). Such cosolute effects can be mapped out more explicitly by plotting _K_eq as a function of PEG 8000 concentration. As shown in Fig. 3_A_, the cosolute crowding effects result in a near-exponential increase in _K_eq as a function of [PEG 8000], with _K_eq = 0.34 (4) in buffered solution now rising by 62-fold to _K_eq = 21 (4) at 16% (wt/wt) PEG 8000. Such an increase corresponds to a preferential stabilization of the folded, more compact tetraloop–receptor state by ΔΔ_G_° ∼ −2.5 kcal/mol (−4.1 RT). By way of theoretical comparison, the dashed gray line in Fig. 3_A_ represents data fits with the simple scaled particle model, the details of which will be discussed later.
Fig. 2.
_E_FRET trajectories comparing 0% and 4% PEG 8000 reveal a significant increase in the frequency of excursions to the high _E_FRET state. Histograms (Left) show an increase in _K_eq by more than sixfold, even for only 4% PEG 8000 solutions.
Fig. 3.
(A) _K_eq is plot as a function of the PEG 8000 solution content. (B) The individual rate constants are shown in a logarithmic plot, highlighting the dramatic increase and much smaller decrease in _k_fold and _k_unfold, respectively.
Solute Crowding Effects on Tetraloop–Receptor Kinetics.
Although cosolute crowding effects on the equilibrium behavior are quite substantial, these data cannot speak to the kinetic origins of this increase. Specifically, the 62-fold increase in _K_eq does not identify whether it arises from cosolute (i) reduced RNA unfolding or (ii) enhanced RNA folding rate constants, or some combination of the two. Considerably more kinetic information for crowding induced stabilization may therefore be gained at the single-molecule level by examining the time domain dynamics of the _E_FRET trajectories themselves (see Fig. S1). As shown in Fig. 2, Upper, the tetraloop–receptor construct in buffer conditions spends most of the time in the unfolded (low _E_FRET) state, with only brief excursions into the folded (high _E_FRET) state. With 4% (wt/wt) PEG 8000 added to the solution, the trajectories change markedly (Fig. 2, Lower). There is a dramatic growth in frequency of excursions into the folded (high FRET) state, yet the mean duration of these excursions in the high _E_FRET state does not change appreciably. Thus, the time trajectories immediately reveal that the primary effect of PEG 8000-based crowding of the GAAA tetraloop–receptor is to increase the folding rates, whereas the unfolding behavior remains essentially constant (see Fig. S1).
The trends in the rate constant dependence on PEG 8000 percentage (wt/wt) are mapped out more completely in Fig. 3_B_. In this plot, _k_fold clearly reveals a superexponential increase [3.4 (0.5) s−1 → 120 (10) s−1] with PEG 8000 concentration for only a less than twofold decrease in _k_unfold [∼10 (1) s−1 → 5.6 (0.8) s−1]. Thus, the dramatic cosolute induced growth in _K_eq is dominated by the increase in _k_fold, with only minor contributions due to a subtle decrease in _k_unfold. This is not an obvious result, as one could have anticipated crowding as a mechanism for suppression of the unfolding process, thereby enhancing the stability of the native tetraloop–receptor tertiary interaction. The better physical explanation consistent with the data would be that the transition state for folding represents a more compact conformation than the unfolded state; thus, the free-energy barrier necessary to achieve the transition state decreases with cosolute crowding. Again, by way of comparison with theory, the dashed gray lines in Fig. 3_B_ represent least-squares fits of the data to a simple scaled particle model, for which the agreement is surprisingly good. Note that these results are fully consistent with kinetic trends predicted from coarse-grained molecular dynamics simulations for folding of the WW peptide domain (24, 25).
Temperature Studies Identify the Predominant Role of Entropy in Molecular Crowding.
