Stabilization of the Transcription Factor Foxp3 by the Deubiquitinase USP7 Increases Treg-Cell-Suppressive Capacity (original) (raw)
. Author manuscript; available in PMC: 2014 Aug 15.
SUMMARY
Stable Foxp3 expression is required for the development of functional regulatory T (Treg) cells. Here, we demonstrate that the expression of the transcription factor Foxp3 can be regulated through the polyubiquitination of multiple lysine residues, resulting in proteasome-mediated degradation. Expression of the deubiquitinase (DUB) USP7 was found to be upregulated and active in Treg cells, being associated with Foxp3 in the nucleus. Ectopic expression of USP7 decreased Foxp3 polyubiquitination and increased Foxp3 expression. Conversely, either treatment with DUB inhibitor or USP7 knockdown decreased endogenous Foxp3 protein expression and decreased Treg-cell-mediated suppression in vitro. Furthermore, in a murine adoptive-transfer-induced colitis model, either inhibition of DUB activity or USP7 knockdown in Treg cells abrogated their ability to resolve inflammation in vivo. Our data reveal a molecular mechanism in which rapid temporal control of Foxp3 expression in Treg cells can be regulated by USP7, thereby modulating Treg cell numbers and function.
INTRODUCTION
Foxp3+ regulatory T (Treg) cells are a specific subset of CD4+ T cells that are crucial for the maintenance of self-tolerance (Khattri et al., 2003; Fontenot et al., 2003). The X-chromosome-encoded transcription factor Foxp3 is essential for both Treg cell development and function. Foxp3 mutations in mice as well as in immune dysregulation polyendocrinopathy, enteropathy, and X-chromosome-linked syndrome (IPEX) patients result in the development of complex autoimmune diseases due to Treg cell deficiency (Khattri et al., 2003). T cells manipulated to ectopically express Foxp3 acquire the Treg cell phenotype (Khattri et al., 2003; Hori et al., 2003). Furthermore, a 90% decrease of Foxp3 protein expression due to destabilizing alterations in the 3′ UTR of the Foxp3 messenger RNA (mRNA), thereby destabilizing mRNA, results in significantly impaired Treg-cell-mediated suppression, demonstrating that the amount of Foxp3 protein directly correlates to Treg cell function (Wan and Flavell, 2007).
Constitutive expression of Foxp3 has been demonstrated to be essential for the maintenance of Treg cell suppressor function (Williams and Rudensky, 2007). Although the precise molecular mechanisms regulating expression of the Foxp3 gene are incompletely understood, it has been reported that TGF-β, IL-2, or T cell receptor (TCR) stimulation of T cells can all result in increased Foxp3 expression (Kim and Leonard, 2007; Yao et al., 2007). This is most likely modulated by the demethylation of the Foxp3 promoter or conserved noncoding regions in the Foxp3 locus (Kim and Leonard, 2007). In addition, multiple transcription factors, including CREB-ATF, Ets-1, Foxo1 and Foxo3, and STAT5 have been demonstrated to regulate Foxp3 transcription (Ouyang et al., 2010; Polansky et al., 2010; Yao et al., 2007; Kim and Leonard, 2007).
Foxp3 expression in Treg cell is not unique, given that in vitro TCR stimulation of CD4+CD25− T cells results in the transient expression of Foxp3 mRNA and protein. However, the vast majority of cells do not exhibit a suppressive phenotype, and it is possible that Foxp3 acts here to prevent T cell hyperactivation (Wang et al., 2007; Gavin et al., 2006). In contrast, a small subpopulation of these TCR-stimulated CD4+CD25− cells expresses both high and stable Foxp3 protein, thus acquiring suppressive capacity (Allan et al., 2005; Passerini et al., 2008). These studies, as well as others, have shown that the persistent expression of Foxp3 is essential for the maintenance of suppressor function.
