Isolation of single-base genome-edited human iPS cells without antibiotic selection (original) (raw)
. Author manuscript; available in PMC: 2014 Sep 1.
Published in final edited form as: Nat Methods. 2014 Feb 9;11(3):291–293. doi: 10.1038/nmeth.2840
Abstract
Precise editing of human genomes in pluripotent stem cells by homology-driven repair of targeted nuclease-induced cleavage has been hindered by the difficulty of isolating rare clones. We developed an efficient method to capture rare mutational events, enabling isolation of mutant lines with single-base substitutions without antibiotic selection. This method facilitates efficient induction or reversion of mutations associated with human disease in isogenic human induced pluripotent stem cells.
Human genetics can be challenging to study in vitro, since the difference between health and disease can be determined by a single point mutation1, 2. Traditional methods to generate mutant cell lines use antibiotic resistance markers, leaving a genetic “scar” that can interfere with studying the resulting phenotype. The advent of human induced pluripotent stem (iPS) cells3, 4 and site-specific nucleases5 has revolutionized our ability to make genetic models that exactly reflect patients' pathological mutations, or to precisely revert pathological mutations to a healthy sequence. A major challenge of nuclease-driven precise mutagenesis is that isolating rare recombinant iPS cell clones is difficult without antibiotic selection. Furthermore, using high levels of nuclease or increasing nuclease activity to attempt to circumvent this problem runs the risk of reducing the fidelity of mutagenesis6-9.
In certain cases the efficiency and fidelity of a nuclease may be so high that direct cloning of the mutant can be done immediately after nuclease treatment. However, we and others have found that many single-base substitutions or deletions often occur at frequencies below 1%10-12, making direct cloning laborious. Furthermore, the expense of iPS cell characterization (including genome sequencing), and the inability to “breed-out” off-target mutations (unlike in model organisms) suggest that lower concentrations of nucleases should be used or that nucleases should be replaced with nickases to avoid off-target mutations13, 14. However, using nickases to improve fidelity also lowers efficiency so that mutants are increasingly rare13,14. Therefore, genome engineering is faced with a logistical challenge: precise mutagenesis with the highest fidelity results in rarer mutagenic events, but isolating a rare mutant cell without antibiotic resistance amid the hundreds of otherwise identical cells is exceedingly difficult.
To help solve this problem, we developed a method that allows efficient detection of a mutation, sib-selection (originally a yeast cloning method)15, and isolation of rare scarless clones with the desired mutation. We used the recently developed droplet digital PCR (ddPCR)16 for this purpose, and adapted iPS cell growth conditions so that rare mutant clones can be isolated with unprecedented efficiency. We used our method to introduce disease-associated point mutations into five genes (PHOX2B, PKP2, RBM20,PRKAG2, and BAG3; see Supplementary Table 1 for details). For each gene, we designed a pair of TALE nucleases (TALENs) or a guide RNA (gRNA) to target sequences close to the desired mutation site and a 60-nt single-stranded oligonucleotide DNA donor containing the mutation.
For our method to work, we needed to efficiently identify a cell harboring a single point mutation among hundreds of parental cells. For this purpose, we combined the TaqMan PCR assay with ddPCR. We designed a common primer pair and allele-specific TaqMan probes conjugated with different fluorophores that distinguish between the wild-type and mutagenized DNA sequences to screen for the mutagenesis events. Only on-target homologous recombination (HR) events are detected with this strategy because the PCR amplicons include the donor sequence and genomic sequences beyond the HR junction (Fig. 1 and Supplementary Fig.1a). We first optimized the PCR annealing and extension temperature (Supplementary Fig. 2). Using the plasmid template mixtures of the wild-type and mutant alleles, we found that this adaptation of ddPCR robustly detects as little as 0.1% of the mutant allele in the wild-type background, a level of sensitivity 100 times greater than that of TaqMan PCR alone (Fig. 1b and Supplementary Fig. 1b-e). ddPCR robustly detected TALEN-induced point mutagenesis using the PHOX2B- and PKP2-targeted TALENs in HEK293T cells, and was more sensitive than the Surveyor assay (Supplementary Fig. 3). ddPCR also detected mutagenesis induced by clustered, regularly interspaced, short palindromic repeats (CRISPR)–CRISPR-associated (Cas) system17, 18 of the RBM20 gene in HEK293T cells (Supplementary Fig. 4).
