Dynamic Reorganization of Metabolic Enzymes into Intracellular Bodies (original) (raw)
. Author manuscript; available in PMC: 2014 Jul 9.
Abstract
Both focused and large-scale cell biological and biochemical studies have revealed that hundreds of metabolic enzymes across diverse organisms form large intracellular bodies. These proteinaceous bodies range in form from fibers and intracellular foci—such as those formed by enzymes of nitrogen and carbon utilization and of nucleotide biosynthesis—to high-density packings inside bacterial microcompartments and eukaryotic microbodies. Although many enzymes clearly form functional mega-assemblies, it is not yet clear for many recently discovered cases whether they represent functional entities, storage bodies, or aggregates. In this article, we survey intracellular protein bodies formed by metabolic enzymes, asking when and why such bodies form and what their formation implies for the functionality—and dysfunctionality—of the enzymes that comprise them. The panoply of intracellular protein bodies also raises interesting questions regarding their evolution and maintenance within cells. We speculate on models for how such structures form in the first place and why they may be inevitable.
Keywords: self-assembly, allosteric regulation, fibers, foci, storage bodies, aggregates, metabolic efficiency
INTRODUCTION
In 2010, three research groups reported the startling discovery that cytidine triphosphate (CTP) synthase, the enzyme that catalyzes the last and rate-limiting step of de novo CTP biosynthesis, can reorganize into extended intracellular fibers in bacterial, fungal, and animal cells (Ingerson-Mahar et al. 2010, Liu 2010, Noree et al. 2010). It is unknown whether these fibers are enzymatically active, if they serve as cytoskeletal elements, or if they fulfill other, as yet undetermined, regulatory or structural roles. However, far from being a unique occurrence, these fibers are just the latest in a growing assortment of intracellular bodies formed by metabolic enzymes.
In fact, observations of such bodies have increased markedly in recent years. The technological capacity to perform large-scale microscopy screens of protein localization has made possible cell-biological studies that focus on particular cellular conditions. Such screens have been further abetted by the availability of extensive libraries of bacterial, yeast, and mammalian cells expressing proteins fused to reporters. In parallel, biologists and bioengineers have begun constructing novel multienzyme complexes to alter or improve metabolic capacity. Thus, the cellular principles underlying the assembly of large protein bodies and their contribution to the dynamic regulation of metabolism are ripe for exploration and exploitation.
In this review, we survey classic and recent examples of intracellular protein bodies, focusing on those formed by metabolic enzymes. We then discuss the current understanding of when and why such bodies form and what their formation implies for the functionality—and dysfunctionality—of the enzymes that comprise them. Finally, we speculate on more general models for how such complex quaternary structures form in the first place and why they may be inevitable.
TYPES OF INTRACELLULAR BODIES
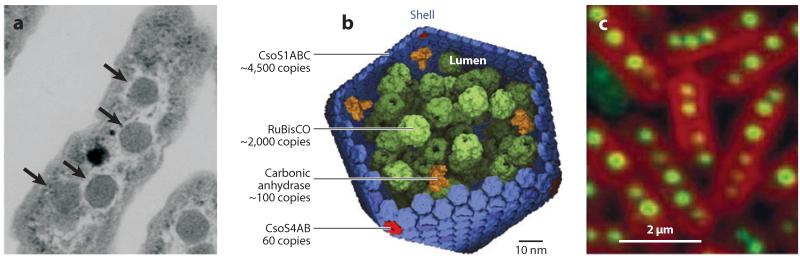
There are a wide variety of intracellular bodies, but they can be classified roughly on the basis of their composition and structures. Bacteria, for the most part (although exceptions exist), do not have membranous compartments, and thus their intracellular bodies tend to be almost exclusively proteinaceous and to serve as subcellular compartments with specialized interiors optimized for their relevant biological roles. Originally called polyhedral bodies when discovered in electron micrographs of Phormidium uncinatum in 1956, bacterial microcom-partments are icosahedrons ~100–200 nm in diameter and delimited by a 3–4-nm-thick protein shell (Figure 1) (Drews & Niklowitz 1956).
Figure 1.
Bacterial microcompartments as exemplified by carboxysomes. (a) Transmission electron micrographs of Halothiobacillus neapolitanus cells showing carboxysomes (arrows) as polyhedral, protein-dense bodies. Adapted from Yeates et al. (2007). (b) The major shell protein (CsoS1ABC in H. neapolitanus) is a hexagonal subunit that oligomerizes into massive sheets that are bent into joined facets at the vertices by a second, pentagonal protein (CsoS4AB) to complete the shell. The interior of the shell is packed with RuBisCO and carbonic anhydrase to maximize CO2 capture. Adapted from Bonacci et al. (2011). (c) Fluorescence microscopy of carboxysomes (green) shows that their in vivo location within cyanobacteria is regulated such that they are centrally aligned and evenly spaced roughly 0.5 μm apart. Adapted from Savage et al. (2010).
In eukaryotes, the formation of membranous compartments is more the norm, and beyond mitochondria and chloroplasts a profusion of specialized intracellular metabolic compartments has been discovered in recent years. These compartments bear a particular relevance to studies of self-assembly of metabolic enzymes, as they frequently exhibit a high level of enzyme self-organization. It can be argued that the crystalline or quasi-crystalline organization of at least some metabolic enzymes within intracellular membranous compartments is a direct result of their increased concentrations within these microbodies.
Finally, beyond the microcompartments and microbodies discussed above, individual metabolic enzymes have been observed to form intracellular fibers and foci. Textbook cases for which the fiber is known to be the enzymatically active form include acetyl-CoA carboxylase and β-glucosidase. Many additional examples of fiber-forming metabolic enzymes have been identified recently, but their functionality is not yet established.
BACTERIAL MICROCOMPARTMENTS
The archetypal bacterial microcompartment, the carboxysome, earned its name and association with carbon fixation in 1973, when the CO2-fixing enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) copurified with polyhedral bodies from the aerobic sulfur bacterium Halothiobacillus neapolitanus (Shively et al. 1973). Initial evidence [such as dispersal upon revival/refeeding (Shively 1974)] supported the idea that carboxysomes were storage bodies rather than the major sites of carbon fixation (Shively 1974). However, studies over the next 20 years showed that the RuBisCO in carboxysomes is active and that carboxysomes also contain carbonic anhydrases to convert bicarbonate into CO2 (Cannon et al. 1991, So et al. 2004).
Crystal structures of carboxysome shell proteins model the wall as a single layer of interlocking hexagonal subunits with a pore in the center of each. The sheets of hexagonal subunits are joined at the vertexes by pentagonal subunits to complete the icosahedrons (Iancu et al. 2007, Kerfeld et al. 2005). The shell seems to provide selective permeability to metabolites while blocking the diffusion of even smaller gas molecules by an unknown mechanism (Dou et al. 2008). The colocalization of enzymes and substrates within the diffusion barrier of the carboxysome shell greatly increases the efficiency of carbon fixation, and carboxysomes are the main sites of CO2 capture in cyanobacteria.
Searches for sequence homology to the major shell proteins have revealed that at least 189 bacterial species contain shell protein orthologs in gene clusters that potentially produce microcompartments (Yeates et al. 2008). Among these, microcompartments with functions other than CO2 capture have begun to be characterized. In Escherichia coli and some Salmonella enterica serovars, orthologs of the carboxysome shell proteins were found in operons containing genes essential for growth on ethanolamine (Stojiljkovic et al. 1995). Acetaldehyde is a toxic and volatile intermediate in this pathway and might otherwise diffuse away rapidly in the absence of the shell proteins. Thus, a portion of the ethanolamine degradation pathway occurs within an ethanolamine-utilizing microcompartment (Brinsmade et al. 2005). A similar 1–2 propanediol-utilizing (PDU) microcompartment was found in an S. enterica serovar (Typhimurium) (Chen et al. 1994). The PDU microcompartment may shield the cell from the toxic intermediate propionaldehyde (Havemann et al. 2002, Havemann & Bobik 2003). The discovery of an N-terminal targeting sequence for loading proteins into PDU microcompartments and the transgenic expression of functional carboxysomes containing fusion proteins in E. coli mark the first steps toward rationally engineering bacterial microcompartments (Bonacci et al. 2011, Fan et al. 2010).
AGGREGATES WITHIN MICROBODIES
It is interesting that protein microcompartments are found primarily in bacteria rather than in eukaryotes, in the same way that it is interesting that bacteriophages primarily have protein coats, whereas their eukaryotic counterparts have membranous coats. It may be that as the complexity of a system scales, the opportunities for inadvertent protein aggregation also scale (a point we touch on later), and that the less precise organization of lipids relative to proteins is a hedge against such aggregates. Conversely, as suggested above, enzyme compartmentalization may provide opportunities for increasing concentrations to the point at which ordered aggregation is possible. Whatever the reason, eukaryotes frequently compartmentalize metabolic enzymes within membranes. We highlight three cases of ordered aggregates within peroxisomes as examples typical of bodies seen in other membrane-bound compartments such as the mitochondria (Polianskyte et al. 2009) and the chloroplast (Englebrecht & Esau 1963, Price et al. 1966).
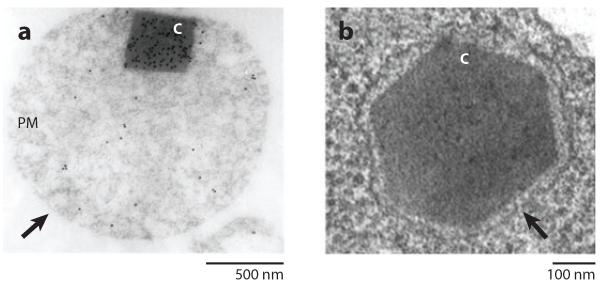
Peroxisomes are eukaryotic microbodies bound by a single lipid membrane. These structures, typically 100 nm to 1 μm across, compartmentalize enzymes and substrates at high concentrations for particular metabolic pathways to improve the rates of catalysis or to shield the rest of the cell from the potential damaging effects of reactive intermediates (Figure 2).
