The emergence of the Middle East Respiratory Syndrome coronavirus (original) (raw)
Abstract
On September 20, 2012, a Saudi Arabian physician reported the isolation of a novel coronavirus from a patient with pneumonia on ProMED‐mail. Within a few days, the same virus was detected in a Qatari patient receiving intensive care in a London hospital, a situation reminiscent of the role air travel played in the spread of severe acute respiratory syndrome coronavirus (SARS‐CoV) in 2002. SARS‐CoV originated in China's Guangdong Province and affected more than 8000 patients in 26 countries before it was contained 6 months later. Over a year after the emergence of this novel coronavirus – Middle East respiratory syndrome coronavirus (MERS‐CoV) – it has caused 178 laboratory‐confirmed cases and 76 deaths. The emergence of a second highly pathogenic coronavirus within a decade highlights the importance of a coordinated global response incorporating reservoir surveillance, high‐containment capacity with fundamental and applied research programs, and dependable communication pathways to ensure outbreak containment. Here, we review the current state of knowledge on the epidemiology, ecology, molecular biology, clinical features, and intervention strategies of the novel coronavirus, MERS‐CoV.
Keywords: MERS‐CoV, coronavirus, epidemiology, molecular biology, intervention strategies
Excellent review on timely and newly emerging infectious diseases.
Introduction
Coronaviruses (family Coronaviridae, subfamily Coronavirinae) circulate in a diverse array of mammalian and avian reservoirs, including humans, bats, pigs, cats, dogs, rodents, and birds (Perlman & Netland, 2009). Coronaviruses (CoV) are classified into four genera (_Alpha_‐, _Beta_‐, _Gamma_‐, and Deltacoronavirus) and are enveloped, positive‐strand RNA viruses between 70 and 120 nm in size (Masters, 2006; de Groot, 2012). The spike glycoproteins that radiate from the virus envelope of the spherical particles are responsible for the characteristic crown‐like appearance of coronaviruses (Fig. 1).
Figure 1.
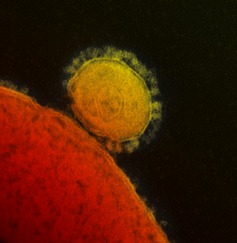
False‐color MERS‐CoV particle visualized by electron microscopy. A MERS‐CoV particle (yellow) attached to the surface of a cell (red). The characteristic MERS‐CoV spike glycoproteins are clearly visible on the surface of the MERS‐CoV particle.
Four coronaviruses continuously circulate in the human population, all of which cause generally mild respiratory disease: HCoV‐229E, HCoV‐NL63 (Alphacoronavirus), HCoV‐OC43, and HKU1 (Betacoronavirus; Hamre & Procknow, 1966; McIntosh et al., 1967; Fouchier et al., 2004; van der Hoek et al., 2004; Woo et al., 2005). In addition, there have been two zoonotic introductions of coronaviruses into the human population over the last decade, both associated with acute respiratory distress syndrome (ARDS) and high case fatality rates: severe acute respiratory syndrome CoV (SARS‐CoV; Drosten et al., 2003; Kuiken et al., 2003) and Middle East respiratory syndrome CoV (MERS‐CoV; Zaki et al., 2012). SARS‐CoV caused the first pandemic of the 21st century, resulting in c. 8400 human cases and an 11% case fatality rate (SARS Epidemiology Working Group, 2003). In addition to the impact of SARS‐CoV on infected individuals and the global public health community, the economic cost of the SARS‐CoV outbreak event was estimated at $16 billion (Brahmbhatt & Dutta, 2008). Although only 163 laboratory‐confirmed cases of MERS‐CoV are currently reported, the high case fatality rate and travel‐related spread across multiple countries are reminiscent of the SARS‐CoV pandemic.
Epidemiology of MERS‐CoV
Virus detection and case definition
The first human case of MERS‐CoV was identified using a pancoronavirus reverse‐transcriptase polymerase chain reaction (RT‐PCR) assay (Zaki et al., 2012). MERS‐CoV‐specific quantitative RT‐PCRs (qRT‐PCR), targeting the region upstream of the E protein gene and the open reading frame 1b, were rapidly developed and have become standards in the laboratory testing and diagnosis of MERS‐CoV (Corman et al.,2012a). Additional qRT‐PCRs targeting the RNA‐dependent RNA polymerase (RdRp) and nucleocapsid (N) genes have been developed as confirmatory assays (Corman et al., 2012b). The World Health Organization (WHO) case definition for MERS‐CoV focuses on patients suffering febrile acute respiratory disease who have a direct epidemiological link to another confirmed case or are residents of or travelers to MERS‐CoV‐source countries (WHO, 2013c). Confirmatory laboratory testing requires a positive qRT‐PCR of at least two specific genomic targets or a single positive target by qRT‐PCR combined with sequencing of a second target (Centers for Disease Control and Prevention, 2013). In instances of inadequate testing or negative tests, a patient with a direct epidemiologic link to a confirmed MERS‐CoV case is determined to be a probable case of MERS‐CoV infection if they present with acute febrile respiratory illness.
Spatial distribution and demographics
While primary cases of MERS‐CoV have been confined to six countries in the Middle East – Saudi Arabia, United Arab Emirates, Qatar, Jordan, Oman and Kuwait – travel‐related cases have been identified in Tunisia the United Kingdom, France, Germany, and Italy (Fig. 2; Bermingham et al., 2012; Buchholz et al., 2013;Gulland 2013a, b; Health Protection Agency, 2013; Hijawi et al., 2013; Mailles et al., 2013;Memish et al., 2013b; Puzelli et al., 2013). Limited secondary transmission occurred after MERS‐CoV introduction in Tunisia, France, and the United Kingdom, while imported cases of MERS‐CoV infection in Germany and Italy did not lead to subsequent confirmed infections (Buchholz et al., 2013; Gulland,2013a, b; Health Protection Agency, 2013; Puzelli et al., 2013). Over 80% of cases of MERS‐CoV have occurred in Saudi Arabia, largely within the Riyadh and Eastern provinces (Centers for Disease Control and Prevention, 2013).
Figure 2.
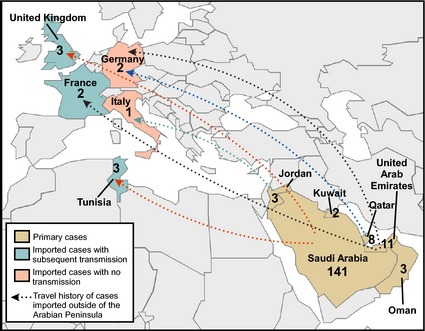
Geographic distribution of the MERS‐CoV outbreak. The geographic distribution of MERS‐CoV cases up to February 1, 2014 is shown. Travel history of cases imported outside of the Arabian Peninsula is indicated with dotted arrows. Countries with primary MERS‐CoV cases are shown in brown, countries with imported MERS‐CoV cases and no confirmed human‐to‐human transmission are shown in pink, and countries with imported MERS‐CoV cases and subsequent human‐to‐human transmission are shown in green.
As of January 20, 2014, there were 178 confirmed cases of MERS‐CoV, 76 (43%) of which were fatal (Fig. 3). Although most cases have been clinically severe, contact surveillance has uncovered at least 27 subclinical or mild infections (Centers for Disease Control and Prevention, 2013). The case fatality ratio of MERS‐CoV (43%) is much higher than that of SARS‐CoV (CFR 11%) (SARS Epidemiology Working Group, 2003). The average age of MERS‐CoV cases is 52 years, with a male‐to‐female ratio of 1.6–1 (The WHO MERS‐CoV Research Group, 2013). Both the case fatality ratio and the male‐to‐female ratio have decreased as the incidence of MERS‐CoV has increased, changes that may be attributed to improved case surveillance (Penttinen et al., 2013). Interestingly, over three quarters of MERS‐CoV cases have occurred in patients with comorbidities (The WHO MERS‐CoV Research Group, 2013). The most common comorbidities for MERS‐CoV cases have been diabetes, hypertension, obesity, cancer, and chronic kidney, heart, and lung disease (Assiri et al., 2013a). While these comorbidities likely affect disease progression and outcome, the strong correlation of chronic disease and MERS‐CoV may be biased by the high rate of these risk factors in the populations of the affected countries. In the Kingdom of Saudi Arabia, for instance, the prevalence of type‐II diabetes across ages is 31.6%, the prevalence of obesity is 31.1% (Al‐Daghri et al., 2011), and one quarter of adult males smoke (WHO, 2013a). Epidemiologic and pathogenesis studies will be necessary to discern how comorbidities impact susceptibility to, and progression of, MERS‐CoV infection.
Figure 3.
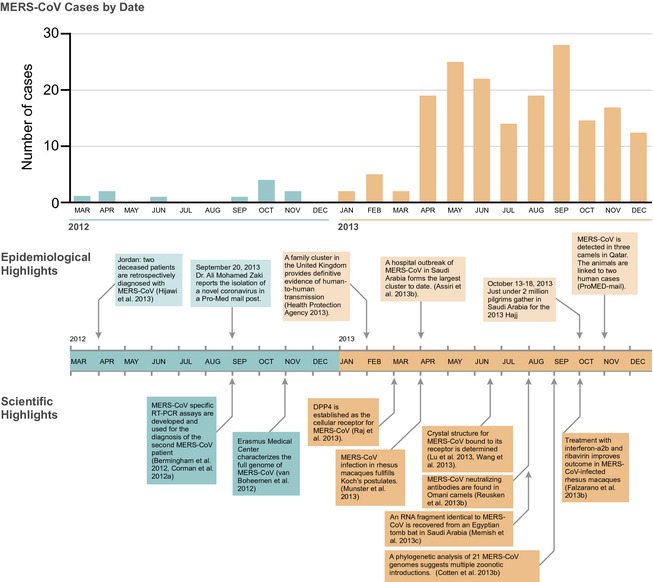
Timeline of the MERS‐CoV outbreak. Temporal distribution of MERS‐CoV cases from March 2012 through December 2013. Significant outbreak events and scientific advances during this time period are highlighted below.
