Rare mutations in SQSTM1 modify susceptibility to frontotemporal lobar degeneration (original) (raw)
Abstract
Mutations in the gene coding for Sequestosome 1 (SQSTM1) have been genetically associated with amyotrophic lateral sclerosis (ALS) and Paget disease of bone. In the present study, we analyzed the SQSTM1 coding sequence for mutations in an extended cohort of 1,808 patients with frontotemporal lobar degeneration (FTLD), ascertained within the European Early-Onset Dementia consortium. As control dataset, we sequenced 1,625 European control individuals and analyzed whole-exome sequence data of 2,274 German individuals (total n = 3,899). Association of rare SQSTM1 mutations was calculated in a meta-analysis of 4,332 FTLD and 10,240 control alleles. We identified 25 coding variants in FTLD patients of which 10 have not been described. Fifteen mutations were absent in the control individuals (carrier frequency <0.00026) whilst the others were rare in both patients and control individuals. When pooling all variants with a minor allele frequency <0.01, an overall frequency of 3.2 % was calculated in patients. Rare variant association analysis between patients and controls showed no difference over the whole protein, but suggested that rare mutations clustering in the UBA domain of SQSTM1 may influence disease susceptibility by doubling the risk for FTLD (RR = 2.18 [95 % CI 1.24–3.85]; corrected p value = 0.042). Detailed histopathology demonstrated that mutations in SQSTM1 associate with widespread neuronal and glial phospho-TDP-43 pathology. With this study, we provide further evidence for a putative role of rare mutations in SQSTM1 in the genetic etiology of FTLD and showed that, comparable to other FTLD/ALS genes, SQSTM1 mutations are associated with TDP-43 pathology.
Electronic supplementary material
The online version of this article (doi:10.1007/s00401-014-1298-7) contains supplementary material, which is available to authorized users.
Keywords: Sequestosome 1, SQSTM1, p62, FTLD, ALS, Rare variants
Introduction
Frontotemporal lobar degeneration (FTLD) represents a heterogeneous group of progressive neurodegenerative dementias, caused by local atrophy of frontal and/or temporal lobes. It is one of the most common forms of early-onset dementia (EOD), with the majority of FTLD patients developing disease between 45 and 65 years. About 15 % of FTLD patients present with a motor neuron disease (MND) syndrome, most commonly amyotrophic lateral sclerosis (ALS). Like FTLD, ALS is a neurodegenerative disorder in which loss of motor neurons leads to progressive weakness of the voluntary muscles. FTLD and ALS show important genetic overlap with mutations identified in the same genes, e.g., the common G4C2 repeat expansion in the chromosome 9 open reading frame 72 gene (C9orf72) and less frequently, mutations in the valosin containing protein (VCP), fused in sarcoma (FUS), TAR DNA-binding protein (TARDBP) and ubiquilin 2 (UBQLN2) genes [5, 8, 30, 31, 37].
Sequencing of the gene coding for sequestosome 1 (SQSTM1) in ALS patients identified several rare mutations [7]. Some of these mutations had been associated with Paget disease of bone (PDB [9, 14, 16]), a localized chronic bone disorder characterized by abnormalities of bone architecture and marrow fibrosis, resulting in an osteodystrophia deformans. Of interest, FTLD, PDB and inclusion body myopathy (IBM) had previously been genetically linked by mutations in VCP [13, 36, 38]. Recently, rare mutations in SQSTM1 were also reported in FTLD patients [17, 28]. The observation of rare mutations (1–3 %) in both FTLD and ALS patients suggested an involvement of the protein SQSTM1, also known as p62, in these pathologies possibly through a common disease pathomechanism. The p62 protein is a stress-responsive ubiquitin-binding protein shown to have a role in degradation of polyubiquitinated proteins via the proteasome pathway or autophagic processes [26]. It is present in neuronal and glial ubiquitin-positive inclusions in different tauopathies and synucleinopathies, including Alzheimer disease, FTLD, dementia with Lewy bodies, Parkinson disease, Huntington disease and multiple system atrophy [15, 20, 23]. Also in FTLD, with or without ALS, p62 co-localizes with TDP-43 and FUS in brain and/or spinal cord [2, 6, 32]. Recently, p62 positive but TDP-43 negative immunoreactivity, extending to the pyramidal cell layer of the hippocampus, basal ganglia and cerebellum, has been recognized as a distinctive feature of _C9orf72_-associated FTLD and ALS [1, 34]. Aggregating dipeptide repeats (DPRs), translated from the expanded GGGGCC repeat, were identified as the main component of these inclusions [21, 22]. In the present study, we aimed at determining the genetic contribution of mutations in SQSTM1 to the etiology of FTLD. Hereto, we analyzed a large study population of 1,808 FTLD patients and compared mutation data to a set of 3,899 European control individuals, as well as 395 European ALS patients.
Materials and methods
The patient and control cohorts under investigation were ascertained through the European Early-Onset Dementia (EU EOD) consortium (Supplementary table 1) [35]. For the present study, DNA and medical/demographic information on 1,808 FTLD patients, originating from Belgium, Italy, Germany, Spain, Portugal, Sweden, Czech Republic, Bulgaria and Austria, was contributed by members of the consortium. From this patient cohort, 1,706 patients were clinically diagnosed with FTLD and 102 with concomitant FTLD and ALS (FTLD-ALS) (for the remainder of the manuscript, the FTLD group will refer to the 1,706 FTLD patients plus the 102 FTLD-ALS patients). The research question of this study was whether genetic variations in SQSTM1 affect FTLD risk. Yet, because of the close relationship with FTLD, the contributed patient cohorts also included a number of ALS patients (n = 395). Patients were evaluated and diagnosed with FTLD according to the Lund and Manchester group criteria [25] and for ALS according to the revised El Escorial criteria [3]. Clinical diagnoses of behavioral variant frontotemporal dementia (bvFTD) were based on the international consensus criteria by Rascovsky et al. [27] and of progressive supranuclear palsy (PSP) on the National Institute of Neurological Disorders and the Society for PSP criteria [18]. Neuropathological examination was performed in 105 autopsied patients, including 67 with FTLD-TDP, 2 FTLD-UPS, 3 FTLD-tau, 1 FTLD-ni, 4 FTLD unspecified, 21 ALS-TDP and 7 ALS unspecified. Genetic mutation profiling of FTLD- and ALS-associated genes was performed for C9orf72 (n = 2,055), GRN (n = 1,024), MAPT (n = 854), VCP (n = 159), CHMP2B (n = 153), TARDBP (n = 272) and FUS (n = 184) and revealed 150 C9orf72 repeat expansion mutations, 24 GRN, 5 MAPT, 2 VCP, 1 CHMP2B, and 2 FUS mutations. A positive family history was defined for index patients with first- or second-degree relatives with symptoms of dementia or MND. Patients were classified as sporadic when no other affected family members were reported. Patients from whom no information on family history could be obtained were classified as ‘family history undocumented’. As control group, we sequenced 1,625 age- and origin-matched Western-European individuals with no personal or family history of neurodegenerative or psychiatric diseases and a Mini Mental State Examination (MMSE) score >26. We further analyzed whole-exome sequencing (WES) data of 2,274 German non-demented individuals [10, 40]. Together, 3,899 control persons were investigated for coding variants in SQSTM1.