Figure 2.
Examples of microbodies visualized by thin-section transmission electron microscopy. (a) Peroxisomes (arrow) within sunflower cotyledon mesophyll cells. Crystalline inclusion bodies are formed from catalase, as shown by the immunogold nanoparticle localization (black dots). Adapted from Tenberge & Eising (1995) and Pavelka & Roth (2010). (b) Aspergillus nidulans showing a Woronin body filled with a HEX-1 protein crystal. Adapted from Yuan et al. (2003). Abbreviations: C, crystalline inclusion bodies; PM, peroxisomal matrix.
Most peroxisomes possess one or more enzymes for purine catabolism or salvage, usually xanthine oxidase and urate oxidase. These proteins are often clustered, even to the extent of forming amorphous and crystalline inclusions. Urate oxidase is a cuproprotein that normally forms homotetrameric rings, but in some mammalian peroxisomes these tetramers stack into fibers, which combine into crystalline cores (Angermüller et al. 1987). These crystalline cores are a common feature of many types of peroxisomes, though their effects on the enzymatic activity are unknown.
In plants, peroxisomes commonly specialize in β-oxidation and contain high concentrations of oxidases and enzymes to degrade hydrogen peroxide produced by the oxidases. Electron microscopy of plant peroxisomes reveals crystalline and amorphous inclusions likely composed of catalase, a homotetrameric enzyme that degrades hydrogen peroxide (Heinze et al. 2000). Biochemical studies comparing crystalline catalase with diffuse catalase found that the crystalline catalase had up to tenfold-less specific activity but greater stability under UV, pH, and temperature stresses (Eising et al. 1998). Although there is a clear loss of function owing to aggregation, the gain of structural stability in a highly oxidizing environment could be an example of adaptive change in catalytic potential mediated by forming larger aggregates.
Specialized types of peroxisomes, called Woronin bodies, are found in filamentous fungi and staunch the flow of cytoplasm from hyphal wounds (Jedd & Chua 2000). The major and essential component of Woronin bodies is an aggregate of HEX protein oligomers, which form the eponymous hexameric crystalline cores. HEX proteins are related most closely in structure and sequence to eIF-5a, though the residues responsible for intersubunit contacts are different, and the precise enzymatic function is unknown (Yuan et al. 2003). Mutation of residues at the oligomerization interface abolishes both the wound-healing function and the classic polymerization phenotype, which shows this to be an example of a functional aggregate (Yuan et al. 2003).
In mammals, the processed form of the β-lactamase-like protein (LACTB) polymerizes into ordered filaments hundreds of micrometers in length in the intermembrane space of mitochondria (Polianskyte et al. 2009). Neither the role these fibers have within mitochondria nor what drove the evolution of fiber formation from the nonfiber-forming bacterial ortholog is known.
FIBERS AND FOCI
Although microcompartments and membranous organelles are complex, highly structured organizing centers of metabolism, metabolic enzymes also self-assemble into a wide array of simpler intracellular bodies, among them fibers and foci. Many such bodies have been discovered; some are clearly functional, whereas the functions of others have yet to be established. The probability of forming functional aggregates arguably scales with the concentrations of the enzymes involved, and thus, perhaps unsurprisingly, many of these fibers and foci seem to be enzymes that support key metabolic processes such as carbon utilization, nitrogen fixation, and nucleotide biosynthesis.
Fibers of Metabolic Proteins: Carbon Utilization
Self-assembling fibers of a single enzyme are perhaps the simplest metabolic bodies observed in cells. The textbook example of a functional fiber is acetyl-CoA carboxylase; it was the first and remains the best characterized of all enzymatic fibers. Most mammals encode acetyl-CoA carboxylase as a single multidomain protein, nominally a homodimer, but one that can assemble into polymers with >50-fold greater activity (Beaty & Lane 1983a, Meredith & Lane 1978). In mammals, there are two isoforms of the enzyme, one localized to the cytoplasm and the other to the mitochondria. Both isoforms carry out the first and rate-limiting step of fatty acid biosynthesis by carboxylating acetyl-CoA to malonyl-CoA.
Although polymerization is dependent on enzyme concentration, many other mechanisms shift the enzymes between inactive monomers and the active polymer form (Beaty & Lane 1983b). Phosphorylation/dephosphorylation will decrease/increase polymerization, respectively; allosteric binding of citrate induces polymerization, whereas excess product (malonyl-CoA) triggers depolymerization; and MIG12 binding also can induce polymerization (Kim et al. 2010). For the mitochondrial isoform of acetyl-CoA carboxylase, MIG12, binding alone is sufficient to drive polymerization and reduces the concentration of citrate needed for polymerization fivefold for the cytoplasmic isoform (Kim et al. 2010).
Gunning (1965) first reported fibers of β-glucosidase in electron micrographs of oat plastids in 1965. This enzyme is nominally a homohexamer that hydrolyzes β1→4 glucose bonds; it also cleaves avenacosides as an antifungal defense. Structural studies 40 years later revealed rings of hexamers stacked into fibers up to 2 μm in length (Kim et al. 2005), and biochemical analyses provided evidence that the fibers might be the active form of the enzyme: longer fibers were more active in hydrolyzing avenacosides and more resistant to inhibitors than were shorter fibers (Kim et al. 2005).
Fibers of Metabolic Proteins: Nitrogen Utilization
Several enzymes at the core of nitrogen metabolism also form fibers. Studies of E. coli glutamine synthase (Class I), which catalyzes the ATP-driven addition of ammonia to glutamate, found that zinc progressively induces the dodecameric rings of the enzyme complex to aggregate into fibers (Miller 1974). Mammals have an eye-specific glutamine synthetase (Class I-like) that is catalytically inactive but essential for proper eye development (Harding et al. 2008). Like crystallins, it is one of many chance adaptations of structured enzyme aggregates to serve as eye proteins. In yeast, glutamine synthetase (Class II) is a decamer that forms foci in vivo under starvation conditions, although its ultrastructure remains to be determined (Narayanaswamy et al. 2009).
When cells are starved of glutamine, they degrade glutamate to α-ketoglutarate and ammonia using the NAD-dependent glutamate dehydrogenase in mitochondria. In mammals, this enzyme normally forms homohexamers but at high concentrations associates into either highly ordered filaments or helical fiber bundles (Josephs & Borisy 1972). It is unclear what effect polymerization has on enzyme activity (Zeiri & Reisler 1978); however, glutamate dehydrogenase has a host of allosteric regulators, including zinc, ADP, and GTP, and those that activate function also promote polymerization and decrease thermal aggregation (Cioni & Strambini 1989, Eisenberg & Reisler 1971, Frieden 1958, Olson & Anfinsen 1952, Sabbaghian et al. 2009). The reverse reaction—glutamate synthesis—can be carried out either by highly related enzymes using NADPH as a cofactor or by glutamate synthases, which catalyze a transamination from glutamine to α-ketoglutarate, forming two molecules of glutamate. In starved yeast, the green fluorescent protein (GFP)-tagged glutamate synthetase forms fibers (Noree et al. 2010); similar to glutamate dehydrogenase, the enzyme forms homohexamers, and the fibers observed by fluorescence microscopy may represent fibrillar bundles of the homohexamers.
Fibers of Metabolic Proteins: Nucleotide Biosynthesis
Cytidine triphosphate synthase
Recently, multiple groups have reported that CTP synthase forms filamentous and ring structures in vivo in fly (Chen et al. 2011, Liu 2010, Noree et al. 2010), bacteria (Ingerson-Mahar et al. 2010), budding yeast (Noree et al. 2010), rat (Noree et al. 2010), and human cells (Carcamo et al. 2011, Chen et al. 2011). In the crescent-shaped bacterium Caulobacter crescentus, CTP synthase forms a single rod lining the inner curvature of the cell. In Drosophila, CTP synthase filaments, termed cytoophidia (meaning “cell snake”), exist in two forms, termed microcytoophidia and macrocytoophidia, the latter being considerably thicker and longer than the former (Figure 3). In yeast, CTP synthase assembles into filaments as cells enter quiescence upon culture growth to stationary phase. In fact, yeast cells express two CTP synthase homologs, Ura7p and Ura8p, that colocalize within the same filaments but not with other known filament-forming proteins (Noree et al. 2010).
Figure 3.
Green fluorescent protein C-terminal cytidine triphosphate (GFP-CTP) synthase fibers within Drosophila egg chamber cells. The fibers exhibit a diverse range of lengths. Adapted from Liu (2010).
Initial data suggest that these fibers may be formed by the self-association of CTP synthase and do not require energy or special machinery (such as chaperones or scaffolds) for formation. Filaments are seen to form upon heterologous expression of C. crescentus CTP synthase in E. coli, and native E. coli CTP synthase forms filaments both in vivo and in vitro (Ingerson-Mahar et al. 2010). That said, other studies now implicate additional components in the fibers (Carcamo et al. 2011).
Although the filaments show distinct intracellular locations, these apparently vary in different organisms. In C. crescentus, CTP synthase filaments colocalize to crescentin along the inner curvature of the cell and apparently help regulate the crescent cell shape (Ingerson-Mahar et al. 2010). In rat neurons, CTP synthase filaments occur in axons but not dendrites (Noree et al. 2010). Fly microcytoophidia associate with Golgi complexes and in some cases with microtubules (Liu 2010). In contrast, human and yeast CTP synthase filaments were not observed to colocalize with microtubules (Carcamo et al. 2011, Noree et al. 2010), and human CTP synthase filaments also were not observed to colocalize to Golgi complexes or centrosomes and were not enriched in actin or vimentin (Carcamo et al. 2011).
