Roberts syndrome: A deficit in acetylated cohesin leads to nucleolar dysfunction (original) (raw)
Abstract
All living organisms must go through cycles of replicating their genetic information and then dividing the copies between two new cells. This cyclical process, in cells from bacteria and human alike, requires a protein complex known as cohesin. Cohesin is a structural maintenance of chromosomes (SMC) complex. While bacteria have one form of this complex, yeast have several SMC complexes, and humans have at least a dozen cohesin complexes alone. Therefore the ancient structure and function of SMC complexes has been both conserved and specialized over the course of evolution. These complexes play roles in replication, repair, organization, and segregation of the genome. Mutations in the genes that encode cohesin and its regulatory factors are associated with developmental disorders such as Roberts syndrome, Cornelia de Lange syndrome, and cancer. In this review, we focus on how acetylation of cohesin contributes to its function. In Roberts syndrome, the lack of cohesin acetylation contributes to nucleolar defects and translational inhibition. An understanding of basic SMC complex function will be essential to unraveling the molecular etiology of human diseases associated with defective SMC function.
Keywords: ribosome, cohesin, cohesinopathies, ribosomal DNA, nucleolus, translation, Roberts syndrome, ESCO2, acetyltransferase
The Cohesin Ring
In order to understand the function of cohesin, it is important to consider its motifs and structure. Cohesin forms a ring, consisting of two long coiled-coil proteins known as SMC proteins, a third component known as the kleisin subunit Rad21/Mcd1/Scc1 (hereafter referred to as Rad21), which closes the complex, and a fourth subunit Scc3/STAG of unclear function (Fig. 1). Smc1 and Smc3 are ATP binding proteins that belong to the structural maintenance of chromosomes (SMC) family, which also includes condensin subunits (Smc2 and Smc4) and Smc5/6 complex components (Smc5 and Smc6). All SMC proteins help modulate chromosome structure and topology. SMC proteins are characterized by ~50 nanometer anti-parallel coiled coils flanked by a globular hinge domain and N and C-terminal domains. The N and C-terminal domains of SMC proteins contain conserved Walker A and Walker B motifs that come together to make an ATPase referred to as the head domain. ATPase activity is important for association of the cohesin complex with the chromosome and relocation of the cohesin ring along DNA.2,3
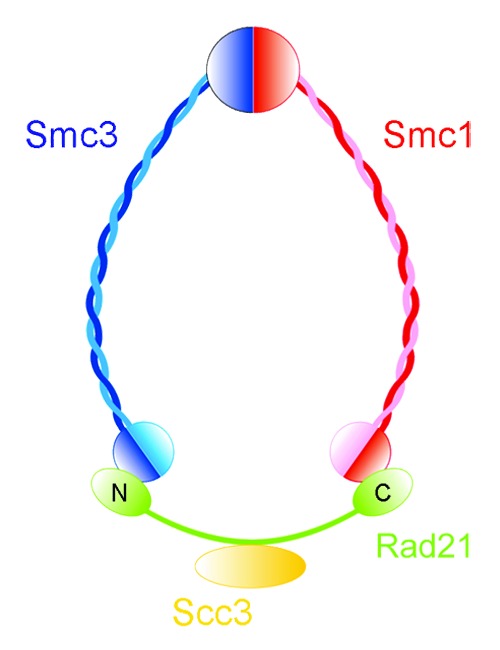
Figure 1. Cohesin subunits are arranged to form a ring. The long coiled-coil Smc1 and 3 subunits are shown in blue and red, respectively, interacting at their hinge domain. The head domains of each subunit form an ATPase. The head domains are held together with the Scc1/Rad21/Mcd1 kleisin subunit (green). The Scc3/STAG subunit (yellow) associates with the kleisin. Adapted from reference 1.