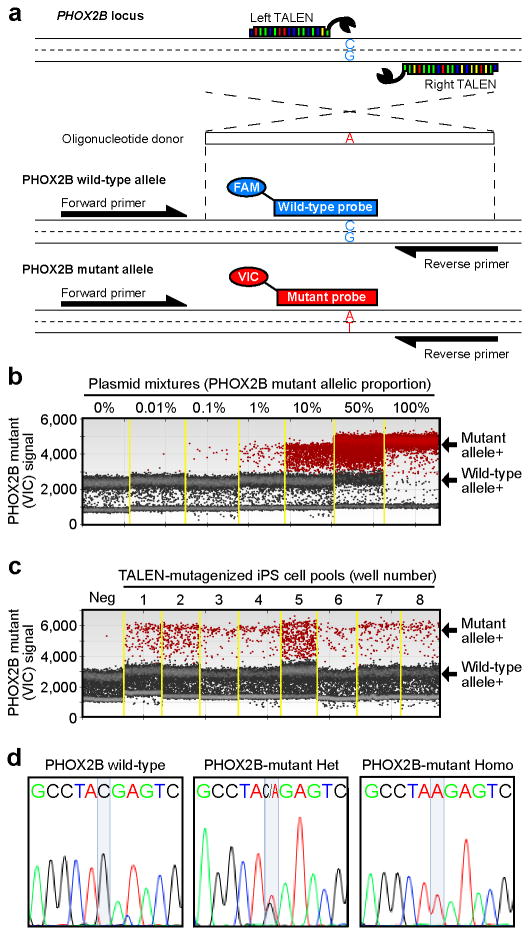
Figure 1. Using ddPCR to detect a point mutation in human iPS cells.
(a) Our mutagenesis and mutation detection strategy for PHOX2B is shown. We designed a pair of TALENs that target the mutation site and a 60-nt oligonucleotide DNA donor with the C→A substitution in the middle. The position of a common primer pair and allele-specific probes conjugated with different fluorophores are indicated. (b) ddPCR with plasmid mixtures. Mixtures containing the indicated concentrations of the mutant allele in a wild-type background were analyzed. Yellow lines indicate borders between different samples. Red dots represent droplets containing the mutant allele. (c) ddPCR with genomic DNA from iPS cells harboring the mutant allele induced by the PHOX2B TALENs. The mutagenized iPS cells were plated in a 96-well plate and 8 wells were analyzed for the mutant and wild-type alleles. An unrelated TALEN pair targeting the PRKAG2 locus was used as a negative control (Neg). (d) Sequencing results of isolated clones after PHOX2B mutagenesis.
Sib-selection is commonly used in yeast genetics to isolate a rare cell type: a population of cells is subdivided and then the ‘mutation-containing’ subpopulation is further subdivided until a rare cell type is purified15. We optimized the conditions for growing iPS cells in 96-well plates. We collected a portion of the cells for genomic DNA analysis and cryopreserved the remaining cells in 96-well format. The quantitative nature of ddPCR enables identification of the well containing the most mutant cells. After recovering cells from the corresponding well in the cryopreserved 96-well plate, we subdivided these cells into another 96-well plate. The stochastic nature of the distribution of mutant cells allowed us to enrich the progeny of mutant cells with each round of selection. Up to ten different iPS cell lines with engineered genomes can be processed at one time (Fig. 2a and Supplementary Table 2).
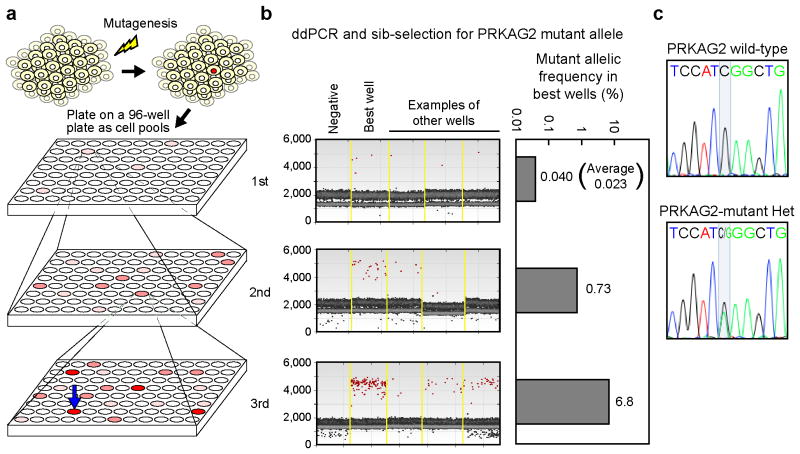
Figure 2. Point mutagenesis in human iPS cells.
(a) Overview of the approach to isolate rare mutants. Cells transfected with a TALEN pair and a donor oligonucleotide harboring the desired point mutation are immediately plated into a 96-well plate. The plates are replicated so that one 96-well plate is cryopreserved, while the other is used for DNA analysis. The mutant allelic frequency is measured from pooled cells from several wells by ddPCR. The well with the highest mutant frequency identifies the well in the cryopreserved plate that is used for re-plating into another 96-well plate for sib-selection. This process can be repeated until the mutants are sufficiently enriched for clonal isolation. iPS cell clones are isolated from the pool with the greatest enrichment of mutated cells and genotyped. (b) Enrichment of PRKAG2 mutant cells by sib-selection. The mutant allelic frequency as measured by ddPCR after the first, second, and third sib-selections is shown. On the left are shown the raw droplet data for the negative controls, the best well, and three random examples. Unrelated PHOX2B TALENs were used as negative controls for the first sib-selection, whereas completely negative “sib” wells are shown as the negative controls for the second and third sib-selections. The calculated mutant allelic frequency from the best wells is shown on the right. The average frequency after the first sib-selection is also shown. (c) Sequencing results for a representative isolated heterozygous clone from the sib-selection screen for the PRKAG2 mutant and a wild-type control.
