APC is an RNA-Binding Protein and its Interactome Provides a Link to Neural Development and Microtubule Assembly (original) (raw)
. Author manuscript; available in PMC: 2015 Jul 17.
SUMMARY
Adenomatous polyposis coli (APC) is a microtubule plus-end scaffolding protein important in biology and disease. APC is implicated in RNA localization, although the mechanisms and functional significance remain unclear. We show that APC is an RNA-binding protein, and identify an RNA interactome by HITS-CLIP. Targets were highly enriched for APC-related functions, including microtubule organization, cell motility, cancer and neurologic disease. Among the targets is β2B-tubulin, known to be required in human neuron and axon migration. We show β2B-tubulin is synthesized in axons and localizes preferentially to dynamic microtubules in the growth cone periphery. APC binds the β2B-tubulin 3'UTR; treatments interfering with this interaction reduced β2B-tubulin mRNA axonal localization and expression, depleted dynamic microtubules and the growth cone periphery, and impaired neuron migration. These results identify APC as a platform binding functionally-related protein and RNA networks, and suggest a self-organizing model for the microtubule to localize synthesis of its own subunits.
INTRODUCTION
Microtubules are fundamental for the spatial organization of the cell, providing structural elements for cell shape, localization of subcellular components, and cell motility. Their function in both static and dynamic cell processes is reflected at the molecular level by the existence of distinct populations of stable and dynamic microtubules. In migrating axons, for example, stable microtubules are bundled in the axon shaft and growth cone central domain, while dynamic microtubules rapidly explore the growth cone peripheral domain which initiates the direction of movement. A key site for regulation of microtubule dynamics is the plus-end, where most growth and shortening occur. Plus-end-tracking (+TIP) proteins are known to regulate microtubule dynamics, although the precise mechanisms by which they operate are generally not well understood (Lowery and Van Vactor, 2009; Dent et al., 2011; Stiess and Bradke, 2011).
Adenomatous Polyposis Coli (APC) is a +TIP protein, which promotes microtubule assembly, and has been studied extensively for its roles in biology and disease. Structurally, APC is a large scaffold protein with binding sites for multiple protein partners (Figure S1A). It was initially identified as a major tumor suppressor, and is mutated in most human colon carcinomas, as well as in brain tumors (Kinzler and Vogelstein, 1996; Aoki and Taketo, 2007; Burgess et al., 2011), and has also been implicated in neurologic disorders including schizophrenia and autism (Cui et al., 2005; Kalkman, 2012). Its normal biological functions include cytoskeletal regulation in cell and axon migration, cell polarity and adhesion (Zhou and Snider, 2006; Aoki and Taketo, 2007; Barth et al., 2008). In addition to its role in microtubule regulation, APC functions in the canonical Wnt signaling pathway, which regulates gene transcription (Clevers and Nusse, 2012). Although Wnt and microtubule pathways were initially studied separately, it has become increasingly clear that they are interconnected, and both are proposed to contribute to effects of APC in biology and cancer (Nathke, 2006; Salinas, 2007).
Both microtubules and APC have been studied extensively in the context of the neuron, a well-suited model due to its highly organized shape and motility. Microtubules play critical roles in axon and neuron migration, axon-dendrite polarity, and synapse formation (Lowery and Van Vactor, 2009; Dent et al., 2011; Stiess and Bradke, 2011). The microtubule has also long been known to have central roles in neurodevelopmental and neurodegenerative diseases (Gerdes and Katsanis, 2005; Manzini and Walsh, 2011). Moreover in recent years, mutations in several individual tubulin genes, including TUBB2B, TUBB3, TUBA1A, and TUBA8 have been found to cause Mendelian disorders in humans resulting in abnormal axon guidance or cortical neuron migration (Tischfield and Engle, 2010). The distinctive phenotypes of these tubulin mutations have supported the multi-tubulin hypothesis, which proposes that the many tubulin isotypes encoded by different genes have distinctive functional properties. However, the mechanisms that contribute to isotype-specific tubulin functions are generally not well understood (Tischfield and Engle, 2010).
RNA-based mechanisms have increasingly been implicated in the development and functioning of the nervous system, including in axon guidance, neuron migration and synapse plasticity, as well as in diseases such as mental retardation (Darnell, 2013; Jung et al., 2014). In principle, important advantages of regulation at the RNA level include the potential to direct protein synthesis to specific subcellular locations, and also the ability to coordinately regulate extensive mRNA networks independent of the structure of the encoded proteins. A major advance in characterizing mRNA networks at the genome-wide level has been the introduction of HITS-CLIP, a technique that uses UV crosslinking to identify RNAs directly bound to a protein of interest in the native context of cells or tissues, followed by high throughput sequencing (Licatalosi et al., 2008; Darnell, 2013), although the understanding of neural protein-RNA regulatory networks remains at an early stage. Intriguingly, a study in fibroblasts found that APC associated, directly or indirectly, with RNA and was required for localization of two RNAs into pseudopodia (Mili et al., 2008). Here we set out to investigate the mechanism of APC interaction with RNA, its genome-wide set of targets, and the biological significance of these interactions.
Our results identify a novel function of APC as an RNA-binding protein. HITS-CLIP in native brain tissue identified 260 high-confidence mRNA targets, encoding proteins strikingly related to APC in function. One of these mRNAs encodes β2B-tubulin / Tubb2b, a tubulin isotype required in cortical neuron migration and axon tract formation in humans (Jaglin et al., 2009; Cederquist et al., 2012; Romaniello et al., 2012). β2B-tubulin protein was found to preferentially localize in dynamic microtubules in the growth cone peripheral domain. Its expression was positively regulated by APC, and treatments to block the interaction of APC with the binding site in the β2B-tubulin mRNA 3’UTR caused loss of dynamic microtubules and the growth cone peripheral domain, and impairment of cortical neuron migration in vivo. These results indicate that the diversity of tubulin isotypes can be explained in part by acquisition of RNA regulatory motifs specifying distinctive subcellular distributions. They also provide a mechanism for control of microtubule dynamics, and suggest a self-organization principle for a polarized cellular structure, the microtubule, to direct the localized synthesis of its own components. The results also show that in addition to APC’s role as a scaffold for a network of proteins, it also binds a functionally-related network of mRNAs, providing a generalizable way to coordinate cellular networks at the protein and RNA levels, with general implications for the actions of APC in biology and disease.
RESULTS
Genome-wide identification of APC target mRNAs in brain
We initially considered the possibility that APC might directly bind RNA for several reasons. APC plays a role in RNA localization in cultured fibroblasts (Mili et al., 2008). Also, APC has an intrinsically unstructured basic region (Figures S1A and S1B) that binds DNA (Deka et al., 1999; Qian et al., 2008). Since unstructured regions are prevalent in both DNA-binding (Vuzman and Levy, 2012) and RNA-binding proteins (Castello et al., 2012), and allow specific binding to more than one type of target (Tompa, 2012), and since other proteins can bind both DNA and RNA, we experimentally investigated the associations of APC with RNA using CLIP. UV crosslinking was carried out in native E14 mouse brain tissue, followed by APC immunoprecipitation, digestion of free RNA, radiolabeling of the protected RNAs, and resolution of protein-RNA complexes on a denaturing gel (Figure 1A). In previous studies using several APC antibodies, the full-length and best-characterized protein isoform migrated at ~300 KDa, with additional shorter isoforms, and here APC-associated RNAs migrated in a comparable pattern (Figure 1A). No protein-RNA complexes were detected in cells bearing truncated APC alleles lacking the region recognized by the APC antibodies, providing a control for antibody specificity (Figure S1C). To further test binding with RNA, the CLIP experiment was repeated using cells transfected with a construct expressing the APC basic domain fused to an HA tag, and the results indicated direct binding of the APC basic domain to RNA (Figure 1B). These data, further confirmed by evidence presented later, identify a new biochemical function for APC as an RNA-binding protein.
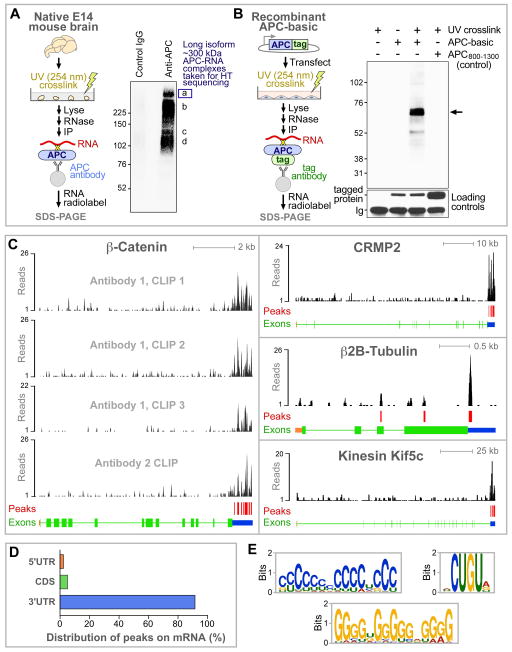
Figure 1. HITS-CLIP identification of RNAs interacting with APC.
