FHIP and FTS proteins are critical for dynein-mediated transport of early endosomes in Aspergillus (original) (raw)
FhipA and FtsA in Aspergillus (human FHIP and FTS homologues) are critical for HookA-dynein–mediated early endosome transport. FhipA and FtsA are required for HookA–early endosome association. FtsA–early endosome association depends on FhipA and HookA, but FhipA is able to localize to early endosomes independently of HookA or FtsA.
Abstract
The minus end–directed microtubule motor cytoplasmic dynein transports various cellular cargoes, including early endosomes, but how dynein binds to its cargo remains unclear. Recently fungal Hook homologues were found to link dynein to early endosomes for their transport. Here we identified FhipA in Aspergillus nidulans as a key player for HookA (A. nidulans Hook) function via a genome-wide screen for mutants defective in early-endosome distribution. The human homologue of FhipA, FHIP, is a protein in the previously discovered FTS/Hook/FHIP (FHF) complex, which contains, besides FHIP and Hook proteins, Fused Toes (FTS). Although this complex was not previously shown to be involved in dynein-mediated transport, we show here that loss of either FhipA or FtsA (A. nidulans FTS homologue) disrupts HookA–early endosome association and inhibits early endosome movement. Both FhipA and FtsA associate with early endosomes, and interestingly, while FtsA–early endosome association requires FhipA and HookA, FhipA–early endosome association is independent of HookA and FtsA. Thus FhipA is more directly linked to early endosomes than HookA and FtsA. However, in the absence of HookA or FtsA, FhipA protein level is significantly reduced. Our results indicate that all three proteins in the FtsA/HookA/FhipA complex are important for dynein-mediated early endosome movement.
INTRODUCTION
Cytoplasmic dynein transports a variety of cargoes, including vesicles/organelles, proteins, and mRNAs along microtubules, and mutations in dynein and its regulators are linked to defects in brain development and neuronal function (Perlson et al., 2010; Ori-McKenney et al., 2011; Franker and Hoogenraad, 2013; Schiavo et al., 2013). What proteins recruit dynein to membranous cargoes is a question of significant interest (Caviston and Holzbaur, 2006; Akhmanova and Hammer, 2010; Tan et al., 2011; Stephens, 2012; Splinter et al., 2012; Yadav et al., 2012; Zhou et al., 2012; Granger et al., 2014). Early endosomes constitute a major class of membranous cargoes of dynein that are transported from the cell periphery inward. Early endosomes undergo long-distance bidirectional movements in filamentous fungi, making these fungi excellent models for studying early endosome movements (Lenz et al., 2006; Abenza et al., 2009; Zekert and Fischer, 2009; Steinberg, 2011, 2014; Egan et al., 2012a; Peñalva et al., 2012; Seidel et al., 2013). Previous studies in fungi and mammalian cells indicate that the p25 component of the dynactin complex (Schroer, 2004), in conjunction with the dynactin complex, plays a critical role in early endosome transport by enhancing dynein–early endosome interaction (Zhang et al., 2011; Yeh et al., 2012). Recently two studies in fungi identified Hook proteins, HookA in Aspergillus nidulans and Hok1 in Ustilago maydis, as proteins that link dynein/dynactin to early endosomes (Bielska et al., 2014; Zhang et al., 2014). HookA and Hok1 both use their C-termini to interact with early endosomes, but it is not clear whether other proteins are required for this interaction.
The prototype Hook was identified in Drosophila as a protein involved in endosome maturation or sorting of endocytic cargoes (Kramer and Phistry, 1996; Sunio et al., 1999). There are three Hook proteins in mammalian cells, and they play distinct cellular roles, including Golgi positioning, centrosome function, aggresome formation, and endocytic cargo sorting (Walenta et al., 2001; Szebenyi et al., 2007a, b; Maldonado-Báez et al., 2013). All three human Hook proteins have been found in the Fused Toes (FTS)/Hook/FHIP (FHF) complex, which contains two additional proteins, FTS, a variant E2 ubiquitin-conjugating enzyme domain–containing protein, and the FTS- and Hook-Interacting Protein called FHIP (Xu et al., 2008). The C-termini of Hook proteins are implicated in targeting Hook to various organelles (Walenta et al., 2001; Bielska et al., 2014; Zhang et al., 2014), and the human Hook C-termini interact with FTS (Xu et al., 2008). The FHF complex interacts with the components of the homotypic vesicular protein-sorting (HOPS) complex, which is known to bind to late endosomes (Xu et al., 2008). Importantly, Hok1 in U. maydis physically associates with the FTS and FHIP proteins, indicating that the FHF complex is conserved from lower to higher eukaryotic systems (Bielska et al., 2014). However, it remains unclear whether FTS and FHIP proteins participate in dynein-mediated early endosome movement in various organisms and/or cell types.
Here we report the identification of the A. nidulans FHIP homologue as a key factor in dynein-mediated early endosome movement. This identification was achieved through a genome-wide screen for early-endosome distribution mutants followed by whole-genome sequencing to identify the causal mutations. This finding further led us to study the function of FtsA, the A. nidulans homologue of the FHIP/Hook-interacting protein FTS, and our results demonstrate that FtsA is also required for early endosome transport.
RESULTS
Identification of FhipA as a protein critical for dynein-mediated early endosome transport in vivo
In filamentous fungi, dynein is required for the distribution of nuclei and other membrane organelles, such as early endosomes and peroxisomes (Plamann et al., 1994; Xiang et al., 1994; Lenz et al., 2006; Abenza et al., 2009; Zekert and Fischer, 2009; Grava et al., 2011; Egan et al., 2012b). The p25 component of dynactin is important for early endosome distribution but not for nuclear distribution (Lee et al., 2001; Zhang et al., 2011). To better understand how dynein interacts with its early endosome cargo, we performed ultraviolet (UV) mutagenesis in A. nidulans to screen for early endosome distribution (eed) mutants that exhibit normal nuclear distribution and accumulation of dynein at the microtubule plus ends (Han et al., 2001; Xiang and Fischer, 2004; Wu et al., 2006). In a previous screen, we identified the eedA gene encoding HookA (Zhang et al., 2014). Here we selected mutant colonies that resemble the _eedA_1 or ∆p25 mutant colonies, which appeared slightly more compact than the wild-type colony (Figure 1A). Subsequently we performed a microscopic screen for mutants that exhibit abnormal accumulation of early endosomes at the hyphal tip but exhibit a normal pattern of nuclear distribution and normal accumulation of dynein at microtubule plus ends, as similarly described recently (Zhang et al., 2014). In the present study, we focused on one eed mutant (#3) that shows a defect in early endosome movement but not in nuclear distribution or dynein localization (Figure 1, B and C).