For all participants, informed consent for participation in the genetic studies was obtained according to sampling protocols that were approved by the Ethics Committee of the respective hospitals. The protocols for the genetic studies were approved by the Ethics Committee of the University of Antwerp, Belgium.
Sample quality control
30 µl at 20 ng/µl of genomic DNA (gDNA) was requested, and concentration and purity were checked spectrophotometrically using the Trinean DropSense96 UV/VIS droplet reader for all consortium samples. Gender and DNA fingerprint were determined for all DNA samples using an in-house developed multiplex polymerase-chain reaction (PCR) genomic DNA Fingerprint panel comprising 13 short tandem repeat (STR) markers distributed over multiple autosomal loci—D20S480, D22S1174, D3S1287, D3S1744, D3S1764, D7S672, D7S2426, D8S1746, D14S1005, D20S866, D10S1237, D20S912, D6S965—and two sex chromosome markers—DXS1187, chrom Y: 2655362–2655672—to enable fast and accurate sample identification and gender determination in a single PCR. After selective amplification of 20 ng gDNA under empirically defined reaction conditions, amplification products were size separated on an ABI 3730 automatic sequencer (Applied Biosystems) using GeneScan-600 LIZ (Applied Biosystems) as internal size standard and genotypes were assigned using in-house developed TracI genotyping software (http://www.vibgeneticservicefacility.be). Duplicate samples, gender mismatches and failed samples due to low DNA quality or contamination were excluded, resulting in the final study population of 1,808 FTLD, 395 ALS, and 1,625 control individuals.
SQSTM1 sequencing
For the 1,808 FTLD, 395 ALS and 1,625 control individuals, the 8 coding exons and intron–exon boundaries were amplified by PCR of gDNA, followed by Sanger sequencing (NM_003900.4, primers available on request). Sequences were analyzed using the software package NovoSNP [39] and confirmed by visual inspection of the DNA sequence traces. Available WES data on an additional 2,274 German controls were checked for SQSTM1 coding variants [10, 40].
Genetic variations were further verified in the Database of Single-Nucleotide Polymorphisms (dbSNP Build ID 137; URL http://www.ncbi.nlm.nih.gov/SNP/); the exome variant server (EVS) of the National Heart, Lung, and Blood Institute GO Exome Sequencing Project (Seattle, WA, USA; URL http://evs.gs.washington.edu/EVS/), and the 1,000 Genomes project (URL http://www.1000genomes.org/). The effects of rare coding variations in SQSTM1 on protein structure and function were predicted using PMUT (http://mmb2.pcb.ub.es:8080/PMut/), SNPs&Go (http://snps.uib.es/snps-and-go//snps-and-go.html) and Provean/SIFT (http://sift.jcvi.org/www/SIFT_enst_submit.html).
Statistical analysis
We determined rare variants as genetic variants with an MAF <0.01 and performed a rare variant burden analysis following a stepwise approach. Alleles of all rare variants were first collapsed across the entire protein. Subsequently, calculations were repeated for rare variants associated with functional domains only. SQSTM1 functional domains were determined according to [11] (Fig. 1b). Finally, rare variant data were calculated for each of the seven protein domains independently.
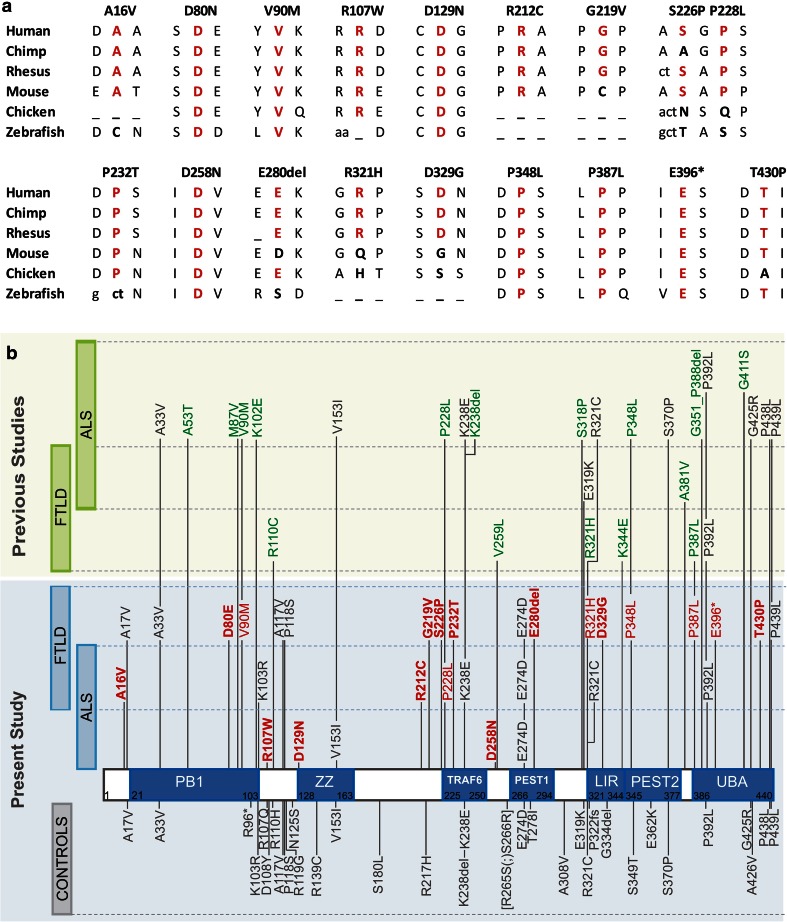
Fig. 1.