(A) APC HITS-CLIP with E14 mouse brain. Protein-RNA complexes were UV-crosslinked in native brain tissue, brains were homogenized, RNAs were trimmed to ~40 nt, APC or control antibodies were used for immunoprecipitation, RNAs were radiolabeled, and RNA-protein complexes were separated by SDS-PAGE and autoradiographed. a–d: RNA-protein complexes reproducibly pulled down with APC antibodies.
(B) Direct interaction between APC basic domain and RNA (arrow). N2A neuroblastoma cells were transfected with a plasmid expressesing APC basic domain fused to an HA tag, followed by UV-crosslinking, lysis, RNA trimming to ~10 nt, immunoprecipitation with HA antibody, RNA radiolabeling, SDS-PAGE and autoradiography. Negative controls are in lanes 1, 2 and 4; APC amino acids 800–1300 provides a control region also predicted to be unstructured (Figure S1B).
(C) Distribution of APC HITS-CLIP sequence reads on representative target genes. Red vertical bars indicate high-confidence APC binding peaks (see Figure S1D and Experimental Procedures).β-Catenin gene is shown starting at exon2 . Exons arein orange (5’UTR), green (protein coding) or blue (3’UTR).
(D) APC binding peaks were predominantly in mRNA 3'UTRs.
(E) Consensus sequence motifs identified within APC binding regions using MEME software.
See also Figure S1 and Table S1.
To perform a genome-wide identification of RNA targets for APC in brain, we next used HITS-CLIP, which combines CLIP with high-throughput sequencing (Figure 1A) (Licatalosi et al., 2008). To reduce potential false positives, two different APC antibodies were used, and for one of them three independent immunoprecipitation replicates were performed, giving a total of four data sets (Figure S1D). Binding peaks were identified on the basis of having multiple sequence reads in all four samples, with height and fold enrichment strongly above background to give a low False Discovery Rate (FDR<0.001; Figure S1D and Experimental Procedures). This procedure identified a set of 631 high-confidence binding peaks in mRNAs from 260 genes.
Figure 1C shows examples of peak distribution for representative target mRNAs encoding proteins well known to have APC-related functions: β-catenin, a central protein in the Wnt signaling pathway; CRMP2, a key regulator of microtubule assembly; β2B-tubulin, a structural component of microtubules; and kinesin Kif5c, a major axonal microtubule motor protein. The great majority of APC binding sites were in mRNA 3’UTRs (582/631 peaks; Figures 1C and 1D). As is common for protein-RNA interactions, no unique binding sequence could be identified. However, three consensus motifs were strongly over-represented (Figure 1E), with 77% of APC target mRNAs carrying at least one of these motifs within APC binding sites.
Global analysis of APC targets in biology and disease
Given the well-known role of APC in microtubule function, we first performed a systematic search for components and regulators of the microtubule cytoskeleton encoded by APC mRNA targets (see Experimental Procedures). This analysis revealed an extensive set of proteins involved in microtubule organization, including tubulins, regulatory proteins that bind microtubules, and key upstream regulatory components (Figure 2A; also Tables S1 and S2). In addition to their relation with microtubules, these proteins bind and regulate one another, forming a highly interconnected protein-protein interaction network. Moreover, APC targets include actin and actin-regulatory factors, consistent with previous studies showing that APC can bind and promote the assembly of actin filaments (Okada et al., 2010), and with the broader concept that microtubules and the actin cytoskeleton are tightly coordinated (Lowery and Van Vactor, 2009; Dent et al., 2011). Together, these observations identify a highly interconnected network of encoded cytoskeletal proteins closely related to known APC functions, suggesting a model where APC function could involve coordinate regulation of both protein and RNA networks.
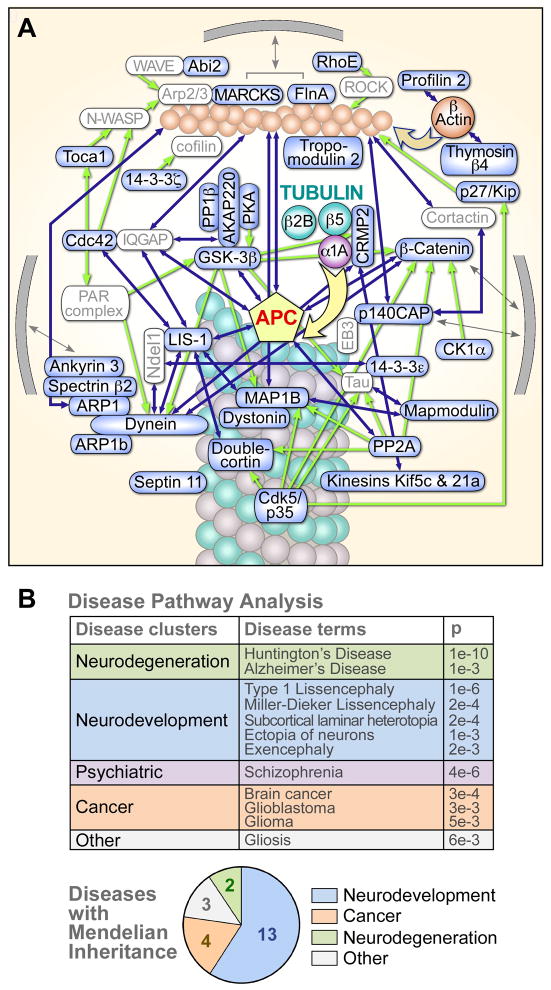
Figure 2. APC RNA interactome in cytoskeleton organization and in disease.
(A) APC RNA interactome encodes a highly interconnected network of cytoskeleton-related proteins. All represented proteins are encoded by APC mRNA targets identified here, except APC itself and icons in gray. Physical interactions are indicated by touching or overlapping icons, or by blue arrows. Green arrows indicate major regulatory interactions; gray arrows indicate association with the plasma membrane. Dync1li2 and Dynll2 are shown as a single dynein icon for simplicity; and Ppp2ca, a subunit of the PP2A complex, as a single PP2A icon. Cytoskeleton-related mRNAs were identified by database searches and manually curated for accuracy (Experimental Procedures and Table S2).
(B) APC target mRNAs encode proteins linked to cancer and neurologic diseases. Upper: Disease terms and clusters associated with APC targets, identified with the IPA package (see Experimental Procedures). Lower: APC mRNA targets corresponding to genes that cause Mendelian disorders in humans.
See also Figure S2, and Tables S1, S2 and S3.
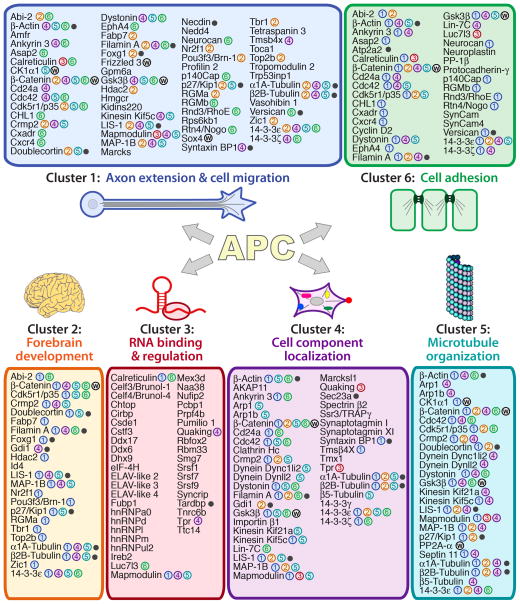
For a more global analysis of APC mRNA targets, we performed gene ontology (GO) analysis of biological processes using the DAVID platform (http://david.abcc.ncifcrf.gov/). The top six clusters are shown in Figure 3 (also Table S4), and notably they all correspond to biological processes in which APC is well known to play key roles. Similar clusters were robustly obtained whether APC targets were compared to a control consisting of the brain transcriptome used here for APC-CLIP (Table S4), or the whole mouse transcriptome (Refseq database)(Table S5). Notably, a substantial proportion of the genes on the APC target list fall into these top six functional categories (128/260 genes), a proportion likely to be an underestimate since not all genes will yet have been tested for these six categories, and because additional APC functions are not represented here (such as Wnt signaling, see below; and synapse formation, the seventh cluster, see Table S4). In addition to their relation with APC, these functional categories are highly related to one another, as reflected by the fact that the majority of the genes are found across more than one category (color coded dots in Figure 3).
Figure 3. APC mRNA targets are functionally related to APC and to one another.