FIGURE 1:
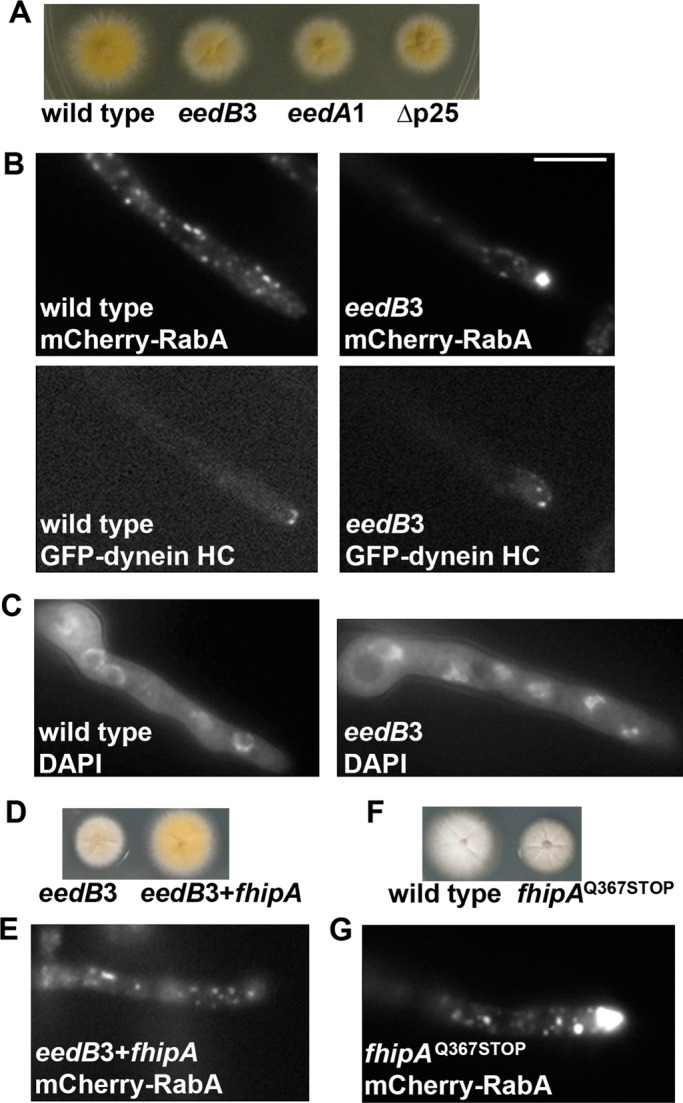
Phenotypic analysis of the _eedB_3/_fhipA_Q367STOP mutant. (A) Colony phenotype of the _eedB_3 mutant in comparison to that of a wild-type control strain, the _eedA_1 mutant (_hookA_L150P, E151K), and the ∆p25 mutant. (B) Microscopic images showing the distributions of mCherry-RabA–labeled early endosomes (mCherry-RabA) and GFP-labeled dynein heavy chain (GFP-dynein HC) in wild type and the _eedB_3 mutant. The same cells are shown for both the mCherry-RabA and GFP-dynein HC images. In the _eedB_3 mutant, dynein-HC proteins form normal comet-like structures, representing their microtubule plus end accumulation. However, abnormal accumulation of the mCherry-RabA signals was observed at ∼71% of the hyphal tips of the _eedB_3 mutant (n = 125), whereas none of the wild-type cells showed the same accumulation (n = 100). (C) Images of nuclei stained by a DNA dye, DAPI, in wild type and the _eedB_3 mutant, indicating that nuclear distribution is normal in the _eedB_3 mutant. (D) Colony phenotypes of the _eedB_3 mutant and the strain in which the wild-type fhipA gene has rescued the mutant. (E) A microscopic image showing the distributions of mCherry-RabA–labeled early endosomes (mCherry-RabA) in the _eedB_3 mutant rescued by the wild-type fhipA gene. In this rescued strain, none of the hyphal tips show the abnormal accumulation of mCherry-RabA signals (n = 100). (F) Colony phenotypes of a wild-type strain and the strain in which the _fhipA_Q367STOP mutation was introduced to the wild-type genome by transformation. (G) A microscopic image showing the distributions of mCherry-RabA–labeled early endosomes (mCherry-RabA) in the _fhipA_Q37STOP mutant. In this strain, ∼73% of the hyphal tips show the abnormal accumulation of mCherry-RabA signals (n = 100). Bar, 5 μm.
Genetic crosses were set up to determine whether the newly identified eed mutation is in the p25 or the hookA gene. In addition, genetic analyses were also done to determine whether the eed mutation is in the nudA gene, encoding cytoplasmic dynein heavy chain (HC), or the nudM gene, encoding the p150 component of dynactin, as some mutations of these genes also affect early endosome distribution more obviously than nuclear distribution (Yao et al., 2012; Tan et al., 2014). Because wild-type progeny were produced from all these crosses, it was apparent that the new eed mutation is not an allele of the nudA (dynein HC), nudM (dynactin p150), dynactin p25 or the eedA (hookA) gene. Thus we named the mutation _eedB_3.
To identify the _eedB_3 mutation, we took a whole-genome-sequencing approach using the genome sequencing and bioinformatic service of Otogenetics (www.otogenetics.com). We also used the software Tablet, version 1.13.12.17 (James Hutton Institute [http://ics.hutton.ac.uk/tablet/]), for visualizing sequence assemblies and alignments (Milne et al., 2013). After eliminating the common single-nucleotide polymorphisms that are not linked to the _eedB_3 phenotype, we focused on 10 candidate mutations for complementation studies. The wild-type DNA fragments covering the positions of the mutations were amplified and transformed into the _eedB_3 mutant. Only one fragment rescued the mutant phenotype (Figure 1, D and E), and this fragment corresponds to the An10801 gene, encoding a protein with 843 amino acids. Remarkably, the protein encoded by AN10801 is homologous to the human FHIP protein, and thus we named it FhipA (standing for FHIP in A. nidulans; Supplemental Figure S1). The mutation we identified in the fhipA gene is CAG to TAG, a nonsense mutation replacing amino acid (aa) 367Q with a stop codon. To confirm that this mutation is causal for the mutant phenotype, we amplified a DNA fragment carrying the mutation and introduced it to the wild type by transformation. We found one transformant (_fhipA_Q367STOP) that shows the same phenotype as the _eedB_3 mutant (Figure 1, F and G), and we crossed it to the original _eedB_3 mutant for genetic linkage analysis. From this cross, 100% of the progeny showed the eedB3 mutant colony phenotype and the abnormal hyphal-tip accumulation of early endosomes, further supporting the notion that fhipA is the gene whose mutation caused the _eedB_3 mutant phenotype.
The human FHIP protein is a component of the FHF complex, which also contains FTS and the human Hook proteins. Because we previously identified the A. nidulans Hook homologue, HookA, as a protein critical for early endosome transport, the present result strongly suggests that the FHF complex functions in dynein-mediated early endosome transport in A. nidulans. To test this idea, we identified the gene encoding FtsA (An0883), the FTS homologue in the A. nidulans genome (Supplemental Figure S2) and made deletion mutants for both the fhipA and ftsA genes. Both the ∆fhipA and ∆ftsA mutants exhibit a colony phenotype and defect in early endosome distribution similar to that of the _eedB_3 mutant (Figure 2, A and B, and Supplemental Movies S1–S3). However, these mutants exhibited normal comet-like structures formed by green fluorescent protein (GFP)–dynein HC and a normal pattern of nuclear distribution without any abnormal clustering of the nuclei (Figure 2, B and C), indicating that neither FhipA nor FtsA is required for the overall function and localization of dynein, but they are essential for dynein-mediated early endosome transport.
FIGURE 2:
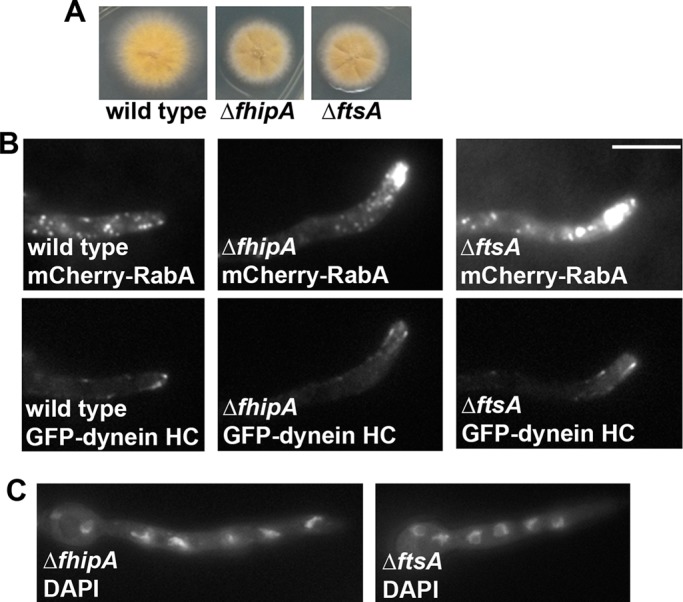
Phenotypes of the ∆fhipA and ∆ftsA mutants. (A) Colony phenotypes of the ∆fhipA and ∆ftsA mutants in comparison to that of a wild-type control strain. (B) Distributions of mCherry-RabA–labeled early endosomes in the ∆fhipA and ∆ftsA mutants. About 79% of the ∆fhipA and 57% of the ∆ftsA hyphal tips exhibited abnormal accumulation of mCherry-RabA signals, whereas none of wild-type hyphal tips shows this accumulation. GFP-dynein HC signals in the same cells are shown (bottom) to indicate that dynein localization is normal in these cells. (C) Images of DAPI-stained nuclei in the ∆fhipA and ∆ftsA mutants. The pattern of nuclear distribution in the mutants is normal, as none of the mutant cells shows any cluster of four or more nuclei when grown under the same condition that allow us to see the hyphal-tip mCherry-RabA accumulation. Bar, 5 μm.