MERS‐CoV clinical features
The clinical manifestations of MERS‐CoV range from subclinical infection to severe respiratory disease. Symptomatic patients often present with fever, myalgia, sore throat, shortness of breath, cough, and occasionally hemoptysis (Albarrak et al., 2012; Assiri et al.,2013a; Drosten et al., 2013; Guberina et al., 2013; Guery et al., 2013; Health Protection Agency, 2013;Memish et al.,2013b, c; Puzelli et al., 2013). Gastrointestinal symptoms such as diarrhea and vomiting are also common (Assiri et al.,2013a; Guery et al., 2013;Memish et al.,2013b). Although coinfections have been limited, MERS‐CoV coinfection with Klebsiella pneumoniae, Staphylococcus aureus, Candida species, and influenza A virus (H1N1), rhinovirus, and herpes simplex virus type‐1 has been reported (Zaki et al., 2012;Assiri et al., 2013a; Drosten et al., 2013; Guery et al., 2013; Health Protection Agency, 2013; Omrani et al., 2013). All clinically described patients have shown abnormal chest radiographs with a spectrum of lower pulmonary infiltrates and consolidation consistent with viral pneumonia (Albarrak et al., 2012; Zaki et al., 2012; Assiri et al.,2013a; Drosten et al., 2013; Guberina et al., 2013; Guery et al., 2013; Memish et al.,2013b). Over 60% of the first 144 MERS‐CoV patients suffered severe disease, requiring intensive care such as mechanical ventilation and extracorporeal membrane oxygenation (The WHO MERS‐CoV Research Group, 2013). Hematological abnormalities reported for clinical cases include thrombocytopenia (36%), lymphopenia (34%), lymphocytosis (11%), and neutrophilia (8%) (Assiri et al.,2013a).
While kidney failure necessitating renal replacement therapy has been reported for a number of MERS‐CoV cases (Albarrak et al., 2012; Zaki et al., 2012; Drosten et al., 2013; Guberina et al., 2013; Guery et al., 2013; Memish et al.,2013b; Omrani et al., 2013), in the absence of postmortem examinations, there is no direct evidence of MERS‐CoV replication in renal tissue. In many cases, renal involvement can be explained by pre‐existing conditions, hemorrhagic complications, and organ stress due to extreme hypoxemia (Albarrak et al., 2012; Drosten et al., 2013; Guery et al., 2013). Long‐term sequelae of acute MERS‐CoV have not yet been described.
Patient care
Clinical treatment for MERS‐CoV patients has centered on intensive care. Drug treatment has consisted of broad‐spectrum antibiotics and often oseltamivir, a drug targeting influenza A virus (Albarrak et al., 2012; Zaki et al., 2012; Assiri et al., 2013a; Drosten et al., 2013; Guery et al., 2013; Memish et al., 2013b; Omrani et al., 2013). Corticosteroids have been used for some patients, and antifungals were administered when necessary (Zaki et al., 2012; Assiri et al.,2013a; Drosten et al., 2013; Guery et al., 2013; Memish et al.,2013b; Omrani et al., 2013). Patients that progressed to severe acute respiratory distress were provided oxygen therapy, mechanical ventilation, or extracorporeal membrane oxygenation (Assiri et al.,2013b; Drosten et al., 2013; Guery et al., 2013; Health Protection Agency, 2013; Memish et al., 2013b; Omrani et al., 2013). Five of the 47 clinically described Saudi patients were treated with ribavirin, and one was given interferon‐α (IFN‐α); a few patients have also been infused with intravenous immunoglobulin (Assiri et al., 2013a). Few clinical case studies and no analyses of MERS‐CoV patients receiving treatments such as ribavirin and immunoglobulin have been published.
The origin of MERS‐CoV
Natural reservoir
Rapid full‐genome sequencing provided the first insight into the origin of MERS‐CoV (van Boheemen et al., 2012;Cotten et al.,2013a). Phylogenetic analysis shows a close genetic relatedness between MERS‐CoV and the group C Betacoronaviruses BtCoV‐HKU4 and BtCoV‐HKU5 detected in insectivorous bats (Woo et al., 2012), although molecular clock analyses suggest they are unlikely to be direct ancestors of MERS‐CoV (Fig. 4; Lau et al., 2013). Since MERS‐CoV's identification in 2012, closely related coronavirus sequences have been detected in bats in Africa, Asia, the Americas, and Eurasia, suggesting a widespread circulation of MERS‐CoV‐related viruses in the order Chiroptera (Annan et al., 2013; Anthony et al., 2013; De Benedictis et al., 2014; Ithete et al., 2013; Lelli et al., 2013; Wacharapluesadee et al., 2013). Investigations of samples from bats roosting in the vicinity of the first MERS‐CoV case in Bisha, Saudi Arabia, revealed the presence of a 190‐nucleotide RNA fragment with 100% match to the RdRp of MERS‐CoV in the feces of an Egyptian tomb bat (Taphozous perforates;Memish et al., 2013c). Unfortunately, the short length of MERS‐like CoV sequences identified in bats limits the strength of phylogenetic analyses and subsequent conclusions about the origin of MERS‐CoV.
Figure 4.
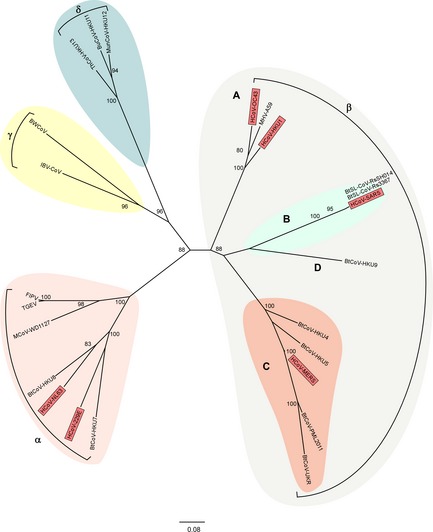
Coronavirus phylogeny. Phylogenetic tree of coronaviruses with representatives of each of the four genera; Alpha (pink), Beta (gray), Delta (blue), and Gammacoronavirus (yellow). Betacoronaviruses are further subdivided into clades A through D, with clade B (green) containing SARS‐CoV and clade C (orange) containing MERS‐CoV. All known human coronaviruses are represented in red. Maximum‐likelihood trees were generated with the MEGA5 software package using a 1231‐nucleotide segment within the RdRp. Trees were visualized using Figtree. Bootstrap values above 75 are shown. CoV isolation origin abbreviations as follows: H: human; Bt: bat; BtSL: bat SARS‐like; BW: beluga whale; IBV: chicken; FIPV: feline; TGEV: swine; M: mink; MHV: murine; Th: thrush; Bu: bulbul; Mun: munia.
Intermediate host
Direct contact between humans and bats is limited, and an intermediate species often plays a role in the transmission of emerging viruses from bats to humans (Field et al., 2001; Luo et al., 2003; Mahalingam et al., 2012; Nel, 2013). Anecdotal evidence of MERS‐CoV patient contact with farm animals has been reported in a few cases (Albarrak et al., 2012; Buchholz et al., 2013; Drosten et al., 2013; The WHO MERS‐CoV Research Group, 2013), and so suspicions about the potential source of MERS‐CoV have focused on livestock common to the Arabian Peninsula, such as goats, sheep, dromedary camels, and cows. The first evidence for the existence of an intermediate animal reservoir was the detection of MERS‐CoV neutralizing antibodies in dromedary camels from Oman and the Canary Islands (Spain) (Reusken et al.,2013b). Subsequent studies have detected MERS‐CoV neutralizing antibodies in dromedary camels from Egypt, Jordan, Saudi Arabia, and importantly, in camel serum collected in 2003 from the United Arab Emirates (Hemida et al., 2013; Perera et al., 2013;Reusken et al.,2013a; Meyer et al., 2014). While MERS‐CoV neutralizing antibodies were not detected in any other species of livestock tested, including chickens, goats, sheep, and cattle, seropositivity among camels passed 90% in every location, even in 2003 (Hemida et al., 2013; Perera et al., 2013;Reusken et al.,2013a, b; Meyer et al., 2014). The high prevalence of neutralizing antibodies across age grades suggests pervasive and early infection of camels with MERS‐CoV or a MERS‐CoV‐like virus. Recently, MERS‐CoV virus was detected by RT‐PCR in nose swabs from three camels in Qatar (Haagmans et al., 2013). The camels were epidemiologically linked to two human cases of MERS‐CoV, and viral fragments sequenced from the camels showed high similarity to sequences from the human cases (Haagmans et al., 2013). While this datum provides more conclusive evidence that dromedary camels form part of the MERS‐CoV outbreak picture, the direction of transmission is still unclear (Fig. 5). Transmission could have occurred from camels to humans, humans to camels, or concurrently from a third source to both humans and camels (Haagmans et al., 2013). Furthermore, the detection of MERS‐CoV neutralizing antibodies in dromedary camels in regions with no reported human cases, such as Egypt and the Canary Islands, raises questions about the extent of MERS‐CoV or MERS‐CoV‐like virus circulation in Africa, the Arabic peninsula, and minor Asia (Perera et al., 2013; Reusken et al.,2013b). Movement of camels between Africa and the Arabian Peninsula is common and could contribute to the spread of MERS‐CoV between regions (Mukasa‐Mugerwa, 1981; Perera et al., 2013).
Figure 5.

Putative MERS‐CoV transmission cycle. The putative transmission cycle for MERS‐CoV. MERS‐CoV likely originated from bats, acting as the natural reservoir. From the natural reservoir, MERS‐CoV spilled either directly over to humans (green arrow) or via an intermediate host (dromedary camels, purple arrow). Currently, the exact route of zoonotic transmission of MERS‐CoV into the human population remains unknown although the presence of MERS‐CoV neutralizing antibodies and the detection of MERS‐CoV in dromedary camels suggest that this species is likely to play a major role in the emergence of MERS‐CoV. Phylogenetic analysis suggests that multiple introductions of MERS‐CoV into the human population have occurred and both zoonotic transmission events and human‐to‐human transmission (blue arrows) drive the current MERS‐CoV outbreak.
