MERS–Related Betacoronavirus in Vespertilio superans Bats, China (original) (raw)
To the Editor: Middle East respiratory syndrome coronavirus (MERS-CoV), a novel lineage C betacoronavirus, was first described in September 2012, and by April 16, 2014, the virus had caused 238 infections and 92 deaths in humans worldwide (1). Antibodies against MERS-CoV in dromedary camels were recently reported (2), as was the full genome of MERS-CoV from dromedary camels (3). Finding the natural reservoir of MERS-CoV is fundamental to our ability to control transmission of this virus to humans (4).
We report a novel lineage C betacoronavirus identified from Vespertilio superans bats in China. The full-length genome of this betacoronavirus showed close genetic relationship with MERS-CoV. Together with other evidence of MERS-CoV–related viruses in bats (5–8), our findings suggest that bats might be the natural reservoirs of MERS-related CoVs.
In June 2013, we collected anal swab samples from 32 V. superans bats from southwestern China. A small proportion of each sample was pooled (without barcoding) and processed by using virus particle–protected nucleic acid purification and sequence-independent PCR for next-generation sequencing analysis with the Illumina (Solexa) Genome Analyzer II (Illumina, San Diego, CA, USA). Redundant reads were filtered, as described (9), from the raw sequencing reads generated by the genome analyzer and then aligned with the nonredundant protein database of the National Center for Biotechnology Information (ftp://ftp.ncbi.nlm.nih.gov/blast/db/) by using BLAST (http://blast.ncbi.nlm.nih.gov). The taxonomy of these aligned reads was parsed by using MEGAN 4 (http://ab.inf.uni-tuebingen.de/software/megan/).
On the basis of the BLAST results, 8,751,354 sequence reads 81 nt in length were aligned with the protein sequences of the nonredundant protein database: 72,084 of the reads were uniquely matched with virus proteins. Of these 72,084 reads, 32,365 were assigned to the family Coronaviridae, primarily to lineage C of the genus Betacoronavirus, and found to share 60%–97% aa identity with MERS-CoV.
The MERS-CoV–related reads were extracted and assembled by using SeqMan software from the Lasergene 7.1.0 program (DNASTAR, Madison, WI, USA), resulting in a draft CoV genome. Reverse transcription PCR selective for the partial RNA-dependent RNA polymerase (RdRp) gene of this novel lineage C betacoronavirus suggested that 5 of the 32 samples (≈16%) were positive for the novel betacoronavirus, and the PCR amplicons shared >98% nt identity with each other. Using a set of overlapped nested PCRs and the rapid amplification of cDNA ends method, we determined the full-length genome of 1 strain of this V. superans bat–derived betacoronavirus (referred to as BtVs-BetaCoV/SC2013, GenBank accession no. KJ473821).
The betacoronavirus strain had a genome length of 30,413 nt, excluding the 3′ poly (A) tails, and a G+C content of 43.1%. Pairwise genome sequence alignment, conducted by the EMBOSS Needle software (http://www.ebi.ac.uk/Tools/psa/emboss_needle/) with default parameters, suggested that the genome sequence of BtVs-BetaCoV/SC2013 showed 75.7% nt identity with that of human MERS-CoV (hCoV-MERS); this shared identity is higher than that for other lineage C betacoronaviruses (from bats and hedgehogs) with full genomes available. hCoV-MERS showed 69.9% nt identity with bat CoV (BtCoV) HKU4-1, 70.1% nt identity with BtCoV-HKU5-1, and 69.6% nt identity with hedgehog CoV EriCoV-2012–174.
Compared with those lineage C betacoronaviruses, which had an 816-bp partial RdRp sequence fragment available, BtVs-BetaCoV/SC2013 shared 96.7 % aa identity with hCoV-MERS. Pipistrellus BtCoVs found in Europe (BtCoV-8-724, BtCoV-8-691, BtCoV-UKR-G17) shared 98.2 % aa identity with hCoV-MERS, and Eptesicus BtCoV found in Italy (BtCoV-ITA26/384/2012) and other lineage C betacoronaviruses shared 96.3 % aa and <95% aa identity, respectively, with hCoV-MERS.