Currently, there is debate as to whether Foxp3+ Treg cells can lose Foxp3 expression and suppressive function and whether they exhibit characteristics of other Th cell subsets. Several independent studies in which Foxp3+ Treg cells were adoptively transferred into lymphopenic mice demonstrated that 10%–50% of the transferred cells lost Foxp3 expression (Gavin et al., 2007; Komatsu et al., 2009; Duarte et al., 2009). Furthermore, Treg cells from both the periphery and the thymus were found to be converted into Th17 cells upon stimulation with anti-CD3, anti-CD28, and IL-6, demonstrating a degree of plasticity (Yang et al., 2008). In addition, Foxp3+ Treg cells have been shown to convert to a Foxp3− Th1 cell phenotype upon Toxoplasma infection (Oldenhove et al., 2009). In contrast, studies with (conditional) Foxp3 GFP-CRE mice that were crossed with ROSA26 reporter mice demonstrated that Foxp3 was remarkably stable and that only a very small subpopulation lost its Foxp3 expression (Rubtsov et al., 2010; Miyao et al., 2012). These differences could potentially be explained by the “pollution” of Teff cells that transiently upregulate Foxp3 without gaining a Treg cell phenotype. In addition, Miyao et al. (2012) demonstrated that Foxp3+ Treg cells could transiently downregulate Foxp3 expression, which was rapidly regained along with suppressive capacity upon activation. Because these studies have all demonstrated that Foxp3 protein expression can be rapidly and, often, transiently lost, we have focused on the molecular mechanism regulating this process.
Protein expression in cells can be regulated by both protein production and degradation rates. Much of the regulated proteolysis in eukaryotic cells is catalyzed by the ubiquitin-proteasome system (Ciechanover and Schwartz, 2002). Covalently attached ubiquitin chains of four or more ubiquitin proteins mark a protein for degradation by the 26S proteasome (Hochstrasser, 2006). Protein ubiquitination is a tightly regulated process modulated by E1, E2, and E3 ligases, which, in a complex, catalyze the addition of ubiquitin to lysine residues of the target protein. Here, the initial ubiquitin serves as an acceptor for additional cycles of ubiquitin modification, resulting in a developing polyubiquitin chain (Eldridge and O’Brien, 2010). Protein deubiquitination is an equally well-regulated process modulated by a large family of deubiquitinating enzymes (DUBs). DUBs catalyze the removal of ubiquitin from specific protein substrates, thereby preventing protein degradation, resulting in increased target protein expression (Nijman et al., 2005).
Our group, as well as others, has previously shown that Foxp3 can be polyubiquitinated; however, the regulation of this process and its modulators has remained elusive (Dang et al., 2011; van Loosdregt et al., 2011; van Loosdregt et al., 2010). Here, we establish that the DUB USP7 (also known as HAUSP) is active in primary Treg cells and associates with Foxp3. Ectopic expression of USP7 specifically decreased Foxp3 polyubiquitination, resulting in increased Foxp3 protein expression. Conversely, knockdown of USP7 resulted in decreased Foxp3 protein. Furthermore, Treg cell function was noticeably decreased when USP7 was knocked down or when DUB activity was inhibited both in vitro and in vivo. The manipulation of Foxp3 ubiquitination provides a molecular mechanism for assuring rapid temporal control of Foxp3 expression in T cells, thereby regulating Treg cell numbers and function.
RESULTS
DUBs Modulate Treg-Cell-Mediated Suppression
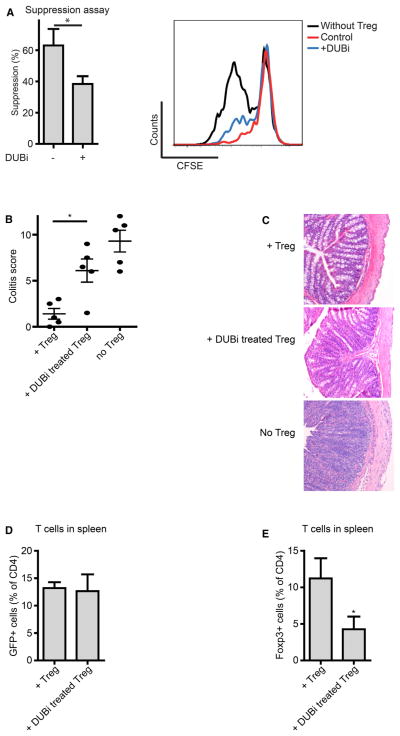
To assess whether ubiquitination could regulate Treg-cell-mediated suppression, we used a pan-DUB inhibitor (DUBi; Figure S1A available online) (Colombo et al., 2010; Patent WO, 2007). Carboxyfluorescein succinimidyl ester (CFSE)-labeled peripheral blood mononuclear cells (PBMCs) were cocultured with sorted human CD4+CD25highCD127low Treg cells that had been pretreated with DUBi separately as described in the Experimental Procedures. Treg-cell-mediated suppression was significantly decreased by preincubating Treg cells with DUBi (Figure 1A). Accordingly, IL-2 mRNA expression was upregulated in Treg cells transiently treated with DUBi both 2 and 7 days after treatment (Figure S1B). Furthermore, treatment with DUBi resulted in the decreased expression of additional Foxp3 transcriptional target genes CD25 and GITR on Foxp3+ Treg cells (Figure S1C).