SQSTM1 mutations identified in FTLD and ALS patient cohorts ascertained with the European EOD consortium. a Sequence alignment for patient-specific mutations showing evolutionary conservation across species. b In the blue panel, SQSTM1 mutations identified in the present study in patients (top) and control individuals (bottom) are presented on the primary structure if the p62 protein indicating known functional domains. Mutations absent from tested and published controls are in red. Mutations not previously associated with FTLD, ALS, or PDB are in red and bold. In the green panel, SQSTM1 mutations reported in previous studies are given [7, 11, 17, 28, 29, 33]. Mutations absent from tested and published controls are in green. Functional domains according to [11]: PB1 = Phox and Bem1p domain; ZZ = zinc finger motif; TRAF6 = TNF receptor–associated factor 6; LIR = LC3 interaction region; PEST1 = proline (P), glutamic acid (E), serine (S), and threonine (T) domain 1; PEST2 = PEST domain 2; UBA = ubiquitin-associated domain
Calculations were performed in our source dataset and in a meta-analysis considering all published datasets generated by full exonic sequencing of SQSTM1 in both FTLD patient and control groups [7, 17, 28] combined with the present study. Overall allele frequencies between patients and control individuals were compared using χ 2 statistics. In the per domain analyses p values were corrected for seven tests, corresponding to the seven functional domains. Rare variant association analysis was limited to the FTLD cohort due to the insufficient power of the ALS cohort to obtain significant association.
Neuropathology of SQSTM1
Formalin fixed, paraffin-embedded tissue blocks from neocortical areas, basal ganglia, thalamus, hippocampus, brainstem and cerebellum were evaluated. In addition to Hematoxylin and Eosin staining, the following monoclonal (mouse) antibodies were used for immunohistochemistry: monoclonal anti-p62 (1:1,000, BD Transduction, Lexington KY, USA), anti-tau AT8 (pS202/pT205, 1:200, Pierce Biotechnology, Rockford, IL, USA), anti-phospho-TDP-43 (pS409/410, 1:2,000, Cosmo Bio, Tokyo, Japan), anti-α-synuclein (1:2,000, clone 5G4, Roboscreen, Leipzig, Germany; specific for disease-associated form), anti-Aβ (1:50, clone 6F/3D, Dako, Glostrup, Denmark). The DAKO EnVision© detection kit, peroxidase/DAB, rabbit/mouse (Dako, Glostrup, Denmark) was used for visualization of the antibody reactions.
Results
Information on family history was available for 76.2 % (1,378/1,808) of the FTLD patients and 83.0 % (328/395) of ALS patients (Supplementary table 1). In the FTLD group, 35.2 % (636/1,808) had a positive family history while 41.2 % (744/1,808) were considered sporadic patients. In the ALS group, 14.9 % (59/395) had a family history of disease while 68.1 % (269/395) were sporadic patients. Onset age distribution was 63 ± 9.9 years in the FTLD group and 58 ± 13.7 years in the ALS group compared to an age at inclusion of 66 ± 12.4 years in the control group (Supplementary table 1).
Mutation screen of SQSTM1 in FTLD
In total we identified 25 rare, heterozygous variations that affected the coding region of SQSTM1, including 23 missense mutations, 1 nonsense mutation and one small in-frame deletion (Tables 1, 2, Supplementary table 2; Fig. 1). Fifteen mutations were present in 16 patients and were absent from 3,899 control individuals (Table 1). The majority of the 15 mutations involved highly conserved amino acid residues in SQSTM1 and were predicted to be pathogenic (Fig. 1a, Supplementary table 3). Among the 16 mutation carriers, 7 had a positive family history of disease and 2 patients had concomitant ALS (Table 1). Nine patients carried a novel mutation not previously associated with FTLD, ALS or PDB (p.Ala16Val, p.Asp80Glu, p.Arg212Cys, p.Gly219Val, p.Ser226Pro, p.Pro232Thr, p.Glu280del, p.Asp329Gly and p.Thr430Pro) (Fig. 1b). The remaining six mutations had been reported in ALS (p.Val90Met, p.Pro228Leu and p.Pro348Leu) [7, 28, 29], FTLD (p.Arg321His, p.Pro387Leu) [17] or PDB (p.Pro387Leu, p.(Glu396*)) [28] (Fig. 1b). Except for p.Ala16Val, p.Arg212Cys and p.Gly219Val, all other mutations were located in a predicted functional protein domain of SQSTM1/p62 (Fig. 1b). In addition to the patient-only mutations, we identified another 10 missense mutations (p.Ala17Val, p.Ala33Val, p.Lys103Arg, p.Ala117Val, p.Pro118Ser, p.Lys238Glu, p.Glu274Asp, p.Arg321Cys, p.Pro392Leu, p.Pro439Leu) that were also present in control individuals at low frequency (Table 2). Except for p.Glu274Asp (MAF of 0.024), all were present in less than 1 % of control individuals. p.Ala17Val was a novel variation not previously reported in FTLD, ALS and/or PDB patients. We identified another 21 variants present in control individuals only (Table 3). When considering all rare variants (MAF <0.01) observed in FTLD patients an overall frequency was calculated of 3.2 % (58/1,808).
Table 1.
SQSTM1 mutations present only in patients and associated clinical phenotypes
| Mutation | Functional domain | Origin | Gender | Clinical diagnosis | Sub-diagnosisb | Family history | Age at onset (years) | Age at death (years) |
|---|---|---|---|---|---|---|---|---|
| FTLD | ||||||||
| p.Ala16Vala | Italian | M | FTLD-ALS | U | 71 | 74 | ||
| p.Asp80Glua | PB1 | Italian | F | FTLD | bvFTD | F | 71 | 85 |
| p.Val90Met | PB1 | Portuguese | F | FTLD | bvFTD | U | 41 | |
| p.Arg212Cysa | Austrian | M | FTLD-ALS | F | 63 | 66 | ||
| p.Gly219Vala | Portuguese | M | FTLD | bvFTD | F | 52 | ||
| p.Ser226Proa | TRAF6 | Spanish | M | FTLD | bvFTD | S | 61 | |
| p.Pro228Leu | TRAF6 | German | M | FTLD | bvFTD | S | 57 | |
| p.Pro232Thra | TRAF6 | Portuguese | F | FTLDc | bvFTD | F | 55 | 68 |
| p.Glu280dela | PEST1 | Italian | F | FTLD | PSP | F | 73 | |
| p.Arg321His | LIR | Italian | F | FTLD | U | 68 | ||
| p.Asp329Glya | LIR | Spanish | M | FTLD | bvFTD | U | 78 | 84 |
| p.Pro348Leu | PEST2 | Italian | M | FTLD | PNFA | F | 74 | |
| p.Pro387Leu | UBA | Italian | F | FTLD | PNFA | F | 65 | |
| p.Pro387Leu | UBA | Italian | M | FTLD | bvFTD | S | 66 | |
| p.(Glu396*) | UBA | Czech | M | FTLD | bvFTD | S | 43 | 47 |
| p.Thr430Proa | UBA | Portuguese | M | FTLD | bvFTD | S | 58 | 63 |
| ALS | ||||||||
| p.Arg107Trpa | Spanish | F | ALS | MND | S | 58 | 62 | |
| p.Asp129Asna | ZZ | Flemish | M | ALS | S | 62 | ||
| p.Asp258Asna | German | F | ALS | F | 52 | 62 |
Table 2.