To optimize the ddPCR assay, we first mutagenized human iPS cells with a highly efficient TALEN pair targeting PHOX2B and then plated the cells evenly within a 96-well plate. We analyzed 8 wells for mutant and wild type alleles and found that the mutant allelic frequency differed in each of the eight wells (Fig. 1c and Supplementary Fig. 5a, b). We isolated two heterozygous and one homozygous mutant PHOX2B iPS clones directly from the pools that had 2-3% mutant cells (Fig. 1d and Supplementary Fig. 5c). When mutant allele frequencies are lower, further rounds of sib-selection can enrich for the desired cells.
We then attempted to use our method to purify an extremely rare population of iPS cells that were mutant at the PRKAG2 locus. We predicted that TALEN-induced recombination at the PRKAG2 site would be low because nonhomologous end-joining (NHEJ) activity was undetectable in the Surveyor assay, even in HEK293T cells (data not shown). Although high concentrations of TALENs would increase HR rates (Supplementary Fig. 6a, b), we chose to use the lower TALEN concentrations in an attempt to avoid dose-dependent off-target DNA damage6-9. After initial transfection of human iPS cells, the induced mutant allelic frequency averaged only 0.023%, representing <1:4000 alleles or <1:2000 heterozygous cells, which was 100-times less efficient than PHOX2B-targeted mutagenesis (Fig. 2band Supplementary Fig. 6c). After three rounds of sib-selection, we obtained a cell pool with a mutant allelic frequency of 6.8% (Fig. 2b and Supplementary Fig. 6c–e), a >290-fold enrichment of the mutated cells. From this enriched cell pool, we isolated 118 iPS cell clones and found that 11 were heterozygous mutants (Fig. 2c and Supplementary Fig. 6f, g). We characterized two of the isolated mutant lines and observed that they maintained normal pluripotent cell morphology, pluripotency marker expression, karyotype (Supplementary Fig. 7a, b), and cardiac differentiation potential (data not shown) after the multiple rounds of sib-selection. We isolated another PRKAG2 heterozygous mutant line from an independent transfection of iPS cells using our method and confirmed that this line also had normal karyotype (Supplementary Fig. 7b).
We sequenced the 10 most likely off-target sites in the five isolated PHOX2B and PRKAG2 mutant lines and saw no mutations (Supplementary Table 3) although complete off-target analysis will require whole genome sequencing. We then used a fourth TALEN pair targeting the BAG3 gene and successfully isolated a BAG3 heterozygous mutant iPS cell line that maintained expression of pluripotency markers and normal karyotype from an initial allelic frequency of 0.045% (∼1:1000 cells) after two rounds of sib-selection (Supplementary Fig. 8). Over the period of 8 months, our lab has isolated 20 independent iPS clones via ddPCR/sib-selection and 90% (18/20) of them had normal karyotype, which is similar to or better than what we obtained during the same 8 months using antibiotic selection and the same culture conditions [82% (9/11) of clones had normal karyotype, unpublished observation]. Interestingly, we did not observe any NHEJ events at the targeted site of any of the 20 clones we have isolated to date (Supplementary Fig. 8c, Fig. 9a, b). The lack of NHEJ events is essential for scarless mutations. This could be due to the lower amounts of nuclease we used, since NHEJ is dose-dependent with zinc-finger nucleases9, and has been reported to be up to 30% when high TALEN-expressing cells are selected10.
The molecular and cellular basis of human phenotypic variation is one of the great challenges of medical research. Advances in human genetics have led to hundreds of single base pair mutations that are hypothesized to cause disease, but we lack an efficient means of testing these mutations. Furthermore, many genetic diseases could be corrected by single-base substitutions without leaving the scar of a selection marker. To meet this challenge, we must have the capability to engineer the human genome, one nucleotide at a time, with tools that are efficient, robust, and accurate. By standard methods, it is extremely difficult to generate otherwise scarless iPS cell lines with precise mutations. The piggyBac transposon system is an elegant method of single base pair mutagenesis19, but it requires an endogenous TTAA transposon insertion site or the creation of it. Furthermore, the piggyBac requires multiple steps of transposon insertion and excision with the potential for transposon re-insertion and imprecise excision (Supplementary Table 2).