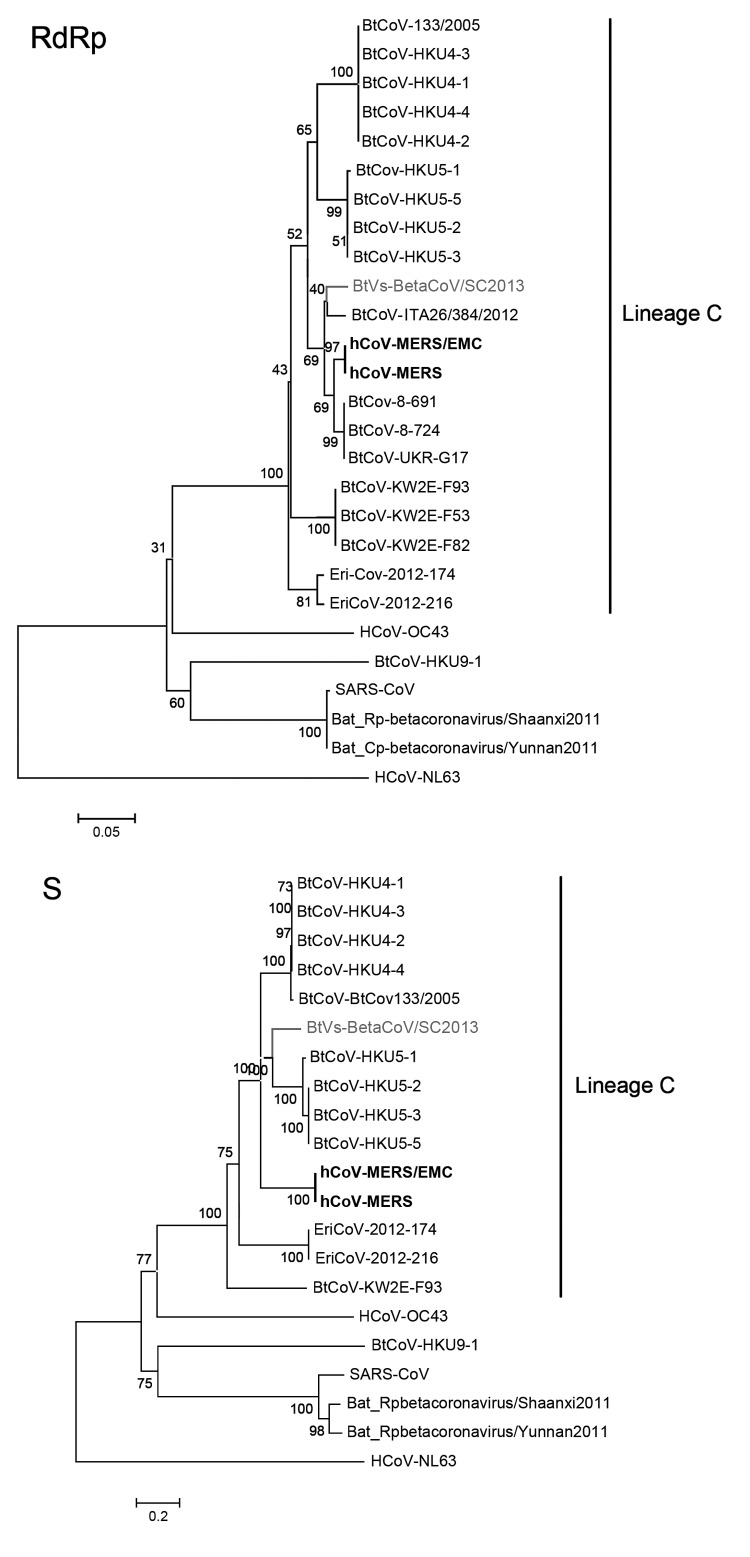
To clarify the evolutionary relationship between BtVs-BetaCoV/SC2013 and other lineage C betacoronaviruses, we performed phylogenetic analyses based on the deduced RdRp and the spike, envelope, membrane, and nucleocapsid proteins by using MEGA5 (http://www.megasoftware.net/) (Figure; Technical Appendix). For RdRp and the envelope, membrane, and nucleocapsid proteins, BtVs-BetaCoV/SC2013 always clustered with hCoV-MERS with short branch lengths, reflecting their high sequence similarities.
Figure.
Phylogenetic trees based on the deduced amino acid sequences of the partial RNA-dependent RNA polymerase (RdRp; an 816-nt sequence fragment corresponding to positions 14817–15632 in human Middle East respiratory syndrome coronavirus [hCoV-MERS; KF192507]) and complete spike (S) protein. The novel virus is shown in gray, and hCoV-MERS is shown in bold. The following coronaviruses were used (GenBank accession numbers are shown in parentheses): severe acute respiratory syndrome coronavirus (SARS-CoV; NC004718), Bat Rp-coronavirus/Shaanxi2011(JX993987), Bat Cp-coronavirus/Yunnan2011(JX993988), Bat coronavirus HKU9-1 (BtCoV-HKU9-1; EF065513), BtCoV-133/2005(NC008315), BtCoV-HKU4-1 (EF065505), BtCoV-HKU4-2 (EF065506), BtCoV-HKU4-3 (EF065507), BtCoV-HKU4-4 (EF065508), BtCoV-HKU5-1 (EF065509), BtCoV-HKU5-2 (EF065510), BtCoV-HKU5-3 (EF065511), BtCoV-HKU5-5 (EF065512), BtCoV-ITA26/384/2012 (KF312399), BtCoV-KW2E-F82 (JX899382), BtCoV-KW2E-F93 (JX899383), BtCoV-KW2E-F53 (JX899384), BtCoV-8–724 (KC243390), BtCoV-8–691 (KC243391), BtCoV-UKR-G17 (KC243392), Human betacoronavirus 2c EMC/2012 (hCoV-MERS/EMC; JX869059), hCoV-OC43 (NC005147), hCoV-NL63 (NC005831), Betacoronavirus ErinaceusCoV/2012-174 (EriCoV-2012-174; KC545383), and EriCoV-2012-216 (KC545386). Scale bar indicates genetic distance estimated by using WAG+G model for the RdRp and WAG+G+F model for the S protein implemented in MEGA5 (http://www.megasoftware.net/).