Figure 1. Ubiquitination Modulates Treg Cell Function.
(A) Sorted human CD4+CD25highCD127low Treg cells were pretreated with 10 μM DUBi for 1 hr, washed, and cocultured with CFSE-labeled PBMCs in anti-CD3-coated wells for 4 days. CFSE dilution of CD4+ cells was analyzed by flow cytometry.
(B) CD4+CD45RBhigh cells were injected into immunodeficient mice. Sorted GFP+ Treg cells from Foxp3-GFP promoter mice were pre-incubated with 10 μM DUBi for 1 hr and injected 3 weeks later. Mice were sacrificed 3 weeks after Treg cell administration. Sections of the colon were analyzed and scored (five mice per group).
(C) Representative hematoxylin- and eosin-stained tissue slides of the colon.
(D) Analysis of CD4+GFP+ cell numbers in the spleen.
(E) Percentage of Foxp3+ CD4+ T cells in the spleen. Data shown are representative of at least three independent experiments, *p<0.05.
Data are represented as mean + SEM. See also Figure S1.
To determine whether ubiquitination could also modulate Treg-cell-mediated suppression in vivo, we used a well-established mouse adoptive transfer colitis model. Here, immunodeficient mice are infused with CD4+CD45RBhigh T cells in order to induce colitis, and disease severity could be reduced by the adoptive transfer of Treg cells (Powrie et al., 1994). Initially, sorted Treg cells isolated from Foxp3-GFP mice were pretreated with DUBi and transferred into _Rag1_−/− mice 3 weeks after the infusion of naïve CD4+ T cells. Mice were sacrificed 3 weeks after Treg cell infusion, and colitis severity was assessed. Treatment with Treg cells noticeably reduced disease scores in comparison to mice that did not receive Treg cells (Figures 1B and 1C). Disease scores of mice receiving Treg cells pretreated with DUBi were significantly increased in comparison to control mice, indicating that the inhibition of DUB activity can significantly abrogate Treg-cell-mediated suppression in vivo. The percentage of GFP+ cells in the spleen was similar in the experimental groups, indicating that Treg cell survival was not affected (Figure 1D). Interestingly, Foxp3 protein expression was significantly reduced in DUBi-treated Treg cells in comparison to untreated Treg cells (Figures 1E and S1D). Given that (poly)ubiquitination is a well-described modulator of protein degradation, these data suggest that Foxp3 protein, and, thus, Treg cell function, may be regulated by ubiquitination.