Both FhipA and FtsA are required for HookA–early endosome interaction
The C-termini of Hook proteins are implicated in cargo binding (Walenta et al., 2001), and the C-termini of fungal Hook proteins are essential for Hook–early endosome interaction (Bielska et al., 2014; Zhang et al., 2014). The human FTS binds to the C-termini of human Hook proteins (Xu et al., 2008), but it remains to be determined whether it affects the cargo-binding ability of Hook proteins. To address this issue, we examined whether loss of FtsA affects HookA–early endosome interaction. To do this, we introduced the ∆ftsA allele into the strain carrying HookA-GFP and mCherry-RabA and observed the fluorescence signals. In the ∆ftsA mutant, HookA-GFP signals are largely diffuse, although mCherry-RabA signals accumulate at the hyphal tip (Figure 3A). Using a similar approach, we found that in the ∆fhip mutant, HookA-GFP no longer colocalizes with mCherry-RabA–labeled early endosomes accumulated at the hyphal tip (Figure 3A). To determine whether the HookA-GFP protein level is affected by the loss of FtsA or FhipA, we performed Western analysis on HookA-GFP in the ∆ftsA and ∆fhip mutant extracts. Our results show that HookA-GFP protein is stably expressed in the mutants (Figure 3B), and thus the localization defects that we observed in the ∆ftsA and ∆fhip mutants are not due to any obvious decrease in HookA protein expression or stability in the absence of FtsA or FhipA. Together these results indicate that both FtsA and FhipA are required for HookA-GFP to associate with early endosomes.
FIGURE 3:
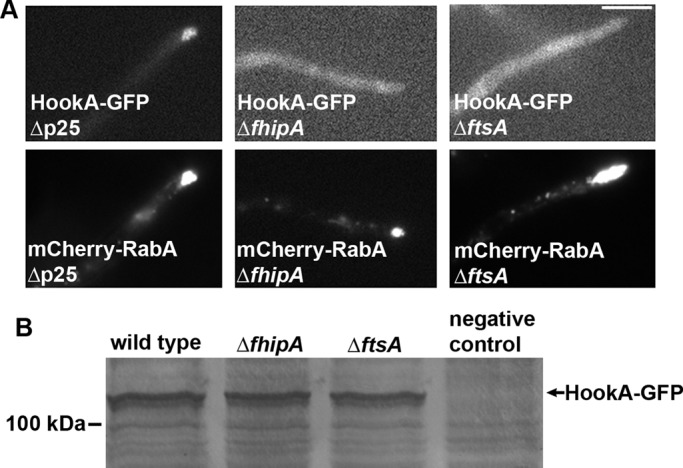
The ∆fhipA and ∆ftsA mutants exhibit a defect in HookA–early endosome interaction. (A) HookA-GFP signals in the ∆p25, ∆fhipA, and ∆ftsA mutants (top), along with the mCherry-RabA signals in the same cells (bottom). In contrast to the hyphal-tip accumulation of HookA-GFP signals in the ∆p25 mutant, representing colocalization with early endosomes, HookA-GFP signals in the ∆fhipA and ∆ftsA mutants do not colocalize with the hyphal tip–accumulated early endosomes. Bar, 5 μm. (B) Because the GFP signals are largely diffuse in the cytoplasm of the ∆fhipA and ∆ftsA mutants, a Western blot is shown to demonstrate that the HookA proteins are expressed and stable in both mutants. The negative control protein sample was from a strain without HookA-GFP.
Both FhipA and FtsA associate with early endosomes
Because the HookA protein is associated with early endosomes and FhipA and FtsA proteins are required for this association, we expected that the FhipA and FtsA proteins are also associated with early endosomes. To examine this in A. nidulans, we constructed FhipA-GFP and FtsA-GFP fusions with GFP fused to the C-termini of these proteins. These fusions were used to replace the endogenous FhipA and FtsA genes and expressed under the control of their endogenous promoters. Both fusions are functional, as evidenced by the normal early endosome distribution in strains containing mCherry-RabA (Figure 4, A and B). FhipA-GFP and FtsA-GFP signals were motile and appeared similar to the HookA-GFP signals that colocalized with early endosomes (Figure 4, A and B; Zhang et al., 2014). To provide further evidence for this notion, we introduced the GFP fusions into the ∆p25 mutant, in which early endosomes fail to move away from the hyphal tip (Zhang et al., 2011). In the ∆p25 mutant background, a dramatic accumulation of FtsA-GFP and FhipA-GFP signals is seen at the hyphal tips where mCherry-RabA–labeled early endosomes also accumulate, and the GFP and mCherry signals largely overlap (Figure 4, A and B). These data suggest that, just like HookA, FhipA and FtsA are associated with early endosomes. This notion is consistent with recent results showing that U. maydis FHIP and FTS proteins colocalize with motile early endosomes (see supplemental data in Bielska et al., 2014).
FIGURE 4:
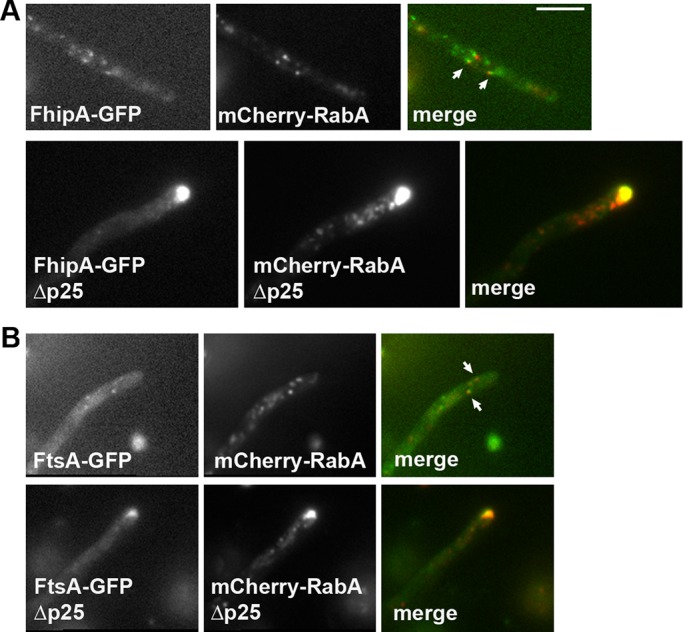
FhipA-GFP and FtsA-GFP colocalize with early endosomes. (A) FhipA-GFP colocalizes with early endosomes. Top, images of FhipA-GFP and mCherry-RabA in the same wild-type cell. Arrows indicate the FhipA-GFP signals that appear to colocalize with the mCherry-RabA signals. Bottom, images of FhipA-GFP and mCherry-RabA in the same ∆p25 mutant cell. Note that in the ∆p25 mutant, FhipA-GFP signals are concentrated at the hyphal tip, where early endosomes accumulate. (B) FtsA-GFP colocalizes with early endosomes. Top, images of FtsA-GFP and mCherry-RabA in the same wild-type cell. Arrows indicate the FtsA-GFP signals that appear to colocalize with the mCherry-RabA signals. Bottom, images of FtsA-GFP and mCherry-RabA in the same ∆p25 mutant cell. Note that in the ∆p25 mutant, FtsA-GFP signals are concentrated at the hyphal tip, where early endosomes accumulate. Bar, 5 μm.