Compounds that modulate CTP synthase protein function modulate fiber formation. For example, CTP synthase inhibitors such as acivicin and the glutamine analogs 6-diazo-5-oxo-L-norleucine (DON) and azaserine (Carcamo et al. 2011, Chen et al. 2011) have dramatic effects on CTP synthase filaments. Interestingly, CTP synthase inhibitors produce different effects in different organisms: DON treatment disrupts CTP synthase filament formation in C. crescentus and also disrupts filaments of heterologously expressed C. crescentus CTP synthase in Schizosaccharomyces pombe or E. coli (Ingerson-Mahar et al. 2010). In contrast, DON and azaserine treatments promote filament formation in fly cells, and DON and acivicin treatments induce filament formation in human cells (Carcamo et al. 2011, Chen et al. 2011). Notably, DON binding induces tertiary (Levitzki et al. 1971) and quaternary (Robertson 1995) structural changes in E. coli CTP synthase. As in the glutamine synthetase case mentioned above, such allosteric conformational changes also alter the conformational state of neighboring subunits coordinately (Levitzki et al. 1971), which is consistent with the hypothesis that differences in intersubunit amino acid contacts might be exposed or hidden by conformational changes. This would help to explain not only the evolution of fiber formation but also how fiber formation is functionally related to regulatory logic.
Filament formation also can vary broadly according to systemic conditions, including cell types, stages, and growth conditions. Ingerson-Mahar et al. (2010) reported that mCherry-CTP synthase filaments were generally shorter in newly formed stalked cells of C. crescentus and then elongated with the progression of the cell cycle; subcellular localization was also dependent on cell cycle. Carcamo et al. (2011) observed the opposite in human cells, in which expression of filaments occurred in all phases of the cell cycle in HEp-2 cells, and Chen et al. (2011) found a similar ubiquity in HeLa cells. However, in other human cells, the filaments varied by cell type and culture conditions. For example, undifferentiated, uninduced human embryonic stem cells contained CTP synthase rings and rods but lost them when stimulated to differentiate by retinoic acid (Carcamo et al. 2011). Similarly, filaments of the yeast CTP synthase Ura7p are induced strongly by growth to saturation or glucose depletion but disappear upon addition of fresh medium. Thus, CTP synthase fibers currently represent something of a quandary: Although widely observed, they show mixed regulatory logic across organisms, and their enzymatic functionality has yet to be established.
Inosine 5[prime]-monophosphate dehydrogenase
In 2011, a study in human cells demonstrated that cytoplasmic filaments consisting of human CTP synthase also contained inosine 5′-monophosphate dehydrogenase (IMPDH) (Carcamo et al. 2011). IMPDH catalyzes the NAD-dependent oxidation of inosine monophosphate (IMP) to xanthosine monophosphate, the first and rate-limiting step for the synthesis of guanosine nucleotides. The predominant isotype, IMPDH type II, is highly expressed in neoplastic and differentiating cells, which makes it an inviting target for antiproliferation drugs such as the immunosuppressive, noncompetitive inhibitor mycophenolic acid (MPA) (Nagai et al. 1991, Zimmermann et al. 1998). MPA induced IMPDH rings and filaments in cultured human cells, reduced the enzyme’s specific activity in cell lysates, and induced purified IMPDH homotetramers of ~15 nm in diameter to form large, disordered aggregates in vitro (Ji et al. 2006). Both MPA-induced rings and fibers in vivo and aggregation in vitro could be dispersed by the addition of GTP at physiological concentrations, with GTP addition restoring the activity of MPA-inhibited IMPDH in cell lysates (Ji et al. 2006). These results are consistent with a hypothesis that MPA stabilizes intertetramer interactions, shifting the equilibrium between active IMPDH tetramers and inactive fibers to favor the inactive form. These results also beg the question of whether cells might naturally form IMPDH fibers to store or regulate the enzyme in an inactive form rather than destroying or recycling it.
Purinosomes
Although sampling may be biased by history, the enzymes of nucleotide metabolism appear to be particularly prone to form intracellular bodies. In particular, the existence of a multienzyme complex consisting of all the members of the de novo purine biosynthesis pathway, termed the purinosome, has been postulated for some time on the basis of the accumulation of a variety of evidence from many experiments. The de novo purine biosynthesis pathway encompasses a ten-step enzymatic reaction converting phosphoribosyl pyrophosphate (PRPP) to IMP. In higher eukaryotes such as mammals, de novo purine biosynthesis is carried out by phosphoribosyl pyrophosphate amidotransferase (PPAT), the trifunctional phosphoribosylglycinamide formyltransferase/phosphoribosylglycinamide synthetase/phosphoribosylaminoimidazole synthetase (GART), phosphoribosylformyl-glycinamidine synthase (FGAMS), the bifunctional phosphoribosylaminoimidazole carboxylase/phosphoribosylaminoimidazole succinocarboxamide synthetase (PAICS), adenylosuccinate lyase (ADSL), and the bifunctional 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC). The de novo purine biosynthesis pathway is upregulated when exogenous hypoxanthine is unavailable (Becker & Kim 1987, Yamaoka et al. 2001).
In higher eukaryotes, these ten enzymes have fused into six polypeptide chains, three of which possess multiple active sites, as listed above. Perhaps the most interesting of these is the trifunctional protein GART, which catalyzes the second, third, and fifth enzymatic steps in the pathway. Early hints that the purinosome might form included coimmuno-precipitation of trifunctional GART and bifunctional PAICS from chicken liver (Caperelli et al. 1980), along with serine hydroxymethyl transferase and the trifunctional methylene tetrahydrofolate dehydrogenase (MTHFD1). These latter two enzymes form a cycle for the production of the labile 10-formyl tetrahydrofolate coenzymes required for steps 3 and 9 of purine biosynthesis. Cross-linking studies in vitro found a physical interaction between GART and MTHFD1, and their interaction increases GART activity (Smith et al. 1980). Fluorescence microscopy of transiently expressed, fluorescently tagged proteins showed colocalization between tetrahydrofolate synthase and both FGAMS and GART.
In 2008, the Benkovic group reported the first direct observation of the purinosome in cells (An et al. 2008). Fluorescent protein constructs of the six mammalian purine biosynthesis enzymes formed punctate intracellular foci when transiently overexpressed in HeLa cells in purine-depleted culture medium. FGAMS-GFP was shown to colocalize with the five other enzymes of the pathway, which suggests the assembly of de novo purine biosynthesis enzymes into purinosomes. FGAMS-GFP foci could be dissolved by exchanging purine-depleted for purine-rich medium, although the addition of the purine hypoxanthine to purine-depleted medium did not dissolve the foci. Intracellular bodies that included PPAT, GART, ADSL, and ATIC were also detected via immunofluorescence confocal microscopy of endogenous enzymes in many human cell types, including primary keratinocytes. The bodies formed and dispersed when culturing cells in purine-depleted and purine-rich medium, respectively (Baresova et al. 2012).
Inhibitor studies have yielded some insights into the mechanism of purinosome formation. Addition of the microtubule-disrupting agent nocodazole reduced purinosome foci formation and decreased overall cellular purine synthesis (An et al. 2010a), which suggests that the formation of these bodies may require cytoskeletally directed active transport. Casein kinase 2 (CK2) inhibitors 4,5,6,7-tetrabromo-1H-benzimidazole and 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole induced foci formation by FGAMS-GFP, GART-GFP, or PPAT-GFP, whereas the subsequent addition of a different CK2 inhibitor, 4,5,6,7-tetrabromobenzotriazole, appeared to reverse or suppress foci formation. In addition, although CK2 inhibitors affected FGAMS-GFP, GART-GFP, and PPAT-GFP foci formation, similar effects were not seen on PAICS-GFP, ADSL-GFP, and GFP-ATIC unless each was cotransfected with one of the former three proteins tagged with orange fluorescent protein (An et al. 2010b). These results suggest a possible, but as yet unclear, role for CK2 in purinosome regulation. One possibility is that phosphorylation by CK2 disperses assemblies, as seen for acetyl-CoA carboxylase (Meggio & Pinna 2003).
The study of the purinosome is also a case study in how the methods used to query intracellular bodies potentially influence outcomes. The visualized purinosome clusters have been observed largely by transient overexpression with fluorescent protein tags. In contrast, previous efforts using cells stably transfected with purinosome constructs did not yield visible purinosome bodies (Gooljarsingh et al. 2001). Overexpression of proteins above their native levels has been shown to result in their aggregation (Cromwell et al. 2006, Klein & Dhurjati 1995, Mayer & Buchner 2004, Yokota et al. 2000, Zhang et al. 2004), and the transiently overexpressed purine biosynthetic enzymes form intracellular bodies to extents that correspond to each enzyme’s predicted aggregation propensity (Zhao et al. 2012). The resultant bodies are marked by ubiquitin and heat-shock chaperones, which suggests that they may represent aggregated protein clusters (Zhao et al. 2012).
The formation of functional purinosomes is not necessarily inconsistent with the observation of aggregated purine biosynthetic enzymes. Indeed, both may be true, depending on the cell state and method of observation. Purinosome bodies appear to be quite heterogeneous, as the penetrance of fluorescent body formation varied broadly for each individual enzyme, ranging from 5% to 77% of the cells (An et al. 2008). A similar broad distribution was observed for CK2 inhibitor-mediated effects (ranging from 15% to 95% penetrance) (An et al. 2010b). Regulatory influences can also be interpreted in terms of either functional or nonfunctional aggregation (or both): although microtubules and CK2 could regulate purinosomes, they have also been implicated in general protein aggregation (Muchowski et al. 2002, Watabe & Nakaki 2011). Nocodazole inhibition of purinosome formation (An et al. 2010a) is consistent with functional assembly requiring microtubules, but nocodazole treatment also inhibits inclusion body formation of aggregated huntingtin (Kaminosono et al. 2008, Muchowski et al. 2002). Future studies will clearly be required to address the key questions of functionality of the purinosome bodies, ideally leading to purification and in vitro characterization, and to assess the relationships, if any, between the intracellular bodies formed by overexpression of purine biosynthetic enzymes and the endogenous forms of those proteins.