Foxp3 Protein Expression Is Regulated by Polyubiquitination
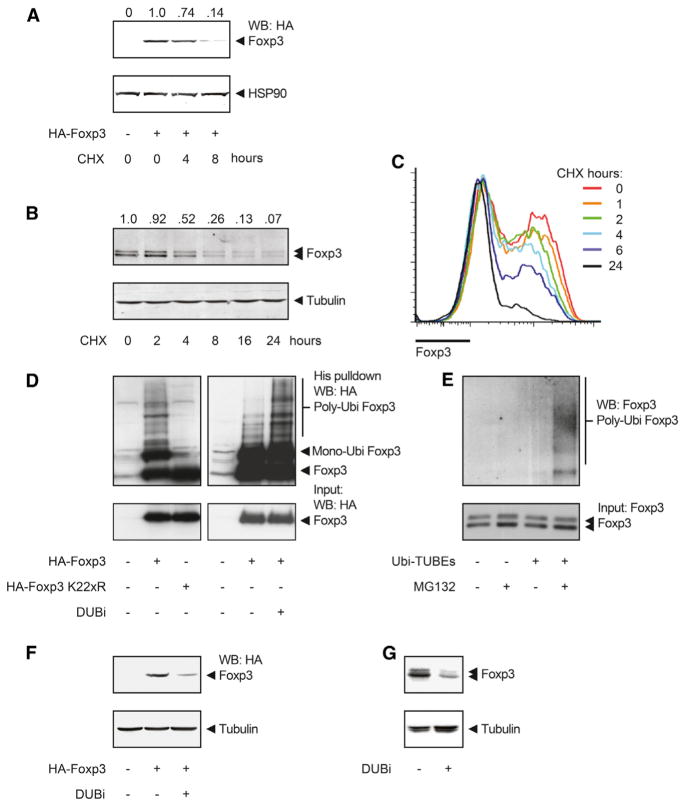
First, to determine whether DUBs can regulate Foxp3 expression, we evaluated Foxp3 protein stability under normal culture conditions. Foxp3-transfected cells or Treg cells were treated with the protein translation inhibitor cycloheximide (CHX), and Foxp3 protein expression was determined. In both transfected human embryonic kidney (HEK) 293T cells (Figure 2A) and human Treg cells (Figure 2B), the inhibition of translation led to a rapid decrease in Foxp3 protein amounts, indicating increased Foxp3 protein degradation. Importantly, Foxp3 mRNA expression in Treg cells was unchanged (Figure S2A). Similar results were obtained when Foxp3 expression was analyzed in human Treg cells by flow cytometry (Figure 2C). Accordingly, protein expression of IL-2Rα (CD25), a direct Foxp3 transcriptional target, also decreased upon CHX treatment (Figure S2B). To further validate that Foxp3 is degraded, we treated Foxp3-transfected HEK 293T cells or natural Treg (nTreg) cells with both CHX and proteasome inhibitor MG132. MG132 treatment impaired the effect of CHX, demonstrating that Foxp3 is indeed degraded by the proteasome (Figures S2C and S2D).
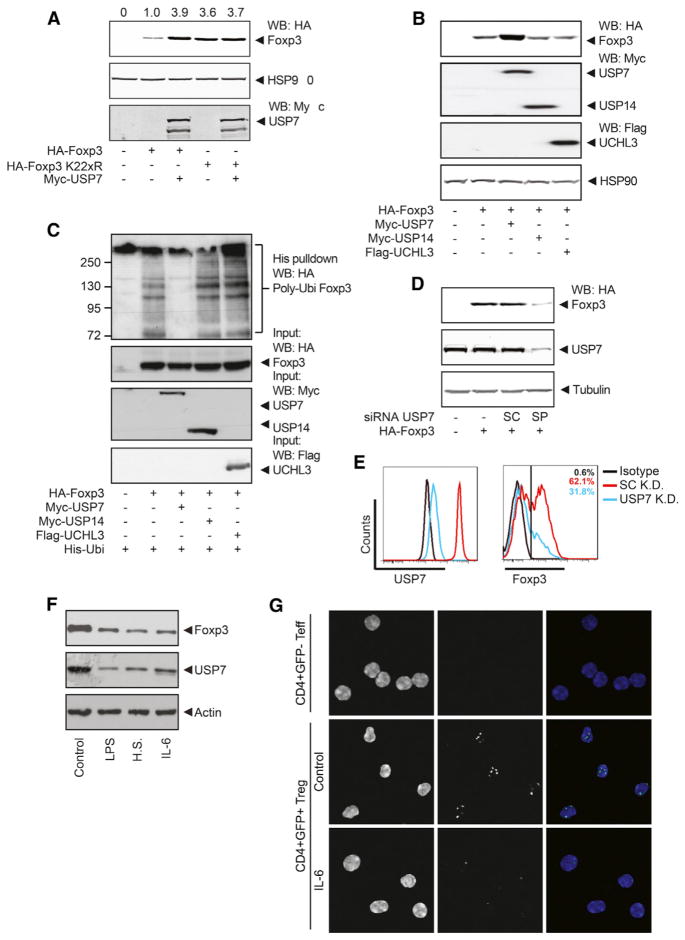
Figure 2. Rapid Turnover of the Foxp3 Protein Is Mediated by Polyubiquitination.
(A) HA-Foxp3-transfected HEK 293T cells were treated with 150 μg/ml cycloheximide (CHX) for 0, 4, and 8 hr. Cell lysates were blotted and analyzed with specific antibodies. Quantification of Foxp3 intensity relative to HSP90 is included.
