Aberrant Topoisomerase-1-DNA Lesions are Pathogenic in Neurodegenerative Genome Instability Syndromes (original) (raw)
. Author manuscript; available in PMC: 2014 Dec 1.
Published in final edited form as: Nat Neurosci. 2014 May 4;17(6):813–821. doi: 10.1038/nn.3715
Abstract
DNA damage is considered a prime factor in multiple spinocerebellar neurodegenerative diseases; however, the DNA lesions underpinning disease etiology are unknown. Here we identify the endogenous accumulation of pathogenic topoisomerase-1-DNA cleavage complexes (Top1cc) in murine models of ataxia telangiectasia and spinocerebellar ataxia with axonal neuropathy 1. We also show that the defective DNA damage response factors in these two diseases cooperatively modulate Top1cc turnover in a non-epistatic and ATM kinase-independent manner. Furthermore, coincident neural inactivation of ATM and DNA single strand break repair factors including tyrosyl-DNA phosphodiesterase-1 or XRCC1 result in increased Top1cc formation and excessive DNA damage and neurodevelopmental defects. Importantly, direct topoisomerase-1 poisoning to elevate Top1cc levels phenocopies the neuropathology of the mouse models above. Our study identifies a critical endogenous pathogenic lesion associated with neurodegenerative syndromes arising from DNA repair deficiency, indicating the essential role that genome integrity plays in preventing disease in the nervous system.
The DNA damage response is essential for maintaining genome integrity and preventing various human diseases, many of which are characterized by pronounced neuropathology1–3. While most components of this signaling pathway have been identified, their tissue-specific function that prevents characteristic disease-related pathology is unclear, as is the precise DNA lesions underpinning the etiology of these syndromes1. The nervous system is particularly at risk from DNA damage4, and endogenous DNA breaks occur spontaneously during development and in the mature brain5–7. A paradigm of defective DNA damage signaling is ataxia telangiectasia (A-T), a neurodegenerative syndrome associated with cancer susceptibility, immunodeficiency and radiosensitivity8–10.
A-T results from dysfunction of ATM (ataxia telangiectasia, mutated), a serine/threonine protein kinase, required for the activation of cell cycle checkpoints, chromatin remodeling, DNA repair or apoptosis after DNA double strand breaks8, 10. This is reinforced by the A-T like neuropathology present when Mre11 is partially inactivated (resulting in A-T like disease, ATLD18, 19); this factor is a component of the ATM-activating Mre11-Rad50-Nbs1 (MRN) DNA double strand break sensor9, 11, 12. ATM activation via the MRN complex is required for apoptosis of immature neural cells after DNA damage via p53 and Chk2 phosphorylation5, 13, and failure to eliminate these damaged neuroprogenitors could predispose mature A-T tissue to later neurodegeneration. However, ATM’s full neuroprotective repertoire in the nervous system remains elusive14.
Two neurodegenerative syndromes similar to A-T, ataxia with oculomotor apraxia (AOA1) and spinocerebellar ataxia with axonal neuropathy (SCAN1), result from defects in the DNA repair enzymes aprataxin (APTX) and tyrosyl-DNA phosphodiesterase 1 (TDP1), respectively. APTX and TDP1 function primarily during DNA single strand break repair15–18; APTX is an adenyl hydrolase that resolves 5′-adenylation intermediates during DNA ligation, while TDP1 cleaves and processes 3′-end covalent topoisomerase-1-DNA intermediates and DNA lesions formed by oxidative damage16–19. To investigate etiologic connections between these diseases and A-T, we considered if ATM function intersects single strand break repair disorders. Because ATM has been implicated in the response to topoisomerase-1 (Top1) adducts20–22 which could increase levels of DNA damage, particularly DNA single strand breaks, we determined if ATM regulates Top1-induced damage in neural tissue. Here we report that a key function of ATM is to avert detrimental DNA lesions in both the developing and mature nervous system by preventing the accumulation of Topoisomerase-1-DNA cleavage complexes. This involves ubiquitination- and sumoylation-mediated turnover of Top1 to resolve Top1cc, and is ATM kinase-independent. Our study further implicates defective Topoisomerase-1 processing and the accumulation of neural DNA damage as causative for neuropathology in multiple neurodegenerative syndromes arising from mutation of DNA damage response factors.
Results
Atm regulates Top1cc in neural tissue
Throughout neural development cells encounter a variety of events that compromise genome integrity, amongst which is endogenous damage via Top1 misfunction during DNA replication and transcription1, 19, 23–25. Top1 alters DNA topology and relaxes DNA supercoiling by breaking and rejoining one strand of DNA, and thousands of these transient Top1cc form during normal cellular function. However, a portion of these can persist in genomic DNA, and when trapped, a Top1cc includes a DNA strand break, which is a direct threat to cell survival19, 23. The anticancer agent camptothecin (CPT) is effective at killing replicating cells because it promotes accumulation of Top1cc that are converted into lethal DNA double strand breaks upon collision with replication forks23, 24. CPT-induced Top1cc and associated DNA breaks during transcription can activate ATM to initiate a DNA damage response (DDR), which is important to prevent genome damage to cells during proliferation21, 22, 26. Therefore, removal of trapped Top1 is critical to avoid DNA damage accumulation and resultant sequela.
Loss of TDP1, whose function is required to repair Top1-DNA complexes via cleavage of the covalent Top1-DNA phosphodiester bond, leads to the neurodegenerative disease, SCAN1. Therefore, we examined brain tissue from Tdp1−/− mice27 to measure the endogenous levels of Top1cc using a modified immune complex of enzyme (ICE) assay28, 29 (Fig. 1a) in a setting where Top1 removal from DNA is compromised. We observed high levels of Top1cc in Tdp1−/− neural tissue compared with control tissue, particularly during early development (Fig. 1). Because of phenotypic similarities between SCAN1 and A-T1, we assessed Top1cc levels in the nervous system of Atm−/− mice. We found that at E12.5, a highly proliferative stage in embryonic brain development, Atm-deficient neural tissue had accumulated substantial Top1cc levels (Fig. 1c). While Top1cc were increased during early brain development in both Tdp1−/− and Atm−/− embryos, they were reduced after E18.5, suggesting these complexes increase during rapid embryonic proliferation rates. Upon birth and into adulthood, elevated Top1cc levels persisted in Atm−/− and Tdp1−/− cerebella compared to WT controls (Fig. 1c, e). At one year of age both Atm−/− and Tdp1−/− cerebella showed higher levels of Top1cc, indicating that loss of either of these factors perturb the steady state levels of Top1-DNA complexes (Fig. 1e). Increased Top1cc did not reflect different Top1 protein levels, as these were equivalent amongst all genotypes (Fig. 1b,d). We also examined Topoisomerase-2α-DNA adducts (Top2cc) and found that unlike Top1, levels of these complexes were similar between control, Tdp1−/− and Atm−/− cerebellum (data not shown). These data show both Atm and Tdp1 are required to prevent Top1cc accumulation in the nervous system.
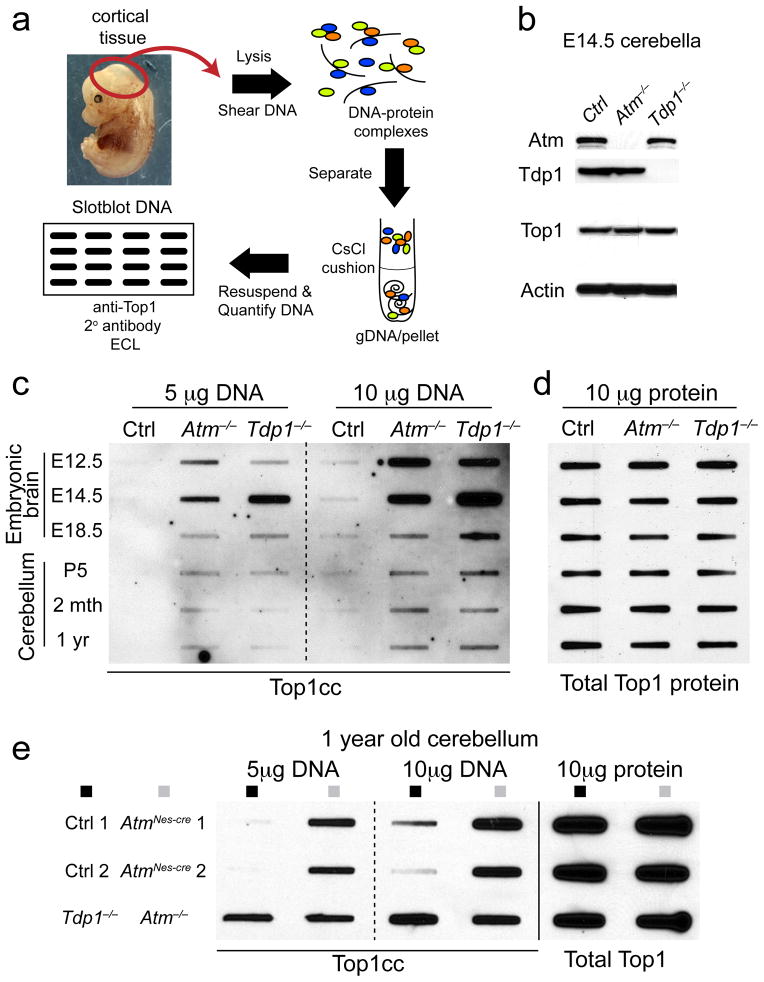
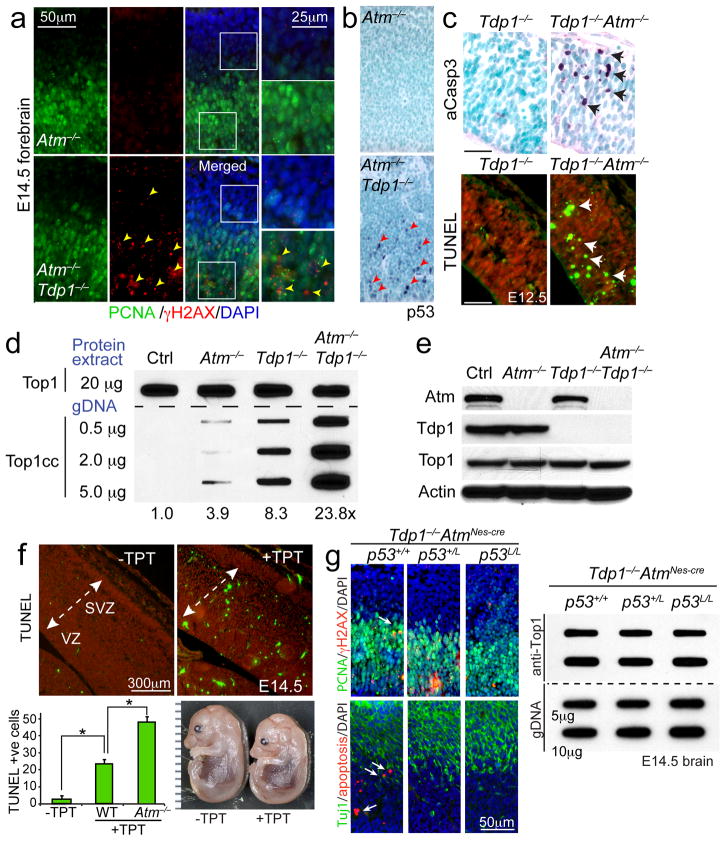
Fig. 1. Atm prevents Top1cc accumulation in vivo.