Transmission of MERS‐CoV
The respective roles of human‐to‐human and zoonotic transmission in the current MERS‐CoV outbreak are not well understood (Fig. 5). Conclusive evidence of human‐to‐human transmission of MERS‐CoV was first reported in a cluster of MERS‐CoV cases in the United Kingdom, when an adult male who had travelled to Saudi Arabia transmitted the virus to two of his family members (Health Protection Agency, 2013). Overall, MERS‐CoV human‐to‐human transmission chains have been self‐limiting and irregular, and more than half of secondary MERS‐CoV cases have originated in a healthcare setting (WHO, 2014). The largest cluster of MERS‐CoV to date has involved 23 patients at three different healthcare facilities in the Eastern Province of Saudi Arabia, highlighting the potential of nosocomial transmission (Assiri et al.,2013b). On the other hand, an early MERS‐CoV patient transferred from Qatar to a specialist lung hospital in Germany was given intensive treatment for almost a month before the hospital learned of his MERS‐CoV diagnosis. Extensive contact investigation and serological analysis of those potentially exposed to the patient revealed no secondary infections (Buchholz et al., 2013). Screening of MERS‐CoV patient contacts has uncovered at least 18 instances of asymptomatic MERS‐CoV infection in healthcare workers and other contacts, although the role these subclinical cases can play in the transmission of infection is unclear (Memish et al.,2013a; The WHO MERS‐CoV Research Group, 2013). Transmission of respiratory viruses is often directly associated with the amount of virus shed. The dynamics of MERS‐CoV shedding throughout the course of disease have not been well characterized, but high viral loads detected in bronchoalveolar lavage samples from infected patients suggests that coughing and exudates from the lower respiratory tract could be important mechanisms of MERS‐CoV human‐to‐human transmission (Drosten et al., 2013; de Sousa et al., 2013).
The zoonotic source of MERS‐CoV continues to play a role in outbreak epidemiology through repeated introductions of virus into the human population (Cotten et al.,2013a, b). The WHO has identified 62 sporadic cases of MERS‐CoV, defined as having occurred with no known human exposure (The WHO MERS‐CoV Research Group, 2013; WHO, 2014). In 14 early clusters, each primary case was an adult male, suggesting that activities unique to adult males in the Arabian Peninsula may expose them to a virus source (Penttinen et al., 2013). Interestingly, the rate of severe disease and death is higher for primary MERS‐CoV patients than for secondary cases, despite a similar prevalence of comorbidities (The WHO MERS‐CoV Research Group, 2013). This could be the result of higher doses of virus exposure among primary patients.
Transmission dynamics of MERS‐CoV
Because of the epidemiologic dynamics described above, the basic reproduction number (R 0) of MERS‐CoV is uncertain. R 0 is a measure of the number of secondary cases generated by one case of disease in a naïve population – an R 0 of < 1 is self‐limiting within a population. Using the epidemiological information from 62 probable cases of MERS‐CoV infection, two different transmission scenarios were used to estimate the R 0 for MERS‐CoV (Breban et al., 2013). One scenario was modeled on a large number of index patients per cluster, each generating a small transmission tree, reflecting the possibility that two epidemiologically linked cases could have been exposed to the same nonhuman source of MERS‐CoV. This scenario yielded an R 0 of 0.60 and a yearly MERS‐CoV introduction rate of 22.3 (Breban et al., 2013). The second scenario, which used a lower rate of introduction with higher human‐to‐human transmission, predicted an R 0 of 0.67 and a yearly introduction rate of 17.1 (Breban et al., 2013). A large‐scale analysis of full‐genome sequences of MERS‐CoV has identified multiple zoonotic introductions of the virus and distinct genomes circulating in the same geographic spaces, providing evidence for multiple zoonotic introductions and transmission dynamics in agreement with a lower R 0 (Cotten et al.,2013b). Furthermore, a comparison of the R 0s predicted for MERS‐CoV and prepandemic SARS‐CoV (R 0 of 0.8) suggests MERS‐CoV has low pandemic potential (Breban et al., 2013). Another transmission model of 111 MERS‐CoV cases predicted a similar R 0 (0.63), but warned that in the absence of control measures, the R 0 could range between 0.8 and 1.3 and allow for self‐sustaining transmission (Cauchemez et al., 2013). Based on extrapolations of the incidence of disease in travelers returning from MERS‐CoV‐source countries, it was predicted that as many as 940 symptomatic cases of MERS‐CoV may have occurred before August 8, 2013 (Cauchemez et al., 2013). Epidemiologic studies will be necessary to assess the prevalence and circulation of MERS‐CoV infection in the human population.
MERS‐CoV biology
The 30 119‐base pair genome of MERS‐CoV consists of at least ten polycistronic open reading frames (ORFs), the organization of which follows that of coronaviruses in general (Masters, 2006; van Boheemen et al., 2012). Over two‐thirds of the 5′ end of the coronavirus genome is composed of the replicase open reading frames ORF1a and ORF1b (Masters, 2006). In MERS‐CoV, these ORFs are translated into two polyproteins, one requiring a ribosomal frame shift, which are eventually cleaved into 16 putative nonstructural proteins (nsps; Sawicki et al., 2007; van Boheemen et al., 2012). The role of these nsps has not been empirically determined for MERS‐CoV, but some function can be predicted based on conserved domains characterized in other coronaviruses. For instance, nsp12 putatively serves as the RdRp, and nsp14 is thought to function as the proofreading exoribonuclease (ExoN; van Boheemen et al., 2012). Interestingly, coronaviruses are the only RNA viruses known to use a specific proofreading enzyme for the maintenance of high‐fidelity viral RNA replication (Smith et al., 2013). Downstream of the two large ORFs are at least nine ORFs encoding the structural proteins – spike, envelope, membrane, and nucleocapsid – and some accessory proteins (van Boheemen et al., 2012). The translation of the downstream ORFs occurs via subgenomic mRNAs, a salient feature of the order to which coronaviruses belong: Nidovirales (Sawicki et al., 2007).
Receptor binding of MERS‐CoV
By granting binding of the virus to the host cell, cellular receptors play an important role in determining the species and tissue tropism of coronaviruses (Thackray & Holmes, 2004; Masters, 2006; Tusell et al., 2007). The MERS‐CoV spike protein is a 1353 amino acid type‐I transmembrane glycoprotein presented as a trimer on the surface of the enveloped virus. After translation, the spike protein is cleaved into two domains: the S1 subunit responsible for receptor binding and the S2 unit that mediates membrane fusion (Ohnuma et al., 2013; Raj et al 2013). The S1 spike glycoprotein binds to the surface enzyme dipeptidyl peptidase 4 (DPP4, also known as CD26). DPP4 is a type‐II transmembrane glycoprotein that catalyzes the cleavage of N‐terminal proline‐containing dipeptides and aids glucose metabolism by proteolytic inactivation of incretins (Engel et al., 2003; Hiramatsu et al., 2003; Lambeir et al., 2003). DPP4 is the third exopeptidase found to act as a receptor for coronaviruses (Raj et al., 2013). Blocking the enzymatic activity of DPP4 does not affect MERS‐CoV susceptibility, and so the significance of these enzymes as receptors for coronaviruses is thought to lie in their widespread expression on endothelial and epithelial tissues (Raj et al., 2013). DPP4 is relatively conserved between mammalian species, allowing MERS‐CoV to bind to species as diverse as bats and humans (Müller et al., 2012; Raj et al., 2013). The receptor‐binding domain (RBD) of MERS‐CoV, the region of the spike protein that attaches to the DPP4 receptor, has been mapped to 240 amino acid residues in the S1 region of the spike protein (Chen et al., 2013; Lu et al., 2013; Mou et al., 2013; Wang et al., 2013). Co‐crystallization of MERS‐CoV spike protein and DPP4 revealed an interaction between the beta‐propeller blades 4 and 5 of DPP4 and beta‐strands 5, 6, 7, and 8 of the MERS‐CoV RBD, also known as the receptor‐binding motif (RBM; Fig. 6; Lu et al., 2013; Wang et al., 2013). The RBD of the spike protein induces neutralizing antibodies, making it an important target for the development of prophylactics and therapeutics (Agnihothram et al., 2013; Du et al., 2013a,b,c; Gierer et al., 2013; Song et al., 2013).
Figure 6.
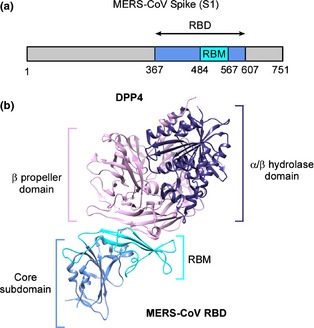
MERS‐CoV spike glycoprotein and DPP4 receptor interaction. (a) The linear organization of the S1 subunit of MERS‐CoV spike glycoprotein with the variable RBD located at amino acid residues 367–607, with a RBM containing the critical amino acid residues for binding at residues 484–567. (b) The crystal structure of the MERS‐CoV RBD coupled with the receptor dipeptidyl peptidase IV (DPP4). DPP4 is structurally divided into an alpha‐ and beta‐hydrolase domain, and a beta‐propeller domain. The beta‐propeller domain of DPP4 (pink) interacts with the RBM region (light blue) of the MERS‐CoV spike protein RBD. The schematic representation of the DPP4 – MERS‐CoV spike protein RBD structure was generated using chimera and protein accession number 4KR0 (Lu et al., 2013).