LARGE-SCALE SCREENS REVEAL MANY ADDITIONAL INTRACELLULAR FOCI AND FIBERS
The frequency at which intracellular bodies have been found during the study of metabolism indicates either that metabolic enzymes are prone to aggregation or, perhaps, that all enzymes are prone to aggregation and, to date, biochemists just happen to have largely studied metabolic enzymes. In fact, large-scale microscopy screens of protein localization dynamics have revealed tens to hundreds of additional enzymes that form intracellular bodies (Figure 4). Using cell microarrays, Narayanaswamy et al. (2009) surveyed large-scale trends in yeast (Saccharomyces cerevisiae) protein reorganization using a genome-wide GFP fusion library (Huh et al. 2003). When ~800 yeast strains expressing normally cytosolic GFP-tagged proteins from their native locus in the genome were grown to stationary phase, 180 proteins involved in intermediary metabolism and stress response were observed to form punctate cytoplasmic foci. The formation of many of these proteins was confirmed also by immunofluorescence and mass spectrometry of untagged proteins, with 33 proteins confirmed by both microscopy and mass spectrometry (Narayanaswamy et al. 2009).
Figure 4.
Hundreds of foci- and fiber-forming proteins have been discovered in systematic protein localization screens; most of these intracellular bodies are still largely uncharacterized. (a,b) Representative foci (ACCase β-subunit)- and fiber (UDP-N-acetylmuramate-alanine ligase)-forming green fluorescent protein (GFP) fusion proteins, respectively, from Caulobacter crescentus. Adapted from Werner et al. (2009). (c,d) Representative foci (Ade4)- and fiber (Pil1)-forming yellow fluorescent protein-fusion proteins, respectively, from Schizosaccharomyces pombe. Adapted from Matsuyama et al. (2006). (e,f) Representative foci (Gln1p)- and fiber (Asn2p)-forming GFP-fusion proteins, respectively, from Saccharomyces cerevisiae. Adapted from Narayanaswamy et al. (2009).
A second screen of a portion of the S. cerevisiae GFP library by Noree et al. (2010) found 27 additional fiber structures. Of these, three were fibers formed by metabolic proteins. One was a CTP synthase fiber (discussed above). The other two fibers, composed of Psa1p and Glt1p, were entirely new. Psa1p is a GDP-mannose pyrophosphorylase essential for building the glycoproteins of the cell wall and is highly conserved across eukaryotes. The other, Glt1p (also discussed above), is an NAD-dependent glutamate synthase, which, along with Gln1p, forms one of the core ammonia incorporation pathways. It is currently unclear whether these fibers are functional and, if so, what role they might serve.
Additionally, several systematic microscopy screens of protein localization have been performed in the fission yeast S. pombe (Ding et al. 2000, Hayashi et al. 2009, Matsuyama et al. 2006, Sawin & Nurse 1996). Although these screens were not searching specifically for new structures, they still describe many proteins as existing as cytoplasmic dots [e.g., CTP synthase and Ade4p, as were seen also in _S. cerevisiae_ (Narayanaswamy et al. 2009)] or in fibrous morphologies (e.g., Pil1p, an essential cell wall peptidoglycan synthetase).
A localization study of ~300 cytoplasmic proteins labeled with mCherry by Werner et al. (2009) identified many proteins that showed nondiffuse localization. Of these, they described 29 proteins’ cellular distributions as foci, 129 proteins as patchy/spotty, and 3 as filament forming. Two of the filament-forming proteins are CTP synthase and an associated structural protein, CreS. The other fiber is formed by UDP-N-acetylmuramate-alanine ligase. Similar to Psa1p, it is an essential enzyme for peptidoglycan synthesis.
These large-scale screens clearly reveal a remarkably extensive assortment of intracellular bodies forming across diverse environmental conditions. It would appear that at least some of these bodies are metabolically inducible and form reversibly, which strongly suggests functionality. For example, the yeast purine biosynthetic enzyme Ade4-GFP formed foci in the absence of adenine, and cycling between punctate and diffuse phenotypes could be controlled by adenine subtraction and addition. Similarly, yeast glutamine synthetase (Gln1-GFP) foci cycled reversibly in the absence and presence of glucose (Narayanaswamy et al. 2009). Finally, there is good evidence for the yeast translation initiation regulatory complexes eIF2 and eIF2B existing as polymerized fibers during log phase growth (Noree et al. 2010). As this is precisely when translation rates are the highest in yeast, it argues the fibers may be functional and regulated. However, as with the purinosome, caution must be used in interpreting whether the bodies form for functional roles or whether regulated changes in concentration lead inadvertently to intracellular aggregates.
Most large-scale localization studies use fusion proteins, and the properties of the tag can affect the solubility and interactions of the tagged protein. One recent study found that a commonly accepted body formed by Clp proteases was entirely dependent on certain fluorescent tags for formation. In the absence of a tag or using a GFP evolved for monomeric expression in E. coli, Clp proteases did not form the bodies. The authors propose that the fluorescent protein tags dimerize, causing homo-oligomeric complexes to assemble into an extended network and producing an intracellular body (Landgraf et al. 2012). If this is a major cause of intracellular bodies, it predicts the set of foci forming proteins should be strongly enriched for oligomers. Thus structures discovered by large-scale fluorescence localization screens need to be confirmed by orthogonal, preferably tag-free, methods to verify their biological relevance.
WHAT’S GOING ON? THREE POTENTIAL ROLES FOR INTRACELLULAR BODIES
The panoply of intracellular protein bodies also raises interesting questions regarding their evolution and maintenance within cells. We conclude by offering additional speculations on several particular aspects of how and why such bodies might evolve.
Case studies such as CTP synthase and the purinosome raise many interesting questions regarding the functionality of intracellular bodies. It is thus useful to consider why metabolic enzymes might assemble into such large intracellular assemblies. In general, metabolic enzymes are notable for often forming complex quaternary structures, some (e.g., pyruvate dehydrogenase) larger than even the largest single proteins. These massive intracellular assemblies may provide functional advantages to the cell, such as catalytic efficiency or improved regulation. Alternatively, these structures might be depots for the storage of functional proteins or the disposal of dysfunctional ones. Although numerous metabolic enzymes clearly form functional and well-characterized metaassemblies, it is not yet clear for many of the most recently discovered intracellular bodies whether they represent functional bodies, storage bodies, or aggregates. Distinguishing these roles represents one of the major challenges for studying these structures.
Catalytic Efficiency and Improved Regulation
Classically, enzymes have been thought to organize into multisubunit assemblies to improve their functionality. Quaternary structures enable the channeling of substrates between active sites on individual subunits, thereby protecting labile intermediates from side reactions in the cell or protecting the cell from toxic reaction intermediates. For example, the first intermediate substrate in de novo purine biosynthesis, 5-phosphoribosylamine, has a cellular half-life of 38 s, and channeling is essential for its subsequent coupling to glycine (Schendel et al. 1988). Also, the addition of CO2 to aminoimidazole ribonucleotide to form 4-carboxy-5-aminoimidazole ribonucleotide occurs without the use of biotin or ATP. In plants, bacteria, and yeast this occurs through two enzymatic steps with N5-CAIR as an unstable intermediate substrate, which may have driven the fusion of the enzymes responsible for these two steps in yeast and plants. Finally, the 5,10-formyl-methylene-tetrahydrofolate coenzyme used in steps 3 and 9 is moderately labile, with a half-life of 30 min (Smith et al. 1980). Such observations underpin the search for the purinosome, which would in principle localize the enzymes within sufficient proximity to prevent the diffusion of unstable intermediates or substrates. Similarly, the peroxisome shields cells from peroxide radicals generated by oxidases in fatty acid and purine catabolism. A related effect is seen in the formation of ethanolamine-utilizing microcompartments that shield cells during growth on ethanolamine by localizing the production and degradation of toxic aldehydes within a protein shell.
Channeling substrates between active sites in a quaternary structure or within a compartment can also improve metabolic efficiency greatly, even when no side reactions are in play. For instance, carboxysomes prevent the diffusive loss of CO2 during carbon fixation and thereby allow unicellular organisms to achieve C4 plantlike efficiencies. In an even grander organization of metabolic machinery, the cellulosome anchors enzymes involved in cellulose production via elaborate, interlocking multidomain protein scaffolds on the cell surface.
Finally, quaternary structure formation allows regulation by cooperative interactions and allosteric effectors. There are, of course, many such known examples of cooperativity in enzyme oligomers. Among these, the dodecameric glutamine synthetase from E. coli is one of the best understood. In addition to two covalent modification enzyme systems, glutamine synthetase has eight direct allosteric inhibitors that, individually, partially inhibit activity and, together, cooperatively inhibit activity (Eisenberg et al. 2000). The allosteric inhibitors bind active sites that are positioned at the interface between the subunits of the two hexameric rings, and in this way binding is transduced into structural changes that can be transmitted between enzymes in both rings. This mode of regulation effectively integrates information about the metabolic state of the cell with overall enzyme activity (Woolfolk 1967), and such changes could in principle modulate the formation of glutamine synthase foci and fibers in vivo, although such an effect has yet to be shown.
Storage Depots
It has also been hypothesized that proteins may assemble into macromolecular depots, in which individual components can be held transiently and released. The advantage of such depots is that proteins need not be resynthesized but are instead retained for potential future use, especially in conditions in which rapid redeployment may be required. For example, in the stationary (quiescent) phase, cells have a remarkable ability to weather extreme stress conditions but can rapidly reenter the cell cycle (Gray et al. 2004). Many changes accompany the transition into stationary phase. Stationaryphase yeast cells, for example, exhibit decreased metabolic rates and increased size and density and in general cease proliferating. The cell walls increase in thickness to provide osmo- and thermotolerance, and the cells accumulate intracellular carbohydrates, including glycogen and trehalose, which may serve to help protect the cells against a variety of stresses (Gray et al. 2004). Although rates of transcription and translation are decreased dramatically in quiescent cells as compared with exponentially growing cells (Choder 1991, Fuge et al. 1994), the quiescent cells must remain able to rapidly restart growth when nutrients do become available. Quiescent cells have been shown to maintain available pools of important cellular components in forms that can be mobilized quickly, including cytoplasmic processing bodies containing mRNAs that can be translated upon restarting growth (Brengues et al. 2005) and actin bodies—localized accumulations of actin that can reassemble into actin fibers and patches as necessary when cellular growth restarts (Sagot et al. 2006). Overall, quiescent cells appear to be rich with dynamic depots that are important for reentry into the mitotic cell cycle. Such a trend is consistent with the tendency for many cellular proteins to be organized–both spatially and functionally–in a manner consistent with the needs of the cell.