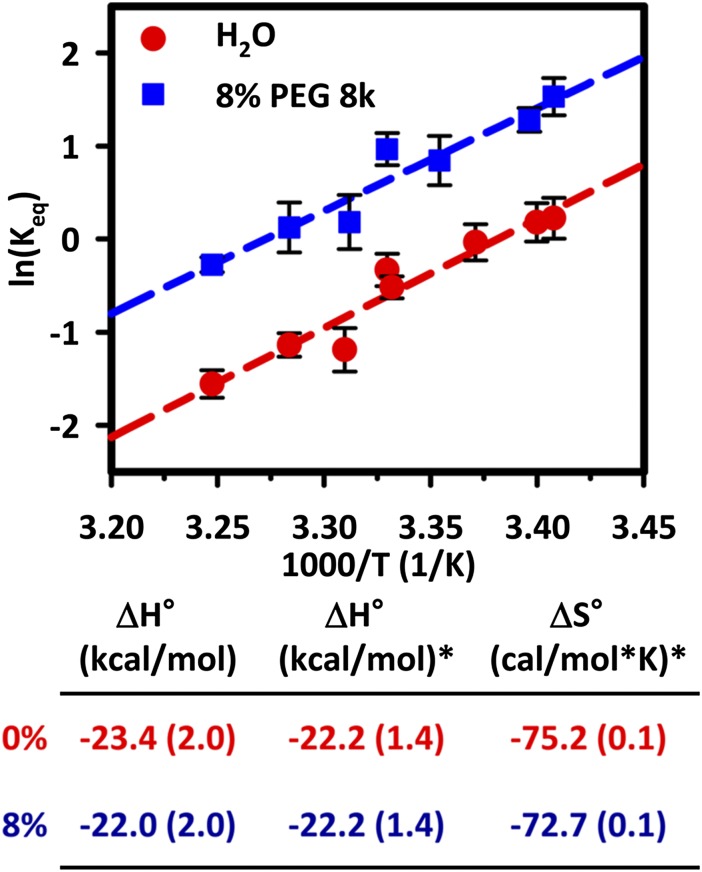
The thermodynamic origin of the observed crowding induced stabilization (ΔΔ_G_° < 0) of the tetraloop–receptor is probed further with temperature-dependent measurements in both aqueous solutions and in an 8% (wt/wt) PEG 8000 solution (Fig. 4). With temperature as an experimental variable, the observed change in the folding free energy [Δ_G_°= −_RT_ln(_K_eq)] can be deconstructed into its enthalpic (Δ_H_°) and entropic (−_T_Δ_S_°) contributions. Fig. 4 displays the results in the form of a plot of ln(_K_eq) vs. 1/T, which for simple Van’t Hoff analysis should yield a straight line with slope and intercept of −Δ_H_°/R and Δ_S_°/R, respectively. By visual inspection, the primary effect of adding PEG 8000 to the solution is to shift the intercepts to higher entropy values, with minimal or no change to the slope. Recast in thermodynamic terms, a parallel upward shift with no slope change corresponds to free energy stabilization driven by a reduction in the entropic cost of folding with no change in reaction enthalpy. In good agreement with this interpretation, the enthalpies measured from independent linear least-squares fits to the two datasets match each other within experimental uncertainty, Δ_H_° = −23.4 (2.0) and −22.0 (2.0) kcal/mol for aqueous solution and 8% (wt/wt) PEG 8000, respectively. Note that these enthalpy values are in excellent agreement with those reported for a similar GAAA-tetraloop construct with a U7 linker (23). To provide more reliable estimates of Δ_S_° and ΔΔ_S_°, the datasets have also been fit assuming a common enthalpy, resulting in entropic folding costs of Δ_S_° = −75.2 (1) and −72.7 (1) cal⋅mol−1·K for aqueous and 8% (wt/wt) PEG 8000, respectively, which yields a positive differential entropy change of ΔΔ_S_° = +2.5 (2) cal⋅mol−1⋅K−1. By itself, this entropy difference translates into ΔΔ_G_° (300 K) = −0.75 kcal/mol, which, compared with room temperature equilibrium data for the same 8% excluded volume fraction reveals that ΔΔ_G_° ∼ −1 kcal/mol. Thus, differential entropy accounts for roughly 75% of the total change in Δ_G_°, clearly providing additional support for an entropically dominated treatment of molecular crowding. Finally, we note that the vanishing of the enthalpic contributions does not imply an absence of interaction between PEG 8000 and the tetraloop–receptor construct, but rather that any additional enthalpic crowding effects are nearly equivalent for the folded and unfolded states. Again, these results are consistent with the entropic stabilization mechanism described for the WW peptide domain and based on a coarse-grained molecular-dynamics model (24).