Host interaction of MERS‐CoV
Preliminary investigations of MERS‐CoV host interactions and innate immune responses have been performed in vitro, ex vivo, and in vivo in a nonhuman primate model. MERS‐CoV has been shown to replicate in vitro in human, bat, and porcine‐derived cell lines, whereas cow, hamster, murine, rat, and canine cell lines were not susceptible (Müller et al., 2012;Chan et al.,2013b; Dijkman et al., 2013; Kindler et al., 2013; Raj et al., 2013; Scobey et al., 2013). While in vitro modeling holds some predictive value for MERS‐CoV susceptibility in vivo, a more complete characterization of the mechanisms of cellular entry is necessary to understand host susceptibility (Leow, 2013). For instance, the species tropism of MERS‐CoV appears to be restricted by variability in the cellular receptor. MERS‐CoV is unable to replicate in mice, hamsters, or ferrets – common small animal models – and the MERS‐CoV spike glycoprotein is unable to bind ferret‐specific DPP4 (Coleman et al., 2013;de Wit et al.,2013a;Raj et al., 2014).
Innate immunity
Infection experiments in human airway epithelial cells have shown that the global transcriptional host response is earlier and more robust after MERS‐CoV infection than after SARS‐CoV infection (Josset et al., 2013). Yet consistent with observations of SARS‐CoV infection, MERS‐CoV infection of human respiratory cells does not lead to a pronounced type‐I IFN response (Kopecky‐Bromberg et al., 2007; Kindler et al., 2013; Zielecki et al., 2013). Type‐I IFNs play a key role in viral immunity, and pathogenic viruses often evade innate immunity through antagonist proteins that disrupt the IFN reaction (García‐Sastre & Biron, 2006; Randall & Goodbourn, 2008; Taylor & Mossman, 2013). In MERS‐CoV, inhibition of type‐I IFNs has been demonstrated by the structural M protein and the accessory proteins encoded by ORF 4a, ORF 4b, and ORF 5 (Yang et al., 2013). The most potent inhibitor, ORF 4a, has been shown to inhibit type‐I IFN activation by blocking the interaction between the RNA helicase sensor MDA5 and viral double‐stranded RNA, a mechanism distinct from that employed by SARS‐CoV (Kopecky‐Bromberg et al., 2007; Niemeyer et al., 2013; Yang et al., 2013).
Efficient MERS‐CoV replication has also been demonstrated in nonciliated bronchial epithelium, alveolar epithelial cells, endothelial cells, and macrophages in human ex vivo organ cultures (Chan et al.,2013c; Zhou et al., 2013). Consistent with the results obtained from human respiratory cell lines, MERS‐CoV infection ex vivo does not lead to a strong type‐I IFN response in these cultures (Chan et al.,2013c ; Zhou et al., 2013). The combined in vitro and ex vivo data suggest that MERS‐CoV actively interacts with and evades innate immune recognition pathways by the host (Kindler et al., 2013).
Ex vivo data support the results of viral dissemination and pathology, and cellular tropism established in the rhesus macaque model (de Wit et al., 2013b). Gene expression analysis in experimentally infected rhesus macaques showed differentially expressed genes in infected lung tissue associated with antiviral immunity, inflammation, and chemotaxis, including IL‐6, chemokine C‐X‐C ligand 1, and matrix metalloproteinase. As expected, type‐I IFNs were not activated (de Wit et al., 2013b). The chemokine IL‐8, a strong recruiter of neutrophils and other granulocytes, was induced in the macaques, perhaps explaining the increased numbers of neutrophils recorded in the blood of infected macaques and some human patients (Zaki et al., 2012; Assiri et al.,2013a; Guery et al., 2013; de Wit et al.,2013b).
MERS‐CoV intervention strategies
Public health measures
Mathematical transmission models highlight two important ways MERS‐CoV can be controlled: reducing the rate of MERS‐CoV introductions into the human population and breaking chains of human‐to‐human transmission. The reduction in the rate of MERS‐CoV introductions in the human population requires a comprehensive understanding of the nonhuman source of MERS‐CoV and the spatial and temporal dynamics of MERS‐CoV circulation in this source. On the other hand, interrupting the human‐to‐human transmission cycle of MERS‐CoV calls for an understanding of the parameters involved in transmission, such as virus shedding, stability, and routes of transmission. Although routes of transmission for MERS‐CoV are not well understood, the spread of MERS‐CoV between people in close contact settings suggests that direct contact and fomite transmission routes are likely to be involved. As stated above, the localization of MERS‐CoV infection in the lower respiratory tract implicates coughing and other exudates as important sources of virus shedding. In addition, the environmental stability of MERS‐CoV provides some information on the potential for fomite transmission. At low temperatures and humidity, both MERS‐CoV and SARS‐CoV virions retain viability on smooth surfaces much longer than many other respiratory viruses, including influenza virus H1N1, HCoV‐229E, and HCoV‐OC43 (Sizun et al., 2000; Chan et al., 2011; van Doremalen et al., 2013). Thus, temperature‐controlled settings such as hospitals may be of particular risk for fomite transmission of MERS‐CoV. Performing high‐risk patient care procedures such as intubation and manual ventilation, along with inconsistent use of surgical masks, was associated with nosocomial transmission of SARS‐CoV to healthcare workers (Ofner‐Agostini et al., 2006; Nishiyama et al., 2008). Healthcare workers are a growing cohort of MERS‐CoV cases, but while routes of exposure may be similar for both viruses, the MERS‐CoV outbreak has not been characterized by the hospital‐based super‐spreader events which defined the epidemiology of SARS‐CoV (Lipsitch et al., 2003).
During infection with SARS‐CoV, viral load in upper respiratory tract secretions remained low for the first 5 days of illness, not peaking in nasopharyngeal aspirates until about 10 days after onset of symptoms (Peiris et al., 2003; Cheng et al., 2004). Thus, SARS‐CoV transmission could be prevented in the general population by basic public health and infection control measures, such as the isolation of patients in negative‐pressure rooms, active surveillance and quarantine of contacts, and the provision of education and protective equipment for healthcare workers (Twu et al., 2003; Svoboda et al., 2004). Estimates of the serial interval of MERS‐CoV infection in the largest cluster of MERS‐CoV to date – 23 cases in a hospital setting – are slightly shorter than those for SARS‐CoV (median 7.6 vs. 8.4 days), suggesting that transmission may occur earlier in the course of illness (Assiri et al.,2013b). Effective implementation of public health measures against the current MERS‐CoV outbreak must integrate knowledge of shedding dynamics, exposure mechanisms, and virus viability.
Therapeutics
Antiviral treatment specific to human coronaviruses has not been developed, and during the short‐lived 2002–2003 SARS‐CoV outbreak, supportive treatment regimens were not optimized (Stockman et al., 2006; Cheng et al., 2013). Commercially available drugs, such as type‐I IFNs, lopinavir, and, at very high concentrations, ribavirin, were shown to inhibit cytopathic effect of SARS‐CoV in in vitro studies, with a synergistic antiviral effect described for type‐I IFNs and ribavirin (Chen et al., 2004; Tan et al., 2004; Birgit et al., 2005). These drugs were used to varying extents on SARS‐CoV patients, often in combination with corticosteroids (Stockman et al., 2006; Cheng et al., 2013). Ribavirin is a structural analog of guanosine with broad‐spectrum mutagenic effects on viruses. Despite its frequent use on SARS‐CoV patients, ribavirin was not shown to be effective for patients, and adverse effects such as hemolytic anemia were common (Stockman et al., 2006; Cheng et al., 2013). Type‐I IFN regimens for SARS‐CoV patients were sometimes used as part of a multi‐drug regimen, and one study of 22 SARS‐CoV patients reported more rapid improvement of radiographic lung pathologies and better oxygenation in patients treated with IFN‐α1 (Loutfy et al., 2003; Stockman et al., 2006; Cheng et al., 2013). Speculations that immunopathology rather than uncontrolled viral replication contributed to lung deterioration during advanced SARS‐CoV infection favored the widespread use of corticosteroids in its treatment (Nicholls et al., 2003; Peiris et al., 2003; Hui & Sung, 2004; Cheng et al., 2013). Retrospective analysis of SARS‐CoV patients showed no benefit of corticosteroid administration except when critical cases were analyzed alone and death‐related variables were adjusted. Among these critical patients, corticosteroid therapy did significantly reduce case fatality (Chen, 2006). Similarly, corticosteroids have been used in some MERS‐CoV patients with severe disease (Assiri et al.,2013a; Guberina et al., 2013; Memish et al.,2013b; Omrani et al., 2013). One previously healthy MERS‐CoV patient who suffered ARDS, multidrug‐resistant Pseudomonas aeruginosa, and suspected concomitant allergic bronchopulmonary aspergillosis improved rapidly when treated with the corticosteroid prednisolone (Guberina et al., 2013). The efficacy of corticosteroid treatment for other MERS‐CoV patients with ARDS has not been reported.
Therapies used during the SARS‐CoV outbreak have shaped investigations of treatment regimens for MERS‐CoV. MERS‐CoV shows greater sensitivity than SARS‐CoV to the antiviral effects of type‐I and type‐III IFNs in vitro and ex vivo (Kindler et al., 2013; Wilde et al., 2013; Zielecki et al., 2013). This greater sensitivity could be explained by mechanistic differences of IFN antagonist accessory proteins between the viruses. For instance, MERS‐CoV lacks a homologue of the SARS‐CoV ORF6 protein, which blocks the STAT1‐activating effects of IFNs and ultimately the transcriptional activation of downstream antiviral genes (Wilde et al., 2013). A comparison of five different IFNs has shown IFN‐β to be the most potent inhibitor of MERS‐CoV (Hart et al., 2013). Although high concentrations of ribavirin were effective in vitro in one study (Falzarano et al.,2013a), another study found no inhibitory effects of ribavirin at a dose translatable to current drug regiments in humans (Hart et al., 2013). When IFN‐α2b and ribavirin were used in combination against MERS‐CoV, however, the 50% effective concentrations (EC50) of the drugs in combination were much lower than for the individual drugs. This additive effect decreased the drug requirements to concentrations potentially achievable in humans (Falzarano et al.,2013a). Interestingly, ribavirin is significantly more effective when the ExoN activity of coronaviruses is knocked out (Smith et al., 2013). Data indicate that in these knockout viruses, ribavirin has a greater ability to inhibit viral RNA synthesis and inosine monophosphate dehydrogenase (IMPDH), and enzyme necessary for the de novo synthesis of guanine nucleotides, which would explain the relatively high ribavirin dose needed to achieve inhibition of viral replication with MERS‐CoV (Smith et al., 2013).