Aggregation of Dysfunctional and/or Unfolded Proteins
In contrast to the above examples of intracellular bodies with active functional roles or storage of functional potential, a third major category of intracellular bodies is now well established: those composed of aggregated and possibly dysfunctional unfolded proteins. Such bodies often are the product of active cellular processes for collecting, sequestering, and disposing of the aggregates (Tyedmers et al. 2010) but also can form when high levels of expression trigger self-association and aggregation (Wickner 1994). Aggregated proteins are commonly sequestered, often via active transport along the cytoskeleton, to specific cellular sites such as aggresomes (Johnston et al. 1998), IPOD, and JUNQ (Kaganovich et al. 2008).
The best-characterized examples of protein aggregates and intracellular aggregation bodies include amyloid fibers and foci as well as inclusion bodies. Amyloid fibers, in particular, are linked to a range of human diseases, including Alzheimer’s, Parkinson’s, and Huntington’s (Ross & Poirier 2004), but they also occur broadly across proteins and organisms (Goldschmidt et al. 2010, Halfmann et al. 2012). The toxicity of protein aggregates is generally attributed not only to the depletion of functional machinery (Stefani & Dobson 2003) but also to the creation of pores inside cell membranes by small oligomers (Ahmed et al. 2010). At the early stages of aggregation,small oligomers of amyloid aggregates have structural similarities to pore-forming bacterial toxins and eukaryotic pore proteins (Hirakura & Kagan 2001), and their insertion into membranes leads to ion loss and cell death (Kourie et al. 2002). Both yeast and human prions constitute a continuous spectrum of aggregation with multiple morphologies (Edskes et al. 2009, Legname et al. 2006), and aggregated prion protein seeds can propagate readily to daughter cells (Krammer et al. 2009).
In contrast to bodies formed by aggregates of dysfunctional proteins, some bodies may be formed by aggregates of functional proteins. For instance recent evidence suggests RNA granules are formed by aggregatation of partially unfolded RNA-binding proteins whose low complexity sequences form hydrogels of amyloid aggregates. However, the proteins retain their RNA-binding capacity, and unlike other amyloid aggregates these show highly dynamic assembly and disassembly rates and can incorporate heterogeneous sequences (Kato et al. 2012). These RNA granules still serve various biological functions despite being formed by amyloid aggregates, which are historically thought to be pathogical and often associated with cell death in neurodegenerative diseases.
SPECULATIONS AND CONCLUSIONS
Are Metabolic Enzymes Intrinsically More Likely to Self-Assemble?
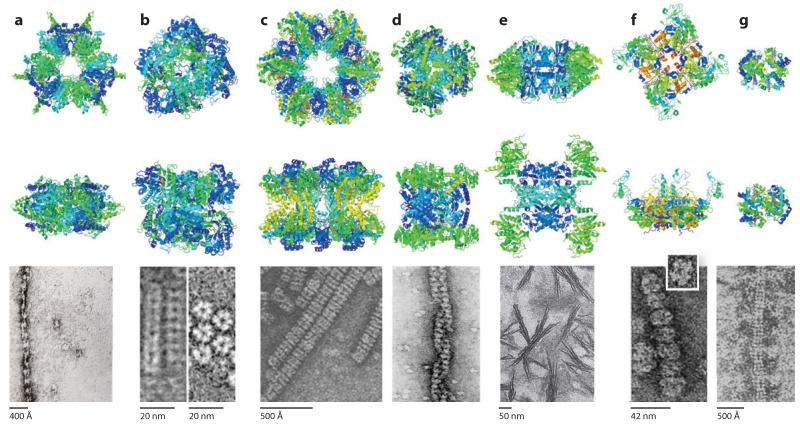
Similar to the large-scale screens of protein localization, microscopy screens have also identified numerous new yeast prions (Alberti et al. 2009). Thus, whether assembling bodies for functional reasons, perhaps in response to metabolic cues, or simply aggregating pathologically, a much larger set of proteins than is broadly appreciated may assemble into bodies or aggregate in vivo, perhaps whenever their abundances exceed tolerated limits (Tartaglia et al. 2007). The breadth of these phenomena raises the interesting possibility that all ordered proteins may exhibit some level of self-aggregation or self-assembly. Because metabolic enzymes often exhibit complex quaternary structures and intersubunit allostery for the purpose of regulation, such quaternary structures might be intrinsically more susceptible to forming intracellular foci and fibers simply as a result of a symmetrical arrangement of enzymes, which would replicate any favorable intersubunit interactions around the structure’s axes of symmetry (Figure 5). For example, stacked E. coli glutamine synthase dodecamers (dimers of hexamers) present six identical interfaces, one between each pair of the six repeated monomers around the dodecamer-dodecamer interface. Any favorable interaction at one such interface is therefore copied six times around the rings, making—via avidity—for a potentially very strong overall interface. Such a mechanism of fiber formation is therefore intrinsically more likely for proteins with complex quaternary structures typical of those found among metabolic enzymes. Figure 6 shows a gallery of metabolic enzyme quaternary structures and fibers thought to be formed by such stacking mechanisms.
Figure 5.
Proteins that assemble into symmetric quaternary structures should in principle have a higher propensity to form fibers, because effectors, whether allosteric, covalent, or mutational, that enhance binding between the oligomeric faces may be multiplied around the axis of symmetry, leading to enhanced fiber stability. The types of effectors and their contribution to fiber stability or destabilization can inform about an enzymatic fiber’s role within cells.
Figure 6.
A sampling of metabolic enzymes that self-assemble into fibers. The quaternary structure of each enzyme is illustrated schematically (top row), following 90° rotation (middle row), and imaged in fiber form by electron microscopy (bottom row). (a) Acetyl-CoA carboxylase: crystal structure of Streptomyces coelicolor acetyl-CoA carboxylase β-subunit, PDB ID: 1XO6 (Diacovich et al. 2004) and electron micrograph of rat liver acetyl-CoA carboxylase (Nelson et al. 2008). (b) β-glucosidase: crystal structure of wheat β-glucosidase, PDB ID: 2DGA (Sue et al. 2006), and electron micrograph of oat β-glucosidase (Kim et al. 2005). (c) Glutamine synthetase: crystal structure of Escherichia coli glutamine synthetase, PDB ID: 1FY (Gill & Eisenberg 2001), and electron micrograph of E. coli glutamine synthetase (Frey et al. 1975). (d) Glutamate dehydrogenase: crystal structure of Clostridium symbiosum glutamate dehydrogenase, PDB ID: 1BGV (Stillman et al. 1993), and electron micrograph of cow liver glutamate dehydrogenase (Josephs & Borisy 1972). Scale not provided. (e) CTP synthase: human CTP synthase 2, PDB ID: 3IHL (M. Moche et al., unpublished data), and electron micrograph of Drosophila CTP synthase (Liu 2010). (f) Inosine monophosphate dehydrogenase: crystal structure of human type II inosine monophosphate dehydrogenase, PDB ID: 1NF7 (D. Risal, M.D. Strickler, B.M. Goldstein, unpublished data), and electron micrograph of human type II inosine monophosphate dehydrogenase (Ingerson-Mahar et al. 2010). (g) Human sickle-cell mutant hemoglobin: crystal structure, PDB ID: 2HBS (Harrington et al. 1997), and electron micrograph human sickle-cell mutant hemoglobin: (Ohtsuki et al. 1977). All crystal structure images created with Jmol.
Given that metabolic enzymes often are also highly expressed and prone to allosteric regulation of alternate conformations, they would seem especially likely cellular candidates for self-assembly into higher-order structures. The resulting fibers might provide regulatory functionality—e.g., providing fine-tuning of enzymatic output by integrating allosteric interactions across the polymeric interfaces. Such appears to be the case for glutamine synthase, at least on the scale of binding of two homopentameric or hexameric rings (Eisenberg et al. 2000). Such fibers or assemblies might also provide stability or rigidity, especially as regards the formation of metabolic enzymes into more extensive structures such as crystals, which we might expect to serve as self-chaperoning structures, decreasing the likelihood for component proteins to unfold and form aggregates. Indeed, lens crystallins are derived frequently from metabolic enzymes (Piatigorsky 1993). Similarly, the crystalline cores of peroxisomes as well as mitochondrial glutamate dehydrogenase fibers are cases in which homo-oligomeric enzymes are highly concentrated in stressful environments.
Protein Aggregation as an Evolutionary Compromise
As we have discussed, the formation of intracellular protein aggregates is widespread, and such aggregates are frequently functional. Therefore, this would seem to be a phenomenon that is not highly selected against and that actually may be beneficial. However, phylogenetic analyses of multiple proteins suggest that a negative correlation exists between the rate of sequence change and the level of expression (Drummond & Wilke 2008, 2009), which has been interpreted to mean that mistranslation of a highly expressed protein is more likely to lead to dysfunctional aggregation than the mistranslation of scarcer proteins. Thus, despite the widespread existence of protein aggregates, the entire proteome is constantly under selection to avoid aggregates. The resolution of this conundrum is seemingly that aggregates are largely unavoidable and that functional organization around aggregates is an evolutionary compromise.
This begs the question of why aggregates are unavoidable, given that evolutionary optimization often grinds genotypes and phenotypes to a very fine degree. The answer must lie in the realm of physical principles that not even evolution can refine, and as described below, sickle-cell hemoglobin remains one of the most illustrative examples. A single mutation that occurred independently several times within a relatively short span of evolutionary time (Wainscoat et al. 1983) leads to polymer formation, which can have the beneficial consequence of increased tolerance to malarial infection. We would argue that such mutations are largely unavoidable, especially in oligomeric proteins that by definition have the opportunity to form multiple, geometrically repeated contact points.