Smc1 and Smc3 interact via their hinge domains. The head domains are held together by a kleisin subunit. Kleisins are a superfamily of bacterial and eukaryotic SMC protein partners.4 The N terminus of Rad21 binds to the head domain of Smc3 while the C terminus binds to the head domain of Smc1,5 which links the head of the SMC proteins. Electron microscopy reveals that the Smc1-Smc3 heterodimer forms a V-shaped structure.5,6 Because Rad21 bridges the heads of Smc1 and Smc3, the three proteins are thought to form a circular complex.5,7,8 The cohesin complex appears to be able to open and close via either the Smc1-Smc3 hinge domain interaction or the junction between the kleisin and the head domain.9 Rad21 is cleaved by a protease prior to the onset of anaphase, allowing the cohesin ring to fall apart and the chromosomes to segregate.10,11
Cohesin associates with hundreds of regions within a genome.12-14 One predominant association site is the pericentromeric region. At this region, cohesin binding opposes the spindle forces and allows chromosomes to orient on the spindle to ensure their accurate segregation. However, cohesin also binds at many other regions in the genome. In metazoans, cohesin is often found at the transcription start site of active genes and at enhancer regions, suggesting cohesin could play a role in transcription. Another region where cohesin is found in both prokaryotes and eukaryotes is the rDNA repeats. These repeats are highly transcribed by RNA polymerase I to make rRNAs that are structural components of the ribosome. While cohesion near centromeres appears to promote chromosome segregation, the role of cohesin at other regions of the genome is less clear, but is likely to influence replication, repair, transcription, and organization of the genome.
Cohesin Acetylation
The opening and closing of the cohesin ring must be regulated in order to control its stability and association with chromatin throughout the cell cycle. Cohesin can interact with DNA in at least two modes, one of which is extremely stable and one of which is more transient. Both acetylation and sumoylation promote stable cohesin-DNA association in budding yeast. Phosphorylation of cohesin subunits can promote or destroy cohesion, depending on the context.1 Both the acetylation mechanism for promoting stable cohesin-DNA interactions and the acetyltransferase are evolutionarily conserved from yeast to human cells. Eco1 (establishment of cohesion) was first discovered in budding yeast as an acetyltransferase required for the establishment of cohesion during S phase.15,16 Eco1 interacts with replication fork components17 and may travel with replication forks to acetylate cohesin complexes during S phase to promote cohesion.18
The target of Eco1 acetylation are lysines 112 and 113 in yeast Smc3.19-21 This acetylation event appears to lock the ring into a stable DNA association mode. When these residues are acetylated, interactions between Smc3 and anti-establishment factors such as Pds5 and Wapl are disrupted, allowing cohesion to be established in S phase.22,23 Deacetylation of Smc3 by Hos1 (known as HDAC8 in human) is also important for recycling Smc3 for the next cell cycle.24-26 In addition to cohesion establishment, acetylation plays an important role in DNA damage repair. Instead of Smc3, Eco1 acetylates the kleisin subunit Rad21 in response to a DNA double-strand break to promote cohesion both at the break site and genome-wide.19,27-31
Budding yeast only contain a single enzyme, Eco1, to acetylate cohesin. Mammalian cells have two enzymes, ESCO1 and ESCO2, both implicated in sister chromatid cohesion.32 This begs the question as to which are the unique and which are the overlapping actions of these two enzymes. Although both are acetyltransferases, knockdown of ESCO1 reduces the acetylation of lysines 105 and 106 on Smc3 more than knockdown of ESCO2,33 suggesting ESCO1 is responsible for the bulk of Smc3 acetylation on these lysines. ESCO1 acetylation of Smc3 is important for cell survival in response to ionizing radiation-induced DNA damage, in association with genome-wide reinforcement of cohesin binding in the G2/M cell cycle phase.34 ESCO2 mutant cells are also sensitive to DNA damage, suggesting ESCO2 also functions as part of the DSB repair response.35 While our current understanding of the functional overlap of these two enzymes is poor, insight into their function will help us understand regulation of cohesin association throughout the genome
Roberts Syndrome
The developmental disorder known as Roberts syndrome (RBS) is caused by mutations in ESCO2 that eliminate its acetyltransferase activity.36,37 RBS patients have growth and mental retardation, limb deformities, and craniofacial defects. Deacetylated cohesin represents the unstable form which is not fully capable of holding sisters together. Metaphase chromosome spreads from RBS patients show heterochromatic repulsion,37 suggesting the possibility that ESCO2 is especially important for cohesion at these regions (Fig. 2). In mouse cells, ESCO2 associates with pericentric heterochromatin and depletion leads to reduced cohesin acetylation, lagging chromosomes, and increased apoptosis.38 In contrast to the case in humans, loss of ESCO2 in mice is lethal. ESCO1 has never been reported to be mutated in association with human disease, possibly because mutations in this gene would be lethal.