SQSTM1 mutations present in patients and control individuals
| Mutation | Functional domain | FTLD n = 1,808 | ALS n = 395 | Controls n = 3,899 |
|---|---|---|---|---|
| p.Ala17Vala | 1 | 1 | ||
| p.Ala33Val | PB1 | 1 | 1 | |
| p.Lys103Arg | PB1 | 1 | 2 | |
| p.Ala117Val | 1 | 3 | ||
| p.Pro118Ser | 1 | 2 | ||
| p.Val153Ile | ZZ | 1 | 3 | |
| p.Lys238Glu | TRAF6 | 17 | 6 | 14 |
| p.Glu274Asp | PEST1 | 109 | 22 | 79 |
| p.Arg321Cys | LIR | 3 | 1 | 1 |
| p.Pro392Leu | UBA | 15 | 3 | 11 |
| p.Pro439Leu | UBA | 2 | 2 |
Table 3.
Descriptives of the SQSTM1 variants found in control individuals only
Mutation screen of SQSTM1 in ALS
For comparison with our FTLD findings, we analyzed the 395 ALS patients that were concurrently ascertained within the EU EOD consortium (Fig. 1). We identified three novel missense mutations present only in the ALS patients (p.Arg107Trp, p.Asp129Asn and p.Asp258Asn) (Table 1) and five other missense mutations (p.Val153Ile, p.Lys238Glu, p.Glu274Asp, p.Arg321Cys and p.Pro392Leu) also present in control individuals at low frequency (Table 2, Supplementary table 2).
Association of rare SQSTM1 variants
We performed a burden analysis collapsing all rare variants with an MAF <0.01 across the whole protein. The same frequency of rare alleles was calculated for FTLD patients 0.016 (58/3,616 rare variant alleles) as for control individuals 0.016 (125/7,798; p value = 0.997, n.s.). We repeated the same analysis for rare variants present in functional domains only, resulting in a shift in allele frequencies of 0.014 (52/3,616) in FTLD versus 0.011 (84/7,798) controls, although not reaching statistical significance (p = 0.098). To increase genetic power, we considered all published datasets that were generated by full exonic sequencing of SQSTM1 in both FTLD patient and control groups [7, 17, 28] and included the data in a meta-analysis with the present study comprising mutant allele frequencies in 4,332 FTLD and 10,240 control alleles. Burden analysis of all rare variants across the protein showed again no increase in patients (70/4,332 = 0.016 in FTLD versus 142/10,240 = 0.014 in controls; p value = 0.291, n.s.), but for the rare variants associated with functional domains a marked significant increase could be calculated of 0.014 (61/4,332 rare variant alleles) in FTLD patients versus 0.009 (97/10,240) in control individuals (relative risk overall (RR) = 1.49 [95 % CI 1.08–2.06]; p value = 0.014).
Subsequently, we investigated which of the functional domains were most contributing to the association. Rare variant data were calculated for each of the seven protein domains. In our source, FTLD cohort significant association was found with the C-terminal ubiquitin-associated (UBA) domain in patients (21/3,616 = 0.006 rare variant alleles) versus control individuals (20/7,798 = 0.003) (RR = 2.27 [95 % CI 1.23–4.20]; p value = 0.007, corrected p value = 0.049). In the meta-dataset, statistically significant clustering in the UBA domain was confirmed in FTLD (23/4,332 = 0.005 rare variant alleles) versus control individuals (25/10,240 = 0.002) (RR = 2.18 [95 % CI 1.24–3.85]; p value = 0.006, corrected p value = 0.042). In addition, also the LC3 interaction region (LIR) domain showed suggestive clustering of rare variants in FTLD (8/4,332 = 0.002) versus control individuals (5/10,240 = 0.0005) (RR = 3.79 [95 % CI 1.24–11.58]; p value = 0.012, corrected p value = 0.084).
Histopathology associated with SQSTM1 mutations
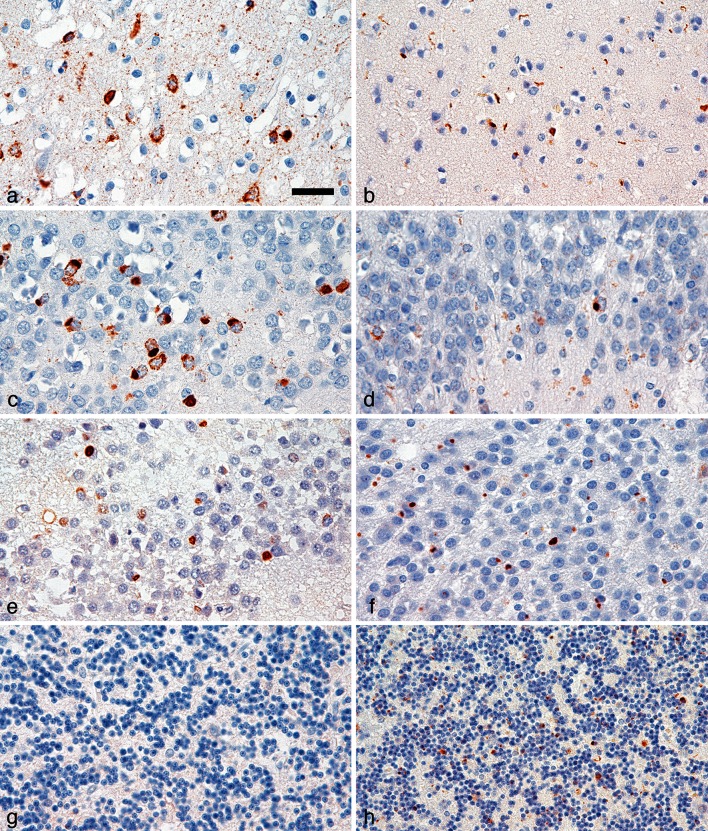
Detailed histopathology was performed on autopsy brain of two carriers of a nonsense mutation p.(Glu396*) located in the UBA domain and a missense mutation p.Arg212Cys in a patient who also carried a C9orf72 repeat expansion. Both cases showed widespread phospho-TDP-43 immunoreactive inclusions in neurons and glial cells. In the p.(Glu396*) carrier, frontal and temporal cortical areas mainly displayed neuronal cytoplasmic inclusions. Neuronal cytoplasmic inclusions were further abundant in the granule cells of the dentate gyrus. In addition, many glial phospho-TDP-43 inclusions were seen in the white matter. Although this brain suffered severe ischemic/hypoxic damage, inclusion bodies were clearly recognized as a distinctive feature. In contrast, neuropathological features of the p.Arg212Cys carrier revealed alterations reminiscent of the C9orf72 pathology, including the characteristic p62-positive but phospho-TDP-43-negative inclusions in the hippocampal pyramidal layer and cerebellar granule cells (Fig. 2).