In a study to identify compounds that inhibit MERS‐CoV, mycophenolic acid demonstrated a particularly high efficacy against MERS‐CoV, with an EC50 of < 10 μM (Chan et al.,2013a,b,c). Mycophenolic acid is an approved drug and, like ribavirin, acts by inhibiting IMPDH. While typically used as an immunosuppressant after tissue transplants, its antiviral activity has been attributed to an inhibition of viral RNA replication (Diamond et al., 2002).
Preliminary in vitro comparisons suggest that mycophenolic acid is a more potent inhibitor of MERS‐CoV than ribavirin (Chan et al.,2013a Falzarano et al.,2013a; Hart et al., 2013). Because of this, effective plasma drug levels may be better achievable for intravenous doses of mycophenolic acid than ribavirin (Chan et al.,2013a). Assumptions about the in vivo usefulness of drugs effective in vitro need to be made with care. Mycophenolic acid, for instance, is a potent inhibitor of T and B lymphocytes and dendritic cell maturation, effectively suppressing antigen presentation and immunoglobulin production (Villarroel et al., 2009). In a well‐characterized animal model, the adverse immunomodulatory effects of the drug may outweigh any reduction in MERS‐CoV virulence.
Animals model for MERS‐CoV
Attempts to establish a small animal model for MERS‐CoV Syrian hamsters, mice, and ferrets have been unsuccessful (Coleman et al., 2013; Raj et al., 2013a, b;de Wit et al.,2013a). However, experimental infection studies in rhesus macaques showed that this animal species was susceptible to MERS‐CoV (Munster et al., 2013). In rhesus macaques, MERS‐CoV causes a lower respiratory tract infection reminiscent of mild to moderate human cases (de Wit et al.,2013b). Clinical signs of MERS‐CoV‐infected macaques included cough and increased respiration rate, and lung samples showed lesions characteristic of mild to marked pneumonia (Munster et al., 2013; de Wit et al.,2013b). Virus was detected throughout the respiratory tract and mediastinal lymph nodes, with viral loads higher earlier during infection. The primary sites of virus replication were type‐I and type‐II pneumocytes, main components of the alveolar architecture. Replication of MERS‐CoV deep in the macaque lower respiratory tract may explain the low potential for transmission of MERS‐CoV. Despite renal failure in some human patients, virus was absent from the kidney tissue of all macaques (de Wit et al.,2013b). A more recent characterization of MERS‐CoV in rhesus macaques reported transient fever in infected monkeys and demonstrated a MERS‐CoV‐specific antibody response in the macaques starting at 7 days postinfection (Yao et al., 2013). The rhesus macaque model of MERS‐CoV infection is the only in vivo model established to date, and development of a small animal model is essential to conduct widespread research on pathogenesis and prophylactic and therapeutic countermeasures.
In vivo testing of antivirals
Of the antiviral and immune modulatory compounds shown effective against MERS‐CoV in vitro, only one treatment option has been examined in vivo. The efficacy of a combination IFN‐α2b and ribavirin treatment regimen was tested in MERS‐CoV‐infected rhesus macaques (Falzarano et al., 2013b). Doses were designed to achieve serum concentrations at or above the EC50 values determined in vitro. Treated animals did not show the clinical signs or hematological changes that developed in the untreated animals, such as breathing difficulties, decreased oxygen saturation levels, and increased neutrophil counts (Falzarano et al., 2013b). Gross pathology of lungs from treated animals was normal, while untreated animals displayed visible lesions. Histopathology revealed mild signs of bronchointerstitial pneumonia in the treated animals, with more abundant alveolar edema and severe lesions seen in the untreated animals. The MERS‐CoV viral load in lung samples was 0.81 log lower for treated animals compared to untreated animals. Furthermore, a lung‐specific host response occurred in untreated macaques but not in treated animals, with increased levels of the cytokines and chemokines IL‐6, IFN‐y, and MCP‐1. Reduced expression of inflammatory genes was observed in the lungs of treated animals (Falzarano et al., 2013b). The significant improvement in clinical score for treated animals despite relatively similar viral loads between the groups suggests that immunopathology may be a factor in the severity of MERS‐CoV infection. However, because the rhesus macaque model only recapitulates mild to moderate disease in humans, the effectiveness of these drugs against MERS‐CoV‐induced ARDS is unclear. Timing is also critical to the efficacy of treatment; most patients do not begin treatment until they are quite ill, whereas drug regimens in the macaques began only 8 h postinoculation. The translation of drug regimens from the laboratory to the clinic must address such discrepancies.
Prophylactics
In the years since the SARS‐CoV outbreak, attempts have been made to prepare for reemergence by establishing vaccines, and the advent of MERS‐CoV has only heightened the need for effective coronavirus vaccines. Inactivated viruses, live‐attenuated viruses, DNA vaccines, virus‐like particles, and viral vector‐based vaccines have all been shown to produce neutralizing antibodies to SARS‐CoV (Chen et al., 2005; Zhao et al., 2005; Graham et al., 2012; Tseng et al., 2012). Unfortunately, SARS‐CoV causes pulmonary immunopathology upon challenge after vaccination in animal models, presumably because they induce an immune response skewed toward T helper 2 (TH2) cell responses (Tseng et al., 2012). While inactivated virus vaccines are particularly prone to inducing this type of TH2‐related hypersensitivity, immunopathology upon challenge has been observed with SARS‐CoV‐based viral vector vaccines (Deming et al., 2006), virus‐like particle vaccines (Tseng et al., 2012), and DNA vaccines (Zhao et al., 2005).
Investigations of the antigenic and serologic relationships of MERS‐CoV to other coronaviruses have confirmed that neutralizing antibodies target the spike protein of MERS‐CoV and are specific to MERS‐CoV, while the N protein induces antibodies cross‐reactive within its coronavirus subgroup (Agnihothram et al., 2013). The spike protein, and especially its RBD, is considered a key component in coronavirus vaccine design (Du et al., 2009, 2013b,c). A replication‐defective vaccinia virus‐based vaccine expressing the full‐length spike protein of MERS‐CoV has been shown to produce high levels of neutralizing antibodies in mice, but virus challenge cannot be applied to the mouse model (Song et al., 2013).Vaccination with recombinant protein containing a truncated RBD of the spike protein (amino acids 377–588) and the Fc of human IgG also induced high titers of neutralizing antibodies. The truncated RBD elicited higher levels of neutralizing antibodies than vaccination with a recombinant protein containing a larger fragment of the spike protein (amino acids 377–662). The authors suggest that non‐neutralizing epitopes within the 588–662 region of the polypeptide may compete with neutralizing epitopes or destabilize the formation of the RBD (Du et al., 2013b). To maximize the protectiveness of neutralizing antibodies across MERS‐CoV strains, the natural variation of MERS‐CoV spike proteins needs to be characterized and considered in the design of vaccines (Graham et al., 2013). Live‐attenuated viruses are another approach to MERS‐CoV vaccine development. A MERS‐CoV mutant lacking the structural E protein has been shown to be replication‐competent but propagation‐defective (Almazán et al., 2013). Development of safe and effective MERS‐CoV vaccines must potentially overcome the challenges that arose for SARS‐CoV. No in vivo testing of vaccines has been performed to date; it remains therefore unclear whether challenge with MERS‐CoV would illicit pulmonary immunopathology upon challenge. Creative approaches, such as the use of adjuvants that promote TH1 cell responses, need to be pursued, and potential vaccines must be rigorously evaluated in animal models (Graham et al., 2013).
Global response
In the year since its identification, MERS‐CoV has not only spread across the Arabic Peninsula, but has been transported to the United Kingdom, France, Germany, Italy, Tunisia, and Spain (Bermingham et al., 2012; Buchholz et al., 2013; Gulland 2013a, b; Health Protection Agency, 2013; Hijawi et al., 2013; Mailles et al., 2013; Memish et al.,2013b; Puzelli et al., 2013). The initial detection of MERS‐CoV was followed by the rapid development of MERS‐CoV molecular and serological diagnostics (Corman et al.,2012a, b). The emergence of MERS‐CoV reminded the infectious disease community of the emergence of SARS‐CoV and called for immediate public health preparedness and response. Research efforts have largely focused on the epidemiology of the outbreak and identification of putative natural and intermediate reservoirs and potential therapeutics. Some controversy has surrounded the identification and sharing of MERS‐CoV isolates – the initial isolate was shared without the consent of the Saudi Arabian government, and material transfer agreements (MTA) of some isolates were said to be unnecessarily restrictive (Butler, 2013). An MTA has several purposes: it defines not only the material to be transferred, but who can use the materials and what the purpose of the transfer is. It also protects the intellectual property rights of each party by defining ownership of the original material and inventions resulting from the use of the material. Lastly, it designates liability of each party and ensures that the receiving party will use the material in a safe way (for instance, at the proper biosafety level). The controversies surrounding the initial detection of MERS‐CoV highlight the importance of creating an international framework for rapid global sharing of virus strains and biologic materials during outbreaks. Similar controversies over ownership have arisen with H5N1 avian influenza (Fidler, 2008). Both the avian flu virus and the MERS‐CoV cases strongly point to the need not only for prenegotiated transfer agreements but also for standardized ‘best practices’ guidelines for highly virulent emerging disease materials where expedited sharing of material and data is of paramount importance.