Because oligomers often are allosterically regulated, and thus by definition assume multiple conformations, there may be unique opportunities for the formation of new mutational contact points. In many cases, new aggregates will be deleterious, as with Alzheimer’s and other prion-based diseases, and evolution will eventually constrain the sequence of the protein in a concentration-dependent manner. In some cases, the aggregates will be neutral, and quaternary structures will form that have the opportunity to eventually benefit the cell—possibly through the adoption of a new regulatory functionality, as in the case of acetyl-CoA carboxylase. CTP synthase remains a tantalizing example of a higher-order structure whose purpose either remains undiscovered or is truly near neutrality, existing solely as a decoration in the cell.
A case study in ambiguity: Are CTP synthase fibers cytoskeletal elements or bacterial sickle-cell disease?
The evolutionary conservation of CTP synthase filaments suggests that filament assembly of CTP synthase may provide a biologically useful purpose for cells, such as enhancing enzymatic regulation, improving CTP synthesis efficiency via compartmentalization, or even forming new cytoskeletal elements. This hypothesis may be supported (weakly) by the observation that yeast CTP synthase isozymes Ura7p and Ura8p colocalize to the same filaments (Noree et al. 2010). However, enzymatic activity has yet to be shown for such filaments. Direct functional evidence for CTP synthase filaments is limited currently to the observation that CTP synthase helps determine the C. crescentus cell curvature (Figure 7) as well as to the observations that the assembly and characteristics of CTP synthase filaments seem to vary according to cell cycle or nutrient availability. However, as other model organisms in which CTP synthase filaments are found do not exhibit curved cell structures, the roles of filaments in those organisms are unresolved. Notably, filament formation and enzymatic roles are at least partially separable; for example, Ingerson-Mahar et al. (2010) separated the filament-forming properties of CTP synthase from one of its two enzymatic activities by showing that catalytically inactive synthetase domain mutants retained the ability to form filaments in C. crescentus. However, a mutation to the glutamine amidotransferase active site abolished filament formation, which implicates this domain in quaternary structure assembly.
Figure 7.
An illustration of the analogous effects on cell morphology by cytidine triphosphate (CTP) synthase and sickle-cell mutant hemoglobin (HbS). (a) Bright field images of Caulobacter crescentus cells depleted of CTP synthase show severe bending, some to the point of circularization. (b) Cells overexpressing CTP synthase are straightened markedly. (c) Transmission electron micrograph of a C. crescentus cell with CTP synthase fibers (arrows) along the cell wall, altering cell morphology. Panels a_–_c adapted from Ingerson-Mahar et al. (2010). (d) Analogous images of red blood cells showing their normal, round morphology when oxygenated, and (e) their highly straightened morphology when the deoxygenated HbS forms into fibers. Adapted from Kaul et al. (1983). (f) Transmission electron micrograph showing HbS fibers (arrows) along the cell walls, altering cell morphology. Adapted from Döbler & Bertles (1968).
Another protein oligomerization event known to disrupt cell shape is interesting to consider in this context—that of sickle-cell mutant hemoglobin (HbS), which arises from normal hemoglobin via a single glutamatic acid–to-valine substitution. Upon deoxygenation, HbS tetramers polymerize into rigid fibers capable of distorting red blood cells (Figure 7), which impedes their passage through capillaries. The structure and assembly of deoxy-HbS fibers have been studied extensively by electron microscopy (Josephs et al. 1976, Lewis et al. 1994, Wang et al. 2000). Although individuals with sickle-cell disease may have drastically reduced quality of life, the mutated gene has been proven to offer resistance against malaria (Ferreira et al. 2011), a lifesaving advantage that may explain its prevalence in the population. Deoxy-HbS fibers thus represent a case of pathological aggregation that nonetheless offers a nonobvious but substantial advantage to individuals carrying the mutation. Hence, it is worth noting that CTP synthase fibers might in fact be generally detrimental but have persisted throughout different phyla owing to a similar but undiscovered benefit to the organisms. CTP synthase and sickle-cell hemoglobin illustrate the difficulties in distinguishing functional roles from aggregation when studying intracellular fibers.
ACKNOWLEDGMENTS
This work was supported by grants from the National Science Foundation, National Institutes of Health, US Army Research (58343-MA), Cancer Prevention Research Institute of Texas, and Welch Foundation (F-1515) to E.M.M. and Welch Foundation grant F-1654 to A.D.E.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Ahmed M, Davis J, Aucoin D, Sato T, Ahuja S, et al. Structural conversion of neurotoxic amyloid-β1–42 oligomers to fibrils. Nat. Struct. Mol. Biol. 2010;17(5):561–67. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137(1):146–58. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Deng Y, Tomsho JW, Kyoung M, Benkovic SJ. Microtubule-assisted mechanism for functional metabolic macromolecular complex formation. Proc. Natl. Acad. Sci. USA. 2010a;107(29):12872–76. doi: 10.1073/pnas.1008451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320(5872):103–6. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- An S, Kyoung M, Allen JJ, Shokat KM, Benkovic SJ. Dynamic regulation of a metabolic multi-enzyme complex by protein kinase CK2. J. Biol. Chem. 2010b;285(15):11093–99. doi: 10.1074/jbc.M110.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermüller S, Bruder G, Völkl A, Wesch H, Fahimi HD, A cytochemical and biochemical study Localization of xanthine oxidase in crystalline cores of peroxisomes. Eur. J. Cell Biol. 1987;45(1):137–44. [PubMed] [Google Scholar]
- Baresova V, Skopova V, Sikora J, Patterson D, Sovova J, et al. Mutations of ATIC and ADSL affect purinosome assembly in cultured skin fibroblasts from patients with AICA-ribosiduria and ADSL deficiency. Hum. Mol. Genet. 2012;21(7):1534–43. doi: 10.1093/hmg/ddr591. [DOI] [PubMed] [Google Scholar]
- Beaty NB, Lane MD. Kinetics of activation of acetyl-CoA carboxylase by citrate. Relationship to the rate of polymerization of the enzyme. J. Biol. Chem. 1983a;258(21):13043–50. [PubMed] [Google Scholar]
- Beaty NB, Lane MD. The polymerization of acetyl-CoA carboxylase. J. Biol. Chem. 1983b;258(21):13051–55. [PubMed] [Google Scholar]
- Becker Ma, Kim M. Regulation of purine synthesis de novo in human fibroblasts by purine nucleotides and phosphoribosylpyrophosphate. J. Biol. Chem. 1987;262(30):14531–37. [PubMed] [Google Scholar]
- Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, et al. Modularity of a carbon-fixing protein organelle. Proc. Natl. Acad. Sci. USA. 2011;109(2):478–83. doi: 10.1073/pnas.1108557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310(5747):486–89. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinsmade SR, Paldon T, Escalante-Semerena JC. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J. Bacteriol. 2005;187(23):8039–46. doi: 10.1128/JB.187.23.8039-8046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon GC, English RS, Shively JM. In situ assay of ribulose-1,5-bisphosphate carboxylase/oxygenase in Thiobacillus neapolitanus. J. Bacteriol. 1991;173(4):1565–68. doi: 10.1128/jb.173.4.1565-1568.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caperelli CA, Benkovic PA, Chettur G, Benkovic SJ. Purification of a complex catalyzing folate cofactor synthesis and transformylation in de novo purine biosynthesis. J. Biol. Chem. 1980;255(5):1885–90. [PubMed] [Google Scholar]
- Carcamo WC, Satoh M, Kasahara H, Terada N, Hamazaki T, et al. Induction of cytoplasmic rods and rings structures by inhibition of the CTP and GTP synthetic pathway in mammalian cells. PLoS ONE. 2011;6(12):e29690. doi: 10.1371/journal.pone.0029690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Zhang J, Tastan OY, Deussen ZA, Siswick MY-Y, Liu J-L. Glutamine analogs promote cytoophidium assembly in human and Drosophila cells. J. Genet. Genomics. 2011;38(9):391–402. doi: 10.1016/j.jgg.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Chen P, Andersson DI, Roth JR. The control region of the pdu/cob regulon in Salmonella typhimurium. J. Bacteriol. 1994;176(17):5474–82. doi: 10.1128/jb.176.17.5474-5482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choder M. A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 1991;5(12A):2315–26. doi: 10.1101/gad.5.12a.2315. [DOI] [PubMed] [Google Scholar]
- Cioni P, Strambini GB. Dynamical structure of glutamate dehydrogenase as monitored by tryptophan phosphorescence: signal transmission following binding of allosteric effectors. J. Mol. Biol. 1989;207(1):237–47. doi: 10.1016/0022-2836(89)90453-1. [DOI] [PubMed] [Google Scholar]
- Cromwell MEM, Hilario E, Jacobson F. Protein aggregation and bioprocessing. AAPS J. 2006;8(3):E572–79. doi: 10.1208/aapsj080366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diacovich L, Mitchell DL, Pham H, Gago G, Melgar MM, et al. Crystal structure of the β-subunit of acyl-CoA carboxylase: structure-based engineering of substrate specificity. Biochemistry. 2004;43(44):14027–36. doi: 10.1021/bi049065v. [DOI] [PubMed] [Google Scholar]
- Ding D-Q, Tomita Y, Yamamoto A, Chikashige Y, Haraguchi T, Hiraoka Y. Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells. 2000;5(3):169–90. doi: 10.1046/j.1365-2443.2000.00317.x. [DOI] [PubMed] [Google Scholar]