a. Schematic of the in vivo complex of enzyme bioassay (ICE assay) following immunodetection with anti-topoisomerase-1 antibody. b. Western analysis indicating the corresponding genotypes of the CNS tissue used for the ICE bioassay. The anti-Top1 immunoblot shows equivalent amounts of Top1 protein amongst cerebella tissue and validates the high-specificity of the antibody. Full-length Western blots are presented in Supplementary Figure 11. c. Genomic DNA was slot-blotted with the indicated amounts (μg) of genomic DNA (a proportion of which is complexed with Top1) isolated from the embryonal brain and postnatal and mature cerebellum. Top1cc were immunodetected with anti-Top1 antibody. Atm−/− embryonic brain and cerebellum showed significant accumulation of Top1cc compared to controls. Top1cc also accumulate in Tdp1−/− neural tissue, due to defects in Top1-DNA cleavage, and serve as a comparative positive control. Embryonic tissue used in ICE assays are derived from gDNA pooled from 3 independent embryos. d. 10 μg of protein extract derived from tissues analyzed in ‘c’ and were immunoblotted to normalize total Top1 protein content. Top1 levels were equivalent amongst WT, Atm−/− and Tdp1−/− tissues at each neurodevelopmental stage. e. One year old ATM-deficient (Atm−/− and AtmNes-cre) cerebella show substantial Top1cc levels, indicating a requirement for Atm in the resolution of Top1-DNA intermediates in the mature nervous system.
Atm−/− neural cells accumulate Top1cc-associated DNA damage
Top1cc involve DNA strand breaks, and persistence of this complex predicts increased amounts of DNA damage23, 30. To determine if Top1cc accumulation is associated with increased DNA strand breaks, we compared DNA repair rates in Atm−/− and Tdp1−/− cells under conditions that promote formation of these complexes, using the Top1 poison, camptothecin (CPT)26, 31. To measure levels of DNA damage we used the alkaline comet assay. We found that quiescent Atm−/− astrocytes accumulated Top1-dependent breaks at levels 3-fold above WT cells after CPT treatment, although at lower levels than Tdp1−/− astrocytes (Fig. 2a). Because cells are non-replicating, DNA breaks from trapped Top1cc are mostly single stranded breaks. In contrast, astrocytes deficient in the ATM-related DNA-dependent protein kinase, catalytic subunit (DNA-PKcs/Prkdc), repaired Top1-induced DNA damage at similar levels to WT astrocytes indicating the relative specificity of the Atm−/− defect (Fig. 2a). Additionally, cells derived from a mouse model of ATLD (Mre11ATLD/ATLD) also failed to show a repair defect after CPT treatment, indicating that the role of Atm in preventing Top1-associated strand breaks is not Mre11-dependent (results not shown). We also confirmed that DNA breaks in these non-replicating cells were transcription-associated Top1 damage21, 22, 32 as the transcription inhibitor 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole abrogated CPT-induced DNA damage (Suppl. Fig. 1).
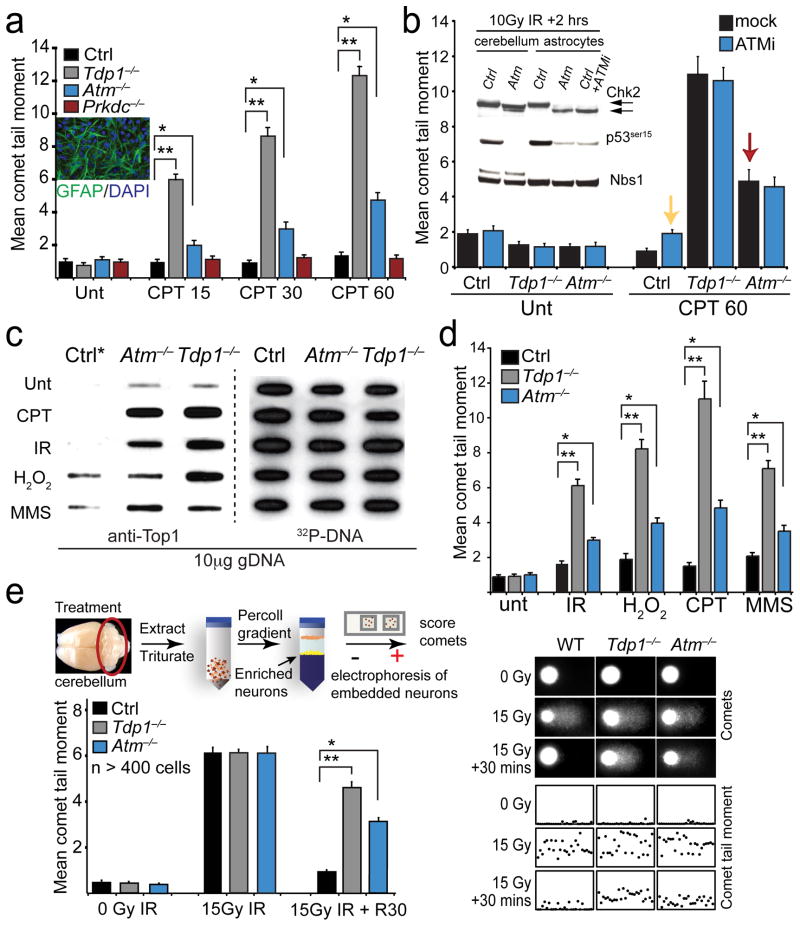
Fig. 2. Atm is required for the normal response to the topoisomerase 1 poison, camptothecin.
a. The repair kinetics of quiescent astrocytes following treatment with the Topoisomerase-1 poison camptothecin (CPT) at the indicated time points is shown. Although Top1-induced DNA breaks are repaired more slowly in Atm−/− astrocytes, deficiency in the related kinase, DNA-dependent protein kinase, catalytic subunit (Prkdc−/−) results in comparable DNA single-strand break repair rates as wild-type (Ctrl) astrocytes. Inset panel shows quiescent GFAP-positive Atm−/− astrocytes. b. Inhibiting Atm kinase activity fails to recapitulate the repair defect observed in Atm−/− cells after CPT. Western blot analysis (inset) of irradiated wild type (Ctrl) astrocytes co-treated with 10 μM ATM inhibitor KU55933 (ATMi) and radiation confirms Atm inhibition by defective DNA damage-induced Chk2 modification (black arrows) and p53 phosphorylation in Atm−/− cerebella and astrocytes. NBS1 was used as a loading control. Comet analysis (bar graph) indicates Atm−/− astrocytes (red arrow) accumulate significantly more CPT-induced DNA damage than ATMi-treated ctrl astrocytes (yellow arrow), indicating ATM kinase-independent repair of Top1-DNA lesions. Full-length Western blots are presented in Supplementary Figure 11. c. ICE analysis of quiescent primary murine astrocytes following treatment with DNA damaging agents can result in accumulation of Top1cc. Treatment conditions were; 14 μm CPT for 60 mins at 37°C; 20Gy IR followed by 60 mins recovery at 37°C; 150 μm H2O2 for 5 mins at 4°C followed by 60 mins recovery at 37°C; 0.20 mg/ml MMS for 10 mins at 37°C. Top1cc were identified by blotting genomic DNA with anti-Top1 and gDNA levels were assessed by re-probing with 32P-labelled mouse ES cell genomic DNA (32P-DNA). d. Alkaline comet analysis of quiescent Atm−/− astrocytes show defective DNA single-strand break repair after treatments listed for ‘c’. For each in vitro comet assay, 100 cells/comet corresponding to each genotype and treatment were analyzed and experiments were performed in triplicate (total of n=300 cells/genotype/treatment). e. In vivo comet analysis comparing relative DNA strand break repair rates amongst ctrl, Atm−/− and Tdp1−/− cerebellar granule cell neurons following ionizing radiation (15 Gy) and a 30 min recovery. Bar graphs represent mean comet tail moments from experiments that were repeated in duplicate (2 mice/treatment) with cells isolated from each cerebella also measured in duplicate (total n=400 independent comet tail moments measured per line/treatment); error bars represent standard error of means (S.E.M.). Scatterplots indicate representative cellular comet tail moments from each corresponding cell/treatment type. For all graphs */** denotes p-values < 0.0001.
ATM has been implicated in the regulation of Tdp1 activity33, 34. Therefore we assessed Tdp1 enzymatic activity in WT and Atm−/− brain. Quantification of the cleavage rate of an oligonucleotide substrate that mimics the Top1 protein-linked DNA 3′-terminus revealed no difference in Tdp1-dependent product formation in Atm−/− cerebellar extracts compared to WT, indicating Atm does not affect Tdp1 enzymatic activity (Suppl. Fig. 2). This suggests that the role of Atm in regulating Top1cc is not via Tdp1.
The kinase activity of ATM is critical for phosphorylation of various substrates that activate cell cycle arrest or apoptosis after DNA damage8–10. Therefore we determined if ATM kinase activity was necessary to prevent Top1cc-associated DNA strand breaks. Using the KU55933 ATM inhibitor35 (ATMi), we compared Atm−/− astrocytes to ATMi-treated WT astrocytes in the presence of CPT. Unlike Atm−/− astrocytes, ATMi-treated WT astrocytes showed normal DNA repair profiles (Fig. 2b). Inhibition of Atm kinase activity in these experiments was confirmed by abrogation of post-translational modification of Chk2 and p53 after DNA damage (Fig. 2b, inset). These data imply that an aspect of the Atm protein, independent of Atm kinase activity, mediates Atm-dependent Top1cc removal to allow DNA repair.