Future perspectives
Despite many advances in our understanding of the MERS‐CoV outbreak, major questions remain. The epidemiology of MERS‐CoV is still poorly understood. Although dromedary camels have been implicated as the most likely intermediate reservoir, more details on the genetic variation of MERS‐CoV viruses circulating in camels and humans are necessary to identify camels as the definitive source of human MERS‐CoV infections. The widespread distribution of dromedary camels across Africa, the Arabic peninsula, and South‐West Asia highlights their potential to facilitate the outbreak's spread (Fig. 7), but information on the spatial and temporal patterns of MERS‐CoV circulation in this species is needed. Because zoonotic introductions continue to play a role in the epidemiology of MERS‐CoV in humans (Cotten et al., 2013b), an elucidation of the mechanisms of zoonotic transmission is essential and intervention strategies should focus on controlling these events.
Figure 7.
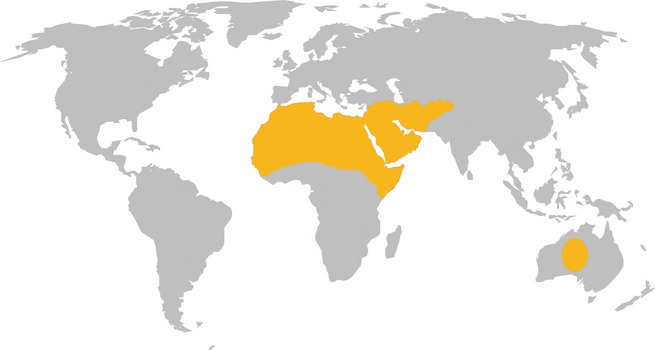
Geographic distribution of dromedary camels. The global distribution of dromedary camels is indicated by yellow shading (Mukasa‐Mugerwa, 1981).
Our clinical understanding of MERS‐CoV infection is based on limited reports (Albarrak et al., 2012; Bermingham et al., 2012; Assiri et al., 2013a; Drosten et al., 2013; Guery et al., 2013;Memish et al., 2013a; Omrani et al., 2013). Of particular interest is the effect of comorbidities on MERS‐CoV pathogenesis and patient outcomes. Preliminary evidence shows a direct relation between underlying comorbidities and disease severity (Assiri et al.,2013a). The observation that primary cases have been more severe than secondary cases, even when controlling for underlying conditions, might suggest that these primary cases were exposed to higher virus doses than secondary cases (The WHO MERS‐CoV Research Group, 2013). This could also indicate that MERS‐CoV is more readily transmissible from the intermediate reservoir to humans than from human‐to‐human and that milder MERS‐CoV cases would be less likely to efficiently transmit MERS‐CoV. Human‐to‐human transmission could increase if MERS‐CoV becomes better adapted to humans. Prolonged MERS‐CoV replication in immune‐compromised patients could increase opportunity for the virus to acquire mutations enabling efficient transmission. The development and testing of MERS‐CoV therapeutics is currently hindered by the absence of small animal models. In addition, phase I–III clinical trials need to be conducted before experimental vaccines and treatment options can be available to humans. Accordingly, research focusing on already‐approved drugs for the treatment of MERS‐CoV with result in faster implementation and wider availability of therapeutics in the clinic.
The increase in emerging infectious disease events over the last decades has made it apparent that a more complete understanding of the ecology, biology, and political economy of infectious disease emergence is necessary. Globalization, climate change, habitat alteration, and wildlife encroachment likely contribute to novel interactions between pathogens and hosts. Adequate preparation for future infectious disease outbreaks requires strong international relationships in research, monitoring and surveillance, and public health response. In 2005, after facing the emergence of SARS‐CoV and avian influenza H5N1, the WHO(2013b) developed International Health Regulations for the coordination of global responses to emerging health threats. The WHO has structured its MERS‐CoV response according to these regulations, forming an emergency committee on MERS‐CoV, creating case definitions of infection, and providing frequent updates of the MERS‐CoV outbreak through the IHR's global alert and response function. The Program for Monitoring of Emerging Diseases (ProMED‐mail), which communicated the first report of MERS‐CoV infection in a human, has also continued to provide the global health and research communities with updates of epidemiological reports and scientific findings. Capacity building, knowledge transfer, and training of the future generation of scientists are key factors in forming multidisciplinary preparedness for future infectious disease outbreaks. In this regard, special attention should be given to prevention and control of emerging infectious diseases in the developing world, the origin of the majority of infectious disease outbreaks.
Acknowledgements
The authors would like to thank Beth Fisher, Anita Mora, Emmie de Wit, Darryl Falzarano, and Alicia Evangelista for their help with the manuscript. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH).
Excellent review on timely and newly emerging infectious diseases.
References
- Agnihothram S, Gopal R, Yount B_et al_ (2013) Platform strategies for rapid response against emerging coronaviruses: MERS‐CoV serologic and antigenic relationships in vaccine design. J Infect Dis [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarrak A, Stephens G, Hewson R & Memish Z (2012) Recovery from severe novel coronavirus infection. Saudi Med J 33: 1265–1269. [PubMed] [Google Scholar]
- Al‐Daghri N, Al‐Attas O, Alokail M, Alkharfy K, Yousef M, Sabico S & Chrousos G (2011) Diabetes mellitus type 2 and other chronic non‐communicable diseases in the central region, Saudi Arabia (Riyadh cohort 2): a decade of an epidemic. BMC Med 9: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazán F, Dediego M, Sola I_et al_ (2013) Engineering a replication‐competent, propagation‐defective Middle East Respiratory Syndrome coronavirus as a vaccine candidate. mBio 4: e00650–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annan A, Baldwin H, Corman V_et al_ (2013) Human betacoronavirus 2c EMC/2012‐related viruses in bats, Ghana and Europe. Emerg Infect Dis 19: 456–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony SJ, Ojeda‐Flores R, Rico‐Chavez O_et al_ (2013) Coronaviruses in bats from Mexico. J Gen Virol 94: 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A, Al‐Tawfiq J, Al‐Rabeeah A_et al_ (2013a) Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East Respiratory Syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis 13: 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A, McGeer A, Perl T_et al_ (2013b) Hospital outbreak of Middle East Respiratory Syndrome coronavirus. N Engl J Med 369: 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham A, Chand M, Brown C_et al_ (2012) Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill 17: 20290. [PubMed] [Google Scholar]
- Birgit M, Martin M, Patrick CB, Hans WD & Jindrich C (2005) Ribavirin and interferon‐β synergistically inhibit SARS‐associated coronavirus replication in animal and human cell lines. Biochem Biophys Res Commun 326: 905–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmbhatt M & Dutta A (2008) On SARS Type Economic Effects During Infectious Disease Outbreaks. The World Bank. [Google Scholar]
- Breban R, Riou J & Fontanet A (2013) Interhuman transmissibility of Middle East Respiratory Syndrome coronavirus: estimation of pandemic risk. Lancet 382: 694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz U, Müller M, Nitsche A_et al_ (2013) Contact investigation of a case of human novel coronavirus infection treated in a German hospital, October–November 2012. Euro Surveill 18: 20406. [PubMed] [Google Scholar]
- Butler D (2013) Tensions linger over discovery of coronavirus. Nat News (online). [Google Scholar]
- Cauchemez S, Fraser C, Van Kerkhove M_et al_ (2013) Middle East Respiratory Syndrome coronavirus: quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infect Dis 14: 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention C (2013) Updated information on the epidemiology of Middle East Respiratory Syndrome coronavirus (MERS‐CoV) infection and guidance for the public, clinicians, and public health authorities, 2012–2013. MMWR Morb Mortal Wkly Rep 62: 793–796. [PMC free article] [PubMed] [Google Scholar]
- Chan K, Peiris J, Lam S, Poon L, Yuen K & Seto W (2011) The effects of temperature and relative humidity on the viability of the SARS coronavirus. Adv Virol 2011: 734690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Chan K‐H, Kao R_et al_ (2013a) Broad‐spectrum antivirals for the emerging Middle East Respiratory Syndrome coronavirus. J Infect 67: 606–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Chan K‐H, Choi G_et al_ (2013b) Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J Infect Dis 207: 1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R, Chan M, Agnihothram S_et al_ (2013c) Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J Virol 87: 6604–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R‐C (2006) Treatment of severe acute respiratory syndrome with glucosteroids*. Chest 129: 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Chan K, Jiang Y_et al_ (2004) In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J Clin Virol 31: 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhang L, Qin C_et al_ (2005) Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J Virol 79: 2678–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Rajashankar K, Yang Y_et al_ (2013) Crystal structure of the receptor‐binding domain from newly emerged Middle East Respiratory Syndrome coronavirus. J Virol 87: 10777–10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Wong D, Tong L_et al_ (2004) Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet 363: 1699–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng VC, Chan J, To K & Yuen K (2013) Clinical management and infection control of SARS: lessons learned. Antiviral Res 100: 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C, Matthews K, Goicochea L & Frieman M (2013) Wild type and innate immune deficient mice are not susceptible to the Middle East Respiratory Syndrome coronavirus. J Gen Virol 95: 408–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V, Eckerle I, Bleicker T_et al_ (2012a) Detection of a novel human coronavirus by real‐time reverse‐transcription polymerase chain reaction. Euro Surveill 17: pii: 20285. [DOI] [PubMed] [Google Scholar]
- Corman V, Müller M, Costabel U_et al_ (2012b) Assays for laboratory confirmation of novel human coronavirus (hCoV‐EMC) infections. Euro Surveill 17: 20334. [DOI] [PubMed] [Google Scholar]
- Cotten M, Lam T, Watson S_et al_ (2013a) Full‐genome deep sequencing and phylogenetic analysis of novel human betacoronavirus. Emerg Infect Dis 19: 736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M, Watson S, Kellam P_et al_ (2013b) Transmission and evolution of the Middle East Respiratory Syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet 382: 1993–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedictis P, Marciano S, Scaravelli D_et al_ (2014) Alpha and lineage C betaCoV infections in Italian bats. Virus Genes 48: 366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R (2012) Family – Coronaviridae Virus Taxonomy (King AMQ, Adams MJ, Carstens EB. & Lefkowitz EJ, eds), pp. 806–828. Elsevier, San Diego, CA. [Google Scholar]