Fig. 4.
Van’t Hoff plot for the tetraloop–receptor equilibrium in aqueous buffer and 8% PEG 8000, where the parallel shift upwards in y intercept reveals that crowding-induced stabilization by PEG 8000 occurs predominantly through reduction in the entropic cost of folding. Left-hand column shows enthalpy changes consistent with ΔΔ_H_° = 0 within experimental error. The second and third columns (*) reflect fits with ΔΔ_H_° = 0 to break parameter correlation and generate a more reliable prediction for entropic lowering of the free-energy difference, i.e., −_T_ΔΔ_S_°.
Discussion
Accelerated Folding Leads to Preferential Stabilization of Compact States.
As a first step, we have explored the isolated GAAA tetraloop–receptor equilibrium behavior as a function of PEG 8000 solution content. Figs. 2 and 3 clearly illustrate that, even under mild crowding conditions [4% (wt/wt) PEG 8000; Fig. 2], there is a significant preferential stabilization by ΔΔ_G_° ∼ −1.1 kcal/mol for folded (high _E_FRET) state of the tetraloop–receptor. Extended to more crowded conditions (Fig. 3_A_), _K_eq is found to be exponentially sensitive to percentage (wt/wt) PEG 8000, resulting in crowding-induced stabilizations up to as much as ΔΔ_G_° ∼ −2.5 kcal/mol. It is worth noting that such effects for this isolated GAAA tetraloop–receptor tertiary motif are remarkably large. By way of comparison, detailed equilibrium thermodynamic studies of DNA crowding effects in PEG 8000 solutions by Knowles et al. (14) found maximal stabilizations only of order ΔΔ_G_° ∼ −1 kcal/mol for formation of 12-nt DNA duplexes.
However, it is the kinetic measurement capabilities of this single-molecule experiment that permit these equilibrium (_K_eq = _k_fold/_k_unfold) changes to be analyzed in more detail. As the PEG 8000 content in the solution is increased, _k_fold increases by ∼40-fold, whereas _k_unfold decreases <2×. Thus, the large crowding-induced shifts observed in _K_eq are dominated by the folding rather than unfolding kinetics of the RNA tertiary structure. This is again not obvious, because an alternative model based on preferential enthalpic stabilization of the folded state would predict a crowding-induced suppression of the unfolding rate.
The increase in rate constant with PEG crowding provides valuable information on the free-energy barrier to folding of the tetraloop–receptor tertiary interaction. In the context of transition state theory, the reactants remain in steady-state equilibrium with the transition state conformation, which, just as for the fully folded vs. unfolded conformations, can be differentially influenced by presence of the cosolute (Fig. 1_B_). In the forward (folding) direction, the rapid increase in _k_fold reveals preferential stabilization of the transition state free energy relative to the unfolded state. In the reverse (unfolding) direction, however, the folded and transition state conformations appear to be stabilized under crowding conditions by similar amounts. The data therefore support a physical picture in which the unfolded and transition states for the tetraloop–receptor represent substantially different levels of compaction, with the transition state more closely approximating the size of the fully folded state.
Furthermore, the temperature-dependent equilibrium constants (Fig. 4) and kinetic rates (Fig. S2) help parse the free-energy shifts into enthalpic and entropic contributions, illustrating a predominantly entropic origin for PEG 8000-induced crowding phenomena in tetraloop–receptor construct. One obvious interpretation would be that the transition state for folding has achieved a similar level of compaction as the completely folded state, i.e., a “late” transition state. However, there must be competing influences, because previous single-molecule thermodynamic studies of the tetraloop–receptor reveal a transition state free energy barrier to folding with predominantly entropic rather than enthalpic contributions, which would imply an “early” barrier under dilute aqueous conditions. One explanation consistent with the data would be that the transition state for achieving the folded tetraloop–receptor tertiary interaction shifts systematically to a more compact conformation as a function of the cosolute crowding conditions. It would be particularly interesting to test these model interpretations with detailed molecular-dynamics simulations and/or more sophisticated statistical theories of RNA folding dynamics as a function of crowding environment. As a first step in this direction, these experimental trends are examined below in the context of a simple hard-sphere statistical model.
Scaled Particle Theory.