Represented here are the top 6 functional clusters of APC targets identified by DAVID analysis. After automated identification of the top six clusters based on gene ontology terms of biological processes, the list of genes for each of these clusters was curated for enhanced accuracy (see Experimental Procedures). Proteins in more than one cluster are cross-referenced with color-coded numbers; those also in the Wnt signaling pathway, marked “w”; and those also causing Mendelian disorders, black dots.
See also Figure S2, and Tables S1, S4 and S5.
When the APC targets were tested for potentially enriched signaling pathways, using the Ingenuity Pathway Analysis (IPA) platform, the Wnt/β-catenin pathway was identified as the most enriched signaling pathway (p<4×10−5; see Table S1 for Wnt pathway components), a striking finding in view of the well-known role played by APC in this pathway. We also investigated the relationship of the APC target list to human disease using the IPA tools (Figure 2B upper panel, also Table S3), and also by a systematic search for genes that cause human disorders with Mendelian inheritance (Figure 2B lower panel, also Figure S2). Both analyses revealed a relationship of APC mRNA targets to neurodevelopmental and neurodegenerative diseases, and cancer, consistent with studies linking these disorders with APC, the Wnt/β-catenin pathway, and microtubules. Taken together, these analyses show that APC binds a highly coherent set of mRNA targets, closely related to its roles in both normal biology and disease, and suggest a model where the action of APC as an RNA-binding protein may play a role in these processes.
β2B-tubulin mRNA as an APC target in the axon
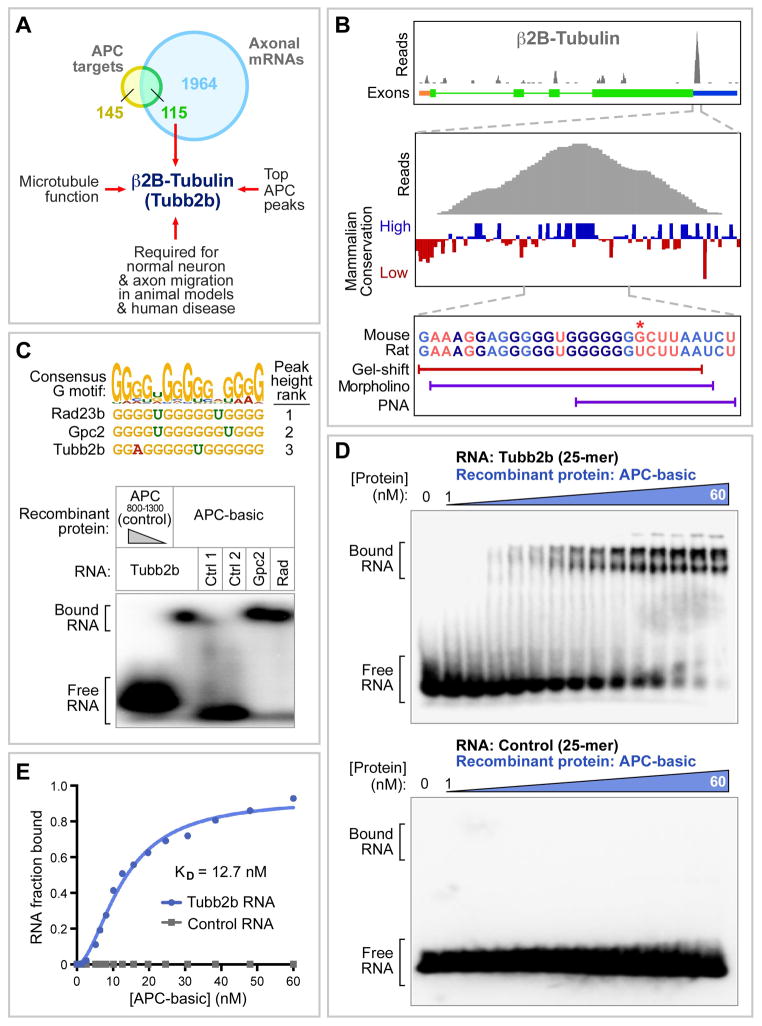
Having obtained a list of mRNAs with APC binding sites, we were interested to experimentally investigate potential functional effects of these binding sites. To identify candidate mRNAs for further study, we compared our target list to a list of mRNAs prsent in dorsal root ganglion (DRG) axons (Gumy et al., 2011), since APC is involved in axon growth, and found that 45% of APC targets were present on the axonally expressed list (Figure 4A, also Tables S1 and S6). Among these, β2B-tubulin emerged as a particularly interesting candidate. β2B-tubulin had a robust APC binding peak in its mRNA 3’UTR (Figures 4B and S3A) and has an obvious role in microtubule assembly. Moreover, in rodent embryos, similar to APC conditional knockout, β2B-tubulin RNAi causes impaired cortical neuron migration (Jaglin et al., 2009; Yokota et al., 2009), while in humans, β2B-tubulin mutations cause asymmetric polymicrogyria, a disorder associated with severe mental retardation, and characterized by defects in cortical neuron migration and axon tracts (Jaglin et al., 2009; Cederquist et al., 2012; Romaniello et al., 2012). Together, these biochemical, developmental and genetic results provided a framework of information to further study β2B-tubulin in the context of migrating axons and neurons.
Figure 4. APC basic domain binds a sequence in the β2B-tubulin mRNA 3’UTR.
**(A)**β2B -tubulin was selected for further study based on the indicated criteria.
(B) APC HITS-CLIP peak in _β2B-tubulin_3’UTR is centered on a conserved G-rich motif. Below the sequence are locations of the RNA probe used for band-shift assay, and the antisense morpholino and PNA oligomers used in subsequent Figures to block the G-motif. Asterisk marks difference between rat (morpholino) and mouse (PNA) sequences. Upper panel is also in Figure 1C.
(C) APC basic domain binds G-rich sequences found within CLIP peaks. Upper: sequences from the top 3 APC target mRNAs containing the G-rich motif (ranked by height of the peak containing the motif). Lower: interactions of APC with these sequences were tested by band-shift. Recombinant APC-basic domain (50 nM) or control APC region (50 nM and 400 nM) was incubated with radiolabeled RNA oligonucleotides (25 or 26mers; see Supplemental information), and run on a native polyacrylamide gel. RNA-protein complexes were observed with probes spanning G-rich motifs from Rad23b, Gpc2, and Tubb2b, but not with control probes (Ctrl 1 contains binding site for the RNA-binding protein HuR; Ctrl 2 is a random 26mer).
(D) RNA-binding affinity was determined by incubating the Tubb2b G-rich motif probe, or control RNA probe, with increasing concentration of APC basic domain.
(E) Binding curve and affinity. Ratio of bound to total (bound plus free) RNA at each protein concentration was derived from the gels in panel D, and fitted with the Hill equation. The Hill coefficient was 1.8, potentially consistent with APC binding as a dimer. KD from repeated experiments was in the 5–15 nM range.
See also Figure S3, and Tables S1 and S6.
To investigate the functional significance of the APC binding site in the β2B-tubulin mRNA, we first wanted to delineate the site in more detail. The HITS-CLIP binding peak in the 3’UTR was centered on a sequence that fitted the consensus G-rich motif, and showed evolutionary conservation (Figures 4B, 4C top, and S3A). To further confirm a direct protein-RNA interaction, we performed in vitro binding assays using purified components. Gel-shift experiments showed that recombinant purified APC basic region bound to short synthetic RNAs containing the G-rich sequence found in the binding peak from β2B-tubulin or other target mRNAs (Figure 4C). The binding had a high affinity, in the range of known RNA-binding proteins interacting with their cognate RNA targets (KD ~10 nM; Figures 4D and 4E). These in vitro binding results, taken together with the CLIP data in cells and in native brain tissue (Figures 1A, 1B and S1C), further confirmed that APC directly binds RNA, and indicate that the G-rich motif in the β2B-tubulin binding peak is a sequence that can bind directly to APC-basic with high affinity.