In addition to the specificity of CPT towards Top1, various DNA lesions including single strand breaks and abasic sites can also result in increased Top1cc that are irreversible23. We compared the relative effects of other genotoxins towards Top1cc accumulation, and found that the loss of Atm or Tdp1 also increased Top1cc formation after ionizing radiation (IR), H2O2 or methylmethane sulphonate (Fig. 2c). Correspondingly, DNA damage in Atm−/− or Tdp1−/− astrocytes was significantly increased compared to WT cells after treatment with these genotoxins (Fig 2d). Atm−/− cerebellar granule neurons also showed an ~3-fold higher level of unrepaired DNA breaks after IR compared to WT counterpart (Fig. 2e). Tdp1−/− granule neurons also showed elevated DNA breaks after IR, although the levels were relatively less than the comparative 3-fold increase observed over Atm−/− astrocytes (Fig. 2d), this possibly reflects intrinsic cell-type differences or cell culture effects. These data indicate that various types of DNA damage including oxidative lesions can increase Top1cc formation, compromising DNA integrity after Atm or Tdp1 loss. Notably, the antioxidant N-acetylcysteine significantly reduced the levels of DNA breaks after CPT in either Atm−/− or Tdp1−/− astrocytes, suggesting oxidative stress-induced DNA breaks can exacerbate damage by trapping Top1cc (Suppl. Fig 3).
Atm regulates DNA damage-dependent Top1 degradation
Removal of trapped Top1cc is associated with proteasome-mediated Top1 degradation, which involves protein modification by ubiquitination and sumoylation, with Tdp1 cleaving the Top1 peptide from DNA21, 23, 30, 36. To determine if ATM modulates Top1cc levels via Top1 degradation, we examined the effect of the proteasome inhibitor MG132 after CPT in Atm deficient and control cells. We found that total Top1 levels were decreased in Ctrl cells after CPT, but less so in Atm−/− cells (Fig. 3a). Consistent with the CPT-induced decrease in Top1 being proteasome dependent, co-treatment with MG132 and CPT elevated Top1 and Top1cc levels in Ctrl cells by 4–5 fold (Fig. 3a). In contrast, Top1 and Top1cc levels in Atm−/− cells were only marginally increased after MG132 suggesting that Atm modulates proteasome-mediated Top1 turnover (Fig. 3a). Accordingly, under these experimental conditions elevated Top1cc levels after MG132 and CPT treatment resulted in a marked increase in DNA breaks in Ctrl astrocytes (Fig. 3b). Notably, MG132 treatment resulted in levels of DNA damage in control cells that were similar to Atm−/− cells. These data imply that Atm has a predominant role in directing proteasome-mediated turnover of Top1 in trapped Top1cc. Additionally, proteasome inhibition also attenuated Atm signaling in WT cells after CPT as indicated by reduced Atm (p-ser-1987) and Kap1 (p-ser-824) phosphorylation (Fig. 3c).
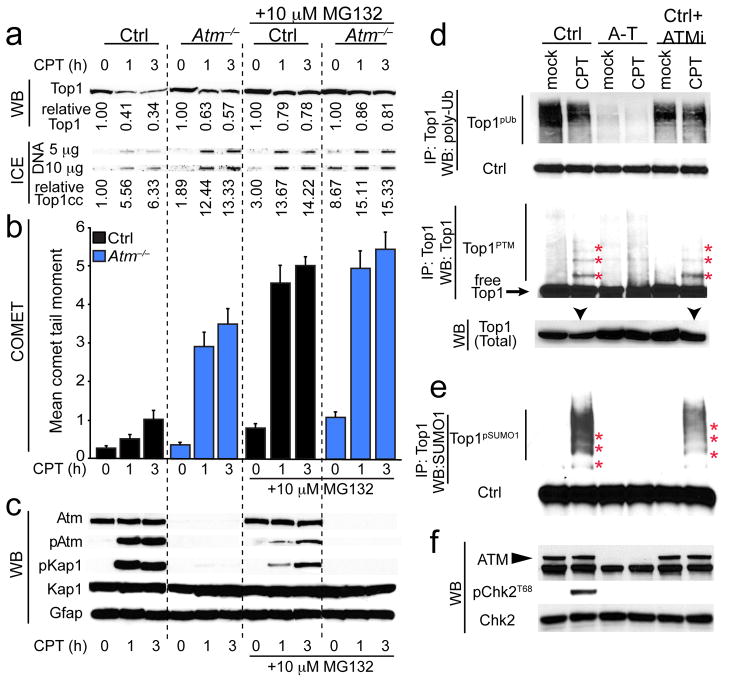
Fig. 3. Atm modulates Top1 turnover after CPT treatment.
a. Top 1 levels are higher in Atm−/− astrocytes compared to WT cells after CPT. The proteasome inhibitor MG132 inhibited Top1 turnover after CPT treatment in control cells. Top1cc levels as determined by the ICE assay correspond to CPT-induced trapping of Top1-DNA and reduced Top1 turnover. Top1/Top1cc quantitation is normalized to untreated control (Ctrl) levels. b. Treatment with MG132 increased the levels of DNA damage, particularly in control cells. c. MG132 also dampened Atm-dependent signaling as shown by decreased Atm and Kap1 phosphorylation. Total Kap1 and Gfap levels show equal protein loading. d. Poly-ubiquitin immunoblots of immunoprecipitated Top1 show that ubiquitination of Top1 is markedly reduced in human A-T lymphoblastoid cells, and that these ubiquitin levels are unaffected by either CPT or KU55933 treatment. Top1 immunoprecipitated from CPT-treated control cells and blotted with Top1 antibodies showed that Top1 migrated as a collection of higher molecular weight species (red asterisks) reflecting Top1 posttranslational modification (Top1PTM). Reduced amounts of post-translationally modified Top1 are found in Top1 immunoprecipitates from ATM−/− lymphoid cells after CPT-treatment. Like CPT-treated control cells, a comparable amount of Top1 is immunoprecipitated from CPT/KU55933 co-treated control lymphoblasts. Immunoblots of extracts prior to immunoprecipitation show reduced total Top1 expression in CPT-treated control cells (black arrows) compared to other cell-types and treatments. e. High molecular weight Top1 bands from Top1 immunoprecipitates are immununoreactive to anti-SUMO1 antibody (red asterisks), thus indicating that Top1 undergoes poly-sumoylation (Top1pSUMO1). In ATM−/− cells, Top1pSUMO1 is reduced after CPT-treatment, while similar to CPT-treated control cells, a comparable amount of Top1pSUMO1 is immunoprecipitated from CPT/KU55933 co-treated control lymphoblasts. f. Western blots indicate absence of ATM in A-T cells, while KU55933 treatment of control cells showed defective CHK2 phosphorylation after CPT treatment, indicating effective ATM inhibition. Full-length Western blots are presented in Supplementary Figure 11.
We also compared the above to human A-T cells. We again found a marked reduction in total Top1 protein in CPT-treated control cells, compared with a ~4-fold higher level of Top1 in the CPT-treated A-T counterparts, consistent with a requirement for Atm in Top1 turnover (Suppl. Fig. 4). Consistent with these results, ICE assays showed increased Top1cc were trapped in A-T cells compared to WT (Suppl. Fig. 4). Co-treatment of control cells with ATMi and CPT resulted in Top1 down-regulation comparable to CPT-only treatment alone, indicating that similar to our earlier data (Fig. 2), Top1 down-regulation is not ATM kinase-dependent (Suppl. Fig 4). Collectively, these data support ATM as a key regulator of Top1 degradation, in a kinase-independent manner after CPT.
To understand how ATM modulates Top1 turnover, we examined ubiquitination and sumoylation status of Top1, as both modifications are important for turnover of this protein21, 36, 37. Analysis of Top1 immunoprecipitates from CPT-treated cells showed ubiquitinated Top1 in control cells (Fig. 3d), but a marked reduction of ubiquitinated Top1 in A-T cells. Further, Top1 immunoprecipitates showed a characteristic Top1-immunoreactive ladder in control cells (Fig. 3d,e; asterisks) reflecting post-translationally modified Top1 (Top1PTM), while in A-T cells this laddering was reduced. These high molecular weight Top1 bands also bind anti-SUMO1 antibody after CPT treatment, and similarly this Top1pSUMO1 immunoreactivity was markedly reduced in A-T cells (Fig. 3e; asterisks). Co-treatment of control cells with ATMi and CPT revealed similar Top1 laddering, confirming that ATM kinase activity is dispensable for these modifications (Fig. 3d–f). We also found that knockdown of ATM, but not TDP1, reduced sumoylation of Top1 after CPT treatment in human cells (Suppl. Fig. 5). Collectively, these results indicate that ATM regulates Top1 degradation in response to DNA damage, in a manner independent of ATM kinase activity or TDP1, to attenuate Top1cc levels thereby preventing DNA damage.
ATM is essential for DNA damage signaling from trapped Top1
Given that a prime role of ATM is as a serine/threonine protein kinase8–10, it was unexpected that this enzymatic activity was dispensable for Top1cc regulation and DNA strand break repair. A prominent feature of DNA damage signaling is the phosphorylation of histone H2AX on serine 139 by ATM or other related kinases (e.g. Prkdc)10, as an early event in establishing the DDR. Therefore, we monitored formation of phosphorylated histone H2AX (γH2AX) in response to CPT and other genotoxins, using quiescent primary cells to avoid replication-associated damage. We found a pronounced defect in the induction of γH2AX after CPT treatment specifically in Atm−/− cells compared to Tdp1−/−, Prkdc−/− or WT counterparts. This indicates that Atm is uniquely required for DNA damage signaling after CPT (Fig. 4a); comparable results using CPT have also been observed in postmitotic neurons22. In contrast to CPT, IR induced similar levels of γH2AX in all genotypes examined (Fig. 4a). CPT-induced γH2AX foci also did not form in Atm−/− MEFs or A-T fibroblasts (Fig. 4b and Suppl. Fig. 6). Additionally, analysis of Atm−/−;Tdp1−/− double knockout cells showed that γH2AX formation in response to CPT is dependent upon Atm, even after Tdp1 inactivation (Fig. 4b). However, CPT-induced γH2AX occurred at similar levels to controls in cells from the A-T-related disease, Nijmegen Breakage Syndrome (NBS; Fig. 4c and Suppl. Fig. 6), and Mre11ATLD/ATLD (not shown), indicating that ATM-dependent phosphorylation of H2AX after CPT is not MRN-dependent.