(B) Human Treg cells were cultured in the presence of 150 μg/ml CHX as indicated. Foxp3 amounts were determined by western blot. Quantification of Foxp3 intensity relative to tubulin is included.
(C) Human Treg cells were cultured in the presence of 150 μg/ml CHX. Foxp3 amounts were determined by flow cytometry.
(D) Ubiquitin pull-down for HA-Foxp3 or HA-Foxp3 K22xR as described in the Experimental Procedures.
(E) Human Treg cells were cultured with 20 μM MG132 for 3 hr, cells were lysed, and ubiquitinated proteins were isolated with TUBE-coupled beads. Ubiquitinated Foxp3 was visualized by western blot with anti-Foxp3.
(F and G) HA-Foxp3-transfected HEK 293T cells (F) or Treg cells (G) were cultured with 5 μM DUBi for 8 hr. Western blots were incubated with antibodies against, HA, Foxp3, or tubulin, as indicated. Data shown are representative of at least three independent experiments. IP, immunoprecipitation; WB, western blot.
See also Figure S2.
Because we had observed that ubiquitination could modulate Treg cell function, we subsequently analyzed the ubiquitination status of Foxp3. HEK 293T cells transfected with both His-tagged ubiquitin and hemagglutinin (HA)-tagged Foxp3 were lysed, and ubiquitinated proteins were isolated with Ni-NTA beads. Foxp3-specific polyubiquitination was determined by immunoblot analysis with anti-HA. A clear polyubiquitination pattern was observed with wild-type (WT) Foxp3 (Figure 2D), and this was abrogated in a Foxp3 mutant in which all lysine residues were mutated (Foxp3 K22xR), indicating that Foxp3 is directly ubiquitinated. Short-term treatment with DUBi increased Foxp3 polyubiquitination, suggesting a continuous deubiquitination of Foxp3 (Figure 2D). Although technically challenging, to confirm that Foxp3 can also be polyubiquitinated in human Treg cells, we lysed cells and incubated them with tandem ubiquitin-binding entities (TUBE)-coupled agarose beads in order to isolate ubiquitinated proteins. Again, Foxp3-specific polyubiquitination was visualized by immunoblot analysis with anti-Foxp3. In the presence of MG132, a clear polyubiquitin pattern was observed, demonstrating that Foxp3 is polyubiquitinated in human Treg cells as well (Figure 2E).
To determine whether Foxp3 polyubiquitination resulted in the regulation of Foxp3 protein expression, we treated HA-Foxp3-transfected cells (Figure 2F) or Treg cells (Figures 2G and S2E) with DUBi, and Foxp3 protein expression was analyzed. Treatment with DUBi decreased Foxp3 protein expression in both cell types. Importantly, DUBi treatment did not affect Foxp3 mRNA expression in Treg cells (Figure S2F). Furthermore, the effect of DUBi on Foxp3 protein expression was dose dependent (Figure S2G). Given that β-catenin protein is heavily dependent on polyubiquitination-mediated degradation, it was used as a positive control for DUBi treatment. As expected, β-catenin amounts were reduced upon treatment (Figure S2H). Foxp3 K22xR expression was not reduced by DUBi treatment because this mutant cannot be polyubiquitinated (Figure S2I). To validate that Foxp3 amounts were decreased because of increased protein degradation, we treated Treg cells with DUBi in the presence of MG132. As expected, treatment with MG132 alone increased Foxp3 expression (Figure S2J), whereas treatment with DUBi resulted in reduced Foxp3 amounts. However, in the presence of MG132, Foxp3 expression was unchanged after DUBi treatment, demonstrating that Foxp3 protein degradation is directly modulated by DUBs. Furthermore, Treg cells were treated with DUBi in the presence of CHX. Again, either DUBi or CHX treatment alone reduced Foxp3 protein expression (Figure S2K). Treatment with both compounds resulted in an even greater reduction in Foxp3 amounts, yet again supporting a role for DUBs in modulating Foxp3 degradation in Treg cells. To determine whether the effect observed of treatment with DUBi was long lasting, we treated Treg cells with DUBi for 1 hr, and Foxp3 protein expression was evaluated either 1 or 7 days after treatment. A single treatment for 1 hr resulted in reduced Foxp3 expression 7 days after treatment (Figure S2L). Taken together, these data show that Foxp3 protein degradation, but not synthesis, is regulated posttranscriptionally, a process that is polyubiquitination dependent and can be rescued by deubiquitination.