- Döbler J, Bertles JF. The physical state of hemoglobin in sickle-cell anemia erythrocytes in vivo. J. Exp. Med. 1968;127(4):711–16. doi: 10.1084/jem.127.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Heinhorst S, Williams EB, Murin CD, Shively JM, Cannon GC. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J. Biol. Chem. 2008;283(16):10377–84. doi: 10.1074/jbc.M709285200. [DOI] [PubMed] [Google Scholar]
- Drews G, Niklowitz W. Cytology of Cyanophycea. II. Centroplasm and granular inclusions of Phormidium uncinatum. Arch. Mikrobiol. 1956;24(2):147–62. [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134(2):341–52. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 2009;10(10):715–24. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, McCann LM, Hebert AM, Wickner RB. Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics. 2009;181(3):1159–67. doi: 10.1534/genetics.108.099929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Gill HS, Pfluegl GMU, Rotstein SH. Structure–function relationships of glutamine synthetases. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2000;1477(1-2):122–45. doi: 10.1016/s0167-4838(99)00270-8. [DOI] [PubMed] [Google Scholar]
- Eisenberg H, Reisler E. Angular dependence of scattered light, rotary frictional coefficients, and distribution of sizes of associated oligomers in solutions of bovine liver glutamate dehydrogenase. Biopolymers. 1971;10(12):2363–76. doi: 10.1002/bip.360101202. [DOI] [PubMed] [Google Scholar]
- Eising R, Heinze M, Kleff S, Tenberge KB. Subcellular distribution and photooxidation of catalase in sunflower. In: Noga G, Schmitz M, editors. Antioxidants Higher Plants: Biosynthesis, Characteristics, Actions and Specific Functions in Stress Defense. Shaker; Aachen, Germany: 1998. pp. 53–66. [Google Scholar]
- Englebrecht AHP, Esau K. Occurrence of inclusions of beet yellows viruses in chlorplasts. Virology. 1963;21:43–47. doi: 10.1016/0042-6822(63)90302-7. [DOI] [PubMed] [Google Scholar]
- Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, et al. Short N-terminal sequences package proteins into bacterial microcompartments. Proc. Natl. Acad. Sci. USA. 2010;107(16):7509–14. doi: 10.1073/pnas.0913199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A, Marguti I, Bechmann I, Jeney V, Chora A, et al. Sickle hemoglobin confers tolerance to Plasmodium infection. Cell. 2011;145(3):398–409. doi: 10.1016/j.cell.2011.03.049. [DOI] [PubMed] [Google Scholar]
- Frey TG, Eisenberg D, Eiserling FA. Glutamine synthetase forms three- and seven-stranded helical cables. Proc. Natl. Acad. Sci. USA. 1975;72(9):3402–6. doi: 10.1073/pnas.72.9.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden C. The dissociation of glutamic dehydrogenase by reduced diphosphopyridine nucleotide (DPNH) Biochim. Biophys. Acta. 1958;27:431–32. doi: 10.1016/0006-3002(58)90364-0. [DOI] [PubMed] [Google Scholar]
- Fuge EK, Braun EL, Werner-Washburne M. Protein synthesis in long-term stationary-phase cultures of Saccharomyces cerevisiae. J. Bacteriol. 1994;176(18):5802–13. doi: 10.1128/jb.176.18.5802-5813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill HS, Eisenberg D. The crystal structure of phosphinothricin in the active site of glutamine synthetase illuminates the mechanism of enzymatic inhibition. Biochemistry. 2001;40(7):1903–12. doi: 10.1021/bi002438h. [DOI] [PubMed] [Google Scholar]
- Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. USA. 2010;107(8):3487–92. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooljarsingh LT, Ramcharan J, Gilroy S, Benkovic SJ. Localization of GAR transformylase in Escherichia coli and mammalian cells. Proc. Natl. Acad. Sci. USA. 2001;98(12):6565–70. doi: 10.1073/pnas.121182998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2004;68(2):187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning BE. The fine structure of the chloroplast stroma following aldehyde osmuim-tetroxide fixation. J. Cell Biol. 1965;24:79–93. doi: 10.1083/jcb.24.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482(7385):363–68. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding RL, Howley S, Baker LJ, Murphy TR, Archer WE, et al. Lengsin expression and function during zebrafish lens formation. Exp. Eye Res. 2008;86(5):807–18. doi: 10.1016/j.exer.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington DJ, Adachi K, Royer WE., Jr. The high resolution crystal structure of deoxyhemoglobin S. J. Mol. Biol. 1997;272(3):398–407. doi: 10.1006/jmbi.1997.1253. [DOI] [PubMed] [Google Scholar]
- Havemann GD, Bobik TA. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 2003;185(17):5086–95. doi: 10.1128/JB.185.17.5086-5095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havemann GD, Sampson EM, Bobik TA. PduA is a shell protein of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 2002;184(5):1253–61. doi: 10.1128/JB.184.5.1253-1261.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi A, Ding D-Q, Da-Qiao D, Tsutsumi C, Chikashige Y, et al. Localization of gene products using a chromosomally tagged GFP-fusion library in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2009;14(2):217–25. doi: 10.1111/j.1365-2443.2008.01264.x. [DOI] [PubMed] [Google Scholar]
- Heinze M, Reichelt R, Kleff S, Eising R. High resolution scanning electron microscopy of protein inclusions (cores) purified from peroxisomes of sunflower (Helianthus annuus L.) cotyledons. Cryst. Res. Technol. 2000;35(6-7):877–86. [Google Scholar]
- Hirakura Y, Kagan BL. Pore formation by beta-2-microglobulin: a mechanism for the pathogenesis of dialysis associated amyloidosis. Amyloid. 2001;8(2):94–100. doi: 10.3109/13506120109007350. [DOI] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, et al. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J. Mol. Biol. 2007;372(3):764–73. doi: 10.1016/j.jmb.2007.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat. Cell Biol. 2010;12(8):739–46. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd G, Chua NH. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2000;2(4):226–31. doi: 10.1038/35008652. [DOI] [PubMed] [Google Scholar]
- Ji Y, Gu J, Makhov AM, Griffith JD, Mitchell BS. Regulation of the interaction of inosine monophosphate dehydrogenase with mycophenolic acid by GTP. J. Biol. Chem. 2006;281(1):206–12. doi: 10.1074/jbc.M507056200. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 1998;143(7):1883–98. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs R, Borisy G. Self-assembly of glutamic dehydrogenase into ordered superstructures: multichain tubes formed by association of single molecules. J. Mol. Biol. 1972;65(1):127–28. doi: 10.1016/0022-2836(72)90496-2. [DOI] [PubMed] [Google Scholar]
- Josephs R, Jarosch H, Edelstein S. Polymorphism of sickle cell hemoglobin fibers. J. Mol. Biol. 1976;102(3):409–26. doi: 10.1016/0022-2836(76)90324-7. [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454(7208):1088–95. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminosono S, Saito T, Oyama F, Ohshima T, Asada A, et al. Suppression of mutant Huntingtin aggregate formation by Cdk5/p35 through the effect on microtubule stability. J. Neurosci. 2008;28(35):8747–55. doi: 10.1523/JNEUROSCI.0973-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, et al. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149(4):753–67. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul DK, Fabry ME, Windisch P, Baez S, Nagel RL. Erythrocytes in sickle cell anemia are heterogeneous in their rheological and hemodynamic characteristics. J. Clin. Investig. 1983;72(1):22–31. doi: 10.1172/JCI110960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld C, Sawaya MR, Tanaka S, Nguyen C, Phillips M, et al. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309(5736):936–38. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- Kim C-W, Moon Y-A, Park SW, Cheng D, Kwon HJ, Horton JD. Induced polymerization of mammalian acetyl-CoA carboxylase by MIG12 provides a tertiary level of regulation of fatty acid synthesis. Proc. Natl. Acad. Sci. USA. 2010;107(21):9626–31. doi: 10.1073/pnas.1001292107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-Y, Kim Y-W, Hegerl R, Cyrklaff M, Kim I-S. Novel type of enzyme multimerization enhances substrate affinity of oat β-glucosidase. J. Struct. Biol. 2005;150(1):1, 10. doi: 10.1016/j.jsb.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Klein J, Dhurjati P. Protein aggregation kinetics in an Escherichia coli strain overexpressing a Salmonella typhimurium CheY mutant gene. Appl. Environ. Microbiol. 1995;61(4):1220–25. doi: 10.1128/aem.61.4.1220-1225.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourie JI, Culverson AL, Farrelly PV, Henry CL, Laohachai KN. Heterogeneous amyloid-formed ion channels as a common cytotoxic mechanism: implications for therapeutic strategies against amyloidosis. Cell Biochem. Biophys. 2002;36(2-3):191–207. doi: 10.1385/CBB:36:2-3:191. [DOI] [PubMed] [Google Scholar]
- Krammer C, Schätzl HM, Vorberg I. Prion-like propagation of cytosolic protein aggregates: insights from cell culture models. Prion. 2009;3(4):206–12. doi: 10.4161/pri.3.4.10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J. Segregation of molecules at cell division reveals native protein localization. Nat. Methods. 2012;9(5):480–82. doi: 10.1038/nmeth.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legname G, Nguyen H-OB, Peretz D, Cohen FE, DeArmond SJ, Prusiner SB. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc. Natl. Acad. Sci. USA. 2006;103(50):19105–10. doi: 10.1073/pnas.0608970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitzki A, Stallcup WB, Koshland DE. Half-of-the-sites reactivity and conformational states of cytidine triphosphate synthetase. Biochemistry. 1971;10(18):3371–78. doi: 10.1021/bi00794a009. [DOI] [PubMed] [Google Scholar]
- Lewis MR, Gross LJ, Josephs R. Cryo-electron microscopy of deoxy-sickle hemoglobin fibers. Microsc. Res. Techn. 1994;27(5):459–67. doi: 10.1002/jemt.1070270512. [DOI] [PubMed] [Google Scholar]