Fig. 2.
Neuropathology observed in SQSTM1 mutation carriers. Immunostaining for phospho-TDP-43 in the temporal cortex (a, b) and in the granule cells of the dentate gyrus (c, d) in a patient with a SQSTM1 p.(Glu396*) mutation (a, c) and a second patient with a SQSTM1 p.Arg212Cys—C9orf72 double mutation (b, d). Notably, p62 immunoreactivity is less in the dentate gyrus (e, f) and lacking in the cerebellar granule cell layer (g, h) in the p.(Glu396*) case (e, g) as compared to the p.Arg212Cys patient with the additional C9orf72 repeat expansion mutation (f, h). Scale bar represents 25 µm for all
Discussion
Previous studies in ALS (total n = 895) [7, 11, 28, 33], and FTLD (total n = 358) [17, 28] estimated the frequency of SQSTM1 coding mutations at 2.42–3.28 % in ALS and 1.76–2.13 % in FTLD. Because of this relatively low prevalence, large patient and control groups are needed to obtain a reliable estimation of the mutation frequency of SQSTM1. Mutations in SQSTM1 were first described in patients with PDB [16]. Up to one-third of patients with familial PDB are explained by a SQSTM1 mutation, with p.Pro392Leu as the most commonly observed mutation [4]. A relationship between PDB and FTLD has been previously demonstrated by the identification of causal mutations in VCP in families segregating the rare syndrome of inclusion body myopathy with Paget disease of bone and frontotemporal lobar degeneration (IBMPFD) [38]. Later studies demonstrated that VCP mutations express significant clinical heterogeneity and patients can present with all, a combination of two or just one of the three core phenotypes of IBMPFD [24, 36]. Also more recently, WES identified a VCP mutation segregating in a family with ALS and subsequent screenings in ALS patients identified additional VCP mutations [12].
In this study, we analyzed a European cohort of 1,808 FTLD patients for mutations in SQSTM1 and identified 25 mutations in the coding sequence of which 15 mutations in 16 patients were absent from 3,899 control persons. This resulted in a mutation frequency for SQSTM1 of 0.9 % overall and 1.1 % in familial FTLD patients. Some of the mutations are listed in dbSNP Short Genetic Variations database, the 1,000 Genomes Project database or the exome variant server (EVS), yet at very low frequencies of MAF ≤0.001 (Supplementary table 2). The SQSTM1 mutation frequency that we calculated in FTLD is lower than in previous studies. This can potentially be explained by the fact that we excluded some of the previously reported patient-only mutations from our calculations since they were present in our extended control group, though at low frequencies <0.01 % (Fig. 1b). This was the case for p.Arg321Cys, p.Ser370Pro, p.Gly425Arg [7]; p.Ala33Val, p.Pro392Leu [7, 17]; p.Lys238Glu, p.Glu319Lys [28]; p.Pro439Leu [11]; and p.Val153Ile [7, 29]. In Table 4, all _SQSTM1_ mutations are listed that have been published [7, 11, 17, 28, 29, 33] and were absent from published and tested control persons. When we considered all rare variants (MAF < 0.01), indifferent of whether or not they appeared in control individuals, we obtained a mutation frequency of 3.2 % in FTLD patients which is the same as calculated in the pooled data analysis of the present and published FTLD cohorts [17, 28]. Overall, no statistical association in patients versus control individuals was observed when pooling all rare _SQSTM1_ variants, not in our study nor in the meta-analysis with all published patient and control datasets [7, 17, 28]. Yet, when considering only domain-associated variants, a trend toward association was observed in our study and a significant increase in patients was reached in the more powerful meta-analysis dataset. Per domain analysis indicated that association was driven by the UBA domain and possibly also the LIR domain. The C-terminal UBA domain, which is primarily affected in PDB, contained significant more variants in FTLD patients when compared to control individuals in both our study and the meta-analysis (0.5 versus 0.2 %) (RRmeta = 2.18 [95 % CI 1.24–3.85]; nominal _p_ valuemeta = 0.006; corrected _p_ valuemeta = 0.042). In the LIR domain through which SQSTM1 binds the autophagy effector protein LC3, 0.2 % of FTLD patients carried a mutation versus 0.05 % of control individuals (RRmeta = 3.79 [95 % CI 1.24–11.58]; nominal _p_ valuemeta = 0.012; corrected _p_ valuemeta = 0.084). In contrast to Rubino and colleagues who sequenced up to 1,700 bp into the _SQSTM1_ promoter and detected 4 variants in 4 out of 170 FTLD patients absent from 145 control individuals (c. −1,221 G>A, c. −1,165 C>T, c. −1,153 C>G, c. −673 T>C) [28], we did not investigate the SQSTM1 promoter for rare variants.
Table 4.
SQSTM1 mutations published in previous studies
| Mutation | Functional domain | FTLD | ALS | Origin | Study |
|---|---|---|---|---|---|
| p.Ala53Thr | PB1 | 1 | Japanese | Hirano et al. [11] | |
| p.Met87Val | PB1 | 1 | French | Teyssou et al. [33] | |
| p.Val90Meta | PB1 | 1 | Japanese | Shimizu et al. [29] | |
| p.Lys102Glu | PB1 | 1 | French | Teyssou et al. [33] | |
| p.Arg110Cys | 2 | 1 | French | Le Ber et al. [17] | |
| p.Pro228Leu | TRAF6 | 1 | Euro-American | Fecto et al. [7] | |
| p.Lys238del | TRAF6 | 1 | Euro-American | Fecto et al. [7] | |
| p.Val259Leu | 1 | Italian | Rubino et al. [28] | ||
| p.Ser318Pro | 1 | Euro-American | Fecto et al. [7] | ||
| p.Arg321His | LIR | 2 | 1 | French | Le Ber et al. [17] |
| p.Lys344Glu | LIR | 1 | Italian | Rubino et al. [28] | |
| p.Pro348Leu | PEST2 | 1 | Italian | Rubino et al. [28] | |
| p.Ala381Val | 1 | 1 | French | Le Ber et al. [17] | |
| p.Pro387Leu | UBA | 1 | French | Le Ber et al. [17] | |
| p.G351_P388del | UBA | 1 | French | Teyssou et al. [33] | |
| p.Gly411Ser | UBA | 1 | Euro-American | Fecto et al. [7] |
The pathogenicity of SQSTM1 mutations, even those that were absent from control individuals, remains unclear at this stage. Most of the patient-only mutations involved highly conserved amino acid residues, with 75 % of the mutations residing in predicted functional domains. In silico prediction programs of amino acid changes indicated that all mutations might be pathogenic by one or more of the in silico programs. Also, the three mutations that resided outside predicted domains were scored as deleterious by one to all four in silico programs. Of course, these in silico predictions are merely indicative and should be interpreted with caution. In contrast to PDB-associated mutations which cluster at the UBA domain [11, 16], FTLD mutations are mostly distributed throughout the protein as also previously reported for ALS, though significantly more mutations seem to be located in the UBA domain. Further, Le Ber and colleagues provided evidence for co-segregation with FTLD for two mutations in the UBA domain, p.Pro387Leu and p.Pro392Leu [17]. The p.Pro387Leu mutation was also present in two unrelated Italian FTLD patients in our study who did not share a common haplotype, suggesting a recurrent de novo mutation. No DNA of relatives of these two mutation carriers was available to test for co-segregation. Three mutation carriers (p.Ala16Val, p.Arg212Cys, p.Gly219Val) also carried a pathogenic C9orf72 repeat expansion. At this stage, we have no clear indication that the co-existence of these mutations influences clinical expression of disease. Onset ages were not different from those in patients carrying only a SQSTM1 mutation. In a Japanese sporadic late-onset ALS patient with pathology confirmed predominant lower motor neuron disease, a compound heterozygous SQSTM1 mutation was detected, p.[Val90Met(;)Val153Ile] [29]. We did not observe compound heterozygous SQSTM1 mutation carriers, though both mutations present in the Japanese patient were also observed in our study, p.Val90Met in one FTLD patient and p.Val153Ile in one other FTLD patient and three control individuals. One could hypothesize that the rare SQSTM1 variants act as high to intermediate penetrant risk alleles and in case of the Japanese patient both had to be present to exert the clinical profile of ALS.
The hallmark lesions of FTLD-TDP are neuronal and glial inclusions with positive immunoreactivity for phospho-TDP-43. In addition to sporadic patients, until now mutations in VCP, GRN and C9orf72 have been associated consistently with TDP-43 pathology, while TARDBP mutations are less frequently observed in FTLD [19]. Here, we provide histopathological evidence that SQSTM1 mutations are associated with TDP-43 pathological inclusions. Interestingly, the patient with only a SQSTM1 mutation (p.Glu396*) showed widespread neuronal and glial phospho-TDP-43 pathology. The patient (p.Arg212Cys) who carried also a C9orf72 mutation showed _C9orf72_-related pathology without other noticeable distinctive features. Potential explanations could be that the C9orf72 mutation masked the effect of the _SQSTM1_mutation, or that the C9orf72 pathology dominates the SQSTM1 pathology. Also the SQSTM1 p.Arg212Cys mutation may well have a lower penetrance and be associated with more subtle pathological changes than the (p.Glu396*) mutation located in the UBA domain. These questions merit further investigation and more _SQSTM1_-positive pathologies will need to be evaluated to resolve these issues. Since mutations in SQSTM1 were first linked to PDB, we carefully reexamined medical records of all carriers of patient-only SQSTM1 mutations for indications of (sub) clinical symptoms of PDB. Only in one Portuguese patient carrying the p.Pro232Thr mutation located in the TRAF6 domain, the diagnosis of possible PDB could be made at hindsight. We, however, cannot exclude that the presence of PDB might have been missed in the other patients since PDB often remains asymptomatic and a diagnosis requires radiography. Likewise we cannot exclude that some of the control individuals, who were selected based on absence of dementia-related symptoms, may have been at risk for/or suffered from PDB, in particular, those with the PDB-associated SQSTM1 p.P392L mutation.
In conclusion, our study represents one of the largest screening efforts of FTLD patients for mutations in SQSTM1. Further, we combined the data from our screening with that of published datasets in a meta-analysis of rare SQSTM1 variants in a total of 4,432 FTLD and 10,240 control alleles. Both our study and the meta-analysis calculated a mutation frequency of 3.2 % in FTLD patients. Also, the meta-analysis suggested that rare mutations clustering in the UBA domain of SQSTM1 may influence disease susceptibility by doubling the risk for FTLD. Histopathology of autopsied brain of SQSTM1 mutation carriers demonstrated a widespread neuronal and glial phospho-TDP-43 pathology. Taken together, our findings provide additional evidence that SQSTM1 is implicated in the pathogenicity of FTLD/ALS spectrum diseases.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors are grateful to the personnel of the Genetic Service Facility for their support of the genetic analyses and to the different neurological centers for their contribution to the diagnosis and sampling of patients. The data generation for this paper was in part funded by the MetLife Award for Medical Research to C.V.B., USA; the Belgian Science Policy Office (BELSPO) Interuniversity Attraction Poles program, the Flemish government support to the European Initiative on Centers of Excellence in Neurodegeneration (CoEN), the Flemish Government initiated Methusalem excellence program, the Alzheimer Research Foundation (SAO/FRA), the Medical Foundation Queen Elisabeth, the Research Foundation Flanders (FWO), the Agency for Innovation by Science and Technology Flanders (IWT) and the University of Antwerp Research Fund, Belgium. The FWO provided a postdoctoral fellowship to J.v.d.Z. and a clinical investigatorship to P.V.D. The Brescia IRCCS Fatebenefratelli site was funded by the Ricerca Corrente, Italian Ministry of Health. R.G. was funded by Fondazione CARIPLO, Grant Number 2009-2633. The Barcelona ACE site thank the patients and controls who participated in this project. We are indebted to Trinitat Port-Carbó and her family who are supporting Fundació ACE research programs. The Lisbon site acknowledges a grant by Grunenthal. The Florence site acknowledges Prin 2010-prot. 2010PWNJXK; Cassa di Rispario di Firenze e Cassa di Risparmio di Pistoia e Pescia. The Stockholm site was financially supported by the Programme in Neuroscience at Karolinska Institutet (StratNeuro); the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet; Swedish Alzheimer Foundation; Swedish Research Council; Karolinska Institutet PhD-student funding; King Gustaf V and Queen Victoria’s Free Mason Foundation; the Gun and Bertil Stohne’s Foundation, Foundation for Old Servants; Clinicians including Dr Vesna Jelic and Anne Börjesson-Hanson. For the Munich Institute of Human Genetics site H.P. was supported by the E-Rare project GENOMIT (01GM1207), and the German Network for mitochondrial disorders (mitoNET 01GM1113C). T.B.H. was supported by the NBIA disorders association. The Barcelona Sant Pau site was in part funded by a grant from the Spanish Ministry of Economy and Competitiveness (Grant Number PI12/01311) and CIBERNED. The Pamplona Center for Applied Medical Research is indebted the UTE project from the Foundation for Applied Medical Research (FIMA) and CIBERNED. The Barcelona Neurological Tissue Bank site is indebted to the Neurological Tissue Bank of the IDIBAPS Biobanc for sample and data procurement. The Neurological Tissue Bank acknowledges all brain donors and relatives for generous brain donation for research and referring physicians. The Czech site was partially supported by grant IGA NT 12094-5 of Grant Agency of the Czech Ministry of Health. The Verona site was in part supported by Fondazione Cariverona (Grant Number 2009.1026 “Cognitive and behavioural disability in dementia and psychosis” to GMF). The Liege site was funded by the FNRS.