Both HookA and FhipA are required for FtsA–early endosome interaction
FtsA is homologous to the FTS protein, which is a variant E2 ubiquitin-conjugating enzyme domain–containing protein (Lesche et al., 1997; Xu et al., 2008). Because FtsA is required for HookA–early endosome interaction (Figure 3A), one possibility is that FtsA may be able to bind to early endosomes independently of HookA but link HookA to early endosomes. However, it is equally possible that the formation of the FtsA-HookA-FhipA complex is important for FtsA–early endosome interaction. To determine whether FtsA–early endosome interaction depends on HookA or FhipA, we introduced FtsA-GFP into the strain background containing the ∆fhipA or ∆hookA allele. Our results showed that instead of associating with the hyphal tip–localized mCherry-RabA signals, FtsA-GFP signals are largely diffuse in these mutants (Figure 5, A and B). This is in sharp contrast to the colocalization of FtsA-GFP and mCherry-RabA signals at the hyphal tip in the ∆p25 mutant (Figure 4B). To determine whether the protein level of FtsA is affected by loss of FhipA or HookA, we performed Western analysis on the protein levels of FtsA-GFP in the mutants. Because it has been shown in mammalian cells that RNA interference of Hook proteins caused a significant decrease in the protein level of FtsA (Xu et al., 2008), we expected that FtsA protein level would be lowered in the ∆hookA mutant. However, our Western analyses showed that the protein level of FtsA is not significantly affected by the loss of either HookA or FhipA (Figure 5C). Thus FhipA and HookA are both required for the early endosome association rather than the expression or stability of FtsA.
FIGURE 5:
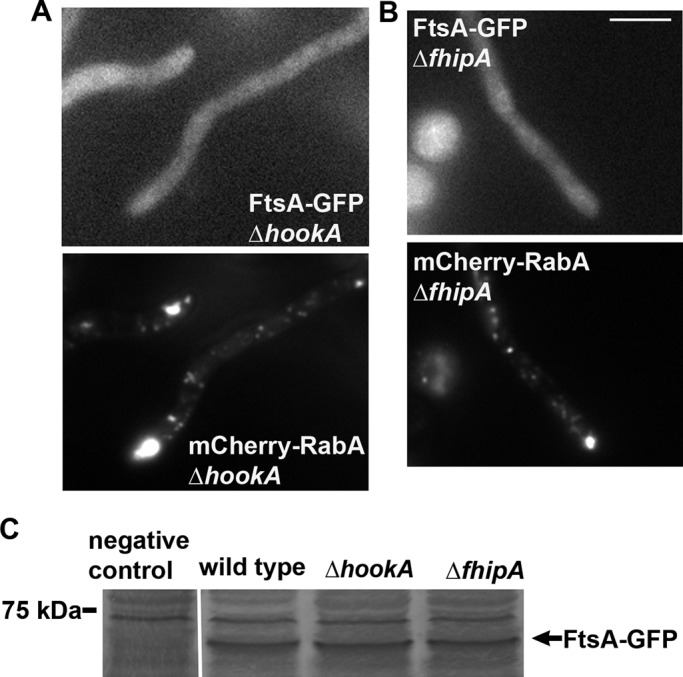
FtsA-GFP does not colocalize with early endosomes in the ∆hookA and ∆fhipA mutants. (A) FtsA-GFP does not colocalize with mCherry-RabA–labeled early endosomes in the ∆hookA mutant. (B) FtsA-GFP does not colocalize with mCherry-RabA–labeled early endosomes in the ∆fhipA mutant. Bar, 5 μm. (C) Because the FtsA-GFP signals are largely diffuse in the cytoplasm, a Western blot is shown to demonstrate that the FtsA proteins are expressed and stable in the ∆hookA and ∆fhipA mutants. The negative control protein sample was from a strain without FtsA-GFP. Bands above the indicated FtsA-GFP band represent proteins that cross-reacted with the anti-GFP antibody.
FhipA-GFP colocalizes with hyphal tip–accumulated early endosomes in the absence of HookA or FtsA
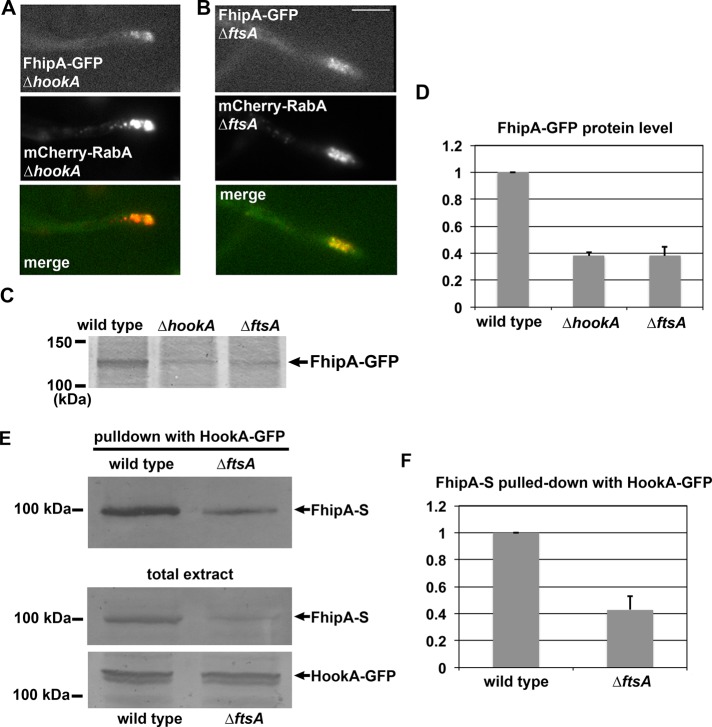
To determine whether FhipA–early endosome interaction depends on FtsA and HookA, we introduced FhipA-GFP into the strain background containing the ∆ftsA or ∆hookA allele. Of interest, we found that Fhip-GFP signals are still concentrated at the hyphal tip in these mutants, suggesting colocalization with the accumulated early endosomes (Figure 6, A and B). However, the intensity of the signals appeared lower than that in the ∆p25 mutant, where FhipA-GFP signals colocalize with hyphal tip–accumulated early endosomes. To examine the protein levels of FhipA-GFP in the ∆hookA and ∆ftsA mutants, we performed Western analyses. Interestingly, in both mutants, the protein levels of Fhip-GFP are significantly decreased (Figure 6, C and D; p < 0.001).
FIGURE 6:
FhipA localization/stability in the ∆hookA and ∆ftsA mutants and FhipA–HookA interaction in the ∆ftsA mutant. (A) FhipA-GFP colocalizes with mCherry-RabA–labeled early endosomes in the ∆hookA mutant. (B) FhipA-GFP colocalizes with mCherry-RabA–labeled early endosomes in the ∆ftsA mutant. Bar, 5 μm. (C) A Western blot showing the FhipA-GFP protein level in the ∆hookA and ∆ftsA mutants. (D) Quantification of the Western results showing that the FhipA-GFP protein level is decreased in the ∆hookA and ∆ftsA mutants (n = 3, p < 0.001 for both mutants). This analysis was done by measuring protein signal intensity on the western blots in relation to protein loading as indicated by Ponceau S staining. The ratio of the FhipA-GFP band intensity to the Ponceau S–stained loading control was calculated. Values presented are relative to the wild-type value, which is set at 1. The mean ± SD values for the Δ_hookA_ and ∆ftsA mutants are shown. (E) A Western blot showing that HookA-GFP is able to pull down FhipA-S from wild-type extract, as well as from ∆ftsA mutant extract. The amount of FhipA-S pulled down is significantly lower from the ∆ftsA extract, which is most likely due to a decrease in the protein level of FhipA-S in the ∆ftsA total extract. A Western blot of HookA-GFP in the ∆ftsA and wild-type extracts is presented as loading control. (F) Quantification of the Western results, showing that the amount of FhipA-S pulled down with HookA-GFP is significantly decreased in the ∆ftsA mutant (n = 3, p < 0.005). Values presented are relative to the wild type value, which is set at 1. The mean ± SD values for the ∆ftsA mutant are shown.