Identification of Multiple Foxp3 Ubiquitination Sites
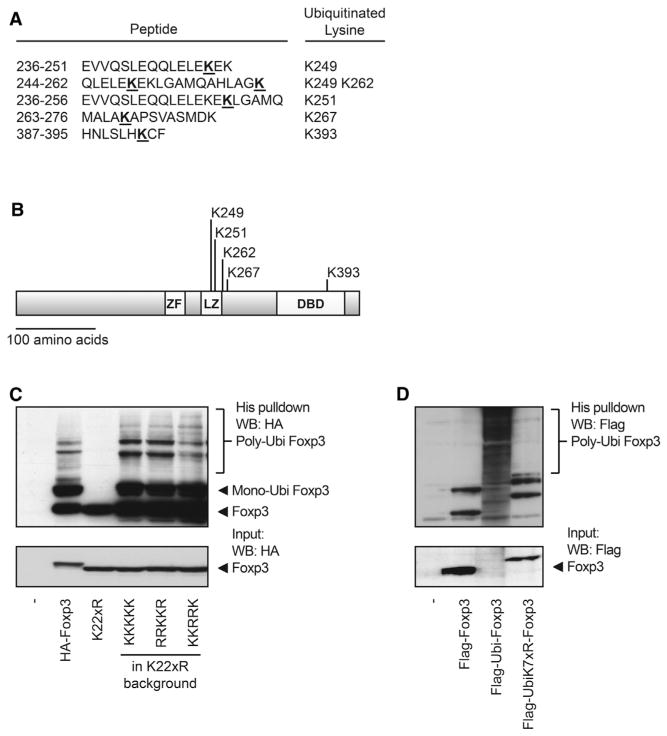
Because we could demonstrate that Foxp3 polyubiquitination results in protein degradation, we examined whether specific lysine residues could be ubiquitinated. Using a mass spectrometry (MS) approach, we found five distinct lysine residues in Foxp3 to be ubiquitinated (K249, K251, K263, K267, and K393; Figures 3A and 3B). Foxp3 expression constructs in which all lysines were mutated into arginines were generated (Foxp3 K22xR) or modified to express combinations of the lysines identified. Subsequently, Foxp3 mutants were analyzed for ubiquitination as previously described. The addition of all five lysine residues that were identified by MS rescued Foxp3 ubiquitination. Similarly, the addition of only lysines 263 and 267 or 249, 251, and 393 restored Foxp3 ubiquitination to amounts similar to WT Foxp3 (Figure 3C).
Figure 3. Identification of Multiple Foxp3 Ubiquitination Sites.
(A) Immunoprecipitated HA-Foxp3 was analyzed for ubiquitinated lysine residues by MS. Peptides continuing ubiquitinated lysine residues are depicted. Ubiquitinated lysine residues are underlined.
(B) A schematic representation of Foxp3 and the lysine residues that are ubiquitinated. ZF, zinc finger motive; LZ, leucine zipper motive; DBD, DNA-binding motive.
(C) Ubiquitin pull-down for HA-Foxp3 or Foxp3 mutants as described in the Experimental Procedures.
(D) FLAG-ubiquitin-Foxp3 or FLAG-ubiquitin-K7xR-Foxp3 fusion constructs (in which all seven lysine residues in ubiquitin are mutated to arginine) were analyzed for ubiquitination as in (C). Data shown are representative of at least three independent experiments. WB, western blot.
See also Figure S3.
To evaluate whether the specific location of modification influences polyubiquitination, we generated a Foxp3 mutant in which ubiquitin was directly fused to the N terminus. The Ubi-Foxp3 fusion protein was highly polyubiquitinated and rapidly degraded, suggesting that polyubiquitination is not dependent on the specific location of lysine residues (Figure 3D). To further verify that the ubiquitin fusion protein was indeed polyubiquitinated, we generated a Ubi-K7xR-Foxp3 fusion construct in which all lysines in ubiquitin, but not in Foxp3, were replaced with arginines. Mutating the ubiquitin fusion protein reduced Foxp3 polyubiquitination back to WT levels. Taken together, these data show that Foxp3 can be polyubiquitinated at multiple lysine residues, which most likely results in rapid proteasome-mediated degradation.