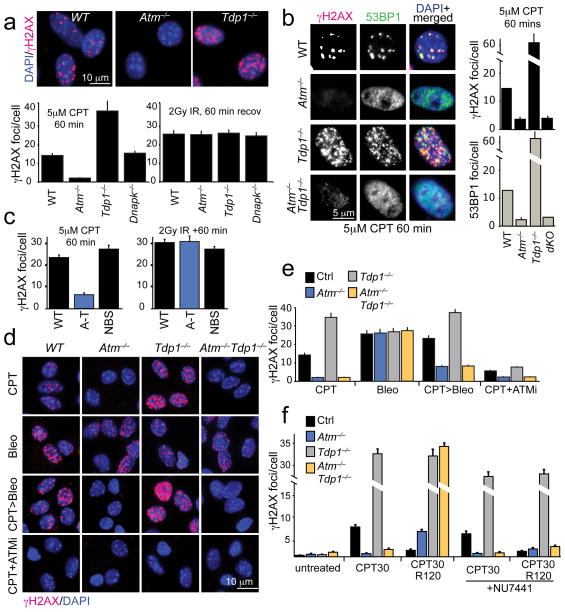
Fig. 4. Atm is essential for DNA damage signaling after CPT treatment.
a. Primary Atm−/− astrocytes form few γH2AX foci after CPT treatment (5 μM for 60 min) while abundant γH2AX foci are seen in WT and Tdp1−/− astrocytes. All genotypes show equivalent levels of γH2AX after IR. Upper panels show immunofluorescence analysis of typical γH2AX foci, which are quantified in the graphs below, as are the foci observed after IR. b. Similar to astrocytes, murine embryonic fibroblasts (MEFs) also exhibit Atm-dependent γH2AX after CPT. 53BP1 foci also show a similar induction after CPT and co-localize with γH2AX foci; these data are quantified in the adjacent graphs. c. In contrast to ATM−/− cells (A-T), the loss of NBS1 does not affect γH2AX foci formation upon CPT treatment. d. While bleomycin treatment (10 μg/ml 30 mins) induces γH2AX foci at similar levels in WT, Atm−/− and Tdp1−/− cells, pre-treatment with CPT prevents bleomycin-induced γH2AX foci formation in Atm−/− and Atm−/−;Tdp1−/− cells. The ATM inhibitor (ATMi) KU55933 prevents CPT-induced γH2AX foci formation indicating that Atm kinase activity is required for H2AX phosphorylation. e. Quantitation of γH2AX foci/cell for the different treatments and genotypes presented in ‘d’ is shown. f. When CPT treated (5μM CPT, 60mins) Atm−/− or Atm−/−;Tdp1−/− cells MEFs are subsequently incubated with CPT-free media, DNA damage signaling is activated in a DNA-dependent protein kinase, catalytic subunit (DNA-PKcs/Prkdc)-dependent manner as γH2AX foci fail to form in the presence of the DNA-PKcs inhibitor, NU7441 (2μM). For all foci quantification experiments, 30 cells for each cell line and corresponding treatment were counted and experiments were repeated in quadruplicate (total n=120 independent cells measured per line/treatment). Bar graphs represent mean cellular foci values of all replicates, error bars represent standard error of means (S.E.M.) and p-value is calculated using student’s unpaired t-test. The y-axis on graphs in ‘b’ and ‘f’ is non-linear to indicate increased foci number in specific genotypes after DNA damage.
We also examined γH2AX formation in Atm−/− cells after dual genotoxins, by co-treating with CPT and bleomycin. Despite bleomycin activating γH2AX in Atm−/− cells, pre-treatment with CPT resulted in minimal bleomycin-induced γH2AX formation implying that CPT-induced damage suppresses DNA damage signaling from ATM-related kinases (Fig. 4d,e). However, after CPT washout (a 30 min CPT pulse followed by a 120 min recovery), γH2AX formed in Atm−/− cells and at higher levels in Atm−/−Tdp1−/− astrocytes, and these foci were suppressed by the Prkdc inhibitor NU7441 (Fig. 4f). Because replication-associated breaks don’t occur in these quiescent cells, the DNA-dependent protein kinase (Prkdc) likely becomes activated as Top1cc lesions are processed. Collectively, our findings indicate a bi-modal response of ATM to CPT-induced DNA damage; ATM kinase activity is essential for DNA damage signaling as shown by γH2AX formation after CPT treatment, while DNA repair (resolution of DNA strand breaks associated with Top1cc) is kinase-independent. Thus these data show that ATM has a unique role, independent of Tdp1, in responding to Top1cc-induced DNA damage.
Synthetic lethality occurs after dual inactivation of Atm and Tdp1
Although the Atm−/− brain accumulates Top1cc, these mice retain apparently normal neurological function and exhibit no overt neuropathology38. A similar situation is found with Tdp1−/− mice27. We reasoned that if Atm and Tdp1 independently regulate Top1cc, intercrossing Atm−/− and Tdp1−/− mice would increase the levels of trapped Top1cc in the brain, and this should be an effective means to assess the in vivo impact of this lesion. Remarkably, we observed a >90% reduction in viability of Atm−/−;Tdp1−/− compound mutant mice (Table 1 and Suppl. Table 1). Analysis during development revealed that most Atm−/−;Tdp1−/− embryos were smaller than controls and lost viability between E13.5-E16.5, a stage at which Top1cc are maximal (Fig. 1c). Since SCAN1 and A-T are neurodegenerative syndromes, we determined the extent to which Atm and Tdp1 loss specifically impacts the nervous system. To do this we used mice in which Atm was inactivated in neural progenitors using Nestin-cre and a conditional Atm allele39 to generate AtmNes-cre mice. AtmNes-cre;Tdp1−/− mice showed a similar phenotype to Atm−/−;Tdp1−/− mice, with premature lethality between E13.5-E16.5 indicating that the lethality upon co-inactivation of Atm and Tdp1 was related to effects upon the nervous system (Table 1 and Suppl. Table 1).
Table 1. Atm loss exacerbates Tdp1 deficiency.
Genetic analysis shows synthetic lethality after coincident loss of Tdp1 and Atm. Compound mutants involving DNA double strand break repair (Lig4 or Prkdc) or MRN (Mre11ATLD/ATLD) deficiency do not affect the expected rate of generation of compound mutant mice. Mouse genetic inheritance ratios were calculated using standard rules for Mendelian genetic inheritance: “Expected” and “Observed” ratios were calculated based upon parental genotypes and the number of live-born mice.
| Genotype | Expected N= | Observed N= | |
|---|---|---|---|
| Atm−/− × Tdp1−/− | Control | 969 | 1083 |
| Mutant | 125 | 11 | |
| AtmNes-cre × Tdp1−/− | Control | 360 | 474 |
| Mutant | 120 | 6 | |
| Mre11ATLD/ATLD × Tdp1−/− | Control | 105 | 108 |
| Mutant | 15 | 12 | |
| Lig4Nes-cre × Tdp1−/− | Control | 284 | 283 |
| Mutant | 9 | 10 | |
| Prkdc−/− × Tdp1−/− | Control | 166 | 166 |
| Mutant | 9 | 10 |
To further understand the basis for the genetic interaction between Atm and Tdp1, we considered if the phenotype of the double mutant mice might actually reflect the established role of Atm in responding to DSBs8–10, rather than a specific function in regulating Top1cc. Therefore we generated compound mutant mice of Tdp1−/− and DNA double strand break repair deficiency, which included DNA Ligase IV deficiency (Tdp1−/−;Lig4Nes-cre) and also the catalytic subunit of the DNA-dependent protein kinase (Tdp1−/−;Prkdc−/−). However, in both cases, the double mutant mice were born at normal Mendelian ratios and did not show any overt phenotype (Table 1 and Suppl. Table 1). We also generated Tdp1−/−;Mre11ATLD/ATLD and Tdp1−/−;Nbs1ΔB/ ΔB double mutant mice to determine if the MRN complex, which is critical for ATM activation8–11, particularly in the nervous system5, was important for Atm activity in the context of Tdp1 inactivation. However, we again found that MRN/Tdp1 double mutants were born at Mendelian frequency without any apparent phenotype (Table 1 and Suppl. Table 1). Collectively, these data indicate that the neuroprotective synergy observed between Atm and Tdp1 likely reflects a role of Atm that is different to its established function in DNA double strand break signaling and activation by the MRN complex.
Atm and Tdp1 are required for CNS development
To understand how dual loss of Atm and Tdp1 affects the nervous system, we examined mutant embryos at various developmental stages. Immunohistochemical analyses of Atm−/−;Tdp1−/− and AtmNes-cre;Tdp1−/− embryos revealed substantial levels of DNA damage as indicated by γ-H2AXpositive cells throughout the developing forebrain, midbrain and cerebellum (Fig. 5a and Suppl. Figs. 7 and 8). Consistent with increased DNA damage, widespread p53 immunostaining and apoptosis was localized primarily in the proliferative ventricular zone (Fig. 5b and Suppl. Figs. 7 and 8). This contrasts WT, Atm−/− or Tdp1−/− embryos, where only sporadic apoptosis and no γH2AX or p53 immunopositive cells were present (Fig. 5a, b and Suppl. Figs. 7 and 8). While the appearance of γH2AX in Atm−/−Tdp1−/− neural tissues seems contradictory to our cellular studies showing Atm is essential for γH2AX formation after CPT (Fig. 4), the in vivo results reflect Top1cc collision with replication forks, which generates DNA double strand breaks, and the subsequent activation of other DNA damage-responsive kinases such as Atr. We also found high levels of apoptosis during early neurogenesis, revealed by active caspase-3 and TUNEL in Atm−/−;Tdp1−/− in E12.5 embryos (Fig. 5c). DNA damage or apoptosis was absent outside of the Atm−/−;Tdp1−/− and AtmNes-cre;Tdp1−/− embryonic nervous system (data not shown).