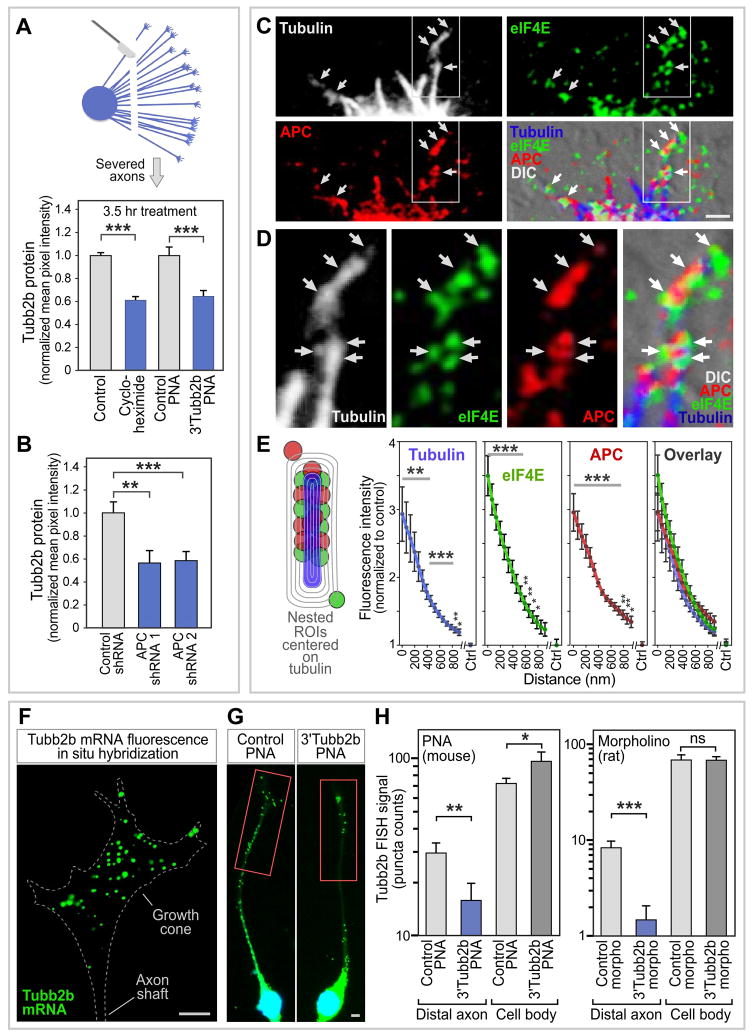
To investigate the regulatory effect of APC binding to the β2B-tubulin mRNA 3’UTR, an antisense oligomer was designed to block the G-motif (3’Tubb2bAPC-site PNA, see Figure 4B, mouse sequence). Peptide-nucleic acids (PNAs) were chosen because they have enhanced binding specificity, and antisense oligos were designed to have multiple mismatches with any other mRNA in the transcriptome (Supplemental Experimental Procedures). Treatment of mouse DRG neurons with the 3’Tubb2bAPC-site PNA reduced β2B-tubulin protein levels in axons (Figure S4C). The presence of β2B-tubulin mRNA in axons, and past evidence for tubulin translation in axons (Eng et al., 1999; Zivraj et al., 2010; Gumy et al., 2011) makes β2B-tubulin a likely candidate for local axonal translation. To confirm local translation in the axon, severed axons in culture were treated with the protein synthesis inhibitor cycloheximide, resulting in a 38% reduction in β2B-tubulin protein expression within 3.5 hours (Figure 5A). To test for a local axonal effect of the APC binding site on β2B-tubulin expression, severed axons were treated with the antisense 3’Tubb2bAPC-site PNA, which had a similar effect to cycloheximide, reducing β2B-tubulin protein expression by 35% within 3.5 hr (Figures 5A, S4A and S4B). These data suggest a model in which APC binding to the β2B-tubulin mRNA would promote β2B-tubulin protein expression. Supporting this model, β2B-tubulin protein levels were reduced when APC was knocked down using small hairpin RNAs (Figures 5B, S4D and S4E), further establishing APC as a positive regulator of β2B-tubulin protein expression.
Figure 5. Axonal synthesis of β2B-tubulin, and localization of translation machinery.
(A) β2B-tubulin protein levels in severed axons are reduced by a protein synthesis inhibitor, or by antisense PNA oligomer to block APC binding to the _β2B-tubulin_3’UTR . β2B-tubulin levels were measured by immunofluorescence. E14 mouse whole DRG explants were cultured for 2 days, then axons were severed and treated as indicated for 3.5 h.
**(B)**Axonal β2B -tubulin protein levels were reduced when APC levels were knocked down using shRNAs in dissociated cultures of DRG neurons.
(C) APC and the translation initiation factor eIF4E cluster at microtubule tips and isolated microtubule fragments (arrows) in axons of dissociated rat DRG neurons. Growth cone periphery is shown here, from central domain at bottom to lamellipodial leading edge at top. Scale bar 2 μm.
(D) Enlargement of boxed area in panel C.
(E) Quantitation of immunolabeling at isolated microtubule fragments in the periphery of DRG growth cones. Isolated microtubule domains were identified and delineated using an automated approach (see Experimental Procedures), and nested regions of interest (ROIs) centered on tubulin were then used to measure the distribution of tubulin, eIF4E and APC labeling, confirming that these markers cluster together at isolated microtubule fragments. Control level was average for peripheral domain.
(F) Fluorescence in situ hybridization (FISH) of β2B-tubulin mRNA in the growth cone of N1E-115 neuronal cell line. Growth cone area is outlined by dashed line. Scale bar 5 μm.
(G) Treatment of N1E-115 cells with the antisense PNA directed against the APC binding site in the _β2B-tubulin_3’UTR reduces β2B-tubulin mRNA FISH signal in the distal axon (boxed in red). Scale bar 5 μm.
(H) Quantitation of the FISH signal in the distal axon, and in the cell body; see panel G. Left: mouse N1E-115 neuronal cell line treated with PNA mouse antisense oligo. Right: rat B35 neuronal cell line treated with morpholino rat antisense oligo. n=8–13.
Error bars, SEM; *p<0.05, **p<0.01, ***p<0.001, t-test.
See also Figure S4.
We were also interested to assess whether APC might localize near components of the translation machinery in growth cones. Immunolocalization showed that markers of translation machinery, including eIF4E and ribosomal protein Rpl19, formed clusters in the growth cone periphery, which were often found in overlapping or contiguous distributions with clusters of APC (Figures 5C–5E, S4F and S4H). The slight offset sometimes seen between APC and eIF4E or Rpl19 puncta is consistent with the large size, oligomerization, and intrinsically unstructured character of APC, which allow it to extend over substantial distances (up to 600 nm per APC molecule) (Minde et al., 2011). APC was similarly found in overlapping or contiguous distributions with sites of newly synthesized protein visualized by pulse labeling with the amino acid analog azidohomoalanine (AHA) (Figure S4G). We were also interested to assess whether APC-translation machinery clusters localize near the microtubule cytoskeleton, focusing on the peripheral domain where microtubules are spread out and can best be observed. Microtubule dynamics in the growth cone involve cycles of polymerization and depolymerization at microtubule filament tips, as well as the presence of small isolated microtubule fragments (Myers et al., 2006; Stiess and Bradke, 2011). APC and markers of translation machinery were seen around a subset of microtubule tips, and in most growth cones showed consistent clustering around isolated microtubule fragments (Figures 5C–5E, S4F and S4H), which may represent either severed or locally nucleated microtubules (Stiess and Bradke, 2011).
We next tested whether the β2B-tubulin mRNA might be found in the growth cone periphery, where dynamic microtubles extend. Using fluorescence in situ hybridization (FISH) we found that β2B-tubulin mRNA is indeed located in the axon including in the growth cone periphery (Figure 5F). When neurons were treated with the antisense 3’Tubb2bAPC-site PNA, β2B-tubulin mRNA levels dropped in the distal axon and increased in the soma (Figures 5G and 5H), indicating that blocking the APC binding site in the _β2B-tubulin_3’UTR inhibits axonal localization of the mRNA.
To further validate the specificity of the antisense approach, these experiments were repeated with antisense morpholinos, another class of nucleic acid analog with enhanced target selectivity (Karkare and Bhatnagar, 2006). Both the 3’Tubb2bAPC-site PNA and morpholino overlap the same G-motif, but the PNA shares only 12/25 residues of the morpholino target sequence. Additionally, the morpholino was designed to be used in a different species, rat, to take advantage of a single-nucleotide mouse/rat difference within the overlapping region (asterisk in Figure 4B), further eliminating the possibility that both antisense oligomers could significantly bind any potential common off-target RNA. As expected, the antisense 3’Tubb2bAPC-site morpholino interfered with the binding of the APC basic domain to the 3’Tubb2bAPC-site RNA oligonucleotide in vitro (Figure S3B) and, confirming the results obtained with the PNA, it inhibited localization of the β2B-tubulin mRNA in the distal axon (Figures 5H and S4I). In contrast, when rat neurons were treated with the 3’Tubb2bAPC-site PNA designed to target the mousesequence, no effect was seen on β2B-tubulin mRNA levels or localization (Figure S4I). Together these results indicate that the APC binding site in the β2B-tubulin mRNA promotes axonal localization of the β2B-tubulin mRNA and axonal expression of β2B-tubulin protein.