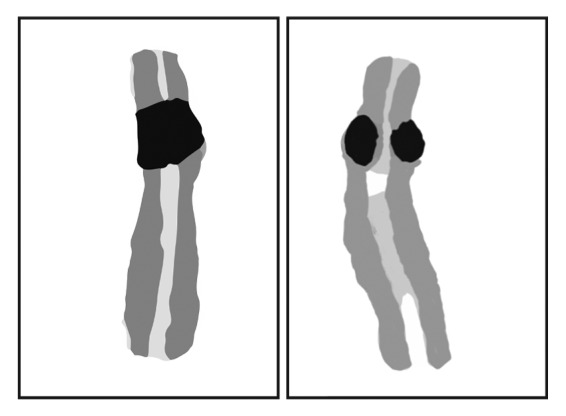
Figure 2. Chromosomes derived from a metaphase spread of a RBS patient cell show repulsion at heterochromatic regions, which includes pericentromeric regions and rDNA. We speculate that the acetylation activity of ESCO2 is necessary for cohesion of these regions. Although both chromosomes are derived from the same metaphase spread presented in Vega et al. (2005),37 the one on the left appears normal while the one on the right shows repulsion.
Because heterochromatic domains are difficult to replicate, it is possible that there is a cohesin acetyltransferase devoted to establishing cohesion in these regions in mammalian cells. The regions with heterochromatic repulsion in human RBS cells include pericentric domains and nucleolar organizing regions (NORs or rDNA). While the repulsion at pericentric domains might be predicted to cause chromosome missegregation and aneuploidy that could contribute to cell death, it remains to be determined if these contribute to the disease etiology. Gene expression is clearly disrupted and contributes to the etiology of both RBS and related cohesinopathies, although the mechanisms behind the changes in gene expression are not clear.39
A budding yeast strain with a mutation in ECO1 that genocopies an RBS allele was found to have defects in nucleolar organization, transcription and cohesion of the rDNA, and translation.40,41 Cohesion within the rDNA appears to promote nucleolar structure and function in budding yeast. In budding yeast the rDNA is present in 100–200 copies at a single locus. At any given moment, about half of the repeats are highly transcribed by RNA polymerase I to produce the rRNAs that become structural components of ribosomes. These RNAs can be limiting for ribosome formation.42 The repeats form loops43 that are reduced in budding yeast bearing an RBS mutation, although RNA polymerase I occupancy remained normal (see Fig. 3 for a model of the role of cohesion at the rDNA). Furthermore, depletion or artificial destruction of cohesion in a single cell cycle was associated with loss of nucleolar integrity.45 Given these observations in budding yeast, we wondered whether nucleolar deficits would accompany ESCO2 mutation in human RBS cells. In human cells there are 5 chromosomes with rDNA repeats. RBS cells have profound defects in transcription of the rDNA and ribosome production.40 Furthermore, the nucleolus is highly fragmented46 (Fig. 4). This leads to global changes in gene expression and cell physiology47 (Xu and Gerton, unpublished).