Fig. 5. Compound Atm−/− and Tdp1−/− mutants are usually lethal during development.
a. Analysis of the E14.5 Atm−/−;Tdp1−/− developing nervous system reveals an accumulation of DNA breaks (γH2AX; arrowheads). In the double-deficient embryonal brain, pronounced γH2AX foci (red signal demarcated by yellow arrowheads) are noted in the forebrain whereas few foci are noted in the Atm−/− control brain. PCNA immunostaining (green) identifies proliferating cells in the ventricular zone of the neocortex. b. Similarly, abundant anti-p53 immunoreactivity (arrowheads) is present in the in the ventricular zone of Atm−/−;Tdp1−/− embryonal forebrain, but are absent in the Atm−/− control. c. Apoptosis occurs early during neurogenesis in the Atm−/− ;Tdp1−/− forebrain, as at E12.5 activated caspase-3 staining and TUNEL (white arrows) is abundant. d. ICE analysis of Top1cc in the E14.5 embryonic brain. 20 μg of whole cell extract derived from E14.5 brain tissue were blotted (first row) to normalize total Topoisomerase-1 protein content amongst the E14.5 CNS tissue. The remaining three rows were blotted with increasing amounts (μg) of genomic DNA isolated from these tissues. Top1cc were immunodetected in genomic DNA (gDNA) using an anti-Top1 antibody. Relative Top1cc levels in Atm−/−;Tdp1−/− embryonal brain compared to controls (Ctrl, Atm−/− and Tdp1−/−) is listed below the blot. Embryonic tissue used in ICE assays are derived from gDNA pooled from 3 independent embryos. e. Western blot analysis of E14.5 Atm−/−;Tdp1−/− neural tissue used for ICE bioassay confirming tissue genotypes and the equivalent amounts of Top1 protein amongst the genotypes for ICE analysis. Full-length Western blots are presented in Supplementary Figure 11. f. Exposure of E12.5 embryos to topotecan results in apoptosis throughout the developing nervous system by E14.5. The graph shows levels of TUNEL positive cells in wild type and the relative increase in apoptosis in Atm−/− after topotecan injections of E12.5 embryos. For quantification, n=2 embryos for each genotype were analyzed. Bar graphs represent mean immunopositive cells for TUNEL or active caspase-3 measured within 0.27mm2 from three representative sections of the neocortex per embryo (total of n=6). Error bars represent standard error of means (S.E.M.) and statistics were calculated using unpaired student’s t-test. g. Disrupted neurogenesis and associated apoptosis after coincident Atm and Tdp1 deletion is prevented when p53 is attenuated via single or dual allele inactivation. Arrows indicate TUNEL positive cells. PCNA immunostaining identifies proliferating cells, while Tuj1 immunostaining identifies differentiating neurons. Top1cc levels in the E14.5 brain are not different in the presence or absence of apoptosis. For all graphs * denotes p-values < 0.001.
Given the synthetic lethality between Atm and Tdp1, and an absence of an overt phenotype after loss of Tdp1 and double strand break repair factors or regulators of Atm activation, we considered it likely that toxic Top1cc accumulation accounted for the Atm−/−;Tdp1−/− phenotype. We found substantially increased Top1cc levels in the double mutants compared to single mutants and controls (Fig. 5d), despite normal levels of Top1 protein in the double mutant embryos (Fig. 5e). As Top1cc are elevated in Atm−/− and Tdp1−/− neural tissues without associated pathology, our data suggest a DNA damage threshold may exist in neurons.
To directly test the pathogenic consequence of elevated Top1cc, we exposed developing embryos to the CPT analog topotecan (TPT). We found a reduction in embryo size and widespread apoptosis throughout the nervous system in response to TPT administration, which was strikingly reminiscent of the Atm−/−;Tdp1−/− phenotype (Fig. 5f and Suppl. Fig. 9). Atm−/− embryos were hypersensitive to topotecan with neural apoptosis being around 2-fold that of wild type embryos (Fig. 5f). Remarkably, despite systemic exposure of the embryos to topotecan and its wide bioavailability, we found that apoptosis was almost exclusively confined to the nervous system, underscoring the sensitivity of neural development to this DNA lesion (Suppl. Fig. 9).
Because apoptosis can potentially result in Top1cc accumulation19, 23, we generated Atm−/− ;Tdp1−/− mice in which p53 was also inactivated as a means to block p53-dependent apoptosis after DNA damage in the nervous system. While loss of one or both copies of p53 effectively blocked apoptosis (Fig. 5g) and rescued lethality (not shown), these triple mutants showed similar Top1cc levels to the Atm−/−;Tdp1−/− mice (Fig. 5g). These data confirm that elevated Top1cc levels in the double mutants result from the inactivation of Atm and Tdp1 and not as an outcome of apoptosis.
Top1cc can accumulate in the nervous system in response to DNA damage
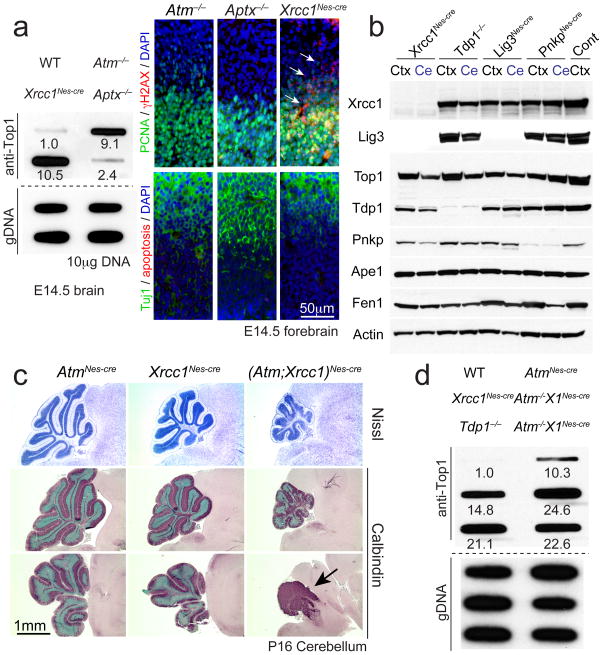
Top1cc can also accumulate when these normally transient complexes encounter various types of DNA lesions including strand breaks and abasic sites19, 23. Therefore we asked if Top1cc accumulation might be a pathologic lesion in other disease-relevant scenarios. To do this we examined mice with defective Xrcc1, a factor central for DNA repair via single strand break/base excision repair (BER)19. Loss of Xrcc1 in the nervous system leads to defective DNA repair and resultant DNA damage accumulation40. Conspicuously, increased Top1cc were present in Xrcc1Nescre proliferative neural tissue at E14.5 reflecting Atm or Tdp1 loss (Fig. 6a), further supporting the notion that DNA damage can result in trapped Top1cc. Importantly, we also examined Aptx−/− embryos, in which the adenyl hydrolase Aptx responsible for preventing AOA1 is inactivated, but failed to find increased Top1cc levels above controls (Fig. 6a). These findings indicate that the type of DNA damage influences Top1cc accumulation. Xrcc1 serves a critical scaffold function during DNA repair and its loss leads to dramatic destabilization and loss of the repair factor DNA ligase III (Lig3). Because TDP1 is known to interact with Xrcc1, we examined levels of the various base excision repair components to determine if Xrcc1 deletion results in destabilization of TDP1 (or other repair factors) as it does Lig3. If so, this could explain elevated Top1cc after Xrcc1 loss. However, while Lig3 was decreased in Xrcc1Nes-cre brain all other BER factors examined showed normal protein levels after Xrcc1 loss, indicating that increased Top1cc in Xrcc1Nes-cre tissue did not result from destabilization of TDP1 (Fig. 6b).
Fig. 6. Top1cc can arise in the nervous system in response to DNA damage.
a. Neural inactivation of Xrcc1 but not Aptx results in elevated Top1cc during development. Values for the ICE assay indicate relative signal compared to wild type (WT). Loss of Xrcc1, but not Atm or Aptx results in increased γH2AX foci, although these were not associated with widespread apoptosis. PCNA immunostaining identifies proliferating cells, while Tuj1 immunostaining identifies differentiating neurons. b. Western blot analysis of base excision repair factors shows that while Xrcc1 loss leads to destabilization of Lig3, other factors such as Tdp1 and Top1 are present at normal levels. Other mutant genotypes serve as controls for protein immuno-detection; Ctx is P5 cortex and Ce is P5 cerebellum. Full-length Western blots are presented in Supplementary Figure 11. c. The P16 (Atm;Xrcc1)Nes-cre cerebellum is markedly affected during neural development (arrow). Nissl staining shows general cerebellar morphology, while calbindin immunostaining identifies Purkinje cells. Bottom panels show a more lateral section of the cerebellum. d. Dual inactivation of Atm and Xrcc1 results in elevated Top1cc levels in the P16 cerebellum. Values for the ICE assay indicate relative signal compared to WT.
We next generated (Atm;Xrcc1)Nes-cre mice to determine the consequences of coincident Atm and Xrcc1 inactivation toward Top1cc occurrence. While Xrcc1Nes-cre mice show a mild reduction in cerebellar size40, dual inactivation of these factors led to marked neural development defects, involving profound cerebellar atrophy and associated Top1cc accumulation in cerebellar tissue (Fig. 6c, d). Levels of Top1cc were higher during neural development than in the P16 cerebellum (Fig. 6d), suggesting that in Xrcc1Nes-cre tissue these lesions are detrimental during development. Accordingly, in (Atm;Xrcc1)Nes-cre neural tissue, we observed pronounced apoptosis that commenced from ~E12.5 onwards (data not shown). Although Top1cc are elevated after Xrcc1 inactivation, it is also likely that the effects of direct DNA strand breaks after Xrcc1 loss additionally exacerbates the (Atm;Xrcc1)Nes-cre phenotypic abnormalities. These data suggest that Top1cc may be a general DNA lesion impacting the nervous system in DNA repair-deficient human diseases.