Roles in growth cone structure and neuron migration
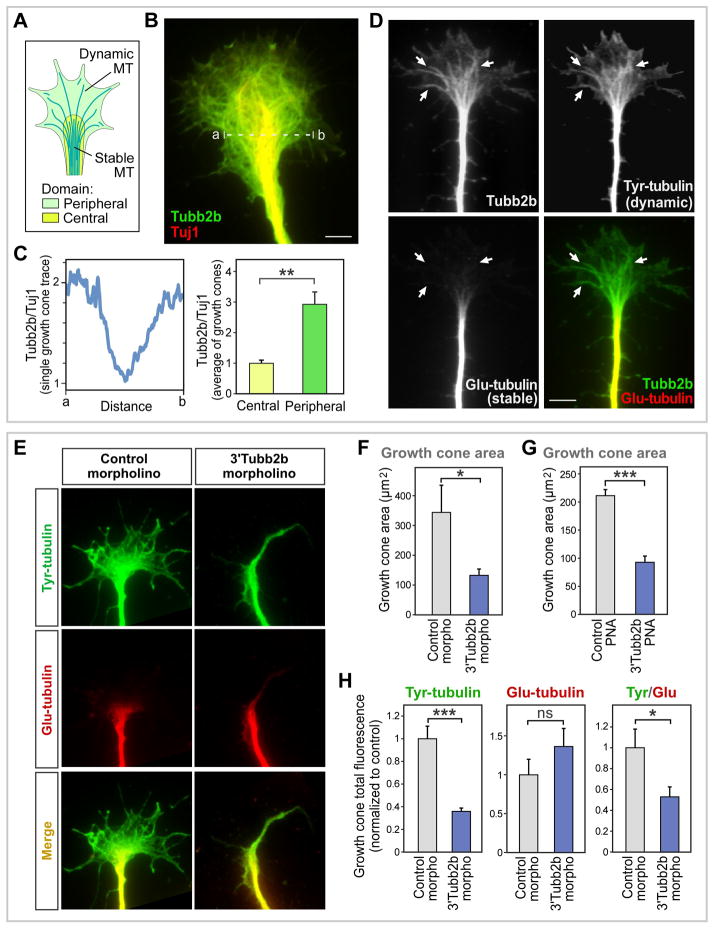
To investigate the functional significance of β2B-tubulin local protein synthesis in axons, we next examined the distribution pattern of this protein in dissociated DRG neurons. β2B-tubulin labeling was seen prominently in axons, and extended well into the peripheral domain of the growth cone, including the actin-rich periphery and filopodia (Figures 6A, 6B and S5A). Similar results were obtained with two different antibodies, and immunolabeling was blocked by siRNAs targeting the β2B-tubulin mRNA (data not shown). When the peripheral to central distribution was assessed quantitatively, compared to β3-tubulin (Tuj1, which is widely used as an axonal marker and was not on our APC target list), there was a strong enrichment of β2B-tubulin labeling in the peripheral domain (2.9-fold enrichment; Figure 6C). A similar result was obtained by comparing to pan-α-tubulin antibodies (data not shown). When β2B-tubulin immunolocalization was compared with tyrosinated (Tyr-tubulin; dynamic) and detyrosinated (Glu-tubulin; stable) microtubules, it was found to overlap with dynamic microtubules in the periphery, where there was little or no labeling for stable microtubules (Figures 6D and S5B). β2B-tubulin labeling was additionally seen in the axon shaft where both dynamic and stable microtubules are present (Figures 6D and S5B). While these results do not exclude the possible presence of β2B-tubulin in stable microtubules (as indeed one might expect if β2B-tubulin were incorporated into dynamic microtubules that subsequently become stabilized), they show prominent overlap of the β2B-tubulin pattern with dynamic microtubules in the growth cone periphery.
Figure 6. Localization and essential role for APC target β2B-tubulin in the growth cone peripheral domain.
(A) Schematic diagram of a growth cone, showing stable microtubules bundled in the central domain, and dynamic microtubules extending into the peripheral domain.
(B) β2B-tubulin is enriched at the growth cone periphery relative to other tubulin isotypes. Dissociated rat DRG neurons were immunolabeled for β2B-tubulin and Tuj1 (β3-tubulin). Scale bar 5 μm.
(C) Left: β2B-tubulin/Tuj1 intensity ratios across the growth cone in B, measured along the dashed line.Right: β2B -tubulin/Tuj1 intensity ratios averaged over multiple growth cones at their minimum (center) and maximum (periphery), normalized to 1 at the center. n=8, **p<0.01.
(D) β2B-tubulin protein distribution overlaps dynamic microtubules. Dissociated rat DRG neurons were immunolabeled for β2B-tubulin, Tyr-tubulin (dynamic microtubules) and Glu-tubulin (stable microtubules). Arrows indicate examples of overlap for β2B-tubulin and dynamic microtubules in the peripheral domain. Scale bar 5 μm.
(E) Growth cone area, and peripheral dynamic microtubules, were reduced by treatment of dissociated rat DRG neurons with 3’Tubb2bAPC-site morpholino that blocks APC-binding site in the β2B-tubulin 3'UTR.
(F) Rat DRG neurons treated with control or 3’Tubb2bAPC-site morpholino were co-electroporated with a GFP plasmid to measure growth cone area. n=37–39, *p<0.05.
(G) Mouse DRG neurons were incubated with control or 3’Tubb2bAPC-site antisense PNA oligomer, and growth cone area was quantified based on actin (phalloidin-rhodamine) fluorescence. n=55–151, ***p<10−14.
(H) Quantitation of growth cone Glu- or Tyr-tubulin fluorescence signals, obtained as described in panel E. n=73–88; *p<0.05, ***p<10−7.
Error bars, SEM; p values by t-test.
See also Figure S5.
Our findings on β2B-tubulin distribution in the axon suggested a functional role in the growth cone periphery. To study the function of APC binding to the _β2B-tubulin_3’UTR , we next analyzed the morphology of DRG neurons treated with the 3’Tubb2bAPC-site antisense oligomers. The 3’Tubb2bAPC-site morpholino had no evident effect on the overall morphology of neurons and their axons, or axon length (data not shown), but growth cone area was strongly reduced (2.6-fold reduction; Figure 6F). Similarly, the 3’Tubb2bAPC-site PNA did not affect overall morphology of the axon or soma, but strongly reduced growth cone area, compared to a non-targeting PNA (2.3-fold reduction; Figure 6G) or compared to a PNA targeted to a neighboring region of the _β2B-tubulin_3’UTR (data not shown). Immunolabeling of stable and dynamic microtubules indicated that the overall loss of growth cone area corresponded to a selective loss of the growth cone peripheral domain and dynamic microtubules, while stable microtubules, found in the central domain, appeared unaffected (Figures 6E and 6H). These results lead to a model in which tubulin synthesis in axons is promoted by APC binding to the G-rich motif in the β2B-tubulin mRNA 3’UTR, and in turn is important for normal extension of peripheral dynamic microtubules and formation of the classic expanded shape of the growth cone.
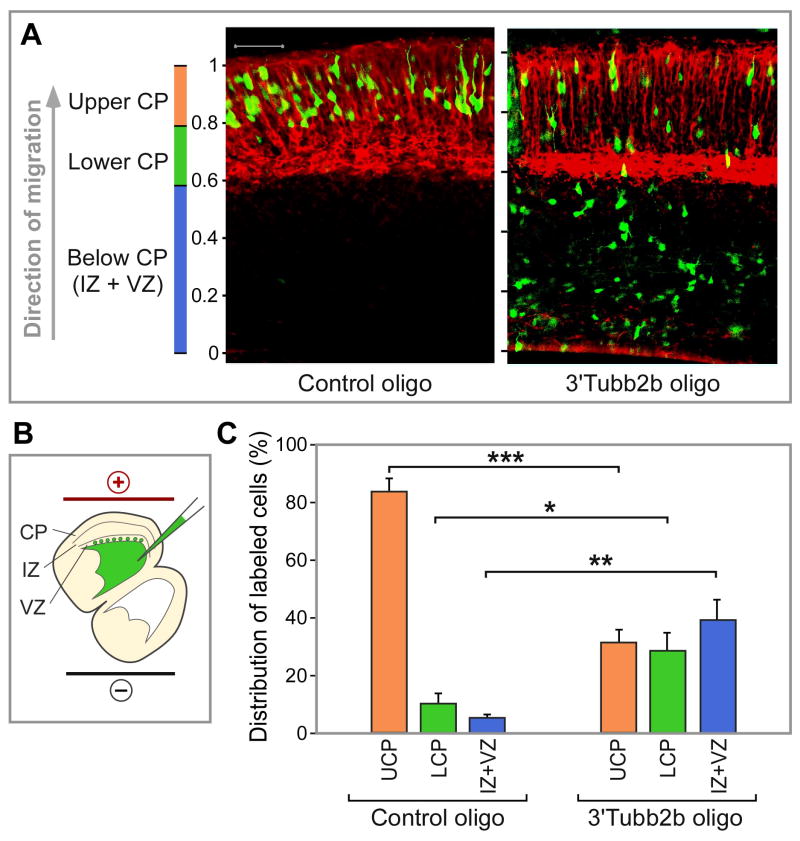
To extend our studies of β2B-tubulin regulation to the developing brain in vivo, we examined cortical neuron migration, for several reasons. Migrating cortical neurons extend a leading process that guides their migration and has a cytoskeletal architecture similar to axons (Marin et al., 2010). Moreover, cortical neuron migration is impaired in both APC- and β2B-tubulin-deficient rodent models (Jaglin et al., 2009; Yokota et al., 2009), while mutations in human β2B-tubulin cause defects in axon tracts and cortical neuron migration (Jaglin et al., 2009; Cederquist et al., 2012; Romaniello et al., 2012). Therefore, based on our data in growth cones, we hypothesized that cortical neuron migration might be impaired by blocking the β2B-tubulin 3’UTR APC binding site. To test this, the 3’Tubb2bAPC-site antisense morpholino was introduced into the brain of E15.5 rat embryos by in utero electroporation. Three days later, cortical neurons electroporated with a control morpholino had migrated all the way to the upper layers of the cortex, while a majority of neurons electroporated with the 3’Tubb2bAPC-site antisense morpholino had not reached the upper cortical plate (Figure 7), showing an impairment of cortical neuron migration in vivo. We also tested the APC basic domain, reasoning that it would have dominant interfering effects on endogenous APC activity, since this domain binds RNAs (Figures 1 and 4), but lacks other regions of APC required for protein-protein interactions and will therefore not localize and function properly (Askham et al., 2000). As predicted, similar to the antisense oligonucleotide against the APC binding site, expressing the basic domain of APC strongly reduced cortical neuron migration in vivo (Figure S7).