- de Sousa R, Reusken C & Koopmans M (2013) MERS coronavirus: data gaps for laboratory preparedness. J Clin Virol 59: 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Prescott J, Baseler L_et al_ (2013a) The Middle East Respiratory Syndrome coronavirus (MERS‐CoV) does not replicate in Syrian hamsters. PLoS ONE 8: e69127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Rasmussen A, Falzarano D_et al_ (2013b) Middle East Respiratory Syndrome coronavirus (MERS‐CoV) causes transient lower respiratory tract infection in rhesus macaques. P Natl Acad Sci USA 110: 16598–16603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming D, Sheahan T, Heise M_et al_ (2006) Vaccine efficacy in senescent mice challenged with recombinant SARS‐CoV bearing epidemic and zoonotic spike variants. PLoS Med 3: e30525/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M, Zachariah M & Harris E (2002) Mycophenolic acid inhibits dengue virus infection by preventing replication of viral RNA. Virology 304: 211–221. [DOI] [PubMed] [Google Scholar]
- Dijkman R, Jebbink MF, Koekkoek SM_et al_ (2013) Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J Virol 87: 6081–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C, Günther S, Preiser W_et al_ (2003) Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348: 1967–1976. [DOI] [PubMed] [Google Scholar]
- Drosten C, Seilmaier M, Corman V_et al_ (2013) Clinical features and virological analysis of a case of Middle East Respiratory Syndrome coronavirus infection. Lancet Infect Dis 13: 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, He Y, Zhou Y, Liu S, Zheng B‐J & Jiang S (2009) The spike protein of SARS‐CoV–a target for vaccine and therapeutic development. Nat Rev Microbiol 7: 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Ma C & Jiang S (2013a) Antibodies induced by receptor‐binding domain in spike protein of SARS‐CoV do not cross‐neutralize the novel human coronavirus hCoV‐EMC. J Infect 67: 348–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Kou Z, Ma C_et al_ (2013b) A truncated receptor‐binding domain of MERS‐CoV spike protein potently inhibits MERS‐CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLoS ONE 8: e81587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Zhao G, Kou Z_et al_ (2013c) Identification of a receptor‐binding domain in the s protein of the novel human coronavirus Middle East Respiratory Syndrome coronavirus as an essential target for vaccine development. J Virol 87: 9939–9942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel M, Hoffmann T, Wagner L_et al_ (2003) The crystal structure of dipeptidyl peptidase IV (CD26) reveals its functional regulation and enzymatic mechanism. P Natl Acad Sci USA 100: 5063–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D, de Wit E, Martellaro C, Callison J, Munster V & Feldmann H (2013a) Inhibition of novel β coronavirus replication by a combination of interferon‐α2b and ribavirin. Sci Rep 3: 1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D, de Wit E, Rasmussen A_et al_ (2013b) Treatment with interferon‐α2b and ribavirin improves outcome in MERS‐CoV‐infected rhesus macaques. Nat Med 19: 1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler D (2008) Influenza virus samples, international law, and global health diplomacy. Emerg Infect Dis 14: 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field H, Young P, Yob J, Mills J, Hall L & Mackenzie J (2001) The natural history of Hendra and Nipah viruses. Microbes Infect 3: 307–314. [DOI] [PubMed] [Google Scholar]
- Fouchier RA, Hartwig NG, Bestebroer TM, Niemeyer B, de Jong JC, Simon JH & Osterhaus AD (2004) A previously undescribed coronavirus associated with respiratory disease in humans. P Natl Acad Sci USA 101: 6212–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Sastre A & Biron C (2006) Type 1 interferons and the virus‐host relationship: a lesson in détente. Science (New York, NY) 312: 879–882. [DOI] [PubMed] [Google Scholar]
- Gierer S, Bertram S, Kaup F_et al_ (2013) The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol 87: 5502–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R, Becker M, Eckerle L, Bolles M, Denison M & Baric R (2012) A live, impaired‐fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat Med 18: 1820–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RL, Donaldson EF & Baric RS (2013) A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol 11: 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guberina H, Witzke O, Timm J, Dittmer U, Müller M, Drosten C & Bonin F (2013) A patient with severe respiratory failure caused by novel human coronavirus. Infection 42: 203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guery B, Poissy J, el Mansouf L_et al_ (2013) Clinical features and viral diagnosis of two cases of infection with Middle East Respiratory Syndrome coronavirus: a report of nosocomial transmission. Lancet 381: 2265–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulland A (2013a) Two cases of novel coronavirus are confirmed in France. BMJ (Clinical Research ed.) 346: f3114. [DOI] [PubMed] [Google Scholar]
- Gulland A (2013b) Novel coronavirus spreads to Tunisia. BMJ (Clinical Research ed.) 346: f3372. [DOI] [PubMed] [Google Scholar]
- Haagmans BL, Al Dhahiry SHS, Reusken CBEM_et al_ (2013) Middle East Respiratory Syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis 14: 140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D & Procknow J (1966) A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med 121: 190–193. [DOI] [PubMed] [Google Scholar]
- Hart B, Dyall J, Postnikova E_et al_ (2013) Interferon‐beta and mycophenolic acid are potent inhibitors of Middle East Respiratory Syndrome coronavirus in cell‐based assays. J Gen Virol 95: 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health Protection Agency UKNCIt (2013) Evidence of person‐to‐person transmission within a family cluster of novel coronavirus infections, United Kingdom, February 2013. Euro Surveill 18: 20427. [DOI] [PubMed] [Google Scholar]
- Hemida M, Perera RA, Wang P_et al_ (2013) Middle East Respiratory Syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Euro Surveill 18: e20659. [DOI] [PubMed] [Google Scholar]
- Hijawi B, Abdallat M, Sayaydeh A_et al_ (2013) Novel coronavirus infections in Jordan, April 2012: epidemiological findings from a retrospective investigation. East Mediterr Health J 19(suppl 1): 8. [PubMed] [Google Scholar]
- Hiramatsu H, Kyono K, Higashiyama Y_et al_ (2003) The structure and function of human dipeptidyl peptidase IV, possessing a unique eight‐bladed beta‐propeller fold. Biochem Biophys Res Commun 302: 849–854. [DOI] [PubMed] [Google Scholar]
- Hui D & Sung J (2004) Treatment of severe acute respiratory syndrome. Chest 126: 670–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithete NL, Stoffberg S, Corman VM_et al_ (2013) Close relative of human Middle East Respiratory Syndrome coronavirus in bat, South Africa. Emerg Infect Dis 19: 1697–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josset L, Menachery V, Gralinski L_et al_ (2013) Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E, Jónsdóttir H, Muth D_et al_ (2013) Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. mBio 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky‐Bromberg S, Martínez‐Sobrido L, Frieman M, Baric R & Palese P (2007) Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81: 548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T, Fouchier R, Schutten M_et al_ (2003) Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362: 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeir A‐M, Durinx C, Scharpé S & De Meester I (2003) Dipeptidyl‐peptidase IV from bench to bedside: an update on structural properties, functions, and clinical aspects of the enzyme DPP IV. Crit Rev Clin Lab Sci 40: 209–294. [DOI] [PubMed] [Google Scholar]
- Lau S, Li K, Tsang A_et al_ (2013) Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East Respiratory Syndrome coronavirus. J Virol 87: 8638–8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelli D, Papetti A, Sabelli C, Rosti E, Moreno A & Boniotti M (2013) Detection of coronaviruses in bats of various species in Italy. Viruses 5: 2679–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow M (2013) Correlating cell line studies with tissue distribution of DPP4/TMPRSS2 and human biological samples may better define the viral tropism of MERS‐CoV. J Infect Dis 208: 1350–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M, Cohen T, Cooper B_et al_ (2003) Transmission dynamics and control of severe acute respiratory syndrome. Science (New York, NY) 300: 1966–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutfy M, Blatt L, Siminovitch K_et al_ (2003) Interferon alfacon‐1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA 290: 3222–3228. [DOI] [PubMed] [Google Scholar]
- Lu G, Hu Y, Wang Q_et al_ (2013) Molecular basis of binding between novel human coronavirus MERS‐CoV and its receptor CD26. Nature 500: 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Li P, Zhang L, Guan Y, Butt K & Wong K (2003) Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302: 276–278. [DOI] [PubMed] [Google Scholar]
- Mahalingam S, Herrero L, Playford E_et al_ (2012) Hendra virus: an emerging paramyxovirus in Australia. Lancet Infect Dis 12: 799–807. [DOI] [PubMed] [Google Scholar]
- Mailles A, Blanckaert K, Chaud P_et al_ (2013) First cases of Middle East Respiratory Syndrome Coronavirus (MERS‐CoV) infections in France, investigations and implications for the prevention of human‐to‐human transmission, France, May 2013. Euro Surveill 18: e20502. [PubMed] [Google Scholar]
- Masters P (2006) The molecular biology of coronaviruses. Adv Virus Res 66: 193–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K, Dees JH, Becker WB, Kapikian AZ & Chanock RM (1967) Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. P Natl Acad Sci USA 57: 933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z, Zumla A & Assiri A (2013a) Middle East Respiratory Syndrome coronavirus infections in health care workers. N Engl J Med 369: 884–886. [DOI] [PubMed] [Google Scholar]
- Memish Z, Zumla A, Al‐Hakeem R, Al‐Rabeeah A & Stephens G (2013b) Family cluster of Middle East Respiratory Syndrome coronavirus infections. N Engl J Med 368: 2487–2494. [DOI] [PubMed] [Google Scholar]