The present single-molecule studies have revealed remarkably strong effects of PEG 8000 crowding on both the equilibrium and kinetic behavior of RNA folding, with the dramatic increase in folding rate constants controlling the overall impact. In conjunction with temperature-dependent studies, the results indicate the crowding-induced changes in the overall free-energy differences for tetraloop–receptor folding to be dominated by entropic contributions, with the negligible enthalpic shifts indicating a nearly balanced cosolute interaction with both folded and unfolded states. These observations suggest that the essential physics of such crowding-induced changes for RNA folding of the tetraloop–receptor interaction can be captured within a minimal statistical framework of noninteracting particles.
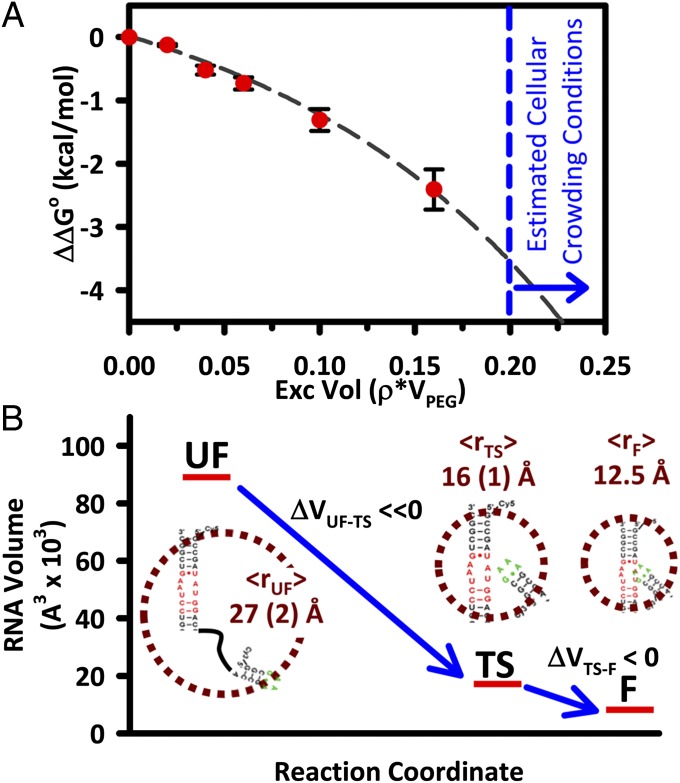
One simple yet powerful statistical treatment of noninteracting cosolutes is so-called “scaled particle theory,” based on the simplifying assumptions of (i) ideal solutions of cosolute particles and (ii) hard-sphere interactions between solute pairs with effective radii for each of the cosolutes. This permits development of analytical expressions for incremental free energies per molecule associated with insertion of a spherical solute of radius r i into a hard-sphere solution of cosolutes of radius r c at a given volume fraction φ. From the equilibrium measurements in Fig. 3_A_, the free-energy change, Δ_G_° = −RT ln(K_eq), as well as ΔΔ_G_° = −_RT ln(_K_eq/_K_°eq) can be calculated for each set of solution conditions. As summarized in Fig. 5_A_, ΔΔ_G_° is a systematically decreasing function of increasing excluded volume fraction, φ=ρV, where ρ and V are the density and effective volume of the PEG solute. In the context of scaled particle theory, this can be recast in terms of:
where ρ c is the number density of the crowding cosolute particles. Because the experimental ΔΔ_G_° values in Fig. 5_A_ are defined relative to aqueous buffered conditions, the plot starts at 0 and decreases with increasing volume fraction. This is consistent with the expectation of preferential stabilization of the folded, more compact state in a crowded solution environment.
Fig. 5.
(A) Stabilization plots for the GAAA tetraloop–receptor folding equilibrium with respect to dilute buffer solution, revealing a nonlinear dependence of ΔΔ_G_° on excluded volume. (B) Fits to the equilibrium and kinetic data reflect the effective radii of the tetraloop–receptor unfolded/folded (UF, F) and transition states (TS), respectively, as well as reveal a large loss of free volume between UF and TS states followed by a much smaller loss between the TS and F states.