Discussion
The precise DNA lesions responsible for neurodegeneration in a spectrum of human genome instability diseases remain unknown. A prime example is that of A-T, a debilitating childhood neurodegenerative syndrome associated with defective DNA damage signaling9. Here, we show that in the nervous system, in addition to its activation by DNA double strand breaks8–10, ATM is required to regulate Top1cc, a transient topoisomerase-DNA intermediate, which occurs frequently during normal cellular function. Our data indicate that ATM functions in a non-epistatic manner to TDP1, to regulate the fate of trapped Top1cc. However, these factors cooperatively function to reduce Top1cc levels, as indicated by the synergistic rise in Top1cc in the absence of both proteins. While Tdp1 has specific enzymatic activity to cleave Top1-DNA adducts, ATM regulates the SUMO/Ubiquitin-mediated turnover of Top1. In this regard, the later onset of neurodegeneration in SCAN1 may indicate that Tdp1 function in vivo is comparatively more critical for preventing transcription-associated Top1cc accumulation in post mitotic tissue.
Aberrant accumulation of Top1cc must be prevented, because the DNA strand break formed during topoisomerase activity is potentially detrimental to a cell. For example, during proliferation, replication fork collision with trapped Top1cc can lead to DNA double strand breaks and cell death. Additionally, Top1cc can be trapped by proximal oxidative DNA breaks that enhance their conversion into DNA damage24, 41, 42. Because oxidative stress potentially results in thousands of DNA single strand breaks per day19, 43, it may be a major contributor towards elevation of Top1cc levels in the nervous system, a tissue that carries a high oxidative load. Thus, DNA lesions associated with Top1cc are pervasive throughout the life of the nervous system, and regulation of this topoisomerase complex is a critical genome maintenance requirement in this tissue.
The canonical role of ATM is regulating the response to DNA double strand breaks9, 10. It is likely that this aspect of ATM operates in parallel with Top1cc regulation (Suppl. Fig. 10). In the nervous system, DNA double strand breaks can result in ATM-dependent apoptosis5, 38, in which ATM activation occurs via the double strand break-sensing MRN complex5, 11. Mutation in the Mre11 component of the MRN complex can lead to ATLD12, a neurodegenerative syndrome with similarity to A-T. Because ATLD reflects the importance of the MRN-ATM axis in responding to DNA double strand breaks, ATM regulation of Top1cc levels implicates an additional prevalent genotoxic lesion in A-T compared with ATLD. The more severe phenotype of A-T compared to ATLD, despite both syndromes having an early onset44, is consistent with the occurrence of this additional detrimental DNA lesion.
Our study points to an important neuroprotective role for regulation of Top1cc in the nervous system, thereby broadening the understanding of the etiologic lesions that contribute to neurodegenerative syndromes. In addition to A-T and SCAN1, this lesion may have a significant pathogenic impact in other syndromes associated with DNA repair deficiency and genome instability. Additionally, given the requirement for other classes of topoisomerases for multiple DNA transactions24, 45, it seems likely defects in these or factors that modulate their activity will impact neural function. For instance, defective topoisomerase activity has been implicated in autism spectrum disorder46, 47, possibly as these enzymes are important for transcriptional regulation of long genes linked to autism48. Furthermore, deficiency in Top3β has recently been identified as an RNA topoisomerase important in the function of the FRMP, a factor that prevents Fragile X mental retardation syndrome49, 50.
FULL ON-LINE METHODS
Mice
Generation of the germline Tdp1−/−, Atm−/−, Mre11ATLD/ATLD, Nbs1ΔB/ΔB, and conditional Atm, Xrcc1, p53 mouse lines have been previously described2, 27, 38, 39. Animals were housed in an AAALAC-accredited facility with a 12 hour day/night cycle. A maximum of 5 adult animals per cage are allowed. All animal experiments were carried out according to NIH regulations and were approved by the SJCRH animal care and use committee.
Isolation of primary cells
Primary astrocytes were prepared from P2 mouse brains as described previously27. Cortices were dissociated by passage through a 5-ml pipette and cells were resuspended in Dulbecco’s modified Eagle’s medium and Ham’s nutrient mixture F-12 (1:1 DMEM/F12, Gibco-BRL) supplemented with 10% fetal bovine serum (v/v), 1× glutamax, 100 U/ml penicillin, 100mg/ml streptomycin and 20 ng/ml epidermal growth factor (EGF; Millipore). Primary astrocytes were established in Primeria T-25 tissue culture flasks (Falcon) at 37°C in a humidified CO2-regulated (5%) incubator.
Neurosphere cultures were prepared using the Neurocult neural stem cell proliferation kit (STEMCELL Technologies) according to the manufacturer’s protocol. Briefly, E14.5 embryos were mechanically dissociated by trituration and then passed through a 40 μm cell strainer. Neural stem cells were then cultured in Neurocult neurobasal media supplemented with proliferation supplements and 20 ng/ml EGF (STEMCELL Technologies) in upright T-25 tissue culture flasks (Falcon) at 37°C in a humidified CO2-regulated (5%) incubator until neurospheres were established. For neurosphere growth assays, whole neurospheres were mechanically dissociated, counted and seeded at 5 × 105 cells/well in a 24-well plate and incubated for five days at 37°C.
Primary mouse embryonic fibroblasts were prepared from E13.5 embryonic mesenchyme. Tissue was minced using dissection scissors, trypsinized and resuspended in Dulbecco’s modified Eagle’s medium supplemented with 10 % fetal bovine serum (v/v), 1× glutamax, 100 U/ml penicillin, 100 μg/ml streptomycin and β-mercaptoethanol and established in T-25 tissue culture flasks (Falcon) at 37° C in a humidified CO2-regulated (5 %) incubator.
Control (Ctrl; GM06889) and A-T (GM01526) human lymphoblastoid cell lines (LCL) and Control (GM01876), A-T (GM05823) and NBS (GM07166) human fibroblast cell lines (HF) were obtained from the Coriell Institute for Medical Research (Camden, NJ, USA). LCLs were maintained in RPMI media supplemented with 15 % fetal bovine serum (v/v), 1× glutamax, 100 U/ml penicillin, 100 μg/ml streptomycin while HFs were maintained in DMEM media supplemented with 15 % fetal bovine serum (v/v), 1× glutamax, 100 U/ml penicillin, 100 μg/ml streptomycin.
Cell survival assays
Mouse embryonic stem cells (ES), neural stem cells (NSCs) and MEFs were plated in replicates of 8 onto 96-well plates (5000 cells/well). Following CPT treatment, WST-1 cell proliferation reagent (10 ml/well) was added, incubated at 37°C for 2 hours and read with a plate-reader at 450 nm (Bio-Rad). Cell survival was tabulated and graphed relative to untreated controls. Experiments were repeated in triplicate.
Cell treatments
For comet analysis, cells were treated with either 150 μM H2O2 (Thermo Fisher) for 10 min on ice, 14 μM camptothecin (CPT, Calbiochem) for various times at 37°C, methyl methanesulfonate (MMS, Sigma) for 10 min at various concentrations at 37°C, bleomycin (Bedford Laboratories) at 40 μg/ml for 30 min at 37°C, or γ–irradiation (20Gy using 137Cs). Following H2O2, bleomycin and IR treatments, cells were incubated for various times in drug-free medium at 37°C prior to analysis whereas following CPT and MMS treatments cells were immediately harvested for analysis. For transcriptional inhibition analysis, cells were preincubated with media containing 50 μM 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB, Sigma) for 30 mins at 37°C followed by replacement of media containing both 14 μM CPT and 50 μM DRB for 60 mins at 37°C. For ATMi analysis, cells were pre-incubated with media containing 10 μM KU55933 (Calbiochem or Selleck chem) for 60 mins at 37°C followed by either irradiation (10 Gy with 2 hr recovery at 37° C; for western analysis) or replacement of media containing both 14 μM CPT and 10 μM KU55933 for 60 mins at 37°C (for comet assay and ICE-T). For foci analysis (γH2AX/53BP1), confluent cells were treated with CPT (5mM; 60 mins, 37°C), IR (2 Gy, 60 mins recovery at 37°C), bleomycin (10 μg/ml, 60 mins at 37°C) or H2O2 (150 mM in PBS on ice, 60 mins recovery in media at 37°C) and underwent immunofluorescence analysis (below). For DNA-PKi analysis, following CPT treatment (5 μM CPT, 30 mins at 37°C), cells were washed 3 times and incubated with 2 μM NU7441 (2 hrs, 37°C) followed by immunofluorescence analysis. For proteasomal analysis, cells were pre-treated with 10 μM MG132 (1 hr at 37°C) prior to CPT co-treatment. Similarly, for NAC analysis, cells were pretreated with 30 mM NAC (1 hr at 37°C) prior to CPT co-treatment.
Alkaline comet analysis
Cells were resuspended in pre-chilled 1× PBS, mixed with an equal volume of 1.2% low-melting point agarose (Invitrogen) maintained at 42°C and immediately layered onto frosted glass slides (Fisher) pre-coated with 0.6% agarose and maintained in the dark at 4°C for all further steps. Slides were immersed in pre-chilled lysis buffer (2.5 M NaCl, 10 mM Tris-HCl, 100 mM EDTA (pH 8.0), 1 % (v/v) Triton X-100, 3 % (v/v) DMSO, pH10) for 1.5 hrs, washed with pre-chilled distilled water (twice for 10 min each) and placed into pre-chilled alkaline electrophoresis buffer (50 mM NaOH, 1mM EDTA, 1% DMSO) for 45 min. Electrophoresis was carried out at 95mA for 25 min, followed by neutralization in 0.4 M Tris-HCl (pH 7.0). Comets were stained with SYBR Green (1:10,000 in 1× PBS, Sigma) for 10 min. A minimum of 100 comet tail moments were measured using the Comet Assay IV system (Perceptive Instruments) coupled to an Axioskop2 plus microscope (Zeiss) at 200× magnification. Experiments were repeated in triplicate and the mean comet tail moments were calculated and graphed (n=3; 300 independent comet tail moments measured per line/treatment). Experimental samples were assigned randomized identity and analysis was performed “blind”.
For in vivo comet analysis, mice were irradiated (individually) with 15 Gy and cerebella were harvested (into cold CMF-PBS) either immediately or after 30 minutes recovery. Granule neurons were isolated via a percoll gradient as previously described (Katyal et al, 2007). Agarose-embedded granule neurons were subjected to comet analysis as described above. Experiments were repeated in duplicate, with each replicate also performed in duplicate (n=4; 400 independent comet tail moments measured per line/treatment). Experimental samples were assigned randomized identity and analysis was performed “blind”.