FhipA is able to interact with HookA in the absence of FtsA
The interactions between the mammalian FTS protein and Hook proteins appear to be direct, and the interactions involve a conserved helix near the C-terminus of Hook and the central β-sheet region of FTS (Xu et al., 2008). However, whether FHIP interacts with Hook directly or via FTS is a question that had not been addressed experimentally. To address this question, we made a strain containing S-tagged FhipA (FhipA-S), and introduced this fusion into a strain containing HookA-GFP and ∆ftsA by genetic crossing. Of interest, HookA-GFP was still able to pull down FhipA-S in the absence of FtsA, although the amount of pulled-down FhipA-S protein was lower from the ∆ftsA mutant extract than that from the wild-type extract (Figure 6, E and F, p < 0.005). The decrease in the level of pulled-down FhipA-S was expected, as the FhipA protein level was reduced in the ∆ftsA mutant (Figure 6, C and D).
DISCUSSION
In this study, by using a classical genetic screen combined with whole-genome sequencing, we found that FhipA, the A. nidulans homologue of the Hook-FTS-interacting protein FHIP, is required for dynein-mediated early endosome transport. We further showed that FtsA, the A. nidulans homologue of FTS, is also important for dynein-mediated early endosome transport. Because the same approach led to the identification of HookA as a dynein–early endosome linker (Zhang et al., 2014), the present results support the idea that HookA, FtsA, and FhipA function as a complex to facilitate dynein-mediated early endosome transport. Moreover, both FhipA and FtsA are important for targeting HookA to early endosomes, and the FtsA–early endosome interaction also requires both HookA and FhipA. Interestingly, FhipA is found to colocalize with early endosomes in the absence of HookA or FtsA, indicating that FhipA is able to associate with early endosomes independently of HookA or FtsA (Figure 7). These results highlight the important roles played by FhipA and FtsA in linking HookA to early endosomes for dynein-mediated transport in vivo.
FIGURE 7:
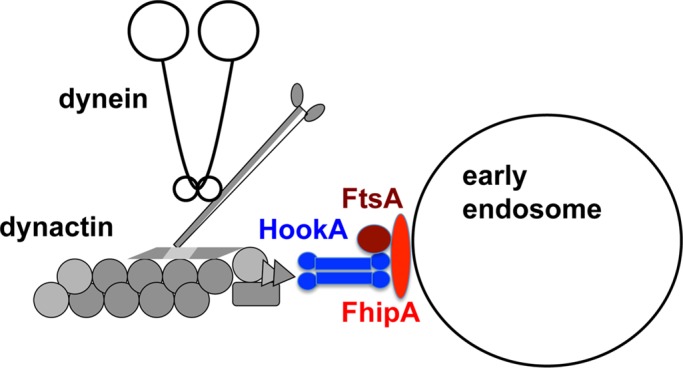
Model showing that the FtsA/HookA/FhipA complex is required for linking HookA to early endosomes. HookA (blue, depicted as a dimer) interacts with dynein–dynactin complexes, as revealed by a previous study (Zhang et al., 2014). The C-terminus of HookA most likely interacts with FtsA (brown) directly, based on the yeast two-hybrid data on human FTS interaction with the C-termini of Hook1 and Hook3 (Xu et al., 2008). FhipA (red) is able to interact with early endosome in the absence of either HookA or FtsA (brown). However, FhipA protein integrity/stability depends on both HookA and FtsA. FhipA-HookA interaction is most likely direct, based on the pull-down data showing this interaction in the absence of FtsA (Figure 6E). The FhipA–HookA interaction may involve the C-terminus of HookA, which is the domain mediating HookA-early endosome interaction (Zhang et al., 2014). In addition, based on data from U. maydis, a region of Hook upstream of the C-terminal early endosome–binding domain may enhance Hook-FTS-FHIP interactions (Bielska et al., 2014).
Our result that HookA–early endosome interaction requires both FhipA and FtsA is surprising, given that the mammalian Hook1 protein interacts directly with endocytic cargo proteins in a yeast two-hybrid assay (Maldonado-Báez et al., 2013). It suggests, however, that the in vivo interaction between HookA and early endosomes may require multiple contact sites involving other proteins. HookA and FhipA are both necessary for the interaction between FtsA and early endosomes. FtsA contains a ubiquitin-conjugating domain (aa 15 to 260) that is catalytically inactive, and thus it belongs to a family of ubiquitin E2 variants (UEVs) just like its human homologue, FTS (Lesche et al. 1997; Xu et al., 2008). However, it is unclear whether FtsA directly contacts early endosomes via binding to cargo proteins containing ubiquitin, a modification implicated in cargo sorting into multivesicular bodies (Tanno and Komada, 2013). A well-known member of the UEV family is Tumor Susceptibility Gene 101 (TSG101), which is the human homologue of the yeast Vacuolar protein sorting 23 (Vps23) protein. TGS101/Vps23 is a component of the ESCRT I complex, which recognizes ubiquitinated cargo proteins during early endosomal sorting of endocytic cargoes (Katzmann et al., 2001; Sundquist et al., 2004). Of interest, whereas a β-sheet region in TSG101 is implicated in binding to ubiquitin, an analogous region in FTS is implicated in binding to the C-terminal helix 1 of Hook proteins (Sundquist et al., 2004; Xu et al., 2008). Thus whether FTS binds to ubiquitin directly is not known. It is possible that unlike TSG101/Vps23, which directly interacts with ubiquitin at the cytoplasmic side of the endocytic cargo proteins, FtsA does not interact with ubiquitin but instead uses the interaction with HookA to fulfill its role in dynein-mediated early endosome movement.
Most interestingly, FhipA appears to be able to interact with early endosomes independently of the FtsA and HookA proteins. However, the mechanism of this interaction is not fully understood. Both FHIP and FhipA contain a retinoid acid–induced protein 16 (RAI-16)–like domain at ∼80–100 aa away from the N-termini. The function of this domain is unclear. FhipA does not contain any transmembrane domain, and thus the interaction between FhipA and early endosome may not be direct. It is likely that additional players are required for FhipA–early endosome interaction. It has been found that the human FHF complex interacts with the components of the HOPS complex, such as Vps18 (Xu et al., 2008). Because Vps18 is also a component of the Rab5 effector class C core vacuole/endosome tethering (CORVET) complex (Balderhaar and Ungermann, 2013), it is possible that FhipA interacts with early endosomes via Vps18. In A. nidulans, Vps18 is required for endosome maturation, which is essential for hyphal growth (Abenza et al., 2012). As a result, the deletion mutant of Vps18 is too sick for us to test whether Vps18 is directly involved in FhipA–early endosome interaction (Abenza et al., 2012). Future work is needed to reveal specifically how FhipA interacts with early endosomes.
It is important to point out that although FhipA appears to be associated with early endosomes in the absence of HookA and FtsA (Figure 6, A and B), the protein level of FhipA is significantly decreased in the absence of HookA and FtsA (Figure 6, C and D). Intriguingly, whereas the physical interaction between HookA and FhipA appears to be independent of FtsA based on the results of the biochemical pull-down assays (Figure 6E), HookA remains largely in the cytosol rather than associates with early endosomes in the absence of FtsA (Figure 3A). One possible explanation for these results is that the amount of FhipA molecules on the early endosomes may be too low to recruit sufficient HookA molecules to early endosomes. It is also possible that formation of the FHF complex may be important for the stable association of FhipA/HookA with early endosomes. Overall our results indicate that the formation of the FhipA/HookA/FtsA complex is important for dynein-mediated early endosome transport. This function of the complex is most likely conserved in filamentous fungi, as U. maydis genes encoding FTS and FHIP have also been identified in a genetic screen for mutants defective in early endosome transport (Gero Steinberg, personal communication). Whether the FHF complex is involved in dynein-mediated transport of cellular cargos in higher eukaryotes will need to be further investigated.
MATERIALS AND METHODS
A. nidulans strains, media, and mutagenesis
A. nidulans strains used in this study are listed in Table 1. For biochemical experiments, yeast extract plus glucose (YG) + UU (or YUU) liquid medium was used. UV mutagenesis on spores of A. nidulans strains was done as previously described (Willins et al., 1995; Xiang et al., 1999). Colonies were grown on YG plus agar (YAG) plates for 2 d at 37°C. For 4′,6-diamidino-2-phenylindole (DAPI) staining of nuclei, cells were incubated in YUU liquid medium for 8 h at 37°C or in liquid minimal medium containing 1% glycerol plus supplements overnight at 32°C. For live-cell imaging experiments, liquid minimal medium containing 1% glycerol plus supplements was used, and cells were cultured at 32°C overnight and observed at room temperature. Colonies were grown on YAG) plates for 2 d at 37°C.