As first demonstrated by Lebowitz and co-workers, the following analytical expression for ΔGidil→crd/RT can be derived:
| ΔGidil→crd/RT=−ln(1−φ)+(φ1−φ)(z3+3z2+z)+(φ1−φ)2(3z3+92z2)+(φ1−φ)3(3z3), | [3] |
|---|
where z = r i/r c is the dimensionless ratio of “crowded” to “crowder” scale lengths (18, 19). If we associate r c with the effective radius of the crowding PEG 8000 solute, and r i with the corresponding radius of a particular folded (_r_fold), unfolded (_r_unfold) or transition state (_r_tst) conformation of the tetraloop–receptor, this yields a similar analytical expression for ΔΔ_G_°/RT, which in turn can be least-squares fitted to experiment results. The connection between the scaled particle theory model and the experimental parameters is given by the following:
| ΔΔGo=ΔGo,Crd−ΔGo=ΔGfolddil→crd−ΔGunfolddil→crd. | [4] |
|---|
For the present fits, the effective radius of PEG 8000 (r c = 15 Å) is taken from literature values for the radius of gyration (26). We can roughly estimate _r_fold for the folded tetraloop–receptor (_r_fold = 12.5 Å) from the crystal structure 1GID by computing the volume enclosed by the solvent accessible surface area (probe radius, 1.4 Å) for nucleotides 147–156 (tetraloop) and 220–229 and 245–253 (receptor) (27). Hard-sphere radius estimates for the unfolded tetraloop–receptor are not available and instead characterized by least-squares analysis of the data. As summarized in Fig. 5, the scaled particle theoretical predictions for ΔΔ_G_° prove to be in surprisingly good agreement with experiment, predicting an effective radius of _r_unfold ∼ 27 (1) Å for the unfolded tetraloop–receptor state. Although the approximate nature of such a hard-sphere model does not warrant rigorous interpretation of length parameters, it is nevertheless worth noting that _r_unfold is comparable to the end-to-end distance (∼24 Å) of the PEG6 linker between the free tetraloop and receptor regions.
We can take this scaled particle treatment one step further and in a similar manner analyze the dependence of the folding/unfolding kinetic rate constants on PEG 8000 molecular crowding (see Fig. S2). Specifically, we start with the simple transition state theory expression:
where Δ_G_‡ represents the free-energy barrier and ν is the attempt frequency along the reaction coordinate to surmount this barrier (22, 23, 28). By scaling the rate constants to the values at zero PEG 8000 concentrations, we can extract Δ[ln(k(T)/ν)] and therefore ΔΔ_G_‡/RT for the tetraloop–receptor folding/unfolding directions as a function of excluded volume fraction φ. This analysis is summarized in Fig. 3_B_, where least-squares fits to _k_fold and _k_unfold keep _r_fold and _r_unfold fixed as determined previously, but allow the effective radius of the transition state conformation (_r_tst) to float. Independent fits to both _k_fold and _k_unfold datasets provide self-consistent results for the transition state radii, with _r_tst (folding) ∼ 16.0 (3) Å and _r_tst (unfolding) ∼ 16.2 (5) Å. The resulting effective hard-sphere volumes for these three states are illustrated along the reaction coordinate in Fig. 5_B_, with the relative radii indicated in the Insets. Once again, despite the obvious approximations implicit in any such hard-sphere model, it is worth noting that the effective radius and volume of the transition state lie much closer to that of the folded tetraloop–receptor construct. This picture is therefore consistent with our previous interpretation of the kinetics, specifically in terms of molecular crowding effects shifting the transition state to a more compact conformation “late” along the reaction coordinate.
Molecular Crowding Model Provides a Physical Basis for Stabilization.