USP7 Associates with and Deubiquitinates Foxp3
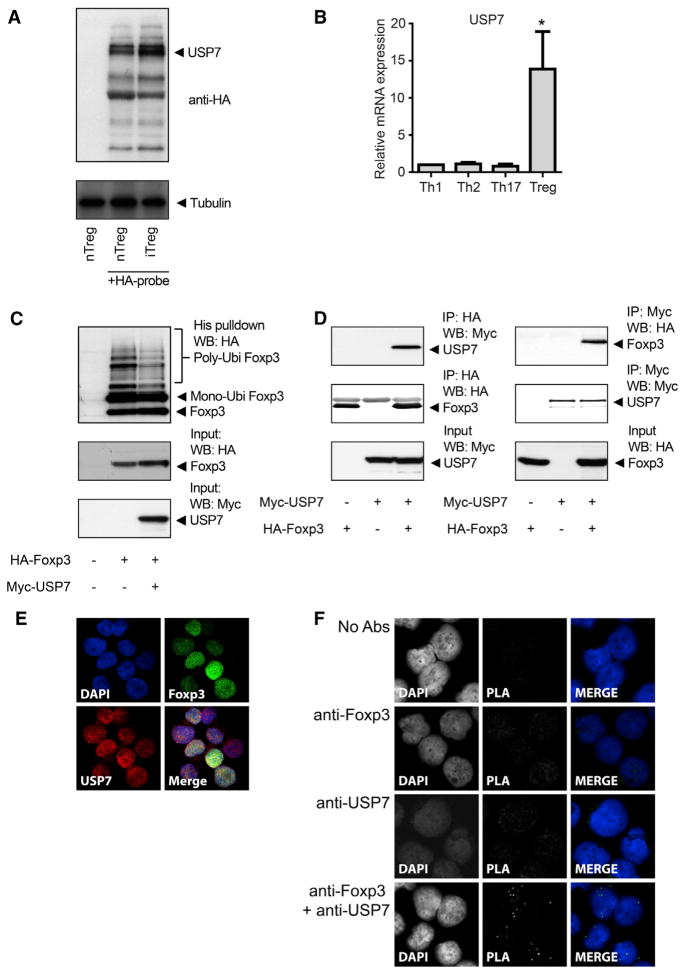
Next, we sought to determine which DUBs could potentially deubiquitinate Foxp3. First, we evaluated which DUBs were both expressed and active in human Treg cells using an HA-tagged probe that covalently binds active DUBs, thus allowing isolation and identification (Borodovsky et al., 2002). Cell lysates from both nTreg and induced Treg (iTreg) cells were incubated with the DUB probe, and active DUBs were immunoprecipitated and separated by SDS-PAGE. Isolated protein bands were analyzed by MS. MS analysis identified the most intense protein band as USP7 (coverage 26%), indicating that this DUB is active in Treg cells (Figures 4A and S3A). Flow cytometric analysis of CD4+CD25highFoxp3+ Treg cells confirmed that USP7 is expressed in Treg cells (Figure S3B). Given that Foxp3 expression is critical during Treg cell development, we analyzed USP7 expression in iTreg cells. Naïve cells were skewed to four different Th cell subsets (Th1, Th2, Th17, and Treg cells; Figure S3C), and USP7 mRNA was analyzed by quantitative RTPCR (qRT-PCR). USP7 mRNA expression was significantly upregulated in iTreg cells but not in other Th subsets, further supporting a role for the USP7 deubiquitination of Foxp3 in Treg cell differentiation (Figure 4B). To determine whether USP7 could also deubiquitinate Foxp3, we analyzed Foxp3 polyubiquitination in transfected HEK 293T cells. Foxp3 polyubiquitination was reduced with ectopic expression of USP7, indicating that USP7 can directly deubiquitinate Foxp3 (Figure 4C). To validate that Foxp3 is a direct USP7 substrate, we analyzed the association of the two proteins by coimmunoprecipitation in HEK 293T cells (Figure 4D). A clear association was observed, indicating that Foxp3 can associate with USP7. To confirm that both proteins interact in human Treg cells, we fixed and permeabilized cells, and both endogenous Foxp3 and USP7 were visualized with specific antibodies. The colocalization of both proteins was observed in the nucleus (Figures 4E and S4A). Similar results were obtained in cells ectopically expressing GFP-tagged USP7 and mKate-tagged Foxp3 (Figure S4B). To further confirm that ectopically expressed or endogenous USP7 and Foxp3 can also directly associate in human Treg cells, we performed an in situ proximity ligation assay (PLA; see the Experimental Procedures). Given that a PLA signal can only be obtained when the proteins of interest are in extremely close proximity, this technique enables the detection of protein-protein interactions in cells. The association of USP7 and Foxp3 was observed, and the interaction was localized specifically to the nucleus (Figures 4F and S4C). Taken together, these data show that USP7, a DUB localized in the nucleus and interacting with Foxp3, is active in both nTreg and iTreg cells and can deubiquitinate Foxp3.