Tdp1 cleavage assay
WT and Atm−/− cerebellar tissue were homogenized (30 strokes with a dounce homogenizer) in cleavage lysis buffer (20 mM Tris–HCl pH 7.5, 10 mM EDTA, 1 mM EGTA, 100 mM NaCl, 1% Triton X-100 and Complete protease inhibitor [Roche]). Soluble cell extracts were recovered by centrifugation at 13 000 r.p.m. for 5 min at 4°C. Protein concentrations were determined by Bradford assay (BioRad). A 3′-linked biotinylated oligo (5′-AGCGCCGAAGGCGCTTCGA-3′) mimicking a Tdp1 substrate containing a phosphotyrosyl bond was end-labeled with 32P using T4 polynucleotide kinase, gel purified and incubated with cerebellar protein extracts, as previously described27. Enzymatic products were separated by 10% SDS-Urea PAGE and visualized and quantified by phosphorimaging analysis (Molecular Dynamics).
Immunodetection of Top1-DNA covalent complexes (ICE assay)
Top1-DNA covalent complexes were isolated using the immunocomplex of enzyme (ICE) bioassay as previously described 28. Briefly, cells or tissues were lysed in 1% sarkosyl with dounce homogenization (10 strokes for cells, 40 strokes for embryonic CNS tissue). DNA was sheered through a 26G needle (10 strokes) and cell lysates were gently layered onto a CsCl cushion and centrifuged in a NVT 90 rotor at 122 000 × g for 20 h at 25°C (Beckman-Coulter). The resulting pelleted covalent DNA-protein complexes were washed, resuspended in TE, and aliquots were diluted with 25 mM sodium phosphate buffer (pH 6.5) and applied to a nitrocellulose membrane by using a slot-blot vacuum manifold (Bio-Rad). Total protein extract was applied to adjacent slots as a control for total Top1 levels amongst tissues. Top1 protein and Top1-DNA complexes were immunodetected using an anti-Top1 polyclonal antibody (rabbit, 1:1000, Bethyl, cat# 302-590A), followed by HRPconjugated anti-rabbit secondary antibody and detection using ECL Prime on X-ray film. ICE blots were subsequently probed with 32P-labeled mouse (ES) or human (293T) genomic DNA (gDNA) to control for relative DNA loading. For astrocyte ICE assays, densitometric analysis of the Top1-DNA signal was performed using Image J. Experiments were performed at least in duplicate.
Western blot analysis
Protein extracts (cells or tissue) were prepared by using lysis buffer (50 mM Tris-HCl, 200mM NaCl, 0.2 % NP-40, 1 % Tween-20 (v/v), 1 mM NaF, 1 mM sodium vanadate, 50 mM β-glycerophosphate, 2 mM PMSF, and protease inhibitor cocktail (Roche) and quantified by Bradford assay (Bio-Rad). Proteins (50 μg per lane) were separated through a 4–12 % (w/v) Bis-Tris SDS polyacrylamide gel (Invitrogen) and transferred onto nitrocellulose membrane (Bio-Rad). Blots were immunostained with the following antibodies: anti-Chk2 (mouse, 1:1000, Upstate, cat# 05-649), anti-phospho-Chk2T68 (rabbit, 1:1000, Cell Signaling, cat# 2661), anti-p53ser15 (rabbit, 1:1000, Cell Signaling, cat# 9284), anti-Nbs1 antibody (rabbit, 1:500, Cell Signaling cat# 3002), anti-ATM (MAT3, mouse, 1:1000, Sigma; D2E2, rabbit, 1:2000, Cell Signaling, cat# 2873), anti-pATMS1981 (10H11.E12, 1:1000, Abcam, cat# ab36810), anti-Tdp1 (mouse, 1:1000, Abnova, cat# H00055775-A01), anti-Topoisomerase 1 (rabbit, 1:1000, Bethyl, cat# 302-590A), anti-KAP1 (rabbit, 1:2000, Abcam, cat# ab10484) and anti-phospho-KAP1S824 (rabbit, 1:1000, Bethyl, cat# A300-767A), followed by appropriate horseradish peroxidase–conjugated secondary antibodies (1:2,000, GE Healthcare) and detected using ECL or ECL Prime chemiluminescence reagent (GE Healthcare). Anti-actin (goat, 1:500, Santa Cruz Biotech, clone I-19, cat# sc-1616) and Ponceau staining of the transferred membrane were used as protein-loading controls. Densitometric analysis of the Top1 signal (following CPT treatment) was performed using ImageJ. Experiments were performed in duplicate.
Top1 immunoprecipitations
Control and A-T human lymphoblasts were used for Top1 immunoprecipitaion as we could not immunoprecipitate Top1 from mouse cells. Cells were pre-incubated with media containing inhibitors (mock/DMSO, 50μM DRB or 10μM KU55933; 60 mins at 37°C) followed by mock (DMSO) or camptothecin treatment (14μM, 60 mins at 37°C). Cells were collected, washed in 1× PBS, lysed (200 mM NaOH, 2 mM EDTA) and neutralized (100 mM HCl, 60 mM Tris-HCl) on ice and underwent sonication (3 × 10 secs bursts, setting 5; 550 sonic dismembrator, Fisher Scientific) in 1 × S7 nuclease buffer (5 mM MgCl2, 5 mM CaCl2, 0.5 mM DTT, 0.1 mM EDTA, 20 mM N-ethylmaleimide, 1× Complete protease inhibitors (Roche). Cell lysates were incubated with 1000 units of S7 nuclease (Roche) on ice for 20 mins and, centrifuged at 10,000 g for 30 mins at 4°C to remove genomic DNA. One milligram of protein extract (resulting supernatant) underwent Top1 immunoprecipitation (2 hours, 4°C) in radioimmunoprecipitation buffer (RIP buffer; 0.5 % NP-40, 0.25% sodium deoxycholate, 0.05 % SDS, 0.5 × PBS, 20 mM N-ethylmaleimide, 1× Complete protease inhibitors, Roche) using 10 μL (neat) anti-Top1 antibody (Bethyl, cat# 302-590A) and 50 μl Protein A/G PLUS agarose beads (Santa Cruz, sc-2003). Immunoprecipitates were washed 3 × RIP buffer and immunoblotted with anti-Top1 (1:1000), anti-SUMO1 (1:1000, Abcam, cat# ab32058) or anti-ubiquitin (1:1000, Santa Cruz, clone P4D1, sc-8017). Experiments were performed in triplicate.
TDP1 overexpression and ATM/TDP1 knockdown analysis
293T cells were transfected (using FuGene 6) with short-hairpin RNA constructs (Sigma) corresponding to human ATM (shATM; 5′-CCGGGCCGTCAACTAGAACATGATACTCGAGTATCATGTTCTAGTTGACGGCTTTTTG-3′ and 5′-CCGGCCTTTCATTCAGCCTTTAGAACTCGAGTTCTAAAGGCTGAATGAAAGGTTTTTG-3′) or a “scrambled” control (shScrambled). 24 hours following transfection, cells underwent puromycin selection (2 μg/ml) with a media change every 24 hours for 10 days. Stable-tranfectants were then re-transfected with shScrambled or shATM constructs along with either pFlag vector control, pFlag-TDP1 or an shRNA targeting human TDP1 (shTDP1; 5′-CCGGCCGATGAATCAAAGTGGTTATCTCGAGATAACCACTTTGATTCATCGGTTTTTG-3′). Following CPT treatment, cells were lysed and underwent Top1 immunoprecipitation to assess levels of Top1 post-translational modification.
Histology
Pregnant female mice underwent transcardial perfusion with 4% (w/v) buffered paraformaldehyde and harvested embryos were cryoprotected in buffered 25% sucrose (w/v) solution. Embryos sectioned sagitally at 10μm using an HM500M cryostat (Microm). Immunostaining was carried out with the antibodies listed below. For colorimetric visualization of positive signals, sections were incubated with antibodies overnight at room temperature after quenching endogenous peroxidase using 0.6% (v/v) H2O2 in methanol. Slides were washed with phosphate-buffered saline (PBS) three times, followed by incubation with biotinylated secondary antibody and avidin-biotin complex (Vectastain Elite kit, Vector Labs). Antibodies were used after citrate buffer–based antigen retrieval. Immunoreactive signals were visualized with the VIP substrate kit (Vector Labs) using the manufacturer’s protocol. Sections were counterstained with 0.1% (w/v) methyl green, dehydrated, and mounted in DPX (Fluka). For fluorescent detection of immunoreactivity, FITC- or Cy3-conjugated secondary antibodies (Jackson Immunologicals) were used and counterstained with 4′6-diamidino-2-phenylindole (DAPI; Vector Laboratories). For immunohistochemistry and immunocytochemistry, we used the following antibodies: γH2AX (Ser-139, rabbit, 1:200, Cell Signaling, cat# 2577), p53 (clone CM5, rabbit, 1:1000, Vector Laboratories, cat# VP-P956), PCNA (clone PC10, mouse, 1:500, Santa Cruz, cat# SC-56), Tuj1/β-tubulin III (mouse, 1:1000, Covance, cat# MMS-435P), Calbindin (mouse, 1:2000, Clone CB955, Sigma cat# C9848). Apoptosis was detected without antigen retrieval using anti-ssDNA antibody (rabbit, 1:300, IBL, cat# 18731). Experiments were performed in triplicate.
For fluorescent labeling of quiescent cortical astrocytes, human fibroblasts or MEFs, cells were grown to confluence on glass coverslips, fixed with 4% PFA in PBS for 10 min and permeabilized for 5 min in 0.5% Triton X-100/PBS. Cells were immunostained with antibodies diluted in PBS/3% BSA; anti-GFAP (rabbit, 1:500; Abcam, cat# 7779), anti-γH2AX (Ser139, rabbit, 1:500; Millipore, cat# JBW301), anti-53BP1 (rabbit, 1:500; Bethyl, cat# A300-272A), followed by Alexa 488/555-conjugated secondary antibodies or Alexa 488-conjugated phalloidin and counterstained with DAPI. Experiments were performed in triplicate.