In the absence of any differential free-energy stabilization of folded vs. unfolded conformations, traditional continuum ideas of friction on molecular length scales (e.g., Kramers’ theory and molecular viscosity) would simply predict a downward shift in the forward and reverse rate constants by precisely the same factor (29). The thermodynamics and kinetics of molecular crowding, therefore, where equilibrium constants can shift by orders of magnitude and forward/reverse rates can be strongly influenced up or down even for ideal noninteracting particles, requires a different physical picture. Fortunately, the ideas of excluded volume and molecular crowding have been explored and are well developed for protein folding systems. Here, we invoke these ideas to explain the experimental observations of our isolated RNA tertiary interaction under crowded conditions (8). From Fig. 3_B_, it is clear that PEG 8000 has the strongest influence on the forward folding free energy barrier, resulting in a large differential increase in _k_fold. With finite excluded volume (Fig. 1_B_), it can be envisioned that the tetraloop–receptor occupies cavities that form between the PEG 8000 molecules. Because forming larger cavities is more entropically costly than smaller ones, RNA conformations with larger effective radii (e.g., unfolded structures) will both perturb and be perturbed to a greater degree with respect to dilute aqueous solutions than more compact (i.e., folded) structures. According to the approximate hard-sphere model fits summarized in Figs. 3 A and B and 5_A_, the unfolded state is characterized by the largest effective radius [_r_unfold ∼ 27 (2) Å], which decreases substantially at the transition state [_r_tst ∼ 16 (1) Å], followed by an additional, but more modest decrease to the final folded state (_r_fold ∼ 12.5 Å). The kinetic data therefore confirm that the greater decrease in free volume for the tetraloop–receptor occurs between the unfolded and transition states, supporting previous claims that the transition state for RNA folding may be relatively compact for similar constructs (22, 23).
As an additional comment, we note that, for all PEG 8000 solutions, the viscosities increase substantially, which in a Kramers’ theory of reaction rates for a continuum solvent model would suggest a decrease of both folding and unfolding rate constants, absent any barrier free-energy changes (29). However, the kinetic trends observed in PEG 8000 solutions clearly illustrate the breakdown of a simple Kramers’ theory description in terms of molecular friction, which motivates the use of an excluded volume model to more accurately describe the experimental results. However, it is worth noting that there must be a smooth transition from continuum behavior to one of molecular crowding, depending on relative sizes of the cosolutes. For example, the scaled particle model clearly must break down in the limit of very small cosolutes (e.g., glycerol), where the size of the particle is now many times smaller than the length scale for folding/unfolding molecular motion.
Interestingly, the transition between these two regimes has recently begun to be mapped out by Knowles et al. for DNA duplex and hairpin constructs (14). They found that PEGs cosolutes with 10 monomer units and higher ( > 456 amu) led to an overall stabilizing influence on DNA hybridization (Δ_G_° < 0), in qualitative agreement with the present PEG 8000 ( = 8,000 amu) results. However, PEGs below 10 monomer units ( < 456 amu) were found to have a slightly destabilizing effect on DNA hybridization (Δ_G_° > 0), whereas Eq. 3 would incorrectly predict the opposite in the limit of small r c. Thus, kinetic crowding effects clearly depend on the ratio of cosolute lengths with respect to the scale of molecular motion, with the observed transition by Knowles et al. around ∼10 PEG monomer units reflecting the specific size of their 12-nt DNA constructs under investigation (14). This result also shows that the approximate nature of the “hard-sphere” picture and that “soft” or chemical interactions likely contribute to the observed thermodynamics to some degree. Additionally, this model has clear limitations for extremely high concentrations of cosolutes, where the RNA can fold/unfold in cavities with significant thermodynamic perturbations to both folded and unfolded structures. Indeed, such effects have been observed in cavity-constrained protein-folding simulations, where the stability of native folded structures are perturbed as well (30). Under such high concentration conditions, however, one would expect to see enthalpic contributions to the thermodynamics of folding stabilization; based on experimental results up to 8% excluded volume, we have apparently not yet reached such a regime.
However, the most important predictions of such a simple model are the estimated ΔΔ_G_° values for excluded volume fractions under cellular conditions, where potential molecular-crowding materials (nucleic acids, proteins, lipids) are thought to occupy ∼20–30% of the total volume (31). The results are shown by the blue dashed line in Fig. 5_A_, which suggest molecular-crowding effects to be on the order of ΔΔ_G_° = −4.5 to −5 kcal/mol. This is a remarkably large effect, as a differential stabilization of the folded state by −5 kcal/mol would correspond to an increase in the folding equilibrium constant by >4,000-fold. Furthermore, because the present study suggests that such free-energy changes arise predominantly from increase in the folding rate constant, this would imply that _k_fold could be increased by as much as factors of 103. Such effects may be especially critical in RNA-based cellular processes where the folding rate of a specific tertiary contact can control the overall rate of enzymatic cleavage, such as in the case of the hammerhead ribozyme (32). For such systems, one might anticipate that molecular crowding could therefore substantially influence both the rates and preferred directions of complex biochemical pathways in the cell (8, 15).