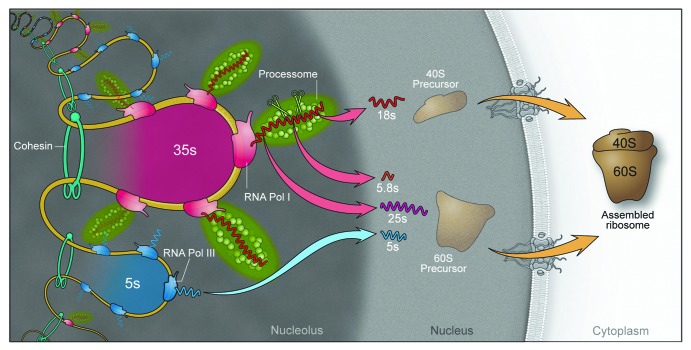
Figure 3. A model for the role of cohesion within the nucleolus. Cohesin binding to the rDNA repeats is evolutionarily conserved from bacteria to human cells. In budding yeast, cohesin binds to the non-transcribed region between repeats44 in such a way that it could trap loops of 35S genes. The 35S gene is transcribed by RNA polymerase I and then cleaved and modified by the processome to make 18S, 5.8S, and 25S rRNAs that are included in the large (60S) and small (40S) ribosomal subunits. RNA polymerase III transcribes the 5S RNA that is included in the large ribosomal subunit. Cohesion at this region could promote transcription by RNA polymerase I as well as the overall structural formation of the nucleolus.
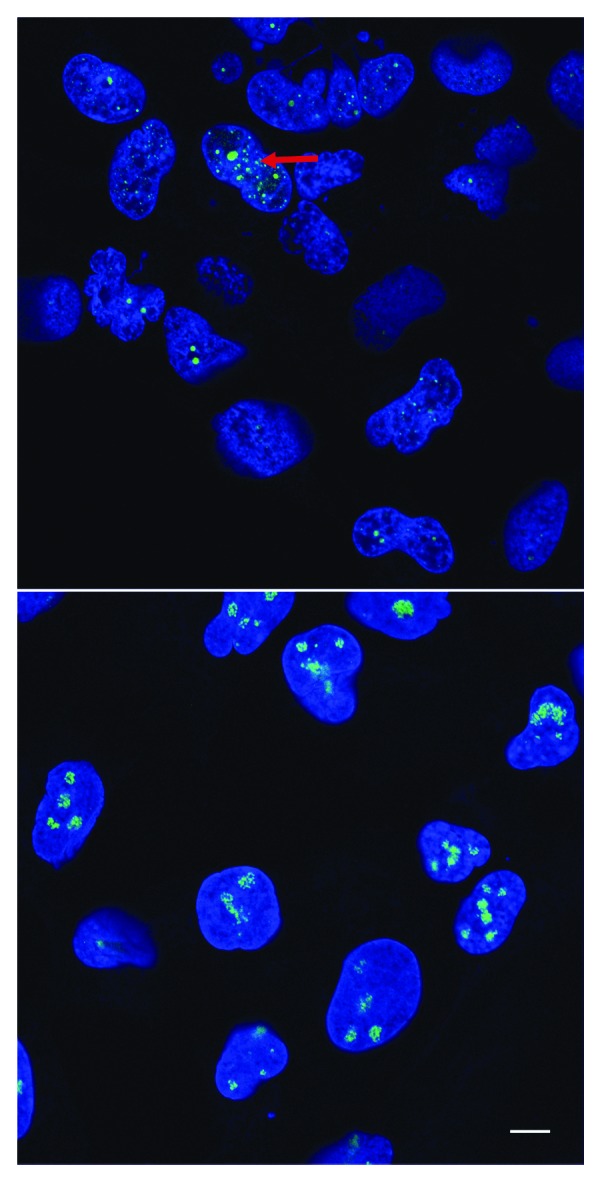
Figure 4. RBS patient cells have highly fragmented nucleoli. Skin cells from patients with RBS display defects in the organization of nucleoli (red arrow), specialized nuclear subdomains dedicated to the production of ribosomes. Fibrillarin, a nucleolar marker stained with an anti-fibrillarin antibody, is shown in green. DNA is stained with DAPI, shown in blue. Colors were adjusted for improved visual presentation. The scale bar is 10 µm. Top, RBS cells; bottom, normal cells.