- Liu J-L. Intracellular compartmentation of CTP synthase in Drosophila. J. Genet. Genomics. 2010;37(5):281–96. doi: 10.1016/S1673-8527(09)60046-1. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2006;24(7):841–47. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- Mayer M, Buchner J. Refolding of inclusion body proteins. Methods Mol. Med. 2004;94:239–54. doi: 10.1385/1-59259-679-7:239. [DOI] [PubMed] [Google Scholar]
- Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17(3):349–68. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- Meredith MJ, Lane MD. Acetyl-CoA carboxylase. evidence for polymeric filament to protomer transition in the intact avian liver cell. J. Biol. Chem. 1978;253(10):3381–83. [PubMed] [Google Scholar]
- Miller R. Zinc-induced paracrystalline aggregation of glutamine synthetase. Arch. Biochem. Biophys. 1974;163(1):155–71. doi: 10.1016/0003-9861(74)90465-2. [DOI] [PubMed] [Google Scholar]
- Muchowski PJ, Ning K, D’Souza-Schorey C, Fields S. Requirement of an intact microtubule cytoskeleton for aggregation and inclusion body formation by a mutant huntingtin fragment. Proc. Natl. Acad. Sci. USA. 2002;99(2):727–32. doi: 10.1073/pnas.022628699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M, Natsumeda Y, Konno Y, Hoffman R, Irino S, Weber G. Selective up-regulation of type II inosine 5′-monophosphate dehydrogenase messenger RNA expression in human leukemias. Cancer Res. 1991;51(15):3886–90. [PubMed] [Google Scholar]
- Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O’Connell JD, et al. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. USA. 2009;106(25):10147–52. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DL, Cox MM. Lehninger Principles of Biochemistry. 4th ed Freeman; New York: 2008. [Google Scholar]
- Noree C, Sato BK, Broyer RM, Wilhelm JE. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J. Cell Biol. 2010;190(4):541–51. doi: 10.1083/jcb.201003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki M, White SL, Zeitler E, Wellems TE, Fuller SD, et al. Electron microscopy of fibers and discs of hemoglobin S having sixfold symmetry. Proc. Natl. Acad. Sci. USA. 1977;74(12):5538–42. doi: 10.1073/pnas.74.12.5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JA, Anfinsen CB. The crystallization and characterization of L-glutamic acid dehydrogenase. J. Biol. Chem. 1952;197(1):67–79. [PubMed] [Google Scholar]
- Pavelka M, Roth J. Functional Ultrastructure. Springer Vienna; Vienna: 2010. [Google Scholar]
- Piatigorsky J. Puzzle of crystallin diversity in eye lenses. Dev. Dyn. 1993;196(4):267–72. doi: 10.1002/aja.1001960408. [DOI] [PubMed] [Google Scholar]
- Polianskyte Z, Peitsaro N, Dapkunas A, Liobikas J, Soliymani R, et al. LACTB is a filament-forming protein localized in mitochondria. Proc. Natl. Acad. Sci. USA. 2009;106(45):18960–65. doi: 10.1073/pnas.0906734106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price WC, Martinez AP, Warmke HE. Crystalline inclusions in chloroplasts of the coconut palm. J. Ultrastruct. Res. 1966;14(5):618–21. doi: 10.1016/s0022-5320(66)80086-2. [DOI] [PubMed] [Google Scholar]
- Robertson JG. Determination of subunit dissociation constants in native and inactivated CTP synthetase by sedimentation equilibrium. Biochemistry. 1995;34(22):7533–41. doi: 10.1021/bi00022a029. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10(Suppl.):S10–17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Sabbaghian M, Ebrahim-Habibi A, Nemat-Gorgani M. Thermal aggregation of a model allosteric protein in different conformational states. Int. J. Biol. Macromol. 2009;44(2):156–62. doi: 10.1016/j.ijbiomac.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Sagot I, Pinson B, Salin B, Daignan-Fornier B. Actin bodies in yeast quiescent cells: an immediately available actin reserve? Mol. Biol. Cell. 2006;17(11):4645–55. doi: 10.1091/mbc.E06-04-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DF, Afonso B, Chen AH, Silver PA. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327(5970):1258–61. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Nurse P. Identification of fission yeast nuclear markers using random polypeptide fusions with green fluorescent protein. Proc. Natl. Acad. Sci. USA. 1996;93(26):15146–51. doi: 10.1073/pnas.93.26.15146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schendel FJ, Cheng YS, Otvos JD, Wehrli S, Stubbe J. Characterization and chemical properties of phosphoribosylamine, an unstable intermediate in the de novo purine biosynthetic pathway. Biochemistry. 1988;27(7):2614–23. doi: 10.1021/bi00407a052. [DOI] [PubMed] [Google Scholar]
- Shively JM, Ball F, Brown DH, Saunders RE. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science. 1973;182(4112):584–86. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- Shively JM. Inclusion bodies of prokaryotes. Annu. Rev. Microbiol. 1974;28:167–87. doi: 10.1146/annurev.mi.28.100174.001123. [DOI] [PubMed] [Google Scholar]
- Smith GK, Mueller WT, Wasserman GF, Taylor WD, Benkovic SJ. Characterization of the enzyme complex involving the folate-requiring enzymes of de novo purine biosynthesis. Biochemistry. 1980;19(18):4313–21. doi: 10.1021/bi00559a026. [DOI] [PubMed] [Google Scholar]
- So AK, Espie GS, Williams EB, Shively JM, Heinhorst S, Cannon GC. A novel evolutionary lineage of carbonic anhydrase (ε class) is a component of the carboxysome shell. J. Bacteriol. 2004;186(3):623–30. doi: 10.1128/JB.186.3.623-630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. 2003;81(11):678–99. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- Stillman TJ, Baker PJ, Britton KL, Rice DW. Conformational flexibility in glutamate dehydrogenase. Role of water in substrate recognition and catalysis. J. Mol. Biol. 1993;234(4):1131–39. doi: 10.1006/jmbi.1993.1665. [DOI] [PubMed] [Google Scholar]
- Stojiljkovic I, Baümler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 1995;177(5):1357–66. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sue M, Yamazaki K, Yajima S, Nomura T, Matsukawa T, et al. Molecular and structural characterization of hexameric β-d-glucosidases in wheat and rye. Plant Physiol. 2006;141(4):1237–47. doi: 10.1104/pp.106.077693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia GG, Pechmann S, Dobson CM, Vendruscolo M. Life on the edge: a link between gene expression levels and aggregation rates of human proteins. Trends Biochem. Sci. 2007;32(5):204–6. doi: 10.1016/j.tibs.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Tenberge KB, Eising R. Immunogold labelling indicates high catalase concentrations in amorphous and crystalline inclusions of sunflower (Helianthus annuus L.) peroxisomes. Histochem. J. 1995;27(3):184–95. [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 2010;11(11):777–88. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- Wainscoat JS, Bell JI, Thein SL, Higgs DR, Sarjeant GR, et al. Multiple origins of the sickle mutation: evidence from beta S globin gene cluster polymorphisms. Mol. Biol. Med. 1983;1(2):191–97. [PubMed] [Google Scholar]
- Wang Z, Kishchenko G, Chen Y, Josephs R. Polymerization of deoxy-sickle cell hemoglobin in high-phosphate buffer. J. Struct. Biol. 2000;131(3):197–209. doi: 10.1006/jsbi.2000.4295. [DOI] [PubMed] [Google Scholar]
- Watabe M, Nakaki T. Protein kinase CK2 regulates the formation and clearance of aggresomes in response to stress. J. Cell Sci. 2011;124(Pt. 9):1519–32. doi: 10.1242/jcs.081778. [DOI] [PubMed] [Google Scholar]
- Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc. Natl. Acad. Sci. USA. 2009;106(19):7858–63. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264(5158):566–69. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Woolfolk C. Regulation of glutamine synthetase III. Cumulative feedback inhibition of glutamine synthetase from Escherichia coli. Arch. Biochem. Biophys. 1967;118(3):736–55. doi: 10.1016/0003-9861(67)90412-2. [DOI] [PubMed] [Google Scholar]
- Yamaoka T, Yano M, Kondo M, Sasaki H, Hino S, et al. Feedback inhibition of amidophosphoribosyl-transferase regulates the rate of cell growth via purine nucleotide, DNA, and protein syntheses. J. Biol. Chem. 2001;276(24):21285–91. doi: 10.1074/jbc.M011103200. [DOI] [PubMed] [Google Scholar]
- Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat. Rev. Microbiol. 2008;6(9):681–91. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]
- Yeates TO, Tsai Y, Tanaka S, Sawaya MR, Kerfeld CA. Self-assembly in the carboxysome: a viral capsid-like protein shell in bacterial cells. Biochem. Soc. Trans. 2007;35(Pt. 3):508–11. doi: 10.1042/BST0350508. [DOI] [PubMed] [Google Scholar]
- Yokota S, Kamijo K, Oda T. Aggregate formation and degradation of overexpressed wild-type and mutant urate oxidase proteins. Quality control of organelle-destined proteins by the endoplasmic reticulum. Histochem. Cell Biol. 2000;114:433–46. doi: 10.1007/s004180000208. [DOI] [PubMed] [Google Scholar]
- Yuan P, Jedd G, Kumaran D, Swaminathan S, Shio H, et al. A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa. Nat. Struct. Biol. 2003;10(4):264–70. doi: 10.1038/nsb910. [DOI] [PubMed] [Google Scholar]
- Zeiri L, Reisler E. Uncoupling of the catalytic activity and the polymerization of beef liver glutamate dehydrogenase. J. Mol. Biol. 1978;124(1):291–95. doi: 10.1016/0022-2836(78)90162-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y-B, Howitt J, McCorkle S, Lawrence P, Springer K, Freimuth P. Protein aggregation during overexpression limited by peptide extensions with large net negative charge. Protein Expr. Purif. 2004;36(2):207–16. doi: 10.1016/j.pep.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Zhao A, Tsechansky M, Swaminathan J, Cook L, Ellington AD, Marcotte EM. Overexpressed purine biosynthetic enzymes form stress bodies. PLoS ONE. 2012 doi: 10.1371/journal.pone.0056203. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann AG, Gu JJ, Laliberté J, Mitchell BS. Inosine-5′-monophosphate dehydrogenase: regulation of expression and role in cellular proliferation and T lymphocyte activation. Prog. Nucleic Acid Res. Mol. Biol. 1998;61:181–209. doi: 10.1016/s0079-6603(08)60827-2. [DOI] [PubMed] [Google Scholar]