Conclusions
In this work, the origins of a large (∼60×), molecular-crowding–based, preferential stabilization of the isolated tetraloop–receptor folded state have been explored at the single-molecule level. Kinetic data reveal a strong acceleration of _k_fold, which accounts for nearly the entire change in _K_eq. The importance of conformational size on these effects is supported by temperature-dependent measurements that illustrate molecular crowding to be primarily entropic rather than enthalpic in both thermodynamic and kinetic origin. To interpret the observed kinetics and equilibria in PEG 8000 solutions, a hard-sphere picture is invoked, for which the presence of the PEG cosolutes restricts the unfolded conformational space of the tetraloop–receptor RNA-plus-PEG cosolute system. These results clearly highlight the need for further single-molecule studies of RNA folding kinetics under more physiologically relevant solution conditions (e.g., with proteins, cellular extracts, or in vivo), to develop and test more sophisticated theoretical models for molecular crowding contributions to folding thermodynamics and kinetics.
Materials and Methods
The SI Text contains full descriptions of all experimental procedures including RNA construct assembly, sample preparation, and single-molecule data acquisition. A detailed discussion of kinetic analysis of single-molecule _E_FRET trajectories is also included (Fig. S1). Additionally, the experimental data and a description of the temperature-dependent kinetic measurements is included.
Supplementary Material
Supporting Information
Acknowledgments
N.F.D. acknowledges the National Research Council for postdoctoral fellowship support. E.D.H. received funding from National Institutes of Health Molecular Biophysics Training Grant T32 GM-065103. Funding for this work was provided by National Science Foundation Grants CHE 1266416 and PHY 1125844 and the National Institute for Standards and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.T. is a guest editor invited by the Editorial Board.
References
- 1.Atkins JF, Gesteland RF, Cech T. RNA Worlds: From Life's Origins to Diversity in Gene Regulation. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2011. [Google Scholar]
- 2.Butcher SE, Pyle AM. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc Chem Res. 2011;44(12):1302–1311. doi: 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- 3.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 4.Sear RP. The cytoplasm of living cells: A functional mixture of thousands of components. J Phys Condens Matter. 2005;17(45):S3587–S3595. [Google Scholar]
- 5.Fiore JL, Hodak JH, Piestert O, Downey CD, Nesbitt DJ. Monovalent and divalent promoted GAAA tetraloop-receptor tertiary interactions from freely diffusing single-molecule studies. Biophys J. 2008;95(8):3892–3905. doi: 10.1529/biophysj.108.134346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Downey CD, et al. Metal ion dependence, thermodynamics, and kinetics for intramolecular docking of a GAAA tetraloop and receptor connected by a flexible linker. Biochemistry. 2006;45(11):3664–3673. doi: 10.1021/bi0520941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodson SA. Metal ions and RNA folding: A highly charged topic with a dynamic future. Curr Opin Chem Biol. 2005;9(2):104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Zhou HX, Rivas GN, Minton AP. Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roque A, Ponte I, Suau P. Macromolecular crowding induces a molten globule state in the C-terminal domain of histone H1. Biophys J. 2007;93(6):2170–2177. doi: 10.1529/biophysj.107.104513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stagg L, Zhang SQ, Cheung MS, Wittung-Stafshede P. Molecular crowding enhances native structure and stability of alpha/beta protein flavodoxin. Proc Natl Acad Sci USA. 2007;104(48):18976–18981. doi: 10.1073/pnas.0705127104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou HX. Protein folding and binding in confined spaces and in crowded solutions. J Mol Recognit. 2004;17(5):368–375. doi: 10.1002/jmr.711. [DOI] [PubMed] [Google Scholar]