Figure 7. Antisense blockade of APC binding site on the β2B-tubulin mRNA impairs cortical neuron migration in vivo (A).
Sections of neocortex from rat embryos electroporated as illustrated in (B), and quantitated in (C). MAP2 immunofluorescence (red) marks the cortical plate. Antisense morpholinos and a GFP-expressing plasmid were co-injected into the ventricles of E15.5 rat embryos in utero, and introduced by electroporation into neurons lining the ventricle. At E18.5, most neurons had reached the upper half of the cortical plate (Upper CP) after treatment with control morpholino, whereas most neurons remained in deeper layers with the antisense 3’Tubb2bAPC-site morpholino. n=3–4 embryos, *p<0.05; **p<0.02; ***p<0.001. Error bars, SEM; p values by t-test. CP: cortical plate; IZ: intermediate zone; VZ: ventricular zone.
See also Figure S7.
DISCUSSION
Here we find that APC, a protein-protein scaffold studied extensively for its important roles in biology and disease, is also an RNA-binding protein, and moreover it associates with a network of mRNAs highly related to its functions. The dual ability of APC to act as a platform for extensive associations with both protein and RNA networks suggests novel mechanisms to regulate biological network function coordinately at the protein and RNA levels. APC is known as a microtubule +TIP protein regulating microtubule assembly, and we find here that APC directs mRNA localization and synthesis of a specific tubulin isotype, β2B-tubulin. Our results suggest a model where APC would promote microtubule polymerization in part by directing the local translation of tubulin in the vicinity of microtubule growing ends, suggesting a self-organizing principle for a polarized cellular structure, the microtubule, to spatially direct the synthesis of its own components.
APC as an mRNA-binding protein
APC has not been previously identified as an RNA-binding protein in part because its sequence does not contain any classical RNA-binding domain. Interestingly, this is consistent with a recent genome-wide screen reporting that approximately half of all proteins that bind RNA do not have any recognizable RNA-binding domain (Castello et al., 2012). Although the APC basic region does not show obvious primary sequence homology with classical RNA-binding domains, it does share properties commonly found in other RNA-binding proteins including a low frequency of acidic residues and a high prediction value for intrinsic disorder (Castello et al., 2012; Dyson, 2012). In addition to being enriched in RNA binding proteins, intrinsically unstructured protein regions are currently of high interest in biology more generally. Such regions allow scaffolding proteins such as APC to sweep a large volume of the cell, and also they have the ability to conformationally adapt to bind more than one type of target molecule with high specificity (Minde et al., 2011; Tompa, 2012).
Regarding recognition motifs, RNA-binding proteins typically bind sequences that are short or highly degenerate, and in many cases it has been difficult to identify any consensus sequences. Here, three enriched consensus motifs were identified within APC target sites, with 77% of the APC target mRNAs containing at least one of them. The ability of RNA-binding proteins to recognize binding sites with a variety of primary sequences can have at least two general explanations. First, structurally flexible RNA-binding domains may be able to bind multiple target motifs (Dyson, 2012). Second, binding sites in RNA often involve secondary structures that can be generated by various primary sequences (Hall, 2002). Our in vitro binding experiments using APC basic domain and short RNA oligonucleotides demonstrated high affinity binding to the G-rich consensus motif. This motif might possibly form a G-quadruplex, a structure recognized by some other nucleic acid-binding proteins (Burge et al., 2006), although the G-quadruplex consensus does not precisely fit the G-motif identified here. Comparable binding to synthetic oligonucleotides containing the other two motifs was not detected (unpublished observations); there could be various reasons for this: the short RNAs used in our assay may not have possessed necessary secondary structures; or additional APC regions outside the basic domain may contribute to RNA binding; or APC might bind cooperatively with other RNA-binding proteins. Indeed, APC has been reported to associate directly or indirectly with RNA binding proteins FMRP and Fus/TLS (Mili et al., 2008; Yasuda et al., 2013), and it would be interesting to investigate whether these interactions contribute to APC mRNA binding specificity.
Isotype specific tubulin regulation
The two main cytoskeletal components at the front of migrating cells and axons are microtubules and actin filaments. Regulation of actin synthesis has been studied extensively over the past two decades. Local actin synthesis at the leading edge is limited to the β-actin isoform, under control of elements in its mRNA 3’UTR (Condeelis and Singer, 2005; Jung et al., 2014). Analogous to actin – indeed substantially more numerous – tubulins exist in multiple isotypes encoded by distinct genes. Although the significance of tubulin isotypes remains little explored, growing evidence indicates that they have distinctive biological functions, confer different dynamic properties to microtubules, and cause different diseases when mutated (Tischfield and Engle, 2010). Our studies on the β2B-tubulin isotype suggest a model where individual tubulin isotypes would have distinct sets of regulatory elements in their 3’UTRs, allowing them to be subject to different posttranscriptional control mechanisms and contributing to their distinctive functions.
One potential reason for RNA-based regulation of specific tubulin isotypes could be to control their differential spatial distribution. This hypothesis is supported by multiple observations in our data. The β2B-tubulin mRNA was localized to axons and locally translated there under control of the APC binding site in its 3’UTR, and we found that APC and translational machinery clustered together near microtubule tips and isolated microtubule fragments in the growth cone peripheral domain. Also, the β2B-tubulin isotype, relative to other tubulins, showed preferential localization to dynamic microtubules in the growth cone periphery. Correspondingly, blocking the APC binding site in the β2B-tubulin 3’UTR caused a strong and selective loss in dynamic microtubules and in area of the growth cone periphery. Consistent with these findings, previous studies have described alteredmicrotubule behavior in APC conditional knockout mice, includinga reduced duration of microtubule growth events, and altered directionality with less frequent growth toward the periphery (Yokota et al., 2009; Chen et al., 2011). Our results all support a model where axonal synthesis of β2B-tubulin contributes to the extension of peripheral dynamic microtubules enriched in β2B-tubulin,and plays an important role in formation of the classical expanded shape of the axonal growth cone. More generally, our data suggest that an advantage of the existence of multiple tubulin isotypes, may have been the evolutionary acquisition of distinct RNA regulatory motifs specifying unique subcellular distributions.
Interconnected networks of APC-bound mRNAs
Regulation at the mRNA level presents unique advantages for biological organization and regulation. One is the ability to direct the synthesis of specific proteins to specific subcellular sites. Consistent with such a role for APC, in addition to β2B-tubulin as discussed above, other proteins encoded by APC mRNA targets – including β-actin, β-catenin, and import in-β – are known to be locally translated at the leading edge of migrating cells or in axons (Hanz et al., 2003; Condeelis and Singer, 2005; Jones et al., 2008).The localization of two mRNAs in fibroblast protrusions was previously reported to involve APC (Mili et al., 2008); these mRNAs were not in our APC RNA interactome, although they were in our mouse brain transcriptome, suggesting either that they interact with APC indirectly, or that there might be brain/fibroblast differences, such as alternative 3’UTRs, or different proteins cooperating with APC. The same study also identified a genome-wide list of mRNAs that move into fibroblast protrusions in response to extracellular cues, although this was not a list of APC-associated mRNAs, and it may be informative to test in future whether all fibroblast protrusion mRNAs are associated with APC (there was little overlap of that list with our brain APC interactome; 2/260 mRNAs). An alternative model, rather than APC being involved in localization of all mRNAs found in the periphery of cells, is that different subsets of mRNAs might be localized by different RNA-binding proteins, a model that may have advantages for biological specificity.