Our method provides a way to capture precise mutagenesis events that are exceedingly rare. For example, with the PRKAG2 TALENs, we initially saw an average mutant allelic frequency of only 0.023% in iPS cells. Without our method, we would have had to isolate >2,000 clones from the initial cell pool to establish one mutant line; with our new method, just 11 clones were required (Fig. 2 and Supplementary Fig. 6). Our method therefore reduces by greater than tenfold the active work time for making scarless iPS cell lines with precise mutations. Since our approach is based on a 96-well plate format, we can easily multiplex mutagenesis so that a single technician would generate 6–10 lines at once. Moreover, subsequent cloning is only attempted when successful HR has occurred, avoiding the potentially wasted effort of directly isolating iPS cell clones even when HR has not occurred (Supplementary Fig. 10 and Supplementary Table 2). Challenges still exist before scarless genome engineering can be routinely used for precise disease models or clinical gene correction. In particular, affordable sequencing of clones for off-target mutations will be essential to determine the proper HR conditions and optimal site-specific nucleases for the highest fidelity. We anticipate that our approach will advance scarless genome engineering by allowing many scientists to use human iPS cells to model human genetics and revert disease mutations with unparalleled precision and efficiency.
Online Methods
Statistical Information
The sample numbers for the experiments to compare ddPCR and canonical genotyping PCR (Supplementary Fig. 1) with the plasmid DNA were technical replicates. Those for the Surveyor assay and ddPCR experiments in HEK293T cells (Supplementary Figs. 3 and 4) were all biological replicates (independent transfections of HEK293T cells). We did not exclude any samples.
TALEN and CRISPR/Cas design and construction
TALEN Targeter (https://tale-nt.cac.cornell.edu/tutorials/talentargeterupdated)20 was used to design the PHOX2B and the PRKAG2 TALENs. ZiFiT (http://zifit.partners.org/ZiFiT/ChoiceMenu.aspx)21, 22 was used to design the PKP2 and BAG3 TALENs. TALENs were constructed using the Voytas laboratory's Golden Gate assembly system provided through Addgene (http://www.addgene.org/TALeffector/goldengateV2/)23 except the backbone vector. We interchangeably used the Goldy24 and the MR015 (a gift from Drs. M. Porteus and M. Rahdar) TALEN backbones for the PHOX2B TALENs. We observed that these two backbones had similar activity in human iPS cells (data not shown). We used the MR015 TALEN backbone for all other TALENs used in this study. The TALEN target sequences were aligned against the reference genome by BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to ensure that they were unique sites. We used the NN repeat variable di-residues (RVDs) for all the TALENs used in this study. For PHOX2B and PKP2, the NN versions are described as the “strong” TALENs. In addition, we made TALENs in which the binding sequence was the same, except that the NN RVDs were substituted with NK RVDs (which induces less frequent recombination25, 26) to create the “weak” TALENs. For the individual TALEN target sequences, see Supplementary Table 4.
For the CRISPR/Cas system, we searched the reference human genome for unique CRISPR target sites (GN20GG) that fit with the Church laboratory's system provided through Addgene (http://www.addgene.org/crispr/church/)18, and found a unique site encompassing the RBM20 mutation site (GGTCTCGTAGTCCGGTGAGCCGG, where the underlined C was mutated to A). We followed the guidelines provided through Addgene to generate the gRNA expression construct.
Design of primers and probes for the TaqMan PCR system and oligonucleotide donors
To design the primers and probes, we used the TaqMan MGB Allelic Discrimination option in Primer Express 3.0 software (Life Technologies). For the PHOX2B mutation, the default setting (minimal amplicon size was 50 bp) was selected, whereas for the PKP2, RBM20, and PRKAG2 mutations, the amplicon size setting was changed to 100 bp to give optimal probe-primer sequences. For the BAG3 mutation, we manually designed the primers and probe by following the instructions of Primer Express 3.0, as the default program did not give us any candidates. The point mutations were inputted as “SNPs for allelic discrimination”, and designed custom TaqMan MGB probes were purchased from Life Technologies. Optimal primer sequences identified by the software were manually checked to ensure that at least one of the primers would anneal outside of the donor sequence. Oligonucleotide donors were manually designed to target the point mutation to the middle of the oligonucleotide sequence. Primers and oligonucleotide donors that underwent standard desalting purification were purchased from Integrated DNA Technologies. For sequences of the primers, probes, and oligonucleotide donors, see Supplementary Table 5.
Culture and transfection of HEK293T cells
HEK293T cells were maintained in Dulbecco's modified Eagle medium with high glucose, sodium pyruvate, and L-glutamine (Life Technologies) supplemented with 10% FetalPlex (Gemini Bio-Products) and 50 units/ml penicillin-streptomycin (Life Technologies). For transfection, 4 x 105 or 2 x 105 cells were plated into a 12- or 24-well plate, respectively. One day after plating, the cells were transfected with DNA with Lipofectamine 2000 (Life Technologies), according to manufacturer's instructions. For point mutagenesis by TALENs, 400 or 200 ng of each TALEN vector and 800 or 400 ng of an oligonucleotide donor DNA were transfected per well in a 12- or 24-well plate, respectively. For point mutagenesis by CRISPR/Cas, 400 or 200 ng of Cas9 vector, 400 or 200 ng of gRNA vector, and 800 or 400 ng of an oligonucleotide donor DNA were transfected per 12- or 24-well, respectively. Genomic DNA was extracted from the cells 3 days after transfection with the DNeasy Blood & Tissue Kit (Qiagen).