TABLE 1:
A. nidulans strains used in this study (markers not confirmed are indicated by question marks).
| Strain | Genotype | Source |
|---|---|---|
| RQ2 | GFP-nudAHC; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB; pyrG89; pyroA4; yA2 | Qiu et al. (2013) |
| RQ54 | argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB; pyrG89; pyroA4; wA2 | Qiu et al. (2013) |
| JZ498 | ∆hookA::Af_pyrG; argB_2_::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB; pyrG89; pyroA4; wA2 | Zhang et al. (2014) |
| JZ500 | HookA_-GFP-Af_pyrG; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB; pyrG89; pyroA4; wA2 | Zhang et al. (2014) |
| JZ556 | HookA_-GFP-Af_pyrG; ∆p25::Af_pyrG_; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB? pyrG89? | Zhang et al. (2014) |
| XX223 | ∆p25::Af_pyrG_; GFP-nudAHC; argB_2::[argB*-alcAp_::mCherry_-RabA]_ | Zhang et al. (2011) |
| XX299 | hookA_L150P, E151K; GFP-nudAHC; argB_2::[argB*-alcAp::mCherry_-RabA]; pantoB100; yA2_ | Zhang et al. (2014) |
| XY110 | eedB_3(FhipAQ367STOP); GFP-nudAHC; argB_2::[argB*-alcAp::mCherry_-RabA];_ ∆nkuA::argB; pyrG89; pyroA4; yA2 | This work |
| XY112 | eedB_3(FhipAQ367STOP)+FhipA; GFP-nudAHC; argB_2::[argB*-alcAp::mCherry_-RabA];_ ∆nkuA::argB; pyrG89; pyroA4; yA2 | This work |
| XY113 | FhipAQ367STOP; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB; pyrG89; pyroA4; wA2 | This work |
| XY114 | ∆fhipA::Af_pyrG;_ GFP-nudAHC; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB; pyrG89; pyroA4; yA2 | This work |
| XY115 | ∆ftsA::Af_pyrG;_ GFP-nudAHC; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB; pyrG89; pyroA4; yA2 | This work |
| XY118 | FhipA-GFP-Af_pyrG; argB_2_::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB; pyrG89; pyroA4; wA2 | This work |
| XY119 | FtsA-GFP-Af_pyrG; argB_2_::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB; pyrG89; pyroA4; wA2 | This work |
| XY123 | HookA-GFP-Af_pyrG_; ∆fhipA::Af_pyrG;_ GFP-nudAHC; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB?; pyrG89?; WA2 | This work |
| XY124 | HookA-GFP-Af_pyrG_; ∆ftsA::Af_pyrG;_ GFP-nudAHC; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB?; pyrG89?; pabaA1 | This work |
| XY125 | FhipA-GFP-Af_pyrG;_ ∆p25::Af_pyrG_; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB? pyrG89? wA2 | This work |
| XY126 | FtsA-GFP-Af_pyrG;_ ∆p25::Af_pyrG_; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB? pyrG89? pantoB100; pyroA4; wA2 | This work |
| XY127 | FhipA-GFP-Af_pyrG;_ ∆hookA::Af_pyrG_; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB? pyrG89? wA2 | This work |
| XY129 | FhipA-GFP-Af_pyrG;_ ∆ftsA::Af_pyrG_; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB? pyrG89? wA2 | This work |
| XY130 | FtsA-GFP-Af_pyrG;_ ∆hookA::Af_pyrG_; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB? pyrG89? pyroA4 | This work |
| XY131 | FtsA-GFP-Af_pyrG;_ ∆fhipA::Af_pyrG_; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB? pyrG89? paba A1; pyroA4 | This work |
| XY132 | FhipA-S-AfpyrG; argB2::[argB*-alcAp::mCherry-RabA]; ∆nkuA::argB; pyrG89; pyroA4; wA2 | This work |
| XY134 | FhipA-S-Af_pyrG;_ HookA-GFP-Af_pyrG; argB_2_::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB? pyrG89? | This work |
| XY135 | FhipA-S-Af_pyrG;_ HookA-GFP-Af_pyrG;_ ∆ftsA::Af_pyrG_; argB_2::[argB*-alcAp_::mCherry_-RabA];_ ∆nkuA::argB? pyrG89? wA2 | This work |
Live-cell imaging and analyses
Fluorescence microscopy of live A. nidulans hyphae was as described (Zhang et al., 2011). All images were captured at room temperature using an Olympus (Center Valley, PA) IX70 inverted fluorescence microscope linked to a PCO/Cooke Corporation (Romulus, MI) Sensicam QE cooled charge-coupled device camera. An UplanApo 100× objective lens (oil) with a 1.35 numerical aperture was used. A filter wheel system with GFP/mCherry-ET Sputtered series with high transmission (Biovision Technologies, Exton, PA) was used. The IPLab software was used for image acquisition and analysis.
Complementation of the eedB3 mutant and recreation of the _fhipA_Q367STOP allele
Complementation of the _eedB_3 (_fhipA_Q367STOP) mutant was done by using the wild-type genomic DNA fragment amplified using the following two oligos: eed3U5 (5′-ACTTTGATATCTCTTCAAGCCATCA-3′), and eed3D3 (5′-GGAGGCAGCCAAGATTGAT-3′). The DNA fragment was used to transform the XY110 strain together with a genomic DNA fragment covering the pyrG gene. To recreate the _fhipA_Q367STOP mutation in a wild-type strain, the same set of oligos was used to amplify the mutant fhipA gene from the _eedB_3 mutant, and the DNA fragment was cotransformed with a linear construct containing pyrG into the RQ54 strain containing ∆nkuA (Nayak et al., 2006) and _pyrG_89.
DNA constructs for generating the ∆fhipA and ∆ftsA mutants
For constructing the ∆fhipA mutant, the following eight oligos were used to make the ∆fhipA construct with the selective marker pyrG from Aspergillus fumigatus, AfpyrG, in the middle of the linear construct (Szewczyk et al., 2006): FPU5 (5′-GCTCGCACATCGCACATCCTA-3′), FPU3 (5′-GCATCCAACGTACAAAATCCAT-3′), FPFus5 (5′-ATGGATTTTGTACGTTGGATGCTGCTCTTCACCCTCTTCGCG-3′), FPFus3 (5′-TCAGGAATCGTCAACTGGCAGCTGTCTGAGAGGAGGCACTG-3′), FPD5 (5′-CTGCCAGTTGACGATTCCTGA-3′), FPD3 (5′-TCACAGGACTATAAGCAGCTCA-3′); FPU5i (5′-GCCAGAGCTTTGCAGCAATACA-3′), and FPD3ii (5′-TAAGCAGCTCAATTGGCAGTC-3′). The deletion construct was obtained via fusion PCRs and used to transform the RQ2 strain containing ∆nkuA and _pyrG_89.
For constructing the ∆ftsA mutant, the following eight oligos were used to make the ∆ftsA construct with the selective marker pyrG from A. fumigatus, AfpyrG, in the middle of the linear construct (Szewczyk et al., 2006): FTSU5 (5′-ATCAGCAATCAGCACTACCATA-3′), FTSU3 (5′-CGTGGACTGAGCATTAAACAA-3′), FTSFus5 (5′-TTGTTTAATGCTCAGTCCACGTGCTCTTCACCCTCTTCGCG-3′), FTSFus3 (5′-AGGAGCAAGTGGAATATCGCGAGCCTGTCTGAGAGGAGGCACTG-3′), FTSD5 (5′-GCTCGCGATATTCCACTTGCTC-3′), FTSD3 (5′-ATCATCAGGAATGGTACAAGCA-3′), FTSU5i (5′-ACAATCTCGACGACTCTGGA-3′), and FTSD3i (5′-ATACGATGTGTTCTTCCAATGTA-3′). The deletion construct was obtained via fusion PCRs and used to transform the RQ2 strain.