Conflict of interest
The authors report no disclosure with regard to the reported findings.
Appendix
BELNEU consortium Jonathan Baets (Department of Molecular Genetics, VIB, Antwerp, Belgium; Institute Born-Bunge, University of Antwerp, Antwerp, Belgium; Antwerp University Hospital, Edegem, Belgium); Dirk Nuytten (Hospital Network Antwerp Stuivenberg, Antwerp, Belgium); Jan De Bleecker (University Hospital Ghent, Ghent, Belgium); Jan Versijpt, Alex Michotte (University Hospital Brussels, Brussels, Belgium); Adrian Ivanoiu (Saint-Luc University Hospital, Brussels, Belgium); Olivier Deryck, Bruno Bergmans (AZ Sint-Jan Brugge, Bruges, Belgium); Christiana Willems (Jessa Hospital, Hasselt, Belgium).
EU EOD consortium Anna Paterlini (Proteomics Unit, IRCCS Istituto Centro San Giovanni di Dio Fatebenefratelli, Brescia, Italy); Elena Alonso (Neurogenetics Laboratory, Division of Neurosciences, Center for Applied Medical Research, Universidad de Navarra, Pamplona, Spain); Manuel Seijo-Martínez (Department of Neurology, Hospital do Salnés, Pontevedra, Spain); Ramon Rene, Jordi Gascon, Jaume Campdelacreu (Department of Neurology, Hospital de Bellvitge, Barcelona, Spain); Madalena Martins, Mafalda Matos, André Janeiro, Ana Verdelho (Faculty of Medicine and Institute of Molecular Medicine, University of Lisbon, Portugal); Irene Piaceri (Department of Neurosciences, Psychology, Drug Research and Child Health (NEUROFARBA) University of Florence, Florence, Italy); Jenny Björkström, Anne Kinhult Ståhlbom, Marie Fallström (Department of Geriatric Medicine, Genetics unit, Karolinska University Hospital, Stockholm, Sweden); Charlotte Forsell, Anna-Karin Lindström, Lena Lilius (Karolinska Institutet, Department of Neurobiology, Care Sciences and Society (NVS), KI-Alzheimer Disease Research Center, Stockholm, Sweden); Inger Nennesmo (Department of Clinical Pathology and Cytology, Karolinska University Hospital, Stockholm, Sweden); Laura Fratiglioni (Aging Research Center, Department of Neurobiology, Care Sciences and Society (NVS), Karolinska Institutet and Stockholm University, Stockholm, Sweden);Tamara Eisele (Department of Psychiatry and Psychotherapy, Technische Universität München, 81675 München, Germany); Thomas Wieland (Institute of Human Genetics, Technische Universität München, 81675 Munich, Germany); Ricard Rojas-García, Marc Suárez-Calvet (Department of Neurology, IIB Sant Pau, Hospital de la Santa Creu i Sant Pau, Universitat Autònoma de Barcelona, Barcelona, Spain; Center for Networker Biomedical Research in Neurodegenerative Diseases (CIBERNED), Madrid, Spain); João Massano (Department of Neurology Centro Hospitalar São João, Portugal; Department of Clinical Neuroscience and Mental Health, Faculty of Medicine University of Porto, Porto, Portugal); Maria Helena Ribeiro (Faculty of Medicine, University of Coimbra, Coimbra, Portugal); Catarina Oliveira (Center for Neuroscience and Cell Biology, University of Coimbra, Coimbra, Portugal); Jose L Molinuevo, Anna Antonell (Alzheimer’s disease and other cogntive disorders unit. Neurology department, Hospital); Robert Rusina, Zdenek Rohan (Department of Pathology and Molecular Medicine, Thomayer Hospital, Prague, Czech Republic); Robert Rusina (Center of Clinical Neurosciences, Department of Neurology, First Medical Faculty, Charles University in Prague, Czech Republic); Tiziana Cavallaro (Department of Neuroscience, AOUI Verona, Verona, Italy).
Footnotes
On behalf of the BELNEU consortium and of the EU EOD consortium are given in Appendix.
The members of BELNEU consortium and EU EOD consortium are given in Appendix.