Besides spatial organization, a second advantage of RNA-based regulation is that it can coordinately regulate large protein networks, independent of the structure of the encoded proteins. Here we find that the APC mRNA interactome encodes protein networks highly interconnected by overlapping functions, regulatory relationships, and physical associations. Although there has been no clear evidence for coordinated assembly of an entire locally synthesized protein network, the proximity of mRNAs during protein synthesis can contribute to formation of protein complexes co-translationally (Chang et al., 2006). The functionally coherent mRNA interactome of APC clearly suggests mechanisms to coordinately regulate an entire protein network temporally, or spatially.
While axonal levels of β2B-tubulin RNA and protein were positively regulated here, it may be interesting to investigate whether APC binding sites on mRNAs might also have negative effects on protein expression, and what are the exact mechanisms involved. The mechanisms of RNA localization, translation and stability are coupled, and it is common for individual RNA-binding proteins to influence more than one of these processes (Dreyfuss et al., 2002). Other aspects that might be interesting for future investigation are the potential relationship with previously described RNA granules (Xing and Bassell, 2012; Yasuda et al., 2013), or cell surface receptors (Tcherkezian et al., 2010), and whether APC is part of a constitutive mechanism of the growth cone machinery or might be regulated by extracellular cues.
Besides its function as a cytoskeletal regulator, APC is well known as a component of the Wnt/β-catenin pathway, where it binds and regulates the degradation of β-catenin protein (Clevers and Nusse, 2012). Interestingly, β-catenin is known to be translated locally at the leading edge of migrating cells (Jones et al., 2008). Here we find that APC binds the mRNA for β-catenin, and several other proteins in the Wnt/β-catenin pathway, leading to a model where APC may influence the expression or localization of the pathway in part via RNA-based mechanisms.
The specific set of mRNAs associated with APC matches not only APC’s functions in normal biology, but also its involvement in disease, including cancer and neurological disorders. Consistent with a potential involvement of RNA-based mechanisms in these diseases, the APC basic domain, found here to bind RNA, is deleted by the APC truncations that are typically associated with cancer, and missense point mutations in this domain have also been identified (Minde et al., 2011). Cancer therapies commonly target microtubules, but toxicity remains a major caveat of this treatment (Pellegrini and Budman, 2005). Therapeutics that act via RNA-based mechanisms may provide greater specificity by targeting the synthesis of specific components of the microtubule regulatory machinery or tubulin isoforms, and the identification here of APC mRNA targets offers tools to test this hypothesis.
APC has been studied extensively for over two decades, yet it has been difficult to identify mechanisms that might coherently unify its diverse array of effects in biology and disease (Burgess et al., 2011; Minde et al., 2011). Our results may help to understand the overall mechanistic basis of APC function, by identifying a function as an RNA-binding protein and delineating an mRNA interactome for APC. These results lead to a model where APC would act as a platform coordinately regulating both protein-protein and protein-RNA interactions. Our initial characterization of the APC RNA interactome indicates that it may provide a link between APC and its broad spectrum of functions in biology and disease.
EXPERIMENTAL PROCEDURES
CLIP, neuron cultures, binding assays, immunolabeling, in vivo migration
Detailed procedures are in supplemental material.
Statistical analysis
All t-tests were two-tailed, unpaired.
Image quantitation
For growth cone area, and tubulin labeling intensities (Figures 6E–H), growth cones were delineated by GFP or phalloidin fluorescence using the MetaMorph auto-trace function for the distal 20 μm. Tubulin labeling intensities in Figures 5A, 5B and S4A–D were measured in axons, including growth cones and axon shafts. For marker intensities in peripheral tubulin clusters (Figures 5C–E), a contour was initially auto-traced in the tubulin (blue) channel, blind to the other channels; concentric ROIs at 1-pixel intervals were then created inside and outside the tubulin contour, and pixel intensities between these lines were averaged for all clusters; control value was labeling intensity averaged over the entire peripheral domain.
For FISH quantitation, neurons were selected blind to the green channel, and green puncta were then counted in the cell body and distal 50 μm of each axon blind to the treatment. Comparable effects of antisense oligomer were seen when the whole axon was counted, and there was no effect on length or total surface area of the axon that could account for the change in FISH signal (data not shown). To confirm results in the cell body, where puncta were dense, average pixel intensity was measured, revealing comparable effects to puncta counting (21% increase, p<0.03, for Tubb2b PNA).
Bioinformatic analysis
To identify CLIP binding site peaks, reads were initially combined from all four replicates; clusters of overlapping reads were identified, and assigned two values: peak height (maximum number of overlapping reads), and median fold enrichment (compared to randomly sampled background from mRNA-seq). To differentiate enriched binding sites from false positives, mock CLIP data sets were generated by randomly sampling the same number of sequence reads as in the APC CLIP samples from the control mRNA-seq data. High confidence peaks were selected on mature mRNAs, based on peak height, fold-enrichment, and presence of multiple reads in all four replicates (Figure S1D; Tables S1 and S7).
GO analysis of biological function was performed using DAVID online tools (http://david.abcc.ncifcrf.gov/). The top six enriched clusters (Figure 3) were determined by automated functional annotation clustering based on GO terms of biological processes (GO_BP_FAT). The individual genes within these top six categories were then curated for enhanced accuracy by systematic search of the GeneCards Compendium (http://www.genecards.org/), which contains a more comprehensive annotation including literature citations; this validated almost all the initial genes, and identified approximately 60 additional genes unambiguously annotated in these categories and cited in the literature.
APC targets related to the cytoskeleton (Figure 2A), were identified by batch query from the Mouse Genome Informatics (MGI) database based on GO annotations related to microtubule or actin cytoskeletons. Interactions among proteins were identified by data mining in GeneCards (http://www.genecards.org), STRING (http://string-db.org), and BioGRID (http://thebiogrid.org), and were then further validated by systematic searching in PubMed and manually selecting those supported by direct experimental evidence in studies of those individual interactions (Table S2).
Disorders and signaling pathways associated with APC targets were identified using IPA tools (http://www.ingenuity.com/); Figure 2B shows disorders corresponding to a specific disease and represented by at least two genes, within the IPA neurological disease category (the next IPA categories are in Table S6). Single genes causing human Mendelian disorders were identified by a systematic manual search in the OMIM database (http://www.ncbi.nlm.nih.gov/omim).
Supplementary Material
01
02
03
04
05
06
07
08
HIGHLIGHTS.
- Microtubule +TIP protein APC is an RNA-binding protein
- HITS-CLIP identifies 260 mRNA targets, which are highly related to APC functions
- APC target β2b-tubulin is axonally synthesized and localized to dynamic microtubules
- Mechanism required for expanded shape of growth cone and for neuron migration in vivo
Acknowledgments
We thank Kyoko Okada, Philippe Roux, David Van Vactor, Peter Park, Xinmin Li, Inke Näthke, Olivier Pertz, Antonina Andreeva and Alexey Murzin for help, reagents, advice, or comments on the manuscript; Jiangwen Zhang, Christian Daly and the Bauer Center for Genomic Research for support with deep sequencing; and Jennifer Waters, Josh Rosenberg, Lauren Piedmont, Lara Petrak, Gaudenz Danuser, Hunter Elliott, Yichao Xu, the Nikon Imaging Center, and the Image and Data Analysis Core at Harvard Medical School for imaging equipment and expertise. This work was supported by grants from the NIH, and a fellowship from the Charles A. King Trust (N.P.).
Footnotes
ACCESSION NUMBERS
Data accompanying this paper are available through NCBI under accession number SRP042131.
AUTHOR CONTRIBUTIONS
N.P. designed, performed and analyzed most experiments, participated in the bioinformatic analysis, and wrote the manuscript. J.Q. designed and performed most bioinformatic analysis, participated in design and interpretation of the experiments, performed the FISH experiments, and contributed to the manuscript. D.W.N., with N.P., initiated the study and performed pilot experiments. M.L.H. performed the cut axon studies and several immunolabeling experiments. J.S. cloned and purified the recombinant APC polypeptides. J.T. performed the AHA experiments. T.L.Y.-P. designed and performed the cortical migration experiments. J.G.F. supervised the study, designed and analyzed it with the other authors, and wrote the manuscript.
Supplemental information includes Extended Experimental Procedures, six figures, and seven tables and can be found with this article online at [to be inserted].