- 12.Denesyuk NA, Thirumalai D. Crowding promotes the switch from hairpin to pseudoknot conformation in human telomerase RNA. J Am Chem Soc. 2011;133(31):11858–11861. doi: 10.1021/ja2035128. [DOI] [PubMed] [Google Scholar]
- 13.Kilburn D, Roh JH, Guo L, Briber RM, Woodson SA. Molecular crowding stabilizes folded RNA structure by the excluded volume effect. J Am Chem Soc. 2010;132(25):8690–8696. doi: 10.1021/ja101500g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knowles DB, LaCroix AS, Deines NF, Shkel I, Record MT., Jr Separation of preferential interaction and excluded volume effects on DNA duplex and hairpin stability. Proc Natl Acad Sci USA. 2011;108(31):12699–12704. doi: 10.1073/pnas.1103382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakano S, Karimata HT, Kitagawa Y, Sugimoto N. Facilitation of RNA enzyme activity in the molecular crowding media of cosolutes. J Am Chem Soc. 2009;131(46):16881–16888. doi: 10.1021/ja9066628. [DOI] [PubMed] [Google Scholar]
- 16.Lambert D, Draper DE. Effects of osmolytes on RNA secondary and tertiary structure stabilities and RNA-Mg2+ interactions. J Mol Biol. 2007;370(5):993–1005. doi: 10.1016/j.jmb.2007.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakano S, Karimata H, Ohmichi T, Kawakami J, Sugimoto N. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J Am Chem Soc. 2004;126(44):14330–14331. doi: 10.1021/ja0463029. [DOI] [PubMed] [Google Scholar]
- 18.Reiss H, Frisch HL, Lebowitz JL. Statistical mechanics of rigid spheres. J Chem Phys. 1959;31(2):369–380. [Google Scholar]
- 19.Lebowitz JL, Helfand E, Praestga E. Scaled particle theory of fluid mixtures. J Chem Phys. 1965;43(3):774. [Google Scholar]
- 20.Minton AP. Macromolecular crowding. Curr Biol. 2006;16(8):R269–R271. doi: 10.1016/j.cub.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 21.Fiore JL, Holmstrom ED, Fiegland LR, Hodak JH, Nesbitt DJ. The role of counterion valence and size in GAAA tetraloop-receptor docking/undocking kinetics. J Mol Biol. 2012;423(2):198–216. doi: 10.1016/j.jmb.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Fiore JL, Holmstrom ED, Nesbitt DJ. Entropic origin of Mg2+-facilitated RNA folding. Proc Natl Acad Sci USA. 2012;109(8):2902–2907. doi: 10.1073/pnas.1114859109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmstrom ED, Fiore JL, Nesbitt DJ. Thermodynamic origins of monovalent facilitated RNA folding. Biochemistry. 2012;51(18):3732–3743. doi: 10.1021/bi201420a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheung MS, Klimov D, Thirumalai D. Molecular crowding enhances native state stability and refolding rates of globular proteins. Proc Natl Acad Sci USA. 2005;102(13):4753–4758. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung MS, Thirumalai D. Effects of crowding and confinement on the structures of the transition state ensemble in proteins. J Phys Chem B. 2007;111(28):8250–8257. doi: 10.1021/jp068201y. [DOI] [PubMed] [Google Scholar]
- 26.Thiyagarajan P, Chaiko DJ, Hjelm RP. A neutron-scattering study of poly(ethylene glycol) in electrolyte-solutions. Macromolecules. 1995;28(23):7730–7736. [Google Scholar]
- 27.Cate JH, et al. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273(5282):1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 28.Fiore JL, Kraemer B, Koberling F, Edmann R, Nesbitt DJ. Enthalpy-driven RNA folding: Single-molecule thermodynamics of tetraloop-receptor tertiary interaction. Biochemistry. 2009;48(11):2550–2558. doi: 10.1021/bi8019788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagen SJ. Solvent viscosity and friction in protein folding dynamics. Curr Protein Pept Sci. 2010;11(5):385–395. doi: 10.2174/138920310791330596. [DOI] [PubMed] [Google Scholar]
- 30.Zhou HX, Dill KA. Stabilization of proteins in confined spaces. Biochemistry. 2001;40(38):11289–11293. doi: 10.1021/bi0155504. [DOI] [PubMed] [Google Scholar]
- 31.Fulton AB. How crowded is the cytoplasm? Cell. 1982;30(2):345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- 32.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10(9):708–712. doi: 10.1038/nsb959. and erratum (2003) 10(10):872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information