In situ cell counts
To determine apoptosis indices in the developing embryonal brain, E12.5 or E14.5 embryos were subjected to quantitative analysis. Two embryos for each genotype were analyzed. Immunopositive signals for TUNEL or active caspase-3 were measured within 0.27mm2 from at least three representative sections of the neocortex per embryo. All statistics were calculated using unpaired student’s t-test. Experiments were performed in duplicate.
Topotecan administration
Topotecan hydrochloride (Hycamtin; Glaxo Smith Kline) is a camptothecin analogue that is widely bioavailable and readily penetrates neural tissue. Topotecan was dissolved in 0.9 % NaCl and diluted to a concentration of 100 μg/ml. Plugged female mice were administered Topotecan via intraperitoneal injection at a dose of 0.5 μg/g body weight at E12.5 and E13.5 and then collected E14.5. The female was perfused with 4% PFA and the embryos were collected in PFA and then cryoprotected and processed for sectioning.
Statistics
Bar and line graphs represent mean values of all replicates, error bars represent standard error of means (S.E.M.) and p-value is calculated using student’s unpaired t-test. For comet assays, a minimum of 100 cells for each treatment were counted and experiments were repeated in triplicate (in vitro comet) or quadruplicate (in vivo comet); exact cell numbers/comets are indicated in their respective figure legends. Mean comet tail moments were calculated and graphed. Experimental samples were assigned randomized identity and analysis was performed “blind”.
For foci quantification experiments, a minimum of 30 cells for each treatment were counted and experiments were repeated in quadruplicate (exact cell numbers/comets are indicated in figure legends). Mean cellular foci values were calculated and graphed. Experimental samples were assigned randomized identity and analysis was performed “blind”.
Mouse genetic inheritance ratios were calculated using standard rules for Mendelian genetic inheritance. Expected and observed genetic ratios were calculated based upon the actual number of live-born mice (indicated by n). No statistical methods were used to pre-determine sample sizes but are similar to those generally employed in the field. Data distribution was assumed to be normal but this was not formally tested.
A methods checklist is available with the supplementary materials.
Supplementary Material
1
10
11
12
13
14
2
3
4
5
6
7
8
9
Acknowledgments
We thank Dr. Eroica Soans and Margarita Mishina for assistance with the ICE bioassay, Bozena Kuzio for general technical assistance, Dr. Fred Alt (Children’s Hospital of Boston) for Prkdc −/− mice and Drs. Keith Caldecott and Sherif El-Khamisy (U. Sussex) and Ramon Klein-Geltink (SJCRH) for helpful discussions and Steven Foster (MSKCC) for help analyzing the mice. We also thank the SJCRH Animal Resource Center and the Transgenic Core Unit for support with mouse work. PJM is supported by the NIH (NS-37956, CA-96832), the CCSG (P30 CA21765) and the American Lebanese and Syrian Associated Charities of St. Jude Children’s Research Hospital. JLN is supported by the NCI (CA52814 and CA82313). JHJP is supported by the NIH (GM59413), the Geoffrey Beene Foundation and the Goodwin Foundation. YSL is supported by the SRC program (2011-0030833). SK is a Neoma Boadway AP Endowed Fellow and is supported by grants from the University of Manitoba, CancerCare Manitoba and a Manitoba Health Research Council (MHRC) Establishment award.
Footnotes
AUTHOR CONTRIBUTIONS.
S.K. and P.J.M. conceived and planned all project experiments and produced the final version of the manuscript. S.K. performed all experiments with contributions from K.N. (in vitro TDP1 cleavage assay), YS.L., M.S. and H.R.R. (generation of the mutant mice and additional technical support), S.D. and Y.L. (processing tissue for ICE bioassay and mouse colony management) and J.Z. (AtmNes-cre and ATMi immunoblotting experiments). J.H.J.P. contributed critical reagents and experimental results. J.L.N. contributed to experimental design and the interpretation of results and the preparation of the final version of the manuscript.
References
- 1.McKinnon PJ. DNA repair deficiency and neurological disease. Nat Rev Neurosci. 2009;10:100–112. doi: 10.1038/nrn2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Driscoll M, Jeggo PA. The role of double-strand break repair - insights from human genetics. Nat Rev Genet. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 4.McKinnon PJ. Maintaining genome stability in the nervous system. Nat Neurosci. 2013;16:1523–1529. doi: 10.1038/nn.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shull ER, et al. Differential DNA damage signaling accounts for distinct neural apoptotic responses in ATLD and NBS. Genes Dev. 2009;23:171–180. doi: 10.1101/gad.1746609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suberbielle E, et al. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-beta. Nature Neuroscience. 2013;16:613–621. doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu T, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 8.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nature reviews. Molecular cell biology. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 9.McKinnon PJ. ATM and the molecular pathogenesis of ataxia telangiectasia. Annual review of pathology. 2012;7:303–321. doi: 10.1146/annurev-pathol-011811-132509. [DOI] [PubMed] [Google Scholar]
- 10.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nature reviews. Molecular cell biology. 2013;14:197–210. [PubMed] [Google Scholar]
- 11.Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nature reviews. Molecular cell biology. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor AM, Groom A, Byrd PJ. Ataxia-telangiectasia-like disorder (ATLD)-its clinical presentation and molecular basis. DNA Repair (Amst) 2004;3:1219–1225. doi: 10.1016/j.dnarep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Takai H, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 2002;21:5195–5205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKinnon PJ. ATM and the molecular pathogenesis of ataxia telangiectasia. Annual review of pathology. 2012;7:303–321. doi: 10.1146/annurev-pathol-011811-132509. [DOI] [PubMed] [Google Scholar]
- 15.Takashima H, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 16.Ahel I, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 17.El-Khamisy SF, et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 18.Date H, et al. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat Genet. 2001;29:184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 19.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 20.Alagoz M, Chiang SC, Sharma A, El-Khamisy SF. ATM deficiency results in accumulation of DNA-topoisomerase I covalent intermediates in neural cells. PLoS One. 2013;8:e58239. doi: 10.1371/journal.pone.0058239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CP, Ban Y, Lyu YL, Desai SD, Liu LF. A ubiquitin-proteasome pathway for the repair of topoisomerase I-DNA covalent complexes. The Journal of biological chemistry. 2008;283:21074–21083. doi: 10.1074/jbc.M803493200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sordet O, et al. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep. 2009;10:887–893. doi: 10.1038/embor.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pommier Y, et al. Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nature reviews. Molecular cell biology. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 25.Wu HY, Liu LF. DNA looping alters local DNA conformation during transcription. J Mol Biol. 1991;219:615–622. doi: 10.1016/0022-2836(91)90658-s. [DOI] [PubMed] [Google Scholar]
- 26.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 27.Katyal S, et al. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007;26:4720–4731. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nitiss JL, Soans E, Rogojina A, Seth A, Mishina M. Topoisomerase Assays. John Wiley & Sons; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian D, Rosenstein BS, Muller MT. Ultraviolet-induced DNA damage stimulates topoisomerase I-DNA complex formation in vivo: possible relationship with DNA repair. Cancer research. 1998;58:976–984. [PubMed] [Google Scholar]
- 30.Heideker J, Prudden J, Perry JJ, Tainer JA, Boddy MN. SUMO-targeted ubiquitin ligase, Rad60, and Nse2 SUMO ligase suppress spontaneous Top1-mediated DNA damage and genome instability. PLoS Genet. 2011;7:e1001320. doi: 10.1371/journal.pgen.1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 32.Sakasai R, Teraoka H, Takagi M, Tibbetts RS. Transcription-dependent activation of ataxia telangiectasia mutated prevents DNA-dependent protein kinase-mediated cell death in response to topoisomerase I poison. J Biol Chem. 2010;285:15201–15208. doi: 10.1074/jbc.M110.101808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiang SC, Carroll J, El-Khamisy SF. TDP1 serine 81 promotes interaction with DNA ligase IIIalpha and facilitates cell survival following DNA damage. Cell Cycle. 2010;9:588–595. doi: 10.4161/cc.9.3.10598. [DOI] [PubMed] [Google Scholar]
- 34.Das BB, et al. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J. 2009;28:3667–3680. doi: 10.1038/emboj.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hickson I, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 36.Lin CP, Ban Y, Lyu YL, Liu LF. Proteasome-dependent processing of topoisomerase I-DNA adducts into DNA double strand breaks at arrested replication forks. The Journal of biological chemistry. 2009;284:28084–28092. doi: 10.1074/jbc.M109.030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao Y, Sun M, Desai SD, Liu LF. SUMO-1 conjugation to topoisomerase I: A possible repair response to topoisomerase-mediated DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herzog KH, Chong MJ, Kapsetaki M, Morgan JI, McKinnon PJ. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science. 1998;280:1089–1091. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]
- 39.Lee Y, et al. ATR maintains select progenitors during nervous system development. The EMBO journal. 2012;31:1177–1189. doi: 10.1038/emboj.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee Y, et al. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nature neuroscience. 2009;12:973–980. doi: 10.1038/nn.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pourquier P, et al. Trapping of mammalian topoisomerase I and recombinations induced by damaged DNA containing nicks or gaps. Importance of DNA end phosphorylation and camptothecin effects. J Biol Chem. 1997;272:26441–26447. doi: 10.1074/jbc.272.42.26441. [DOI] [PubMed] [Google Scholar]
- 42.Pourquier P, et al. Effects of uracil incorporation, DNA mismatches, and abasic sites on cleavage and religation activities of mammalian topoisomerase I. J Biol Chem. 1997;272:7792–7796. doi: 10.1074/jbc.272.12.7792. [DOI] [PubMed] [Google Scholar]
- 43.Beal MF. Oxidatively modified proteins in aging and disease. Free Radic Biol Med. 2002;32:797–803. doi: 10.1016/s0891-5849(02)00780-3. [DOI] [PubMed] [Google Scholar]
- 44.Palmeri S, et al. Clinical Course of Two Italian Siblings with Ataxia-Telangiectasia-Like Disorder. Cerebellum. 2013 doi: 10.1007/s12311-013-0460-4. [DOI] [PubMed] [Google Scholar]
- 45.Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.King IF, et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501:58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoll G, et al. Deletion of TOP3beta, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat Neurosci. 2013;16:1228–1237. doi: 10.1038/nn.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu D, et al. Top3beta is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat Neurosci. 2013;16:1238–1247. doi: 10.1038/nn.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
10
11
12
13
14
2
3
4
5
6
7
8
9