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aoki K, Taketo MM. Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. J Cell Sci. 2007;120:3327–3335. doi: 10.1242/jcs.03485. [DOI] [PubMed] [Google Scholar]
- Barth AI, Caro-Gonzalez HY, Nelson WJ. Role of adenomatous polyposis coli (APC) and microtubules in directional cell migration and neuronal polarization. Semin Cell Dev Biol. 2008;19:245–251. doi: 10.1016/j.semcdb.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess AW, Faux MC, Layton MJ, Ramsay RG. Wnt signaling and colon tumorigenesis--a view from the periphery. Exp Cell Res. 2011;317:2748–2758. doi: 10.1016/j.yexcr.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Cederquist GY, Luchniak A, Tischfield MA, Peeva M, Song Y, Menezes MP, Chan WM, Andrews C, Chew S, Jamieson RV, et al. An inherited TUBB2B mutation alters a kinesin-binding site and causes polymicrogyria, CFEOM and axon dysinnervation. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Shav-Tal Y, Trcek T, Singer RH, Goldman RD. Assembling an intermediate filament network by dynamic cotranslation. J Cell Biol. 2006;172:747–758. doi: 10.1083/jcb.200511033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Tian X, Kim WY, Snider WD. Adenomatous polyposis coli regulates axon arborization and cytoskeleton organization via its N-terminus. PLoS One. 2011;6:e24335. doi: 10.1371/journal.pone.0024335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Condeelis J, Singer RH. How and why does beta-actin mRNA target? Biol Cell. 2005;97:97–110. doi: 10.1042/BC20040063. [DOI] [PubMed] [Google Scholar]
- Cui DH, Jiang KD, Jiang SD, Xu YF, Yao H. The tumor suppressor adenomatous polyposis coli gene is associated with susceptibility to schizophrenia. Mol Psychiatry. 2005;10:669–677. doi: 10.1038/sj.mp.4001653. [DOI] [PubMed] [Google Scholar]
- Darnell RB. RNA protein interaction in neurons. Annu Rev Neurosci. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deka J, Herter P, Sprenger-Haussels M, Koosch S, Franz D, Muller KM, Kuhnen C, Hoffmann I, Muller O. The APC protein binds to A/T rich DNA sequences. Oncogene. 1999;18:5654–5661. doi: 10.1038/sj.onc.1202944. [DOI] [PubMed] [Google Scholar]
- Dent EW, Gupton SL, Gertler FB. The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Dyson HJ. Roles of intrinsic disorder in protein-nucleic acid interactions. Mol Biosyst. 2012;8:97–104. doi: 10.1039/c1mb05258f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng H, Lund K, Campenot RB. Synthesis of b-tubulin, actin, and other proteins in axons of sympathetic neurons in compartmented cultures. J Neurosci. 1999;19:1–9. doi: 10.1523/JNEUROSCI.19-01-00001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes JM, Katsanis N. Microtubule transport defects in neurological and ciliary disease. Cell Mol Life Sci. 2005;62:1556–1570. doi: 10.1007/s00018-005-5007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Yeo GS, Tung YC, Zivraj KH, Willis D, Coppola G, Lam BY, Twiss JL, Holt CE, Fawcett JW. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17:85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall KB. RNA-protein interactions. Curr Opin Struct Biol. 2002;12:283–288. doi: 10.1016/s0959-440x(02)00323-8. [DOI] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, et al. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- Jaglin XH, Poirier K, Saillour Y, Buhler E, Tian G, Bahi-Buisson N, Fallet-Bianco C, Phan-Dinh-Tuy F, Kong XP, Bomont P, et al. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat Genet. 2009;41:746–752. doi: 10.1038/ng.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KJ, Korb E, Kundel MA, Kochanek AR, Kabraji S, McEvoy M, Shin CY, Wells DG. CPEB1 regulates beta-catenin mRNA translation and cell migration in astrocytes. Glia. 2008;56:1401–1413. doi: 10.1002/glia.20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Gkogkas CG, Sonenberg N, Holt CE. Remote control of gene function by local translation. Cell. 2014;157:26–40. doi: 10.1016/j.cell.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkman HO. A review of the evidence for the canonical Wnt pathway in autism spectrum disorders. Mol Autism. 2012;3:10. doi: 10.1186/2040-2392-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkare S, Bhatnagar D. Promising nucleic acid analogs and mimics: characteristic features and applications of PNA, LNA, and morpholino. Appl Microbiol Biotechnol. 2006;71:575–586. doi: 10.1007/s00253-006-0434-2. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, Van Vactor D. The trip of the tip: understanding the growth cone machinery. Nat Rev Mol Cell Biol. 2009;10:332–343. doi: 10.1038/nrm2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzini MC, Walsh CA. What disorders of cortical development tell us about the cortex: one plus one does not always make two. Curr Opin Genet Dev. 2011;21:333–339. doi: 10.1016/j.gde.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Valiente M, Ge X, Tsai LH. Guiding neuronal cell migrations. Cold Spring Harb Perspect Biol. 2010;2:a001834. doi: 10.1101/cshperspect.a001834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453:115–119. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minde DP, Anvarian Z, Rudiger SG, Maurice MM. Messing up disorder: how do missense mutations in the tumor suppressor protein APC lead to cancer? Mol Cancer. 2011;10:101. doi: 10.1186/1476-4598-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KA, He Y, Hasaka TP, Baas PW. Microtubule transport in the axon: Re-thinking a potential role for the actin cytoskeleton. Neuroscientist. 2006;12:107–118. doi: 10.1177/1073858405283428. [DOI] [PubMed] [Google Scholar]
- Nathke I. Cytoskeleton out of the cupboard: colon cancer and cytoskeletal changes induced by loss of APC. Nat Rev Cancer. 2006;6:967–974. doi: 10.1038/nrc2010. [DOI] [PubMed] [Google Scholar]
- Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189:1087–1096. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini F, Budman DR. Review: tubulin function, action of antitubulin drugs, and new drug development. Cancer Invest. 2005;23:264–273. doi: 10.1081/cnv-200055970. [DOI] [PubMed] [Google Scholar]
- Qian J, Sarnaik AA, Bonney TM, Keirsey J, Combs KA, Steigerwald K, Acharya S, Behbehani GK, Barton MC, Lowy AM, et al. The APC tumor suppressor inhibits DNA replication by directly binding to DNA via its carboxyl terminus. Gastroenterology. 2008;135:152–162. doi: 10.1053/j.gastro.2008.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniello R, Tonelli A, Arrigoni F, Baschirotto C, Triulzi F, Bresolin N, Bassi MT, Borgatti R. A novel mutation in the beta-tubulin gene TUBB2B associated with complex malformation of cortical development and deficits in axonal guidance. Dev Med Child Neurol. 2012;54:765–769. doi: 10.1111/j.1469-8749.2012.04316.x. [DOI] [PubMed] [Google Scholar]
- Salinas PC. Modulation of the microtubule cytoskeleton: a role for a divergent canonical Wnt pathway. Trends Cell Biol. 2007;17:333–342. doi: 10.1016/j.tcb.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Stiess M, Bradke F. Neuronal polarization: The cytoskeleton leads the way. Developmental Neurobiology. 2011;71:430–444. doi: 10.1002/dneu.20849. [DOI] [PubMed] [Google Scholar]
- Tcherkezian J, Brittis PA, Thomas F, Roux PP, Flanagan JG. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell. 2010;141:632–644. doi: 10.1016/j.cell.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield MA, Engle EC. Distinct alpha- and beta-tubulin isotypes are required for the positioning, differentiation and survival of neurons: new support for the 'multi-tubulin' hypothesis. Biosci Rep. 2010;30:319–330. doi: 10.1042/BSR20100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci. 2012 doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Vuzman D, Levy Y. Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Mol Biosyst. 2012;8:47–57. doi: 10.1039/c1mb05273j. [DOI] [PubMed] [Google Scholar]
- Xing L, Bassell GJ. mRNA Localization: An Orchestration of Assembly, Traffic and Synthesis. Traffic. 2012 doi: 10.1111/tra.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Zhang H, Loiselle D, Haystead T, Macara IG, Mili S. The RNA-binding protein Fus directs translation of localized mRNAs in APC-RNP granules. J Cell Biol. 2013;203:737–746. doi: 10.1083/jcb.201306058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Kim WY, Chen Y, Wang X, Stanco A, Komuro Y, Snider W, Anton ES. The adenomatous polyposis coli protein is an essential regulator of radial glial polarity and construction of the cerebral cortex. Neuron. 2009;61:42–56. doi: 10.1016/j.neuron.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FQ, Snider WD. Intracellular control of developmental and regenerative axon growth. Philos Trans R Soc Lond B Biol Sci. 2006;361:1575–1592. doi: 10.1098/rstb.2006.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivraj KH, Tung YC, Piper M, Gumy L, Fawcett JW, Yeo GS, Holt CE. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J Neurosci. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05
06
07
08