Surveyor assay
The Surveyor assay was carried out according to manufacturer's instructions (Transgenomics). Using Lipofectamine 2000, 800 ng or 400 ng of each TALEN vector was transfected into HEK293T cells plated into a 12- or 24-well plate, respectively. To PCR-amplify the target genomic regions, we used Phusion High-Fidelity DNA Polymerase (New England Biolabs) and the following primers: PHOX2B FW 5′-CTCCAGCCACCTTCTCCATA-3′, PHOX2B RV 5′-CGCTGAGAAAGCTGAAGGTC-3′, PKP2 FW 5′-CAAACTCAAGAATCTCATGATAACAGAA-3′, PKP2 RV 5′-ACAAACCATCAAACAAACTGTG-3′. The digested DNA fragments were analyzed by 15% acrylamide gel electrophoresis. The intensities of the bands were quantified by ImageJ (http://rsbweb.nih.gov/ij/). Quantification of the frequency of NHEJ was calculated using the formula; 100 x {1 – [1 – a/(a + b)]1/2 }, where a = the integrated intensity of both cleavage product bands, and b = the integrated intensity of the uncleaved PCR product band.
Culture and transfection of iPS cells
For iPS cells, UCSF Committee on human research #10-02521 approved the study protocol. The human iPS cell lines used in this study were generated from a healthy male patient, WTC10 and WTC1127, using the episomal reprogramming method28. We successfully applied our ddPCR/sib-selection method to another iPS cell line, WTB6, which was generated from a healthy female patient using episomal reprogramming method28 (data not shown). Informed consent was obtained for this procedure. iPS cells were maintained on Matrigel (BD Biosciences) in Essential 8 medium (Life Technologies), which was exchanged every other day. To sparsely populated wells (i.e., passaged wells), we added 10 μM Y-27632, a Rho-associated kinase (ROCK) inhibitor (Millipore), to promote cell survival29.
For TALEN and donor transfections, we used the Human Stem Cell Nucleofector Kit-1 and a Nucleofector 2b Device (Lonza). WTC11 was used for the BAG3 mutagenesis, whereas WTC10 was used for the mutagenesis of other genes. When cells were 60–70% confluent in six-well plates, the medium was changed to Essential 8 with 10 μM Y-27632 at least 1 hour before transfection. The medium was then transferred into a conical tube to be used as conditioned medium. Cells were washed with 2 ml/well phosphate-buffered saline (PBS) and treated with 500 μl/well Accutase (Millipore) at 37°C for 5–10 minutes. Cells were then resuspended in PBS and counted using a Countess Automated Cell Counter (Life Technologies).
For each transfection, 2 million cells were then transferred into a conical tube and spun down at 1,500 rpm for 5 minutes. The supernatant was aspirated, the cells were resuspended in 100 μl/transfection of Human Stem Cell Solution 1, and 3 μg of each TALEN vector (10 μg of each TALEN vector was used only in Supplementary Fig. 6b) and 6 μg of an oligonucleotide donor were added. The total volume of the DNA solution was less than 10 μl. The suspension was transferred to a cuvette and electroporated using program A-23. Conditioned medium (500 μl/transfection) was added to the transfected cells, which were then transferred to a conical tube with 9.5 ml of conditioned medium. The cells were plated into a Matrigel-coated 96-well plate using a multichannel micropipetter.
Detection of point mutations by ddPCR
The composition of the pre-mixtures of allele-specific TaqMan probes and primers was 5 μM of an allele-specific FAM or VIC TaqMan MGB probe, 18 μM of a forward primer, and 18 μM of a reverse primer in water. For optimization of the PCR annealing and extension temperatures, we mixed the following reagents in 1.5 ml tubes: 9 μl water, 12.5 μl 2x ddPCR Supermix for Probes (Bio-Rad), 1.25 μl FAM probe and primer pre-mixture, 1.25 μl VIC probe and primer pre-mixture, and 1 μl 0.5 pg/μl 1:1 plasmid mixture of a wild-type allele and a mutant allele (25 μl total volume). Droplet generation with a QX100 Droplet Generator was performed according to the manufacturer's instructions (Bio-Rad), and the reaction was transferred into a 96-well PCR plate for standard PCR on a C1000 Thermal Cycler (Bio-Rad). The thermal cycling program conducted was:
- Step 1: 95°C 10 minutes;
- Step 2: 94°C 30 seconds;
- Step 3: 50°C–58 or 60°C gradient 1 minute, go back to Step 2 39 times;
- Step 4: 98°C 10 minutes.
After the PCR was complete, the droplets were analyzed using a QX100 Droplet Reader (Bio-Rad) with the “absolute quantification” option. We first checked whether a probe set could properly discriminate between negative, FAM-positive, VIC-positive, and double-positive populations. We then chose the highest annealing temperature that gave the best separation of the four distinct populations as the optimal temperature (see also Supplementary Fig. 2). The optimal temperatures differed between different probe-primer sets. For example, for the PHOX2B mutation shown in Supplementary Fig. 2, the optimal annealing and extension temperature was 51°C. For the PKP2-, RBM20-, PRKAG2-, and BAG3- mutations, the optimal annealing temperatures were 57°C, 57°C, 51°C, and 55°C, respectively (data not shown).
For ddPCR to detect point mutagenesis, we mixed the following reagents in 0.2 ml PCR 8-tube strips: 9 μl (for HEK293T cells) or 4 μl (for iPS cells) water, 12.5μl 2x ddPCR Supermix for Probes (Bio-Rad), 1.25 μl FAM probe and primer pre-mixture, 1.25 μl VIC probe and primer pre-mixture, and 1 μl (for HEK293T cells, 100 ng) or 5 μl (for iPS cells, 50-150 ng) genomic DNA solution (25 μl total volume). Droplets were generated, followed by PCR at the optimal temperatures, and the droplets were analyzed as described for the temperature optimization to calculated mutant allelic frequencies in a wild-type allelic background. For iPS cells, a multichannel micropipetter was used to directly transfer genomic DNA extracted from 96-well plates to reaction mixtures, and then to transfer the reaction mixtures to a cartridge for droplet generation.
Freezing iPS cells and genomic DNA extraction from iPS cells on a 96-well plate for sib-selection
When iPS cells (plated in 96-well plates) were 80–90% confluent, the cells were washed with 100 μl/well PBS and treated with 30 μl/well Accutase at 37°C for 5–10 minutes. Half of the detached cells were placed into a 96-well plate for genomic DNA extraction, and the other half left in the original 96-well plate were frozen at −80 °C for future sib-selection. For genomic DNA extraction, 50 μl of genomic DNA lysis buffer [10 mM Tris pH 7.5, 10 mM EDTA pH 8.0, 10 mM NaCl, 0.5%N-lauroylsarcosine, and 1 mg/ml proteinase K (added fresh)] was aliquoted into each well on another 96-well plate. We transferred 15 μl/well of the cell suspension (in Accutase) to the 96-well plate containing the lysis buffer, and the mixture was combined using a multichannel micropipetter. The plate was then incubated at 55°C overnight in a plastic container with a small amount of water (to prevent drying). Then, 100 μl 75 mM NaCl in ethanol that had been stored at -80°C was added to each well, the solution was mixed, and the plate was incubated at room temperature for 2 hours. The solution was discarded by inverting the plate (the DNA remained adhered to the plate), and each well was washed with 100 μl 70% ethanol twice. The plate was then air-dried for 30–45 minutes to remove all residual alcohol. After drying, DNA in each well was dissolved in 30 μl TE buffer, and the DNA concentration was measured. For most wells, we obtained DNA concentrations of 10–30 ng/μl.
For cell freezing, 75 μl freezing medium [HyClone Fetal Bovine Serum (Thermo Scientific) with 10% dimethyl sulfoxide (Sigma)] was added to cells in each well of the 96-well plate and mixed by pipetting up and down, taking care to avoid creating too many bubbles. Then, 75 μl of mineral oil (Sigma, embryo culture grade) was layered onto the cell suspension. The 96-well plate was sealed with Parafilm and frozen at -80°C in a Styrofoam container. When thawing the cells, the plate was placed in a 37°C CO2 incubator for 10–15 minutes. The cell suspension was transferred to a 1.5-ml tube, mixed with 0.5 ml Essential 8 medium with 10 μM Y-27632, and the number of cells was counted using a Countess Automated Cell Counter. An aliquot of the cell suspension was then removed and placed into a conical tube containing Essential 8 medium supplemented with 10 μM Y-27632 and mixed well. Cells were then plated into Matrigel-coated 96-well plates at the appropriate cell density.
For sib-selection, cell pools containing mutated cells were plated at a density of 500 cells/well into a 96-well plate, fed with Essential 8 medium with 10 μM Y-27632 every other day for 6 days, and then fed with Essential 8 every other day for the next 4 days. After culturing cells in these conditions, they were 80–90% confluent and ready for the next round of sib-selection or cloning.
Isolation of iPS cell clones
The process of thawing a cell pool from which iPS clones were isolated was the same as described for sib-selection (above). For isolating iPS cell clones, the cells were plated at limiting dilutions for an average density of 100 cells/well into 96-well plates. The cells were fed with Essential 8 medium with Y-27632 every other day for 10 days, and then fed with Essential 8 every other day for the next 8 or 10 days. By 18 or 20 days after plating, cells in only 10–30% of wells survived. Wells containing cells with pluripotent morphology that were likely to have been derived from a single clone were chosen for clonal analyses. We are aware that iPS cells purified via limiting dilution may be the progeny of more than one cell (mixed colony). However analysis via ddPCR allowed us to rapidly determine whether a putative clone is genetically homogenous by quantifying the alleles. After the initial purification we also frequently recloned cells to be sure that populations are homogenous (see Genotyping PCR and expansion of isolated clones and Supplementary Fig. 6g). These wells were washed in 100 μl/well PBS, and cells were detached with 30 μl/well Accutase. Cells were divided into two even pools for genomic DNA extraction and freezing as described above, except that the transferred cells were strategically arranged in wells of new 96-well plates. In this way, the genomic DNA samples could be readily analyzed by genotyping PCR on a 384-well format, as described below. The concentration of genomic DNA extracted from the clones was 3–10 ng/μl, which was sufficient for genotyping PCR. Some investigators prefer cloning human iPS cells on feeder cells, and this process also works well with our method. However we have found that using feeder free culture conditions to be more compatible with higher throughput production and analysis in a 96-well plate format. However it is essential to confirm homogeneity of the alleles via ddPCR and/or subsequent subcloning.
Genotyping PCR and expansion of isolated clones
The following reagents were mixed in each well of a 384-well plate: 2.5 μl 2x TaqMan Genotyping Master Mix (Life Technologies), 0.125 μl FAM probe and primer pre-mixture (the same pre-mixture used for ddPCR), 0.25 μl VIC probe and primer pre-mixture (the same pre-mixture used for ddPCR), 1.625 μl water, and either 0.5 μl extracted genomic DNA from iPS clones, 0.5 μl 0.5 pg/μl mutant allele plasmid/wild-type allele plasmid/1:1 mixture of both plasmids, or 0.5 μl water (no template control) (5 μl total). We made a master mix of these reagents, excluding the templates, aliquoted 4.5 μl of the master mix into each well on a 384-well plate, and added 0.5 μl of each template to the relevant wells. After mixing, the plate was centrifuged at 1,300 rpm for 5 minutes. PCR genotyping was carried out using the Applied Biosystems 7500 or 7900 Fast Real-Time PCR System (Life Technologies) using the SDS2.4 software (Life Technologies). Plate documents of Allelic Discrimination (AD) and the Standard Curve (AQ) for the samples were created according to the manufacturers' instructions. Pre-Read was performed to measure background signals of each well with the AD plate document, a thermal cycling was performed with the AQ plate document, and finally Post-Read and genotyping were done with the AD plate document (according to manufacturer's instructions). The thermal cycling parameters for the default condition of the AQ plate document were as follows:
- Step 1: 50°C 2 minutes;
- Step 2: 95°C 10 minutes;
- Step 3: 95°C 15 seconds;
- Step 4: 60°C 1 minute, go back to Step 3 39 times.
During thermal cycling, both the FAM and VIC signals were monitored.
The target genomic regions were amplified by conventional PCR with Phusion High-Fidelity DNA Polymerase from the genomic DNA of clones that were genotyped as positive for mutant alleles, and sequence-verified for the mutations. The 96-well plates that had the positive clones were then thawed in the same way as for sib-selection (described above). The cell suspension was mixed with 0.5 ml Essential 8 medium with 10 μM Y-27632 in a 1.5-ml tube, and the cells were spun down at 6,000 rpm for 5 minutes. Using a micropipetter, the surface oil layer followed by the medium was carefully removed. The cells were then resuspended in Essential 8 medium with 10 μM Y-27632 and plated into Matrigel-coated 6- or 12-well plates. Wells containing iPS cells that partially differentiated during the cloning step were passaged at a 1:10 ratio 2–3 times, to remove these cells and recover pools that are fully pluripotent.
We confirmed that the mutant allele: wild-type allele ratio was 1:1 and 1:0 for heterozygous and homozygous mutant lines, respectively, by ddPCR to verify that they were pure clones. The results for the PRKAG2-mutant Het are shown in Supplementary Fig. 6g as an example.
Fluorescent staining of iPS cells
iPS cells cultured in 24-well plates were washed with 500 μl/well PBS, fixed with 250 μl/well 4% paraformaldehyde at room temperature for 20 minutes, and then washed with 500 μl/well PBS three times. The cells were blocked and permeabilized with 250 μl/well PBS with 0.1% Triton-X (PBS-T) supplemented with 5% bovine serum albumin (BSA) at room temperature for 1 hour, and then in 200 μl/well primary antibodies diluted 200 times in PBS-T with 5% BSA at 4°C overnight. The primary antibodies (all purchased from Abcam) used in this study were SOX2 (ab59776), OCT4 (ab19857), SSEA4 (ab16287), and TRA-1-81 (ab16289). The cells were washed with 500 μl/well PBS-T three times, incubated with 200 μl/well secondary antibodies diluted 500 times in PBS-T with 5% BSA at room temperature for 1 hour, and then washed with 500 μl/well PBS three times. The secondary antibodies used were Alexa Fluor 488 Goat Anti-Mouse IgG (A-11001) and Alexa Fluor 488 (A-11008) or 594 (A-11012) Goat Anti-Rabbit IgG (Life Technologies). Finally, the cells were mounted with 75 μl/well VECTASHIELD Mounting Medium with DAPI (H-1200) (Vector Laboratories). The pictures were taken by using BZ-9000 microscope (Keyence).
Karyotyping
Karyotyping was carried out by Cell Line Genetics (Madison, WI).