mTORC1 is a protein kinase that senses cellular stress, such as amino acid deprivation, and responds by shutting down protein translation. Given the translation defects in RBS cells, we examined the mTOR pathway. Based on several key effector proteins, we found that the TOR pathway was inhibited and p53 was activated in RBS cells. A zebrafish model for RBS showed that depletion of ESCO2 was associated with disruption of the cell cycle and high levels of apoptosis.48 Using zebrafish RBS models, we found that the mTOR and p53 pathways were inhibited and activated, respectively, as observed in human RBS cells.46 Stimulation of the TOR pathway with l-leucine helped to rescue both protein synthesis and cell division and block cell death.46 At the organismal level, l-leucine had a substantial rescue effect on the development of zebrafish RBS embryos, suggesting that many of the transcriptional changes could be rescued by boosting translation (Fig. 5). The fact that l-leucine can push the TOR pathway to work harder also suggests that the basic machinery needed to make ribosomes and carry out translation is intact in RBS cells.
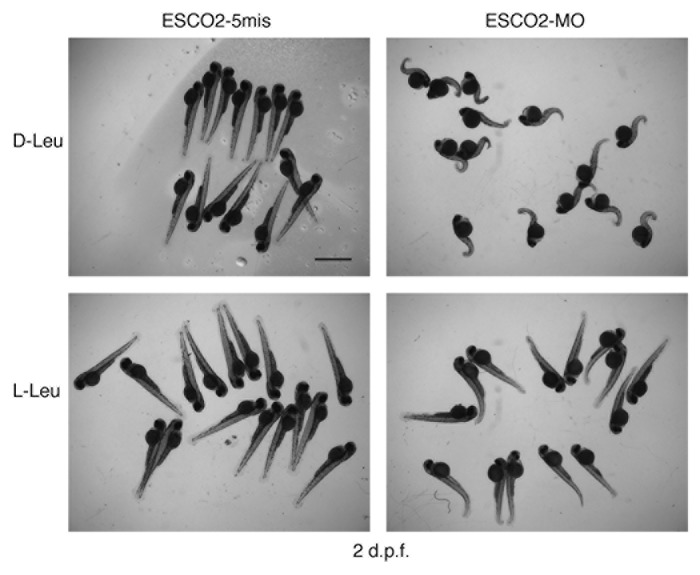
**Figure 5.**l-leucine treated RBS zebrafish show improved development. A morpholino to ESCO2 (ESCO2-MO) or a control morpholino (ESCO2–5mis) was used for knock down in wild type embryos at the 1–2 cell stage. Following microinjection, embryos were incubated with 4 mM L-glutamine plus either 10 mM d-leucine, which is biologically inactive, or 10 mM l-leucine for 2 d post fertilization (dpf) and then photographed. L-glutamine assists in uptake of leucine. The scale bar is 2 mm.
In the future, it will be interesting to quantify the extent to which translational changes drive differential gene expression in RBS.49 An experiment performed in eco1 mutant budding yeast in which poor rRNA production was corrected by deletion of FOB1 reveals that the differential expression of hundreds of genes is also corrected.50 This experiment suggests that a substantial portion of the differential gene expression associated with a mutation in ECO1 in budding yeast is due to defects at the rDNA locus and reduced translation.
Cornelia de Lange Syndrome and Cancer
Cornelia de Lange syndrome (CdLS) is a genetically dominant disorder caused by the mutation of one copy of NIPBL, SMC1, SMC3, or HDAC8.51-57 A related disorder is caused by mutation of RAD21.58 These mutations affect the cohesin ring (SMC1, SMC3, RAD21), the deacetylation of the ring (HDAC8), or the loading of the ring (NIPBL). CdLS is characterized by growth and mental retardation, limb deformities, and craniofacial defects. The severity of CdLS can range from a mild disorder of the nervous system to a severe multisystem disorder. About 60% of CdLS cases are caused by mutations in NIPBL, a loading factor for the cohesin complex. While gene expression is clearly disrupted and contributes to the etiology of CdLS, the underlying mechanism is unclear.59 Many models posit that cohesin binding at a given locus has direct effects on its transcription. Because cohesin binds at many sites in the genome, a mutation in cohesin has the potential to affect the transcription of hundreds of genes. However, translational changes such as those associated with RBS would also be expected to affect the transcription and translation of many genes.
We speculate that mutations in ESCO2 may affect a particular pool of cohesin, the cohesin at heterochromatic regions. When cohesion at the rDNA is reduced, nucleolar structure is disrupted and nucleolar function is less efficient, with an accompanying deficit in the TOR pathway. In contrast, mutations that cause CdLS may affect cohesin throughout the genome, unless the mutation compromises protein-protein or protein-DNA interactions that occur at specific cohesin associated regions. These mutations could have anywhere from undetectable to severe effects on the pool of cohesin at the rDNA and on nucleolar function. In budding yeast, a mutation in SMC1 that genocopies a mutation associated with CdLS causes nucleolar phenotypes that are milder than the ECO1 mutation,40,41 suggesting some CdLS mutations may be associated with mild defects in nucleolar function. In zebrafish models for CdLS, some mutations appear to be associated with more growth delay and apoptosis than others;48,60-63 nucleolar function has not been analyzed. However, if nucleolar function is affected by non-ESCO2 mutations, this could potentially contribute to the gene expression changes associated with CdLS. Furthermore, if translation is not operating at full capacity, l-leucine might be able to stimulate mTOR to help rescue translation and some of the differential gene expression associated with CdLS.
Translational defects are associated with both human developmental disorders and cancer.64,65 A recent study found that 13% of acute myeloid leukemia (AML) cases were caused by mutations in cohesin genes, including RAD21, STAG2, SMC1, and SMC3.66 In general, AML is not associated with abnormal karyotypes, raising the possibility that the cohesin mutations are causing transcriptional or translational changes that lead to AML, and emphasizing the need to understand the molecular origin of these changes. If we can understand the contribution of translational mechanisms and particular key regulatory pathways, such as p53 and mTOR, in Roberts syndrome and CdLS, we might also understand how cohesin mutations lead to myeloid neoplasms.66,67
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
We thank Richard Shrock and Mark Miller for assistance in manuscript and figure preparation.
References
- 1.Xiong B, Gerton JL. Regulators of the cohesin network. Annu Rev Biochem. 2010;79:131–53. doi: 10.1146/annurev-biochem-061708-092640. [DOI] [PubMed] [Google Scholar]
- 2.Hu B, Itoh T, Mishra A, Katoh Y, Chan KL, Upcher W, Godlee C, Roig MB, Shirahige K, Nasmyth K. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr Biol. 2011;21:12–24. doi: 10.1016/j.cub.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr Biol. 2003;13:1941–53. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 4.Schleiffer A, Kaitna S, Maurer-Stroh S, Glotzer M, Nasmyth K, Eisenhaber F. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol Cell. 2003;11:571–5. doi: 10.1016/S1097-2765(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 5.Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–88. doi: 10.1016/S1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 6.Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156:419–24. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–77. doi: 10.1016/S0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 8.Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Löwe J. Structure and stability of cohesin’s Smc1-kleisin interaction. Mol Cell. 2004;15:951–64. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 9.Nasmyth K. Cohesin: a catenase with separate entry and exit gates? Nat Cell Biol. 2011;13:1170–7. doi: 10.1038/ncb2349. [DOI] [PubMed] [Google Scholar]
- 10.Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–3. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 11.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–86. doi: 10.1016/S0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 12.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 13.Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–8. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–19. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tóth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–33. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenna MA, Skibbens RV. Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol Cell Biol. 2003;23:2999–3007. doi: 10.1128/MCB.23.8.2999-3007.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell. 2006;23:787–99. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 19.Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–9. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 20.Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–6. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 21.Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouët F, Underwood P, Metson J, Imre R, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell. 2009;33:763–74. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 22.Terret M-E, Sherwood R, Rahman S, Qin J, Jallepalli PV. Cohesin acetylation speeds the replication fork. Nature. 2009;462:231–4. doi: 10.1038/nature08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–6. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 24.Xiong B, Lu S, Gerton JL. Hos1 is a lysine deacetylase for the Smc3 subunit of cohesin. Curr Biol. 2010;20:1660–5. doi: 10.1016/j.cub.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Borges V, Lehane C, Lopez-Serra L, Flynn H, Skehel M, Rolef Ben-Shahar T, Uhlmann F. Hos1 deacetylates Smc3 to close the cohesin acetylation cycle. Mol Cell. 2010;39:677–88. doi: 10.1016/j.molcel.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Beckouët F, Hu B, Roig MB, Sutani T, Komata M, Uluocak P, Katis VL, Shirahige K, Nasmyth K. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol Cell. 2010;39:689–99. doi: 10.1016/j.molcel.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heidinger-Pauli JM, Unal E, Koshland D. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol Cell. 2009;34:311–21. doi: 10.1016/j.molcel.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ström L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjögren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–5. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- 29.Ström L, Lindroos HB, Shirahige K, Sjögren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–15. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 30.Heidinger-Pauli JM, Unal E, Guacci V, Koshland D. The kleisin subunit of cohesin dictates damage-induced cohesion. Mol Cell. 2008;31:47–56. doi: 10.1016/j.molcel.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 32.Hou F, Zou H. Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol Biol Cell. 2005;16:3908–18. doi: 10.1091/mbc.E04-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell. 2008;31:143–51. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Kim BJ, Li Y, Zhang J, Xi Y, Li Y, Yang T, Jung SY, Pan X, Chen R, Li W, et al. Genome-wide reinforcement of cohesin binding at pre-existing cohesin sites in response to ionizing radiation in human cells. J Biol Chem. 2010;285:22784–92. doi: 10.1074/jbc.M110.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Lelij P, Godthelp BC, van Zon W, van Gosliga D, Oostra AB, Steltenpool J, de Groot J, Scheper RJ, Wolthuis RM, Waisfisz Q, et al. The cellular phenotype of Roberts syndrome fibroblasts as revealed by ectopic expression of ESCO2. PLoS One. 2009;4:e6936. doi: 10.1371/journal.pone.0006936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordillo M, Vega H, Trainer AH, Hou F, Sakai N, Luque R, Kayserili H, Basaran S, Skovby F, Hennekam RC, et al. The molecular mechanism underlying Roberts syndrome involves loss of ESCO2 acetyltransferase activity. Hum Mol Genet. 2008;17:2172–80. doi: 10.1093/hmg/ddn116. [DOI] [PubMed] [Google Scholar]
- 37.Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagihara I, Yamada M, van Gosliga D, Kayserili H, Xu C, Ozono K, et al. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet. 2005;37:468–70. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 38.Whelan G, Kreidl E, Wutz G, Egner A, Peters JM, Eichele G. Cohesin acetyltransferase Esco2 is a cell viability factor and is required for cohesion in pericentric heterochromatin. EMBO J. 2012;31:71–82. doi: 10.1038/emboj.2011.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Krantz ID. Cohesin and human disease. Annu Rev Genomics Hum Genet. 2008;9:303–20. doi: 10.1146/annurev.genom.9.081307.164211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bose T, Lee KK, Lu S, Xu B, Harris B, Slaughter B, Unruh J, Garrett A, McDowell W, Box A, et al. Cohesin proteins promote ribosomal RNA production and protein translation in yeast and human cells. PLoS Genet. 2012;8:e1002749. doi: 10.1371/journal.pgen.1002749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gard S, Light W, Xiong B, Bose T, McNairn AJ, Harris B, Fleharty B, Seidel C, Brickner JH, Gerton JL. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J Cell Biol. 2009;187:455–62. doi: 10.1083/jcb.200906075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laferté A, Favry E, Sentenac A, Riva M, Carles C, Chédin S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–40. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayan M, Aragón L. Cis-interactions between non-coding ribosomal spacers dependent on RNAP-II separate RNAP-I and RNAP-III transcription domains. Cell Cycle. 2010;9:4328–37. doi: 10.4161/cc.9.21.13591. [DOI] [PubMed] [Google Scholar]
- 44.Laloraya S, Guacci V, Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol. 2000;151:1047–56. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris B, Bose T, Lee KK, Wang F, Lu S, Ross RT, Zhang Y, French SL, Beyer AL, Slaughter BD, et al. Cohesion promotes nucleolar structure and function. Mol Biol Cell. 2013 doi: 10.1091/mbc.E13-07-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu B, Lee KK, Zhang L, Gerton JL. Stimulation of mTORC1 with L-leucine rescues defects associated with Roberts syndrome. PLoS Genet. 2013;9:e1003857. doi: 10.1371/journal.pgen.1003857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J, Zhang Z, Bando M, Itoh T, Deardorff MA, Clark D, Kaur M, Tandy S, Kondoh T, Rappaport E, et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7:e1000119. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mönnich M, Kuriger Z, Print CG, Horsfield JA. A zebrafish model of Roberts syndrome reveals that Esco2 depletion interferes with development by disrupting the cell cycle. PLoS One. 2011;6:e20051. doi: 10.1371/journal.pone.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerton JL. Translational mechanisms at work in the cohesinopathies. Nucleus. 2012;3:520–5. doi: 10.4161/nucl.22800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu S, Lee K, Harris B, Xiong B, Bose T, Saraf A, Hattem G, Florens L, Seidel C, Gerton J. The cohesin acetyltransferase Eco1 coordinates rDNA replication and transcription. EMBO R. doi: 10.1002/embr.201337974. Forthcoming 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodríguez C, Arnedo M, Loeys B, Kline AD, et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80:485–94. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–30. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- 53.Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–5. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–41. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 55.Deardorff MA, Bando M, Nakato R, Watrin E, Itoh T, Minamino M, Saitoh K, Komata M, Katou Y, Clark D, et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–7. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Braunholz D, Hullings M, Gil-Rodríguez MC, Fincher CT, Mallozzi MB, Loy E, Albrecht M, Kaur M, Limon J, Rampuria A, et al. Isolated NIBPL missense mutations that cause Cornelia de Lange syndrome alter MAU2 interaction. Eur J Hum Genet. 2012;20:271–6. doi: 10.1038/ejhg.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lightfoot J, Testori S, Barroso C, Martinez-Perez E. Loading of meiotic cohesin by SCC-2 is required for early processing of DSBs and for the DNA damage checkpoint. Curr Biol. 2011;21:1421–30. doi: 10.1016/j.cub.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 58.Deardorff MA, Wilde JJ, Albrecht M, Dickinson E, Tennstedt S, Braunholz D, Mönnich M, Yan Y, Xu W, Gil-Rodríguez MC, et al. RAD21 mutations cause a human cohesinopathy. Am J Hum Genet. 2012;90:1014–27. doi: 10.1016/j.ajhg.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J, Krantz ID. Cornelia de Lange syndrome, cohesin, and beyond. Clin Genet. 2009;76:303–14. doi: 10.1111/j.1399-0004.2009.01271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deardorff MA, Wilde JJ, Albrecht M, Dickinson E, Tennstedt S, Braunholz D, Mönnich M, Yan Y, Xu W, Gil-Rodríguez MC, et al. RAD21 mutations cause a human cohesinopathy. Am J Hum Genet. 2012;90:1014–27. doi: 10.1016/j.ajhg.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muto A, Calof AL, Lander AD, Schilling TF. Multifactorial origins of heart and gut defects in nipbl-deficient zebrafish, a model of Cornelia de Lange Syndrome. PLoS Biol. 2011;9:e1001181. doi: 10.1371/journal.pbio.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ghiselli G. SMC3 knockdown triggers genomic instability and p53-dependent apoptosis in human and zebrafish cells. Mol Cancer. 2006;5:52. doi: 10.1186/1476-4598-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horsfield JA, Anagnostou SH, Hu JK, Cho KH, Geisler R, Lieschke G, Crosier KE, Crosier PS. Cohesin-dependent regulation of Runx genes. Development. 2007;134:2639–49. doi: 10.1242/dev.002485. [DOI] [PubMed] [Google Scholar]
- 64.Stumpf CR, Ruggero D. The cancerous translation apparatus. Curr Opin Genet Dev. 2011;21:474–83. doi: 10.1016/j.gde.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–69. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson G, et al. Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R, et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet. 2013;45:1232–7. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]