DNA constructs for generating strains containing FhipA-GFP, FhipA–S-tag, and FtsA-GFP
For constructing the FhipA-GFP fusion, we used the following eight oligos to amplify genomic DNA and the GFP-AfpyrG fusion from the plasmid pFNO3 (deposited in the Fungal Genetics Stock Center [FGSC] by Steve Osmani; Yang et al., 2004; McCluskey et al., 2010): HPOrfF (5′-GAAGTAGGGCACCTACTGACA-3′), HPOrfR (5′-GGAATCGTCAACTGGCAGAGA-3′), HPFusF (5′-TCTCTGCCAGTTGACGATTCCGGAGCTGGTGCAGGCGCTGGAG-3′), HPFusR (5′-GCCGCTTAGGCATAGTTTCACTGTCTGAGAGGAGGCACTGATG-3′), HPUtrF (5′-TGAAACTATGCCTAAGCGGC-3′), HPUtrR (5′-GCCGTAGATGATTCACCTCAT-3′), HPOrfFii (5′-ACGTTAGACCAGTCATTCGATA-3′), and HPUtrRii (5’-TGAAGCTGACTGGATTCTCAACA-3’). The fusion PCRs generated the FhipA-GFP-Af_pyrG_ fragment that we used to transform into RQ54. For making the strain containing S-tagged FhipA, the same strategy and the same oligos were used, except that the pAO81 plasmid (instead of pFNO3) containing the coding sequence for the S-tag was used.
For constructing the FtsA-GFP fusion, we used the following six oligos to amplify genomic DNA and the GFP-AfpyrG fusion from the plasmid pFNO3 (deposited in the FGSC by Steve Osmani; Yang et al., 2004; McCluskey et al., 2010): FTSOrfF (5′-CGAGTATTGTGCCAAGCTCCA-3′), FTSOrfR (5′-TTCTGACGCCTGCTCCGGAGGA-3′), FTSFusF (5′-ATTCCTCCGGAGCAGGCGTCAGAAGGAGCTGGTGCAGGCGCTGGAG-3′), HKFusR (5′-GAGCAAGTGGAATATCGCGAGCTTACTGTCTGAGAGGAGGCACTGATG-3′), FTSUtrF (5′-TAAGCTCGCGATATTCCACTTGC-3′), FTSUtrR (5′-GAACTGATCAACTTTGCAGGTTA-3′), FTSOrfFi (5′-ACCAACTGATCCAGGAGCAGTA-3′), and FTSUtrRi (5′-TTGGATGCTGGCTCATCTACATA-3′). The fusion PCRs generated the FtsA-GFP-Af_pyrG_ fragment that we used to transform into RQ54.
Analyses of protein–protein interactions and Western analysis
The μMACS GFP-tagged protein isolation kit (Miltenyi Biotec, San Diego, CA) was used to determine whether GFP-tagged HookA pulls down S-tagged FhipA. This was done as described in Qiu et al. (2013). Strains were grown overnight in liquid-rich medium YG. About 0.4 g of hyphal mass was harvested from overnight culture for each sample, and cell extracts were prepared using a lysis buffer containing 50 mM Tris-HCl, pH 8.0, and 10 μg/ml protease inhibitor cocktail (Sigma-Aldrich). Cell extract was centrifuged at 8000 × g for 20 min and then 16,000 × g for 10 min at 4°C, and supernatant was used for the pull-down experiment. To pull down GFP-tagged protein, 25 μl of anti-GFP MicroBeads was added to the cell extracts for each sample and incubated at 4°C for 30 min. The MicroBeads/cell extract mixture was then applied to the μColumn, followed by gentle wash with the lysis buffer and the wash buffer 2 provided in the kit (Miltenyi Biotec). Preheated (95°C) SDS–PAGE sample buffer was used as elution buffer. Antibodies against GFP and S-tag used for Western analyses were from Clontech (Mountain View, CA) and Cell Signaling Technology (Danvers, MA), respectively. Western analyses were performed using the alkaline phosphatase (AP) system, and blots were developed using the AP color development reagents from Bio-Rad (Hercules, CA). Quantitation of the protein band intensity was done using the IPLab software as described previously (Yao et al., 2012; Qiu et al., 2013).
Supplementary Material
Supplemental Materials
Acknowledgments
We thank Jun Zhang and Rongde Qiu for sharing strains and reagents. We thank Berl Oakley for the ∆nkuA strain, Miguel Peñalva for the mCherry-RabA strain, and the Fungal Genetic Stock Center for the pFNO3 and pAO81 plasmids and Steve Osmani for depositing them. We also thank Gero Steinberg for communicating unpublished results. Services for primer synthesis and DNA sequencing were provided by the Biomedical Instrumentation Center of the Uniformed Services University. This work was funded by National Institutes of Health Grant RO1 GM097580 (to X.X.) and a Uniformed Services University Intramural Grant (to X.X.).
Abbreviations used:
FHIP
FTS- and Hook-interacting protein
FTS
Fused Toes
Footnotes
REFERENCES
- Abenza JF, Galindo A, Pinar M, Pantazopoulou A, de los Rios V, Peñalva MA. Endosomal maturation by Rab conversion in Aspergillus nidulans is coupled to dynein-mediated basipetal movement. Mol Biol Cell. 2012;23:1889–1901. doi: 10.1091/mbc.E11-11-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abenza JF, Pantazopoulou A, Rodriguez JM, Galindo A, Peñalva MA. Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic. 2009;10:57–75. doi: 10.1111/j.1600-0854.2008.00848.x. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Hammer JA., 3rd Linking molecular motors to membrane cargo. Curr Opin Cell Biol. 2010;22:479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes—coordinators of endosome and lysosome fusion. J Cell Sci. 2013;126:1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- Bielska E, Schuster M, Roger Y, Berepiki A, Soanes DM, Talbot NJ, Steinberg G. Hook is an adapter that coordinates kinesin-3 and dynein cargo attachment on early endosomes. J Cell Biol. 2014;204:989–1007. doi: 10.1083/jcb.201309022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston JP, Holzbaur EL. Microtubule motors at the intersection of trafficking and transport. Trends Cell Biol. 2006;16:530–537. doi: 10.1016/j.tcb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Egan MJ, McClintock MA, Reck-Peterson SL. Microtubule-based transport in filamentous fungi. Curr Opin Microbiol. 2012a;15:637–645. doi: 10.1016/j.mib.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MJ, Tan K, Reck-Peterson SL. Lis1 is an initiation factor for dynein-driven organelle transport. J Cell Biol. 2012b;197:971–982. doi: 10.1083/jcb.201112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franker MA, Hoogenraad CC. Microtubule-based transport—basic mechanisms, traffic rules and role in neurological pathogenesis. J Cell Sci. 2013;126:2319–2329. doi: 10.1242/jcs.115030. [DOI] [PubMed] [Google Scholar]
- Granger E, McNee G, Allan V, Woodman P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol. 2014;31:20–29. doi: 10.1016/j.semcdb.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grava S, Keller M, Voegeli S, Seger S, Lang C, Philippsen P. Clustering of nuclei in multinucleated hyphae is prevented by dynein-driven bidirectional nuclear movements and microtubule growth control in Ashbya gossypii. Eukaryot Cell. 2011;10:902–915. doi: 10.1128/EC.05095-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G, Liu B, Zhang J, Zuo W, Morris NR, Xiang X. The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr Biol. 2001;11:719–724. doi: 10.1016/s0960-9822(01)00200-7. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Kramer H, Phistry M. Mutations in the Drosophila hook gene inhibit endocytosis of the boss transmembrane ligand into multivesicular bodies. J Cell Biol. 1996;133:1205–1215. doi: 10.1083/jcb.133.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IH, Kumar S, Plamann M. Null mutants of the Neurospora actin-related protein 1 pointed-end complex show distinct phenotypes. Mol Biol Cell. 2001;12:2195–2206. doi: 10.1091/mbc.12.7.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz JH, Schuchardt I, Straube A, Steinberg G. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 2006;25:2275–2286. doi: 10.1038/sj.emboj.7601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesche R, Peetz A, van der Hoeven F, Ruther U. Ft1, a novel gene related to ubiquitin-conjugating enzymes, is deleted in the Fused Toes mouse mutation. Mamm Genome. 1997;8:879–883. doi: 10.1007/s003359900604. [DOI] [PubMed] [Google Scholar]