- Memish Z, Mishra N, Olival K_et al_ (2013c) Middle East Respiratory Syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis 19: e131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BMM, Corman VM, Reusken CBEM, Ritz D & Godeke GD (2014) Antibodies against MERS Coronavirus in Dromedary Camels, United Arab Emirates, 2003 and 2013. Emerg Infect Dis 20: e131746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H, Raj V, van Kuppeveld F, Rottier P, Haagmans B & Bosch B (2013) The receptor binding domain of the new Middle East Respiratory Syndrome coronavirus maps to a 231‐residue region in the spike protein that efficiently elicits neutralizing antibodies. J Virol 87: 9379–9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukasa‐Mugerwa E 1981. The Camel (Camelus dromedarius): A Bibliographical Review. International Livestock Research Institute (ILCA), Addis Ababa. [Google Scholar]
- Müller M, Raj V, Muth D_et al_ (2012) Human coronavirus EMC does not require the SARS‐coronavirus receptor and maintains broad replicative capability in mammalian cell lines. mBio 3: e00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V, de Wit E & Feldmann H (2013) Pneumonia from human coronavirus in a macaque model. N Engl J Med 368: 1560–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel L (2013) Discrepancies in data reporting for rabies, Africa. Emerg Infect Dis 19: 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls J, Poon L, Lee K_et al_ (2003) Lung pathology of fatal severe acute respiratory syndrome. Lancet 361: 1773–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer D, Zillinger T, Muth D_et al_ (2013) Middle East Respiratory Syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol 87: 12489–12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Wakasugi N, Kirikae T_et al_ (2008) Risk factors for SARS infection within hospitals in Hanoi, Vietnam. Jpn J Infect Dis 61: 388–390. [PubMed] [Google Scholar]
- Ofner‐Agostini M, Gravel D, McDonald L_et al_ (2006) Cluster of cases of severe acute respiratory syndrome among Toronto healthcare workers after implementation of infection control precautions: a case series. Infect Control Hosp Epidemiol 27: 473–478. [DOI] [PubMed] [Google Scholar]
- Ohnuma K, Haagmans B, Hatano R_et al_ (2013) Inhibition of Middle East Respiratory Syndrome coronavirus (MERS‐CoV) infection by anti‐CD26 monoclonal antibody. J Virol 87: 13892–13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omrani A, Matin M, Haddad Q, Al‐Nakhli D, Memish Z & Albarrak A (2013) A family cluster of Middle East Respiratory Syndrome coronavirus infections related to a likely unrecognized asymptomatic or mild case. Int J Infect Dis 17: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J, Chu C, Cheng VC_et al_ (2003) Clinical progression and viral load in a community outbreak of coronavirus‐associated SARS pneumonia: a prospective study. Lancet 361: 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penttinen P, Kaasik‐Aaslav K, Friaux A_et al_ (2013) Taking stock of the first 133 MERS coronavirus cases globally – is the epidemic changing?. Euro Surveill 18: e20596. [DOI] [PubMed] [Google Scholar]
- Perera R, Wang P, Gomaa M_et al_ (2013) Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill 18: e20574. [DOI] [PubMed] [Google Scholar]
- Perlman S & Netland J (2009) Coronaviruses post‐SARS: update on replication and pathogenesis. Nat Rev Microbiol 7: 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzelli S, Azzi A, Santini M_et al_ (2013) Investigation of an imported case of Middle East Respiratory Syndrome coronavirus (MERS‐CoV) infection in Florence, Italy, May to June 2013. Euro Surveill 18: e20564. [DOI] [PubMed] [Google Scholar]
- Raj V, Mou H, Smits S_et al_ (2013) Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus‐EMC. Nature 495: 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj VS, Smits SL, Provacia LB_et al_ (2014) Adenosine deaminase acts as a natural antagonist for dipeptidyl peptidase 4 mediated entry of the Middle East Respiratory Syndrome coronavirus. J Virol 88: 1834–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall R & Goodbourn S (2008) Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol 89: 1–47. [DOI] [PubMed] [Google Scholar]
- Reusken C, Ababneh M, Raj VS_et al_ (2013a) Middle East Respiratory Syndrome coronavirus (MERS‐CoV) serology in major livestock species in an affected region in Jordan, June to September 2013. Euro Surveill 18: e20662. [DOI] [PubMed] [Google Scholar]
- Reusken C, Haagmans B, Müller M_et al_ (2013b) Middle East Respiratory Syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis 13: 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SARS Epidemiology Working Group (2003) Consensus Document on the Epidemiology of Severe Acute Respiratory Syndrome. World Health Organization, Geneva. [Google Scholar]
- Sawicki S, Sawicki D & Siddell S (2007) A contemporary view of coronavirus transcription. J Virol 81: 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobey T, Yount B, Sims A_et al_ (2013) Reverse genetics with a full‐length infectious cDNA of the Middle East Respiratory Syndrome coronavirus. P Natl Acad Sci USA 110: 16157–16162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizun J, Yu M & Talbot P (2000) Survival of human coronaviruses 229E and OC43 in suspension and after drying onsurfaces: a possible source ofhospital‐acquired infections. J Hosp Infect 46: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Blanc H, Vignuzzi M & Denison M (2013) Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog 9: e1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Fux R, Provacia L_et al_ (2013) Middle East Respiratory Syndrome coronavirus spike protein delivered by modified vaccinia virus Ankara efficiently induces virus‐neutralizing antibodies. J Virol 87: 11950–11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockman L, Bellamy R & Garner P (2006) SARS: systematic review of treatment effects. PLoS Med 3: e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda T, Henry B, Shulman L_et al_ (2004) Public health measures to control the spread of the severe acute respiratory syndrome during the outbreak in Toronto. N Engl J Med 350: 2352–2361. [DOI] [PubMed] [Google Scholar]
- Tan E, Ooi E, Lin C‐Y, Tan H, Ling A, Lim B & Stanton L (2004) Inhibition of SARS coronavirus infection in vitro with clinically approved antiviral drugs. Emerg Infect Dis 10: 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K & Mossman K (2013) Recent advances in understanding viral evasion of type I interferon. Immunology 138: 190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackray L & Holmes K (2004) Amino acid substitutions and an insertion in the spike glycoprotein extend the host range of the murine coronavirus MHV‐A59. Virology 324: 510–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The WHO MERS‐CoV Research Group (2013) State of knowledge and data gaps of Middle East Respiratory Syndrome Coronavirus (MERS‐CoV) in humans. PLoS Curr 5, ecurrents.outbreaks.0bf719e352e7478f8ad85fa30127ddb8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C‐T, Sbrana E, Iwata‐Yoshikawa N_et al_ (2012) Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS ONE 7: e35421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusell S, Schittone S & Holmes K (2007) Mutational analysis of aminopeptidase N, a receptor for several group 1 coronaviruses, identifies key determinants of viral host range. J Virol 81: 1261–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twu S‐J, Chen T‐J, Chen C‐J_et al_ (2003) Control measures for severe acute respiratory syndrome (SARS) in Taiwan. Emerg Infect Dis 9: 718–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boheemen S, de Graaf M, Lauber C_et al_ (2012) Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3: e00473–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L, Pyrc K, Jebbink MF_et al_ (2004) Identification of a new human coronavirus. Nat Med 10: 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N, Bushmaker T & Munster V (2013) Stability of Middle East Respiratory Syndrome coronavirus (MERS‐CoV) under different environmental conditions. Euro Surveill 18: e20590. [DOI] [PubMed] [Google Scholar]
- Villarroel M, Hidalgo M & Jimeno A (2009) Mycophenolate mofetil: an update. Drugs Today (Barc) 45: 521–532. [DOI] [PubMed] [Google Scholar]
- Wacharapluesadee S, Sintunawa C, Kaewpom T_et al_ (2013) Group C betacoronavirus in bat guano fertilizer, Thailand. Emerg Infect Dis 19: 1349–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Shi X, Jiang L_et al_ (2013) Structure of MERS‐CoV spike receptor‐binding domain complexed with human receptor DPP4. Cell Res 23: 986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2013a) Risk Factors: Tobacco by Country. World Health Organization, Geneva. [Google Scholar]
- WHO (2013b) Alert, Response, and Capacity Building Under the International Health Regulations. World Health Organization, Geneva. [Google Scholar]
- WHO 2013c. Revised Interim Case Definition for Reporting to WHO – Middle East Respiratory Syndrome Coronavirus (MERS‐CoV). World Health Organization Global Alert and Response (GAR) Geneva. [Google Scholar]
- WHO (2014) Middle East Respiratory Syndrome Coronavirus (MERS‐CoV) Summary and Literature Update. WHO, Global Alert and Response, Geneva. [Google Scholar]
- Wilde AHD, Raj VS, Oudshoorn D_et al_ (2013) MERS‐coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon‐ treatment. J Gen Virol 94: 1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P, Lau S, Chu C‐M_et al_ (2005) Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 79: 884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PCY, Lau SKP, Li KSM, Tsang AKL & Yuen K‐Y (2012) Genetic relatedness of the novel human group C betacoronavirus to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5. Emerg Microbes Infect 1: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhang L, Geng H_et al_ (2013) The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East Respiratory Syndrome coronavirus (MERS‐CoV) are potent interferon antagonists. Protein Cell 4: 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Bao L, Deng W_et al_ (2013) An animal model of MERS produced by infection of rhesus macaques with MERS coronavirus. J Infect Dis 209: 236–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A, van Boheemen S, Bestebroer T, Osterhaus A & Fouchier R (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367: 1814–1820. [DOI] [PubMed] [Google Scholar]
- Zhao P, Cao J, Zhao L‐J_et al_ (2005) Immune responses against SARS‐coronavirus nucleocapsid protein induced by DNA vaccine. Virology 331: 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Chu H, Li C_et al_ (2013) Active MERS‐CoV replication and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielecki F, Weber M, Eickmann M_et al_ (2013) Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J Virol 87: 5300–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]