References
- 1.Al-Sarraj S, King A, Troakes C, et al. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- 2.Arai T, Nonaka T, Hasegawa M, et al. Neuronal and glial inclusions in frontotemporal dementia with or without motor neuron disease are immunopositive for p62. Neurosci Lett. 2003;342:41–44. doi: 10.1016/S0304-3940(03)00216-7. [DOI] [PubMed] [Google Scholar]
- 3.Brooks BR, Miller RG, Swash M, Munsat TL. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;32:505–514. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 4.Chung PYJ, Beyens G, Guañabens N, et al. Founder effect in different European countries for the recurrent P392L SQSTM1 mutation in Paget’s disease of bone. Calcif Tissue Int. 2008;83:34–42. doi: 10.1007/s00223-008-9137-2. [DOI] [PubMed] [Google Scholar]
- 5.Cruts M, Gijselinck I, Van Langenhove T, et al. Current insights into the C9orf72 repeat expansion diseases of the FTLD/ALS spectrum. Trends Neurosci. 2013;36:450–459. doi: 10.1016/j.tins.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Deng HX, Zhai H, Bigio EH, et al. FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Ann Neurol. 2010;67:739–748. doi: 10.1002/ana.22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fecto F, Yan J, Vemula SP, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- 8.Gelpi E, van der Zee J, Turon Estrada A, et al. TARDBP mutation p.Ile383Val associated with semantic dementia and complex proteinopathy. Neuropathol Appl Neurobiol. 2014;40:225–230. doi: 10.1111/nan.12063. [DOI] [PubMed] [Google Scholar]
- 9.Gennari L, Gianfrancesco F, Di Stefano M, et al. SQSTM1 gene analysis and gene-environment interaction in Paget’s disease of bone. J Bone Miner Res. 2010;25:1375–1384. doi: 10.1002/jbmr.31. [DOI] [PubMed] [Google Scholar]
- 10.Haack TB, Kopajtich R, Freisinger P, et al. ELAC2 mutations cause a mitochondrial RNA processing defect associated with hypertrophic cardiomyopathy. Am J Hum Genet. 2013;93:211–223. doi: 10.1016/j.ajhg.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirano M, Nakamura Y, Saigoh K, et al. Mutations in the gene encoding p62 in Japanese patients with amyotrophic lateral sclerosis. Neurology. 2013;80:458–463. doi: 10.1212/WNL.0b013e31827f0fe5. [DOI] [PubMed] [Google Scholar]
- 12.Johnson JO, Mandrioli J, Benatar M, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimonis VE, Fulchiero E, Vesa J, Watts G. VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia: review of a unique disorder. Biochim Biophys Acta. 2008;1782:744–748. doi: 10.1016/j.bbadis.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Kurihara N, Hiruma Y. Mutation of the sequestosome 1 (p62) gene increases osteoclastogenesis but does not induce Paget disease. J Clin Invest. 2007;117:133–142. doi: 10.1172/JCI28267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuusisto E, Salminen A, Alafuzoff I. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport. 2001;12:2085–2090. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- 16.Laurin N, Brown JP, Morissette J, Raymond V. Recurrent mutation of the gene encoding sequestosome 1 (SQSTM1/p62) in Paget disease of bone. Am J Hum Genet. 2002;70:1582–1588. doi: 10.1086/340731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Ber I, Camuzat A, Guerreiro R, Campion D. SQSTM1 mutations in French patients with frontotemporal dementia or frontotemporal dementia with amyotrophic lateral sclerosis. JAMA Neurol. 2013;70:1403–1410. doi: 10.1001/jamaneurol.2013.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Litvan I, Agid Y, Calne D, et al. Clinical research criteria for the diagnosis of progressive supranuclear palsy (Steele-Richardson-Olszewski syndrome): report of the NINDS-SPSP international workshop. Neurology. 1996;47:1–9. doi: 10.1212/WNL.47.1.1. [DOI] [PubMed] [Google Scholar]
- 19.Mackenzie IRA, Neumann M, Baborie A, et al. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 2011;122:111–113. doi: 10.1007/s00401-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizuno Y, Amari M, Takatama M, et al. Immunoreactivities of p62, an ubiqutin-binding protein, in the spinal anterior horn cells of patients with amyotrophic lateral sclerosis. J Neurol Sci. 2006;249:13–18. doi: 10.1016/j.jns.2006.05.060. [DOI] [PubMed] [Google Scholar]
- 21.Mori K, Arzberger T, Grässer FA, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013;126:881–893. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- 22.Mori K, Weng S-M, Arzberger T, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 23.Nakaso K, Yoshimoto Y, Nakano T, et al. Transcriptional activation of p62/A170/ZIP during the formation of the aggregates: possible mechanisms and the role in Lewy body formation in Parkinson’s disease. Brain Res. 2004;1012:42–51. doi: 10.1016/j.brainres.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 24.Nalbandian A, Donkervoort S, Dec E, et al. The multiple faces of valosin-containing protein-associated diseases: inclusion body myopathy with Paget’s disease of bone, frontotemporal dementia, and amyotrophic lateral sclerosis. J Mol Neurosci. 2011;45:522–531. doi: 10.1007/s12031-011-9627-y. [DOI] [PubMed] [Google Scholar]
- 25.Neary D, Snowden JS, Gustafson L, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. doi: 10.1212/WNL.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 26.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 27.Rascovsky K, Hodges JR, Knopman D, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134:2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubino E, Rainero I, Chiò A, et al. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology. 2012;79:1556–1562. doi: 10.1212/WNL.0b013e31826e25df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimizu H, Toyoshima Y, Shiga A, et al. Sporadic ALS with compound heterozygous mutations in the SQSTM1 gene. Acta Neuropathol. 2013;126:453–459. doi: 10.1007/s00401-013-1150-5. [DOI] [PubMed] [Google Scholar]
- 30.Synofzik M, Born C, Rominger A, et al. (2013) Targeted high-throughput sequencing identifies a TARDBP mutation as a cause of early-onset FTD without motor neuron disease. Neurobiol Aging 35:1212.e1–5. doi:10.1016/j.neurobiolaging.2013.10.092 [DOI] [PubMed]
- 31.Synofzik M, Maetzler W, Grehl T, et al. Screening in ALS and FTD patients reveals 3 novel UBQLN2 mutations outside the PXX domain and a pure FTD phenotype. Neurobiol Aging. 2012;33(2949):e13–e17. doi: 10.1016/j.neurobiolaging.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Tanji K, Zhang H-X, Mori F, et al. p62/sequestosome 1 binds to TDP-43 in brains with frontotemporal lobar degeneration with TDP-43 inclusions. J Neurosci Res. 2012;90:2034–2042. doi: 10.1002/jnr.23081. [DOI] [PubMed] [Google Scholar]
- 33.Teyssou E, Takeda T, Lebon V, et al. Mutations in SQSTM1 encoding p62 in amyotrophic lateral sclerosis: genetics and neuropathology. Acta Neuropathol. 2013;125:511–522. doi: 10.1007/s00401-013-1090-0. [DOI] [PubMed] [Google Scholar]
- 34.Troakes C, Maekawa S, Wijesekera L, et al. An MND/ALS phenotype associated with C9orf72 repeat expansion: abundant p62-positive, TDP-43-negative inclusions in cerebral cortex, hippocampus and cerebellum but without associated cognitive decline. Neuropathol Off J Japanese Soc Neuropathol. 2011;32:505–514. doi: 10.1111/j.1440-1789.2011.01286.x. [DOI] [PubMed] [Google Scholar]
- 35.van der Zee J, Gijselinck I, Dillen L, et al. A Pan-European study of the C9orf72 repeat associated with FTLD: geographic prevalence, genomic instability, and intermediate repeats. Hum Mutat. 2013;34:363–373. doi: 10.1002/humu.22244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Zee J, Pirici D, Van Langenhove T, et al. Clinical heterogeneity in 3 unrelated families linked to VCP p.Arg159His. Neurology. 2009;73:626–632. doi: 10.1212/WNL.0b013e3181b389d9. [DOI] [PubMed] [Google Scholar]
- 37.Van Langenhove T, van der Zee J, Van Broeckhoven C. The molecular basis of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum. Ann Med. 2012;44:817–828. doi: 10.3109/07853890.2012.665471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watts GDJ, Wymer J, Kovach MJ, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 39.Weckx S, Del-Favero J, Rademakers R, et al. novoSNP, a novel computational tool for sequence variation discovery. Genome Res. 2005;15:436–442. doi: 10.1101/gr.2754005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimprich A, Benet-Pagès A, Struhal W, et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet. 2011;89:168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.