- Maldonado-Baez L, Cole NB, Kramer H, Donaldson JG. Microtubule-dependent endosomal sorting of clathrin-independent cargo by Hook1. J Cell Biol. 2013;201:233–247. doi: 10.1083/jcb.201208172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey K, Wiest A, Plamann M. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J Biosci. 2010;35:119–126. doi: 10.1007/s12038-010-0014-6. [DOI] [PubMed] [Google Scholar]
- Milne I, Stephen G, Bayer M, Cock PJ, Pritchard L, Cardle L, Shaw PD, Marshall D. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013;14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori-McKenney KM, McKenney RJ, Vallee RB. Studies of lissencephaly and neurodegenerative disease reveal novel aspects of cytoplasmic dynein regulation. In: King SM, editor. Dyneins: Structure, Biology and Disease. Maryland Heights, MO: Elsevier; 2011. pp. 441–453. [Google Scholar]
- Peñalva MA, Galindo A, Abenza JF, Pinar M, Calcagno-Pizarelli AM, Arst HN, Pantazopoulou A. Searching for gold beyond mitosis: mining intracellular membrane traffic in Aspergillus nidulans. Cell Logist. 2012;2:2–14. doi: 10.4161/cl.19304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlson E, Maday S, Fu MM, Moughamian AJ, Holzbaur EL. Retrograde axonal transport: pathways to cell death. Trends Neurosci. 2010;33:335–344. doi: 10.1016/j.tins.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann M, Minke PF, Tinsley JH, Bruno KS. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu R, Zhang J, Xiang X. Identification of a novel site in the tail of dynein heavy chain important for dynein function in vivo. J Biol Chem. 2013;288:2271–2280. doi: 10.1074/jbc.M112.412403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavo G, Greensmith L, Hafezparast M, Fisher EM. Cytoplasmic dynein heavy chain: the servant of many masters. Trends Neurosci. 2013;36:641–651. doi: 10.1016/j.tins.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- Seidel C, Moreno-Velasquez SD, Riquelme M, Fischer R. Neurospora crassa NKIN2, a kinesin-3 motor, transports early endosomes and is required for polarized growth. Eukaryot Cell. 2013;12:1020–1032. doi: 10.1128/EC.00081-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D, et al. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol Biol Cell. 2012;23:4226–4241. doi: 10.1091/mbc.E12-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G. Motors in fungal morphogenesis: cooperation versus competition. Curr Opin Microbiol. 2011;14:660–667. doi: 10.1016/j.mib.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Steinberg G. Endocytosis and early endosome motility in filamentous fungi. Curr Opin Microbiol. 2014;20:10–18. doi: 10.1016/j.mib.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DJ. Functional coupling of microtubules to membranes—implications for membrane structure and dynamics. J Cell Sci. 2012;125:2795–2804. doi: 10.1242/jcs.097675. [DOI] [PubMed] [Google Scholar]
- Sundquist WI, Schubert HL, Kelly BN, Hill GC, Holton JM, Hill CP. Ubiquitin recognition by the human TSG101 protein. Mol Cell. 2004;13:783–789. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- Sunio A, Metcalf AB, Kramer H. Genetic dissection of endocytic trafficking in Drosophila using a horseradish peroxidase-bride of sevenless chimera: hook is required for normal maturation of multivesicular endosomes. Mol Biol Cell. 1999;10:847–859. doi: 10.1091/mbc.10.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebenyi G, Hall B, Yu R, Hashim AI, Kramer H. Hook2 localizes to the centrosome, binds directly to centriolin/CEP110 and contributes to centrosomal function. Traffic. 2007a;8:32–46. doi: 10.1111/j.1600-0854.2006.00511.x. [DOI] [PubMed] [Google Scholar]
- Szebenyi G, Wigley WC, Hall B, Didier A, Yu M, Thomas P, Kramer H. Hook2 contributes to aggresome formation. BMC Cell Biol. 2007b;8:19. doi: 10.1186/1471-2121-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Tan K, Roberts AJ, Chonofsky M, Egan MJ, Reck-Peterson SL. A microscopy-based screen employing multiplex genome sequencing identifies cargo-specific requirements for dynein velocity. Mol Biol Cell. 2014;25:669–678. doi: 10.1091/mbc.E13-09-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SC, Scherer J, Vallee RB. Recruitment of dynein to late endosomes and lysosomes through light intermediate chains. Mol Biol Cell. 2011;22:467–477. doi: 10.1091/mbc.E10-02-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno H, Komada M. The ubiquitin code and its decoding machinery in the endocytic pathway. J Biochem. 2013;153:497–504. doi: 10.1093/jb/mvt028. [DOI] [PubMed] [Google Scholar]
- Walenta JH, Didier AJ, Liu X, Kramer H. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J Cell Biol. 2001;152:923–934. doi: 10.1083/jcb.152.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willins DA, Xiang X, Morris NR. An alpha tubulin mutation suppresses nuclear migration mutations in Aspergillus nidulans. Genetics. 1995;141:1287–1298. doi: 10.1093/genetics/141.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Xiang X, Hammer JA., 3rd Motor proteins at the microtubule plus-end. Trends Cell Biol. 2006;16:135–143. doi: 10.1016/j.tcb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Xiang X, Beckwith SM, Morris NR. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc Natl Acad Sci USA. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Fischer R. Nuclear migration and positioning in filamentous fungi. Fungal Genet Biol. 2004;41:411–419. doi: 10.1016/j.fgb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Xiang X, Zuo W, Efimov VP, Morris NR. Isolation of a new set of Aspergillus nidulans mutants defective in nuclear migration. Curr Genet. 1999;35:626–630. doi: 10.1007/s002940050461. [DOI] [PubMed] [Google Scholar]
- Xu L, Sowa ME, Chen J, Li X, Gygi SP, Harper JW. An FTS/Hook/p107(FHIP) complex interacts with and promotes endosomal clustering by the homotypic vacuolar protein sorting complex. Mol Biol Cell. 2008;19:5059–5071. doi: 10.1091/mbc.E08-05-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Puthenveedu MA, Linstedt AD. Golgin160 recruits the dynein motor to position the Golgi apparatus. Dev Cell. 2012;23:153–165. doi: 10.1016/j.devcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ukil L, Osmani A, Nahm F, Davies J, De Souza CP, Dou X, Perez-Balaguer A, Osmani SA. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot Cell. 2004;3:1359–1362. doi: 10.1128/EC.3.5.1359-1362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Zhang J, Zhou H, Wang E, Xiang X. In vivo roles of the basic domain of dynactin p150 in microtubule plus-end tracking and dynein function. Traffic. 2012;13:375–387. doi: 10.1111/j.1600-0854.2011.01312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TY, Quintyne NJ, Scipioni BR, Eckley DM, Schroer TA. Dynactin's pointed-end complex is a cargo-targeting module. Mol Biol Cell. 2012;23:3827–3837. doi: 10.1091/mbc.E12-07-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekert N, Fischer R. The Aspergillus nidulans kinesin-3 UncA motor moves vesicles along a subpopulation of microtubules. Mol Biol Cell. 2009;20:673–684. doi: 10.1091/mbc.E08-07-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Qiu R, Arst HN, Jr, Peñalva MA, Xiang X. HookA is a novel dynein-early endosome linker critical for cargo movement in vivo. J Cell Biol. 2014;204:1009–1026. doi: 10.1083/jcb.201308009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yao X, Fischer L, Abenza JF, Peñalva MA, Xiang X. The p25 subunit of the dynactin complex is required for dynein-early endosome interaction. J Cell Biol. 2011;193:1245–1255. doi: 10.1083/jcb.201011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Cai Q, Xie Y, Sheng ZH. Snapin recruits dynein to BDNF-TrkB signaling endosomes for retrograde axonal transport and is essential for dendrite growth of cortical neurons. Cell Rep. 2012;2:42–51. doi: 10.1016/j.celrep.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials