Actin filament severing by cofilin dismantles actin patches and produces mother filaments for new patches (original) (raw)
. Author manuscript; available in PMC: 2014 Aug 14.
Published in final edited form as: Curr Biol. 2013 May 30;23(13):1154–1162. doi: 10.1016/j.cub.2013.05.005
Summary
Background
Yeast cells depend on Arp2/3 complex to assemble actin filaments at sites of endocytosis, but the source of the initial filaments required to activate Arp2/3 complex is not known.
Results
We tested the proposal that cofilin severs actin filaments during endocytosis in fission yeast cells using a mutant cofilin defective in severing. We used quantitative fluorescence microscopy to track mGFP-tagged proteins, including early endocytic adaptor proteins, activators of Arp2/3 complex and actin filaments. Consistent with the hypothesis, actin patches disassembled far slower in cells depending on severing-deficient cofilin than wild type cells. Even more interesting, actin patches assembled slowly in these cofilin mutant cells. Adaptor proteins End4p and Pan1p accumulated and persisted at endocytic sites more than 10 times longer than in wild type cells, followed by slow put persistent recruitment of activators of Arp2/3 complex, including WASP and myosin-I. Mutations revealed that actin filament binding sites on adaptor proteins Pan1p and End4p contribute to initiating actin polymerization in actin patches.
Conclusions
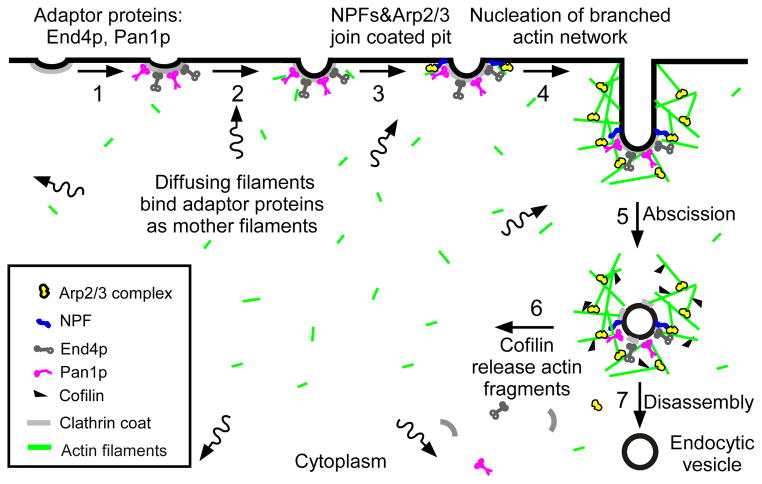
We propose a “sever, diffuse and trigger” model for the nucleation of actin filaments at sites of endocytosis whereby cofilin generates actin filament fragments that diffuse through the cytoplasm, bind adapter proteins at nascent sites of endocytosis and serve as mother filaments to initiate the autocatalytic assembly of the branched actin filament network of each new patch. This hypothesis explains the source of the “mother filaments” that are absolutely required for Arp2/3 complex to nucleate polymerization.
Keywords: cofilin, actin patch, endocytosis, adaptor protein, Arp2/3 complex
Introduction
Assembly of actin plays a key role in clathrin-mediated endocytosis in yeast by providing both scaffold for the membrane invagination and force for scission of the vesicle against turgor pressure [1–3](for review see [4, 5]). The sites of endocytosis are called actin patches.
Genetics and quantitative fluorescence microscopy provided many details about the molecular composition and assembly pathway of actin patches including evidence that Arp2/3 complex nucleates the actin filaments [6–8], but questions remain about every stage of the process. Biochemical studies showed that Arp2/3 complex must bind both a nucleation promoting factor and a pre-existing filament to initiate a new filament [9]. However, the source of these mother filaments is unclear in fungi [7]. An immediately adjacent actin patch can be a source of actin filaments [10], but most patches form in regions of cortex apparently devoid of actin filaments [11]. However, electron microscopy of animal cells suggested that actin filaments may be present at sites of endocytosis earlier than indicated by fluorescence microscopy [12]. Questions also remain about disassembly of the actin coat. Simulations of a mathematical model of fission yeast endocytic patches demonstrated that dissociation of subunits from the ends of actin filaments is too slow to account for disappearance of the filaments in 10 s, unless the filaments are severed into fragments small enough to diffuse out of the patch and disassemble elsewhere [7]. Dissociation of Arp2/3 complex from the branched actin filaments has been proposed as a mechanism [13], and while cofilin stimulates debranching [14], this reaction was not necessary to account for the turnover of filaments in patches [7].
The small protein cofilin severs actin filaments [15–17] and contributes to many processes including endocytosis, cell motility and cytokinesis [18–20]. Cofilin concentrates in yeast endocytic actin patches [21–23] and cofilin mutations cause endocytic defects [19, 21, 23].
Here we used a mutant cofilin with a selective defect in severing [18] to study how actin filament severing contributes to endocytosis in fission yeast. Endocytic actin patches disassembled very slowly in mutant cells as anticipated by studies in budding yeast [21, 23]. Surprisingly, actin patch assembly took longer in cofilin mutant cells, with prolonged accumulation of both membrane adaptor proteins and activators of Arp2/3 complex before assembly of actin. Depletion of the actin monomer pool could not explain these defects in cofilin mutant cells. Instead, we present evident to support a feedback mechanism where severing by cofilin generates short actin filaments that interact with endocytic adapter proteins End4p and Pan1p and Arp2/3 complex to initiate actin polymerization at sites of endocytosis.
Results
Cofilin concentrates in actin patches
We studied the localization of cofilin in fission yeast by expressing cofilin tagged on the N-terminus with mGFP (Figure S1A and 1A). The protein is the product of the adf1+ gene, but we use the common name, cofilin or mGFP-cofilin, because adf1, actin depolymerizing factor 1, is a misleading name for a protein that does not depolymerize actin filaments [15–17]. This fusion protein must be over expressed from either the adh1 (alcohol dehydrogenase 1) or the strong 3nmt1 promoter to complement a Δ_adf1_ null mutation [18, 22]. The adf1::Padh-mGFP-adf1 cells expressed about 10 fold more mGFP-Adf1p than Adf1p in the adf1+ cells (Figure S1B). These cells grew normally at both 25°C and 30°C but slightly slower at 36°C compared to wild type cells (Figure S1C). After correcting for excluded volume [24]the total fluorescence in adf1::Padh-mGFP-adf1 cells corresponded to a cytoplasmic concentration of 200 μM mGFP-cofilin.
We used two-color fluorescence microscopy to compare mGFP-cofilin with either End4p-mCherry, an early endocytic adaptor protein, or Fim1p-mCherry, the fission yeast homolog of the actin filament binding protein fimbrin (Figure 1B and S1D). Actin patches in cells depending on over expressed mGFP-cofilin assembled and disassembled Fim1p-mCherry normally (Figure S1E), and the lifetime of End4p increased only slightly from 36 s to 39 s (n = 20). mGFP-cofilin appeared at sites of endocytosis 5 s before actin patches began to move from the cell surface (defined as time zero), peaked at 6,000 molecules at time +10 s and then gradually dissipated over the next 10 s (Figure 1A). The mGFP-cofilin fluorescence peaked after both End4p and Fim1p and persisted after both proteins dissociated from moving patches (Figure 1B and S1D).
Figure 1.
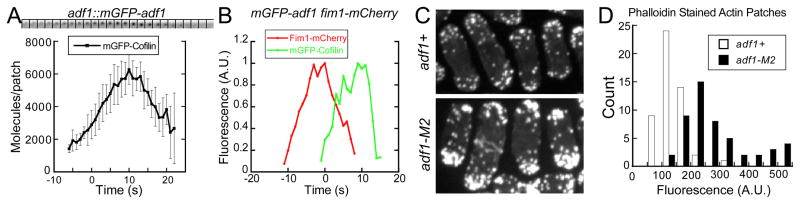
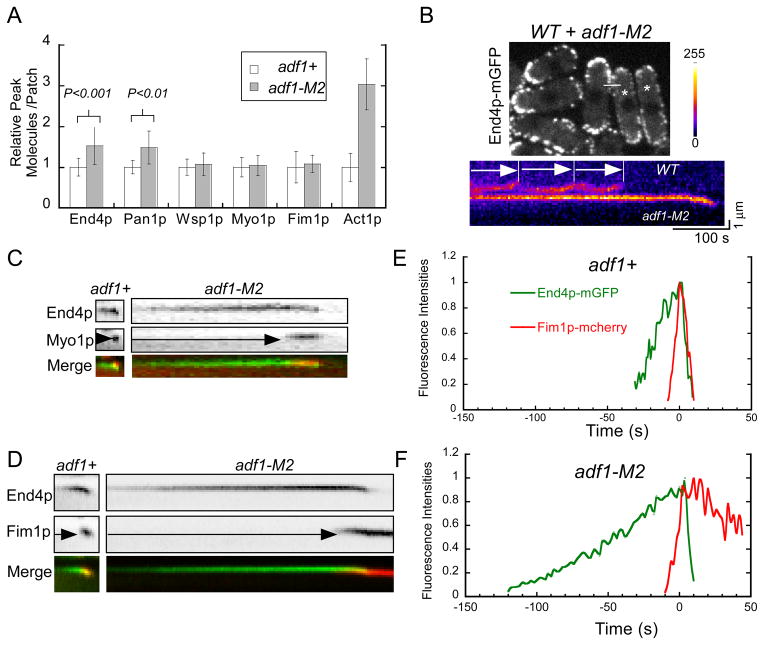
Cofilin localization in endocytic actin patches of fission yeast cells and effects of a cofilin mutation that reduces severing on actin filament accumulation in patches. (A) Time course of mGFP-cofilin in actin patches. Upper panel: a time series of negative contrast fluorescence micrographs of an actin patch at 1 s intervals. Images are sums of 5 median slices of the cell. Lower panel: graph of the average number of molecules of mGFP-Adf1p per patch (± SD) over the time. (B) Time courses of normalized fluorescence intensities of mGFP-cofilin (green) and Fim1p-mCherry (red) in one actin patch. Also see Figure S1. (C–D) Comparisons of actin patches in wild type adf1+ and cofilin mutant adf1-M2 cells. (C) Maxim intensity projections of Z-series of fluorescence micrographs of adf1+ cells and adf1-M2 cells fixed and stained with rhodamine-phalloidin for actin filaments. (D) Histogram of fluorescence intensities of 50 actin patches from adf1+ (gray) and adf1-M2 (black) cells. See also Figure S1.
Endocytic defects in cofilin mutant adf1-M2 cells
To study the role of cofilin during endocytosis, we used cells depending on mutant cofilin-M2 that binds and severs actin filaments far slower than the wild type cofilin [18]. Fission yeast adf1-M2 cells have larger diameters than the wild type cells, similar to many other endocytic mutants [25]. Calcofluor white stained the cell walls more intensely in these mutant cells than in wild type cells (data not shown), a defect observed in other endocytic mutants. The average fluorescence intensity of actin patches stained by rhodamine-phalloidin was 2 times greater in adf1-M2 cells (281 ± 107, n = 50) than wild type cells (139 ± 42, n = 50) (Figure 1C–D). During interphase, these bright actin patches packed more densely at the poles of cofilin mutant cells than in wild type cells (Figure 1C).
We looked for endocytic defects in the adf1-M2 cells using two approaches: localization of the fission yeast homolog of the SNARE protein synaptobrevin Syb1p (Figure S2F); and pulse chase experiments with the fluorescent lipophilic dye FM4-64 (Figure S2G). After exocytosis Syb1p recycles from the plasma membrane through endocytic pathway, a process compromised by many endocytic mutations in both budding and fission yeast [10, 26]. GFP-Syb1p concentrated in numerous cytosolic puncta in both wild type and adf1-M2 cells, but associated mostly with the plasma membrane in adf1-M2 cells (Figure S2F), consistent with defects in endocytosis. Pulse-chase experiments confirmed that adf1-M2 cells internalized FM4-64 very slowly. Wild type cells took up FM4-64 from the plasma membrane into numerous cytoplasmic puncta in <8 min (Figure S2G), but after 16 min most of the dye remained on the surface of adf1-M2 cells with very few fluorescence puncta in the cytoplasm (Figure S2G).
Defects in actin patch assembly and disassembly in cofilin adf1-M2 mutant cells
We used quantitative fluorescence microscopy to compare actin patch dynamics of wild type and adf1-M2 cells expressing 6 endocytic proteins tagged with mGFP (Figure 2 and S2A). We aligned time courses of all actin patches based on their initial inward movement (time zero). As in budding yeast [4, 8] fission yeast initiate endocytosis by sequentially recruiting clathrin, adaptor proteins, nucleation promoting factors for Arp2/3 complex and finally Arp2/3 complex itself [6]. We tracked two adaptor proteins End4p/Sla2p (a homolog of Hip1r [27]) and Pan1p (an Eps15 protein [28]). We also tracked nucleation promoting factors (NPFs), the homolog of Wiskott-Aldrich syndrome protein (Wsp1p) and myosin-I (Myo1p), which stimulate Arp2/3 complex to initiate the explosive assembly of actin filaments [7, 25, 29, 30] in cooperation with the F-BAR proteins [31]. We tracked actin filaments with either Fim1p-mGFP expressed from its genomic locus or mGFP-act1 (mGFP-actin) expressed at a low level from the leu1 locus under the control of a suppressible 41nmt1 promoter in cells with a wild type actin gene.
Figure 2.
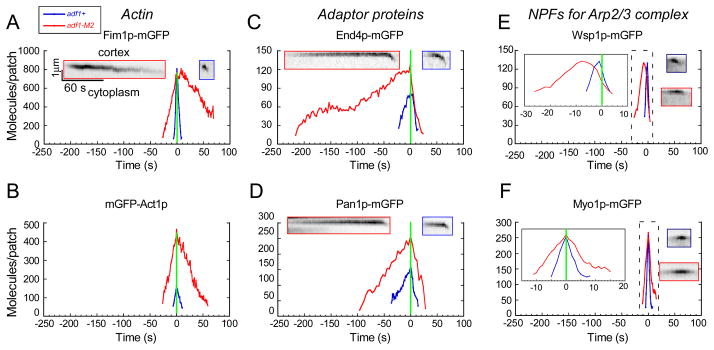
Time courses of the accumulation and disappearance of fluorescent proteins in actin patches of (blue lines) wild type adf1+ cells and (red lines) cofilin mutant adf1-M2 cells. Graphs of the average number of molecules at each point in time. Data from individual patches were aligned at the time they started to move (time zero, green line). Inserts are kymographs of fluorescence micrographs of individual actin patches in (blue outline) wild type cells or (red outline) adf1-M2 cells. Inserts in (E) and (F) are expanded time scales for data in dashed boxes. (A) Fim1p-mGFP, (B) mGFP-Act1p, (C) End4p-mGFP, (D) Pan1p-mGFP, (E) mGFP-Wsp1p and (F) mGFP-Myo1p. See also Figure S2.
Actin patches in adf1-M2 cells accumulated mGFP-Act1p at the same rate as wild type cells, but the accumulation phase lasted 3 times longer (30 s vs. 10 s) (Figure 2B), so the peak number of mGFP-actin molecules per actin patch was 3 fold higher in cofilin mutant cells than wild type cells (Figure 2B), confirming the more intense staining with rhodamine-phalloidin (Figure 1C). Nevertheless, these actin patches in adf1-M2 cells accumulated the normal amount of the actin binding protein Fim1p-mGFP but slower than wild type cells (Figure 2A). After time zero mGFP-actin and Fim1p-mGFP took about 10 s to disappear from patches in wild type cells (Figure 2A), but both proteins persisted far longer in adf1-M2 cells, taking as long as 180 s to completely disappear from some patches (Figure 2A). In many cases, these persisting actin patches in adf1-M2 cells moved from their initial locations and occasionally appeared to merge with nearby actin patches.
Quantitative microscopy revealed that the early stages of actin patch assembly were prolonged in adf1-M2 cells (Figure 2C–D). Two adaptor proteins, End4p-mGFP and Pan1p-mGFP, appeared at actin patches around time −30 s in wild type cells, but both appeared before −100 s in adf1-M2 cells and accumulated to higher levels than in wild type cells (Figure 2C–D). Prolonged accumulation of End4p and Pan1p resulted in ~1.5-fold more of these adapters in actin patches (Figure 3A) at the time NPFs arrived prior to actin polymerization. Imaging actin patches in adf1-M2 and wild type cells side by side confirmed the presence of more End4p-mGFP in the long-lived actin patches of the mutant cells (Figure 3B) as observed by quantitative fluorescence imaging. The NPFs Wsp1p and Myo1p also accumulated slower in the actin patches of adf1-M2 cells than wild type cells (Figure 2E–F), but the cofilin mutation did not alter their peak numbers (Figure 3A). Both membrane adaptor proteins dissociated from actin patches in 20 s in adf1-M2 cells, slightly slower than wild type cells (10 s) (Figure 2C–D). The movements of patches away from the cell surface were indistinguishable in adf1-M2 and wild type cells (Figure S2B). Similarly, both NPFs disappeared slightly slower than normal from patches in adf1-M2 cells. We used two-color fluorescence microscopy of live cells expressing a tagged adaptor protein and either Myo1p or Fim1p to confirm the timing of these events (Figure 3C–F).
Figure 3.
Accumulation of endocytic adaptor proteins in actin patches of wild type and cofilin mutant cells before actin assembly. (A) Histogram of peak numbers of proteins per actin patch in wild type adf1+ cells (open bars) and cofilin mutant adf1-M2 cells (shaded bars). Mean numbers ±SD are normalized against wild type actin patches. P values are from two-tailed student t-tests. (B) Side by side comparison of actin patches in wild type (asterisk) and adf1-M2 cells expressing End4p-mGFP. (Top) Fluorescence micrograph (sum of 5 middle Z slices of the cells) of a mixed population of cells. Wild type cells were identified by expression of Fim1p-mCherry. Data for the kymograph were collected along the white line. (Bottom) Kymograph of pseudo colored fluorescence micrographs of actin patches in a pair of neighboring cells showing (top) three actin patches in the wild type cell and (bottom) one prolonged actin patch in the adf-M2 cell. (C–D) Kymographs of fluorescence micrographs of actin patches in wild type adf1+ (left) and adf1-M2 (right) cells expressing pairs of fluorescent proteins, either End4p-mYFP and mCFP-Myo1p or End4p-mGFP and Fim1p-mCherry. Bottom panels are merged images with End4 in green and Myo1p/Fim1p in red. (E–F) Time courses of normalized fluorescence intensities of End4p-mGFP (green) and Fim1p-mCherry (red) in actin patches of (E) wild type adf1+ and (F) adf1-M2 cells. See also Figure S2.
Thus, defective actin filament severing in adf1-M2 cells not only slowed disassembly of actin patches (as expected) but also prolonged the accumulation of adaptor proteins and NPFs, delaying the initiation of actin filament assembly in actin patches, a defect that we confirmed in cells depending on the mutant cofilin-M3 with a slightly milder defect in severing actin filaments (Figure S2C and D) [18]. We considered two mechanisms that might explain these assembly defects: depletion of the cytoplasmic pool of actin monomers; and a deficiency of actin filament fragments to stimulate Arp2/3 complex in adjacent actin patches.
Depletion of actin monomers does not mimic the defects in actin patch assembly of cofilin mutant cells
We investigated whether slow severing and depolymerization of actin filaments in adf1-M2 cells might deplete the cytoplasmic pool of actin monomers, leading secondarily to defects in actin patch assembly. A shortage of actin monomers would not only slow nucleation and elongation of actin filaments but might also compromise binding of Arp2/3 complex to mother filaments [32].
Quantitative fluorescence microscopy of cells expressing mGFP-actin from the 41nmt1 promoter in the leu1 locus showed that the cytoplasmic mGFP-actin fluorescence was 10% lower in a_df1-M2_ cells than wild type cells (Figure S2E). To test whether this slightly lower pool of cytoplasmic actin might cause endocytic defects in adf1-M2 cells, we studied actin patches in wild type cells treated with Latrunculin A (LatA), a small molecule that binds actin monomers with high affinity and inhibits polymerization [33–35]. At 8 μM LatA increased the cytoplasmic fluorescence of mGFP-actin, ~30% (Figure S2F). We interpret this change to be sequestered mGFP-actin bound to LatA. This effect of LatA on the pool of unpolymerized actin available to assemble actin patches should be sufficient to mimic the 10% smaller amount of cytoplasmic actin in adf1-M2 cells.
Low concentrations of LatA (0–8 μM) prolonged the accumulation of both End4p-mYFP and mCFP-Myo1p in patches (Figure S2F, G and H). However, LatA only led to a small delay in the interval between the appearance of End4p and Myo1p in actin patches of wild type cells (Figure S2F, G and H) as observed in adf1-M2 cells (Figure 3). Furthermore, low concentrations of LatA inhibited the inward movement of actin patches, a defect not observed in cofilin mutant cells. For example, most actin patches (9/10) failed to move inward in cells treated with 8 μM LatA. Therefore, sequestering a fraction of the pool of monomeric actin with LatA did not replicate the phenotype of cofilin mutant cells.
Roles of End4p and Pan1p in initiating actin polymerization in actin patches
A second hypothesis to explain the defects in actin patch assembly in cofilin mutant cells is that the process depends on short actin filaments normally severed by cofilin from endocytic patches to serve as mother filaments to activate Arp2/3 complex. We also considered the possibility that proteins recruited early in the endocytic pathway might tether these filaments and enhance their impact. Among the proteins that join clathrin early in the endocytic pathway [6, 8], Pan1p and End4p are known to bind actin filaments [36, 37] making them candidates to retain diffusing actin filaments at endocytic sites before the arrival of NPFs and Arp2/3 complex. Eps15 homolog Pan1p [28] is essential in fission yeast (Figure S3A). End4p is a nonessential homolog of budding yeast Sla2p and animal Hip1r [27, 38]. Actin patches in Δend4 cells formed elongated tails as they moved from the cell surface (Figure S3D), similar to Δ_sla2_ deletion mutations in budding yeast [8].
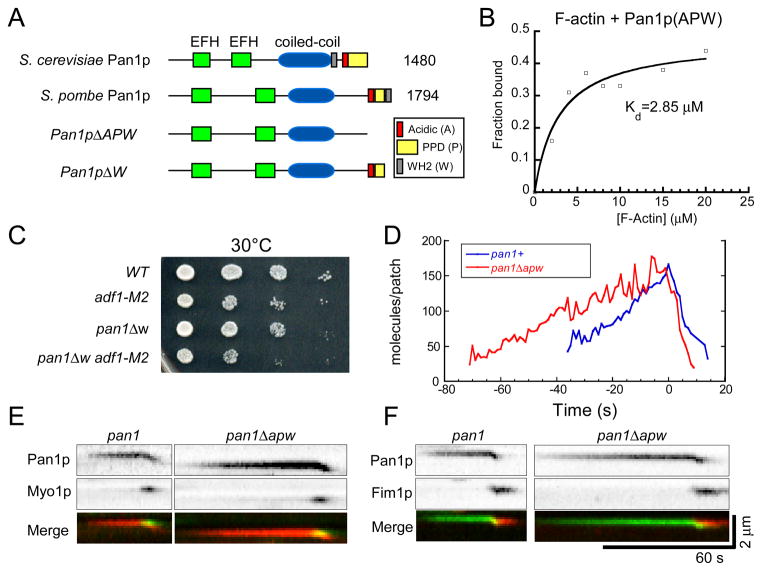
Biochemical experiments showed that fission yeast Pan1p binds actin filaments like budding yeast Pan1p [39]. Pan1p has a conserved WH2 (W) motif, located at the very C-terminus after acidic (A) and polyproline (PP) motifs (Figure 4A and S3B). A purified C-terminal fragment of Pan1p (Pan1p1633-1974) including the A, PP and W domains bound muscle actin filaments with a Kd of about 3 μM (Figure 4B), although only 40% of the GST tagged protein was active.
Figure 4.
Analysis of Pan1p binding to actin filaments and effects of deleting the actin binding region of Pan1p on actin patches. (A) Domain structures of budding yeast and fission yeast Pan1p and Pan1p truncation mutants. (B) Graph of the dependence of the fraction of Pan11633 -1794 pelleted on the concentration of actin filaments. Samples of 2 μM Pan11633 -1794 with 0–20 μM actin filaments were incubated at room temperature for 30 min in KMEI buffer before pelleting at 212,300 g for 40 min. A bimolecular binding isotherm fit the data well with a Kd of 2.9 μM. Only 40% of GST-Pan11633 -1794 (APW) was depleted from the supernatant with 20 μM actin indicating that much of the protein was inactive. (C) Viability of strains with the pan1Δw truncation mutation, adf1-M2 mutation or both mutations measured by growth of dilution series for 2 days at 30°C. (D) Time course of average fluorescence intensities of actin patches marked with Pan1p-mGFP (n = 10) or Pan1pΔW-mGFP (n = 12). (E–F) Kymographs of fluorescence micrographs of actin patches from cells expressing wild type or mutant Pan1p. (E) Pan1pΔAPW-tdTomato and mGFP-Myo1p. (F) Pan1pΔAPW-mGFP and Fim1p-mcherry. See also Figure S3.
We tested the effects on actin patch dynamics of two Pan1p truncation mutants designed to compromise binding to actin filaments. We deleted either the W domain alone in pan1_Δ_1743-1794 (pan1Δw), or the A, PP and W domains altogether in pan1Δ1643-1794 (pan1Δapw) (Figure 4A). Cells expressed the pan1 truncation mutant proteins at levels similar to full length Pan1p (Figure S3C). Both mutations had negative genetic interactions with the adf1-M2 mutation, with double mutants growing slower than the wild type cells at 30°C (Figure 4C). Pan1pΔAPW-mGFP took twice as long as full length Pan1p-mGFP to accumulate to normal levels in actin patches but dissociated from actin patches over the same time course as full length Pan1p-mGFP (Figure 4D). The behavior of Pan1pΔW-mGFP was similar (data not shown). Strains depending on these pan1 truncation mutants took ~40 s longer than wild type cells to recruit Myo1p to actin patches (Figure 4E), so actin polymerization was delayed (Figure 4F). Similarly the interval between the appearances of End4p and Myo1p was longer in actin patches of pan1 truncation mutants than wild type cells (data not shown). Therefore, the WH2 domain of Pan1p contributes to initiation of actin patch assembly but is not essential.
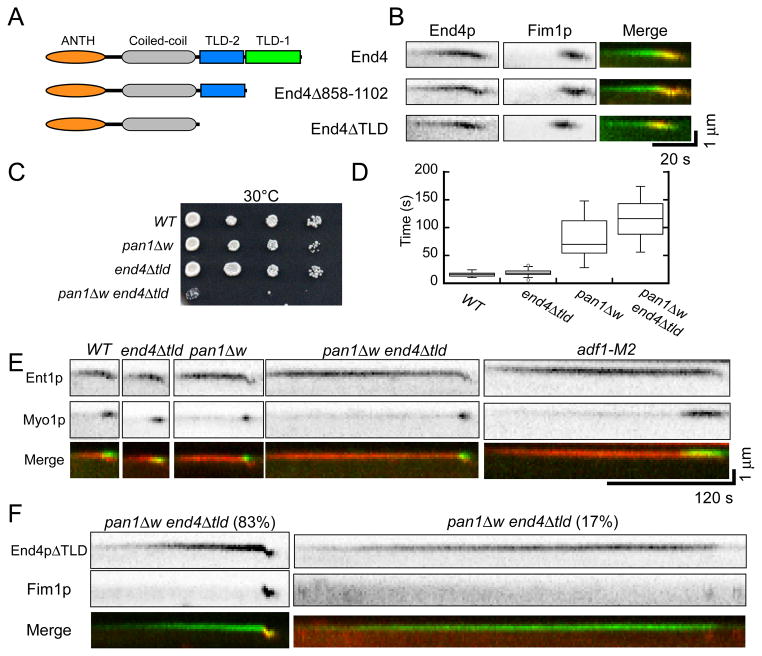
We tested the effects on actin patch dynamics of two End4p truncation mutants designed to compromise binding to actin filaments. The C-terminal talin-like (TL) domain (also called an I/LWEQ domain) of budding yeast End4p can bind actin filaments [36, 40], but removing the TL domain had no detectable effect on the assembly of actin patches [40]. Using homology based structural prediction (Protein Homology/analogY Recognition Engine, http://www.sbg.bio.ic.ac.uk/phyre2) we found that the End4p sequence between the central coiled-coil and C-terminal TL domain has a high degree of sequence homology to the actin filament binding domain of talin (Figure 5A). This previously unrecognized TL domain may have compensated for deletion of the C-terminal TL domain in previous work. To compromise actin filament binding of End4p, we deleted either the C-terminal TL domain (end4Δ858-1102) or both TL domains (end4_Δ_663-1102/end4Δtld) (Figure 5A).
Figure 5.
Actin filament binding domains of End4p and Pan1p cooperate to initiate actin polymerization in endocytic patches**. (A–B)** Deletion of one or both talin-like domains from End4p. (A) Domain structure of End4p and the two truncation mutants. (B) Kymographs of fluorescence micrographs of actin patches in cells expressing either End4p or End4p truncation mutants tagged with mGFP and Fim1p-mcherry. (C) Viability of strains with the pan1Δw mutation, end4Δtld mutation and both mutations (pan1Δw end4Δtld) measured by growth of dilution series for 2 days at 30°C. (D) Box plot of the time interval between the appearances of Ent1p and Myo1p in actin patches of wild type, end4Δtld, pan1Δw and end4_Δ_tld pan1_Δ_w cells. Horizontal lines in the boxes are average values. The boxes include ± 25% from the average values. Vertical lines mark minimum and maximum values. (E) Kymographs of fluorescence micrographs of actin patches in the wild type, end4Δtld, pan1Δw, end4_Δ_tld pan1_Δ_w double mutant and adf1-M2 cells expressing both Ent1p-Tdtomato (top panels) and mGFP-Myo1p (bottom panels). The bottom panels are merged kymographs with Ent1p in red and Myo1p in green. (F) Kymographs of fluorescence micrographs of actin patches in end4Δtld pan1ΔW cells expressing (top panels) End4pΔTLD-mGFP and (bottom panels) Fim1p-Tdtomato. Bottom panels are merged kymographs with End4pΔTLD in green and Fim1p in red. (Left) Recruitment of Fim1p (left)was delayed in 83% of patches, and (right) failed in 17% of patches. See also Figure S3 and S4.
Both end4 TL domain deletion mutations had negative, temperature sensitive interactions with the cofilin mutation adf1-M2 (Fig S3E). mGFP-End4p lacking one or both TL domains was expressed at the same level as the full length protein (Figure S3F) and accumulated in and disappeared from actin patches normally (Figure S3G and H). Both end4 mutant strains initiated actin assembly without delay (Figure 5B).
The end4 mutants lacking TL domains had strong negative genetic interactions with pan1 mutants lacking the WH2 motif (Figure 5C). The double mutant pan1Δw end4Δtld was temperature sensitive, growing very slowly at 30°C (Figure 5C) and dead at 36°C. On the other hand, the function of the newly identified actin filament binding motif of Epsin (Ent1p) [41] did not overlap with those of Pan1p (Figure S4 and Supplemental Results). The pan1Δw end4Δtld cells took up FM4-64 much slower than wild type cells, even at the permissive temperature 25°C. Wild type cells took up most of the cell surface FM4-64 in less than 9 min, but most of the dye remained on the cell surface of double mutant cells after 15 min (Figure S3I).
We observed endocytic adaptor protein Ent1p-tdTomato and mGFP-Myo1p to confirm that serious defects in actin patch assembly compromise endocytosis in pan1Δw end4Δtld double mutant cells (Figure 5D and E). Ent1p is the fission yeast homolog of epsin that normally appears in actin patches ~30 s before initiation of actin assembly. The interval between the appearances of Ent1p and Myo1p in actin patches was 15.6 ± 3.9 s (n = 20) in wild type cells, 19.6 ± 5.8 s (n = 30) in end4Δtld cells, 81.9 ± 35.2 s (n = 30) in pan1Δw cells and 122.3 ± 55.8 s (n = 31) in end4Δtld pan1Δw double mutant cells at the permissive temperature (Figure 5D). Actin patches in adf1-M2 cells also recruited Myo1p slowly (Figure 3C and 7E). Actin assembly followed with Fim1p-mcherry was severely compromised in the double mutant cells with 17% (n = 30) of actin patches failing to recruit Fim1p-mcherry and initiate actin polymerization (Figure 5F). The patches in the double mutant cells that polymerized actin did so with an average of 140 ± 62 s (n = 25) between the appearance of End4p and Fim1p-mcherry compared with just 25 s in wild type cells (Figure 5F).
Although we did not detect small actin filaments diffusing from patches in cells expressing mGFP-Lifeact (explained in Supplemental Results), we found that the depletion of actin filaments in cells by LatA inhibited recruitment of Arp2/3 complex and Myo1p at sites of endocytosis without preventing the accumulation of both End4p and Wsp1p (Figure S5 and Supplemental Results).
Discussion
Our experiments with a cofilin mutant defective in severing actin filaments confirmed that actin filament severing is not only essential for disassembling actin filaments in endocytic patches, but also contributes to the assembly of actin patches. We attribute the endocytic defects of adf1-M2 cells to slow severing by the mutant cofilin. Although the affinity of cofilin-M2 for actin filaments is five-fold lower than wild type cofilin, cooperative binding allows 2 μM cofilin-M2 to saturate 1 μM actin filaments [18], so cofilin-M2 will bind actin filaments but sever them slowly in the mutant cells. The effects of truncation mutations of two endocytic adapter proteins support the proposal that cofilin produces short actin filaments that bind adapter proteins and serve as primers to initiate actin polymerization at site of endocytosis.
Actin filament severing by cofilin disassembles endocytic actin patches
Disassembly of endocytic actin patches depends on cofilin in budding yeast [8, 21, 23], and our experiments demonstrate that the serving activity of cofilin is required (step 6, Figure 6). Slow binding of the mutant cofilin to actin filaments should also slow dissociation of branches [14] and may contribute to the slow turnover of patches. Since the cofilin mutation had little effect on the movement of actin patches (Figure S2B), neither scission of endocytic vesicles from the plasma membrane (step 5) nor vesicle movements appear to depend on disassembly of actin filaments. Membrane adaptor proteins and NPFs dissociated normally from actin patches (step 7), so these events are also independent of actin filament severing.
Figure 6.
“Sever, diffuse and trigger” model for actin filament turnover in actin patches. The 7 steps (numbers next to the arrows) are (1) clathrin coated pits bind adaptor proteins End4p and Pan1p, (2) short, diffusing actin filaments bind to End4p and Pan1p associated with coated pits, and (3) Arp2/3 complex interacts with these mother filaments and nucleation promoting factors to (4) initiate branching nucleation of actin filaments that promote elongation of the endocytic tubule. (5) After abscission of the vesicle, (6) cofilin severs actin filaments to generate a pool of short, diffusing actin filaments, some of which return to the cycle at step 2. See also Figure S5.
A “sever, diffuse and trigger” hypothesis explains the cofilin requirement for actin patch assembly
Our most important finding is that the early steps in the assembly of actin patches depend on both actin filament severing by cofilin and actin filament binding by early endocytic adaptor proteins. These two seemingly disparate features suggest a “sever, diffuse and trigger” hypothesis (Figure 6): cofilin generates actin filament fragments that diffuse through the cytoplasm (step 6), bind adapter proteins at nascent sites of endocytosis (step 2) and serve as mother filaments (step 3) to initiate the autocatalytic assembly of the branched actin filament network of each new patch (step 4). This hypothesis offers one feasible explanation for the source of mother filaments that are absolutely required for Arp2/3 complex to nucleate polymerization [9] but appear to be absent from the cortex of yeast [11, 42]. We recognize other potential sources of mother filaments including the possibility that Wsp1p/Las17p may nucleate actin filaments in patches [43].
Our hypothesis is an extension of a proposal that actin patches can provide mother filaments to initiate the assembly of an adjacent new patch through an “touch and trigger” mechanism [10]. Actin patches were observed to trigger actin filament assembly at a nearby site of endocytosis by direct contact. Although “touch and trigger” events were rare in wild type cells, they were frequent in the dip1 mutant, which has defects in nucleation of actin filaments in patches [10]. The ”sever, diffuse and trigger” mechanism can act over longer distances to explain actin assembly at sites of endocytosis physically separated from other actin patches. Our hypothesis does not preclude the possibility that the actin filament binding domains of adaptor proteins have separate roles at a later stage after the assembly of actin patches, as suggested by studies in budding yeast [1, 41]. Indeed, our studies suggest that the TLD domain of End4p could complement the ABD domain of Ent1p in a function separate from initiating the assembly of actin patches, perhaps coupling the actin filament network to the membrane of endocytic vesicles (step 5).
Simulations of actin patch dynamics [7] estimated that severing releases short actin filaments at a prodigious rate of ~30 per second from each >100 actin patches. The cytoplasmic concentration of actin in such fragments would rapidly reach the micromolar range, even if further severing and polymerization turn over the fragments in 10 s. A filament of 30 subunits has a translational diffusion coefficient of 5.2 μm2/s (see Experimental Procedures) and shorter fragments move even faster, so they diffuse rapidly throughout the cytoplasm. These short, rapidly diffusing filaments [7, 44] are difficult to distinguish from cytoplasmic actin monomers in cells with a small fraction of the total actin tagged with mGFP. Severing of filaments in actin cables nucleated by formin For3p may provide a second source of actin filament fragments [45].
Animal cells may also have small diffusing actin filaments that contribute to endocytosis. Fluorescence correlation spectroscopy (FCS) detected short (~80 subunits) actin filaments diffusing in the cortex of tissue culture cells [46], and electron microscopy of Hela cells showed actin filaments associated with early endocytic invaginations that were not detected by correlated fluorescence microscopy [12]. Cofilin may generate these filaments, perhaps explaining how cofilin contributes to actin polymerization at the leading edge of animal cells [20].
Actin filament binding sites on adaptor protein Pan1p and End4p are important to initiate actin nucleation in patches. The affinities of the APW motif of Pan1p and the TLD domain of End4p for actin filaments (Kd = ~3 μM) are higher than alpha-actinins (Kd = 20 μM), which bind actin filaments in physiological settings. Given 240 molecules of End4p and Pan1p on the tiny surface of a coated pit (12000 nm2, assuming a diameter of 50 nm [47]), the density of actin filament binding sites is 1 per 50 nm2. This high density of actin filament binding sites on cytoplasmic surface of a coated pit may favor multi-valent binding of short filaments.
Why are actin filament severing and actin filament binding by adaptor proteins required to terminate the accumulation of adaptor proteins and recruit NPFs?
Slow severing of actin filaments by cofilin and deletion of actin binding sites from the adapter proteins End4p and Pan1p cause similar defects, prolonged recruitment of both adaptors (step 3, Figure 6) at sites of endocytosis and delayed recruitment of the NPFs Wsp1p and Myo1p. Our experiments show that the actin filament binding sites on End4p and Pan1p are complementary, since deletion of both sites cause more severe defects than deletion of the individual sites. Interestingly, a budding yeast pan1 mutant is synthetic lethal with a clathrin light chain (clc1) truncation mutant that is missing its sla2/end4 binding motif [48].
These features suggest that recruitment of actin filaments by adaptor proteins is part of a negative feedback loop that limits the accumulation of the adaptor proteins, as well as a positive signal for binding NFPs. Actin filaments are not required for adaptor proteins to concentrate at nascent sites of endocytosis, since both accumulate in the presence of LatA. However nothing is known about how binding of actin filaments to these adaptor proteins provides negative feedback to limit their accumulation. Similarly, the LatA experiments showed that actin filaments are not required for Wsp1p to concentrate with the adaptor proteins at sites of endocytosis. Nevertheless, when severing is compromised or the adaptor proteins lack actin filament binding sites, Wsp1p binds only after the adaptor proteins have accumulated to much higher levels. Thus actin filaments must directly or indirectly favor Wsp1p binding.
On the other hand, LatA reduced the recruitment of both Myo1p and Arp2/3 complex and eliminated actin filament polymerization. Verprolin (Vrp1p) may recruit a small number of Myo1p molecules to patches by linking to Wsp1p [25], but direct interactions with actin filaments are likely to recruit most Myo1p to actin patches [25, 29]. Since Myo1p appears in patches prior to Arp2/3 complex [6, 24], the initial binding sites for Myo1p may be actin filament fragments bound to End4p and Pan1p.
Experimental Procedures
Fission yeast strains, cell culture and microscopy
Table 1 lists the strains used in this study. Standard yeast culture techniques were used. Cells were imaged with an Olympus IX71 microscope equipped with a confocal spinning disk unit. The Supplemental Experimental Procedures provide details.
Construction of the adf1::GFP-adf1 strain
We amplified the strong ADH promoter (Padh) sequence from fission yeast genomic DNA by PCR and subcloned into the pFA6a-KanMX6-41nmt1-mGFP vector [49] in place of the 41nmt1 promoter. We amplified the KanMX6-Padh-mGFP fragment for targeting to the 5′ end of endogenous adf1 coding sequence by homologous recombination [49].
Tracking and counting the numbers of fluorescent fusion proteins in actin patches
We tracked actin patches semi-manually with Image J plug-ins. We selected actin patches in time lapse fluorescence micrographs with maximum fluorescence intensities within the middle 3 slices of a 5-slice Z-series. We measured the fluorescence intensity of each actin patch as the sum of the fluorescence in 3 slices within a circle with a diameter of 0.67 μM (5 pixels), after subtracting the background cytosolic fluorescence based on the fluorescence surrounding the patch. Final values of the fluorescence intensities of the patches were corrected for exposure time, camera noise, uneven illumination and photobleaching [6, 31]. We converted the fluorescence intensities of actin patches into the numbers of mGFP fusion proteins with a standard curve based on the total cellular intensities of seven standard proteins [6, 24].
Protein purification and binding assay
GST-Pan11633-1794 was expressed in E. coli and purified by passing through affinity, gel filtration and ion exchange columns. GST-Pan11633-1794 was bound to actin filaments at room temperature and centrifuged at 213,300 g for 40 min. For details, please see Supplemental Experimental Procedures.
Supplementary Material
Supplemental data
Highlights.
- Cofilin severs actin filaments during disassembly of endocytic actin patches
- Defective actin filament severing also delays early steps in actin patch assembly
- Actin binding domains of adaptor proteins help initiate actin polymerization
- We propose a “sever, diffuse and trigger” model for activation of Arp2/3 complex
Acknowledgments
This work was supported by NIH research grant GM-025338 to TDP. The authors thank Julien Berro for providing Image J plug-ins, Rajesh Arasada for providing strains and helpful discussions. The authors also thank Jian-Qiu Wu and Masaki Edamatsu for providing strains.
References
- 1.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 2.Newpher TM, Smith RP, Lemmon V, Lemmon SK. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Aghamohammadzadeh S, Ayscough KR. Differential requirements for actin during yeast and mammalian endocytosis. Nat Cell Biol. 2009;11:1039–1042. doi: 10.1038/ncb1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinberg J, Drubin DG. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 2012;22:1–13. doi: 10.1016/j.tcb.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- 6.Sirotkin V, Berro J, Macmillan K, Zhao L, Pollard TD. Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol Biol Cell. 2010;21:2894–2904. doi: 10.1091/mbc.E10-02-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berro J, Sirotkin V, Pollard TD. Mathematical modeling of endocytic actin patch kinetics in fission yeast: disassembly requires release of actin filament fragments. Mol Biol Cell. 2010;21:2905–2915. doi: 10.1091/mbc.E10-06-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 9.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basu R, Chang F. Characterization of dip1p reveals a switch in Arp2/3-dependent actin assembly for fission yeast endocytosis. Curr Biol. 2011;21:905–916. doi: 10.1016/j.cub.2011.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulholland J, Preuss D, Moon A, Wong A, Drubin D, Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins A, Warrington A, Taylor KA, Svitkina T. Structural organization of the actin cytoskeleton at sites of clathrin-mediated endocytosis. Curr Biol. 2011;21:1167–1175. doi: 10.1016/j.cub.2011.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin AC, Welch MD, Drubin DG. Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat Cell Biol. 2006;8:826–833. doi: 10.1038/ncb1443. [DOI] [PubMed] [Google Scholar]
- 14.Chan C, Beltzner CC, Pollard TD. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr Biol. 2009;19:537–545. doi: 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. J Mol Biol. 2007;365:1350–1358. doi: 10.1016/j.jmb.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Q, Pollard TD. Actin filament severing by cofilin is more important for assembly than constriction of the cytokinetic contractile ring. J Cell Biol. 2011;195:485–498. doi: 10.1083/jcb.201103067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lappalainen P, Drubin DG. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 21.Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J Cell Biol. 2007;178:1251–1264. doi: 10.1083/jcb.200703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano K, Mabuchi I. Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol Biol Cell. 2006;17:1933–1945. doi: 10.1091/mbc.E05-09-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin MC, Galletta BJ, Sept D, Cooper JA. Overlapping and distinct functions for cofilin, coronin and Aip1 in actin dynamics in vivo. J Cell Sci. 2010;123:1329–1342. doi: 10.1242/jcs.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 25.Sirotkin V, Beltzner CC, Marchand JB, Pollard TD. Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast. J Cell Biol. 2005;170:637–648. doi: 10.1083/jcb.200502053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HR. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engqvist-Goldstein AE, Kessels MM, Chopra VS, Hayden MR, Drubin DG. An actin-binding protein of the Sla2/Huntingtin interacting protein 1 family is a novel component of clathrin-coated pits and vesicles. J Cell Biol. 1999;147:1503–1518. doi: 10.1083/jcb.147.7.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wendland B, Emr SD. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee WL, Bezanilla M, Pollard TD. Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp. J Cell Biol. 2000;151:789–800. doi: 10.1083/jcb.151.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lechler T, Shevchenko A, Li R. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J Cell Biol. 2000;148:363–373. doi: 10.1083/jcb.148.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arasada R, Pollard TD. Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast. Curr Biol. 2011;21:1450–1459. doi: 10.1016/j.cub.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ti SC, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci U S A. 2011;108:E463–471. doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coue M, Brenner SL, Spector I, Korn ED. Inhibition of actin polymerization by latrunculin A. FEBS Lett. 1987;213:316–318. doi: 10.1016/0014-5793(87)81513-2. [DOI] [PubMed] [Google Scholar]
- 34.Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morton WM, Ayscough KR, McLaughlin PJ. Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat Cell Biol. 2000;2:376–378. doi: 10.1038/35014075. [DOI] [PubMed] [Google Scholar]
- 36.McCann RO, Craig SW. The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc Natl Acad Sci U S A. 1997;94:5679–5684. doi: 10.1073/pnas.94.11.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toshima J, Toshima JY, Martin AC, Drubin DG. Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nat Cell Biol. 2005;7:246–254. doi: 10.1038/ncb1229. [DOI] [PubMed] [Google Scholar]
- 38.Iwaki T, Tanaka N, Takagi H, Giga-Hama Y, Takegawa K. Characterization of end4+, a gene required for endocytosis in Schizosaccharomyces pombe. Yeast. 2004;21:867–881. doi: 10.1002/yea.1134. [DOI] [PubMed] [Google Scholar]
- 39.Duncan MC, Cope MJ, Goode BL, Wendland B, Drubin DG. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]
- 40.Wesp A, Hicke L, Palecek J, Lombardi R, Aust T, Munn AL, Riezman H. End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:2291–2306. doi: 10.1091/mbc.8.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skruzny M, Brach T, Ciuffa R, Rybina S, Wachsmuth M, Kaksonen M. Molecular basis for coupling the plasma membrane to the actin cytoskeleton during clathrin-mediated endocytosis. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1207011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodal AA, Kozubowski L, Goode BL, Drubin DG, Hartwig JH. Actin and septin ultrastructures at the budding yeast cell cortex. Mol Biol Cell. 2005;16:372–384. doi: 10.1091/mbc.E04-08-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urbanek AN, Smith AP, Allwood EG, Booth WI, Ayscough KR. A novel actin-binding motif in Las17/WASP nucleates actin filaments independently of Arp2/3. Curr Biol. 2013;23:196–203. doi: 10.1016/j.cub.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 44.Young ME, Cooper JA, Bridgman PC. Yeast actin patches are networks of branched actin filaments. J Cell Biol. 2004;166:629–635. doi: 10.1083/jcb.200404159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin SG, Chang F. Dynamics of the formin for3p in actin cable assembly. Curr Biol. 2006;16:1161–1170. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 46.Gowrishankar K, Ghosh S, Saha S, CR, Mayor S, Rao M. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell. 2012;149:1353–1367. doi: 10.1016/j.cell.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 47.Kukulski W, Schorb M, Kaksonen M, Briggs JA. Plasma Membrane Reshaping during Endocytosis Is Revealed by Time-Resolved Electron Tomography. Cell. 2012;150:508–520. doi: 10.1016/j.cell.2012.05.046. [DOI] [PubMed] [Google Scholar]
- 48.Boettner DR, Friesen H, Andrews B, Lemmon SK. Clathrin light chain directs endocytosis by influencing the binding of the yeast Hip1R homologue, Sla2, to F-actin. Mol Biol Cell. 2011;22:3699–3714. doi: 10.1091/mbc.E11-07-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data