Sequencing of potential off-target genomic sites
Potential off-target sites for the PHOX2B and PRKAG2 TALENs were predicted by a recently reported bioinformatic tool, TALENoffer30. We set the distance between two TALEN binding sites from 12 to 24 bp, and other parameters were the default settings for the prediction. For both TALEN pairs, the top candidates were their on-targets, indicating that the prediction was properly done. We chose top 10 off-target sites for each TALEN pair and designed primers that amplify 400–1100-bp genomic regions around the off-target sites. The genomic regions were amplified by using Phusion High-Fidelity DNA Polymerase from the PHOX2B-mutant Het, PHOX2B-mutant Homo, PRKAG2-mutant Het, PRKAG2-mutant Het 2, and Independent PRKAG2-mutant Het lines, as well as the parental iPS cell line, WTC10. The amplicons were sequenced from both 5′ and 3′ ends by using the primers that amplified them. All the sequencing results from the mutant lines were compared to those from WTC10. We did not see any mutations in the mutant lines (see Supplementary Table 3 for the details).
Supplementary Material
1
10
11
12
2
3
4
5
6
7
8
9
Acknowledgments
We thank members of the Conklin laboratory for technical assistance and critical reading of the manuscript. We thank B. G. Bruneau, D. Srivastava, S. Yamanaka, K. Tomoda, Y. Hayashi, K. E. Eilertson (Gladstone Institutes), B. S. Moriarity, D. A. Largaespada (University of Minnesota), and W. A. Weiss (University of California, San Francisco) for valuable discussions and advice. We thank A. K. Holloway (Gladstone Institutes) for generating the list of unique CRISPR target sites. We thank K. Carver-Moore (Gladstone Institutes) for her technical assistance in iPS cell culture on 96-well plates. We are also grateful to M. Porteus and M. Rahdar (Stanford University) for providing us with the TALEN backbone plasmid MR015. We also thank the Roddenberry Stem Cell Core (Gladstone Institutes) for providing a stimulating environment. Y.M. is a recipient of a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad and the Uehara Memorial Foundation Research Fellowship. L.M.J. is supported by a Postdoctoral Fellowship, TG2-01160 from the California Institute of Regenerative Medicine. M.H. is supported by a Postdoctoral Fellowship, PF-13-295-01 – TBG from the American Cancer Society. B.R.C. received support from the Gladstone Institutes, US National Heart, Lung, and Blood Institute, National Institutes of Health, U01-HL100406, U01-GM09614, R01-HL108677, U01-HL098179 and R01-HL060664.
Footnotes
Competing Financial Interests: The authors declare no competing financial interests.
Author Contributions: Y.M. and B.R.C. conceived and designed the experiments. Y.M. and A.H.C. conducted most of the experiments. L.M.J. and J.Y. performed the BAG3 mutagenesis, and J.Y. constructed the PKP2 TALENs. M.H. designed and constructed the PHOX2B TALENs and helped the Surveyor assays. T.D.N. helped to conceive of ddPCR experiments, and with the construction of TALENs. P.P.L. conducted immunofluorescence staining. Y.M., P.L.S., and B.R.C. wrote the manuscript with support from all authors.
References
- 1.da Cunha Santos G, Shepherd FA, Tsao MS. Annual review of pathology. 2011;6:49–69. doi: 10.1146/annurev-pathol-011110-130206. [DOI] [PubMed] [Google Scholar]
- 2.Moore JR, Leinwand L, Warshaw DM. Circulation research. 2012;111:375–385. doi: 10.1161/CIRCRESAHA.110.223842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi K, et al. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, et al. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 5.Gaj T, Gersbach CA, Barbas CF., 3rd Trends in biotechnology. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Y, et al. Nature biotechnology. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA. Nucleic acids research. 2011;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu PD, et al. Nature biotechnology. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Nature methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding Q, et al. Cell stem cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen F, et al. Nature methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soldner F, et al. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mali P, et al. Nature biotechnology. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ran FA, et al. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormick M. Methods in enzymology. 1987;151:445–449. doi: 10.1016/s0076-6879(87)51036-9. [DOI] [PubMed] [Google Scholar]
- 16.Hindson BJ, et al. Analytical chemistry. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cong L, et al. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mali P, et al. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yusa K, et al. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyle EL, et al. Nucleic acids research. 2012;40:W117–122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sander JD, et al. Nucleic acids research. 2010;38:W462–468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. Nucleic acids research. 2007;35:W599–605. doi: 10.1093/nar/gkm349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cermak T, et al. Nucleic acids research. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bedell VM, et al. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christian ML, et al. PloS one. 2012;7:e45383. doi: 10.1371/journal.pone.0045383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Nature communications. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kreitzer FR, et al. American journal of stem cells. 2013;2:119–131. [PMC free article] [PubMed] [Google Scholar]
- 28.Okita K, et al. Nature methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe K, et al. Nature biotechnology. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 30.Grau J, Boch J, Posch S. Bioinformatics. 2013;29:2931–2932. doi: 10.1093/bioinformatics/btt501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
10
11
12
2
3
4
5
6
7
8
9