In the spike protein phylogenetic tree, BtVs-BetaCoV/SC2013 clustered with a clade defined by BtCoV-HKU5, with which it shares 74.8% aa identity. The spike proteins of hCoV-MERS form a sister clade of the clade defined by HKU5 BtCoVs and BtVs-betaCoV/SC2013, and the spike proteins share 69.0% aa identity with BtVs-betaCoV/SC2013. Spike proteins of BtVs-BetaCoV/SC2013, HKU5 BtCoVs, HKU4 BtCoVs, and hCoV-MERS, rather than EriCoV-2012-174, EriCoV-2012-216, and BtCoV-KW2E-F93, form a super clade. Spike protein is the critical factor for receptor recognition, binding, and cellular entry of CoVs in different host species (10), which may explain why the spike proteins in our study were relatively conserved within the same host species.
We identified a novel lineage C betacoronavirus from a V. superans bat and determined its full-length genome sequence. This novel betacoronavirus represents one of the most MERS-like CoVs that have been identified in bats as of the end of March 2014. The full-length genome sequence of the novel virus showed a closer genetic relationship with hCoV-MERS and camel MERS-CoV than with any other fully sequenced lineage C betacoronaviruses previously identified in bats or hedgehogs. Further studies of CoVs from more bat species worldwide may, therefore, help provide additional clues to the origins of pathogenic hCoV-MERS.
Technical Appendix
Phylogenetic trees of a novel Middle East respiratory syndrome–related coronavirus, human Middle East respiratory syndrome coronaviruses, severe acute respiratory syndrome virus, and various other coronaviruses.
Acknowledgments
This work was supported by the National S&T Major Project, “China Mega-Project for Infectious Disease” (grant nos. 2011ZX10004-001 and 2014ZX10004001) from the Chinese government.
Suggested citation for this article: Yang L, Wu Z, Ren X, Yang F, Zhang J, He G, et al. MERS–related betacoronavirus in Vespertilio superans bats, China [letter]. Emerg Infect Dis. 2014 July [_date cited_]. http://dx.doi.org/10.3201/eid2007.140318
1
These authors contributed equally to this article.
References
- 1.MERS-CoV—Eastern Mediterranean. ProMED-mail 2014. Apr 16 [cited 2014 Apr 16]. http://www.promedmail.org/, archive no. 20140416.2406647.
- 2.Meyer B, Muller MA, Corman VM, Reusken CB, Ritz D, Godeke GJ, et al. Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg Infect Dis. 2014;20:552–9. 10.3201/eid2004.131746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemida MG, Chu DKW, Poon LLM, Perera RAPM, Alhammadi MA, Ng HY, et al. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg Infect Dis. [Epub ahead of print]. 2014. Jul [cited 2014 Apr 19]. [DOI] [PMC free article] [PubMed]
- 4.Corman VM, Kallies R, Philipps H, Gopner G, Muller MA, Eckerle I, et al. Characterization of a novel betacoronavirus related to middle East respiratory syndrome coronavirus in European hedgehogs. J Virol. 2014;88:717–24. 10.1128/JVI.01600-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Benedictis P, Marciano S, Scaravelli D, Priori P, Zecchin B, Capua I, et al. Alpha and lineage C betaCoV infections in Italian bats. Virus Genes. 2014;48:366–71. 10.1007/s11262-013-1008-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Annan A, Baldwin HJ, Corman VM, Klose SM, Owusu M, Nkrumah EE, et al. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis. 2013;19:456–9. 10.3201/eid1903.121503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis. 2013;19:1819–23. 10.3201/eid1911.131172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wacharapluesadee S, Sintunawa C, Kaewpom T, Khongnomnan K, Olival KJ, Epstein JH, et al. Group C betacoronavirus in bat guano fertilizer, Thailand. Emerg Infect Dis. 2013;19:1349–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z, Ren X, Yang L, Hu Y, Yang J, He G, et al. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J Virol. 2012;86:10999–1012. 10.1128/JVI.01394-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–50. 10.1038/nrmicro2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Technical Appendix
Phylogenetic trees of a novel Middle East respiratory syndrome–related coronavirus, human Middle East respiratory syndrome coronaviruses, severe acute respiratory syndrome virus, and various other coronaviruses.