Figure 4. USP7 Deubiquitinates Foxp3.
(A) Human iTreg and nTreg cells were lysed and incubated with an HA-DUB probe for 30 min. Using anti-HA-coupled beads, we immunoprecipitated active DUBs were, and proteins were separated on gel and analyzed by MS.
(B) Naïve human CD4+ T cells were differentiated to Th1, Th2, Th17, and Treg cells. mRNA expression of USP7 was analyzed by quantitative RT-PCR. mRNA expression was normalized for the housekeeping gene GAPDH.
(C) Ubiquitin pull-down for HA-Foxp3 or HA-Foxp3 K22xR as described in the Experimental Procedures.
(D) Cell lysates of HA-Foxp3- and Myc-USP7-transfected cells were immunoprecipitated with anti-HA- or anti-Myc-coupled beads. Immunoblots were analyzed with anti-HA or anti-Myc.
(E) Representative confocal microscopy images of human Treg cells. Endogenous USP7 (red) and Foxp3 (green) were visualized with specific antibodies, and DAPI was used to visualize the nuclei (blue).
(F) Foxp3-USP7 association was visualized in human Treg cells with an in situ proximity ligation assay as described in the Experimental Procedures. Punctate staining (green) indicates a Foxp3-USP7 interaction as detected by the assay, and DAPI was used to visualize the nuclei(blue). *p < 0.05. IP, immunoprecipitation; WB, western blot.
Data are represented as mean + SEM. See also Figures S3 and S4.
USP7-Mediated Foxp3 Deubiquitination Results in Increased Foxp3 Protein Expression
To determine whether USP7-mediated deubiquitination results in increased Foxp3 protein amounts, we assessed Foxp3 expression in HEK 293T cells that were cotransfected with Foxp3 and USP7. Ectopic expression of USP7 with Foxp3 noticeably increased Foxp3 protein amounts (Figure 5A). Foxp3 K22xR protein amounts were not increased by the cotransfection of USP7, indicating that USP7 stabilizes Foxp3 expression by direct deubiquitination. To verify these data, we treated HEK 293T cells transfected with Foxp3 and USP7 with CHX in order to determine the effect of USP7 on Foxp3 half-life. Indeed, USP7 decreased the rate of degradation of Foxp3 (Figure S5A). Furthermore, a pulse chase experiment was performed to determine the half-life of Foxp3 or Foxp3 K22xR in the presence or absence of USP7 (Figure S5B). We observed a Foxp3 half-life of approximately 4 hr; however, Foxp3 protein amounts were very stable in the presence of ectopically expressed USP7 or when all lysines were mutated to arginines. Accordingly, Foxp3 half-life was similar in a Foxp3-expressing Jurkat T cell line after treatment with CHX, whereas Foxp3 K22xR was very stable (Figure S5C). To investigate whether these observations were specific for USP7, we analyzed the effect of additional DUBs. Using a global DUB probe analysis, we found that both USP14 and UCH-L3 were active in Treg cells (data not shown). In contrast to USP7, neither USP14 nor UCH-L3 influenced Foxp3 protein expression or Foxp3 polyubiquitination (Figures 5B and 5C). In addition, a USP7 knockdown was performed in cells ectopically expressing Foxp3 with a pool of four separate small interfering RNAs (siRNAs; Figure S5D). USP7 knockdown resulted in markedly reduced Foxp3 protein expression in comparison to control (Figure 5D). To verify these data in primary human Treg cells, we performed USP7 knockdown with a USP7 small hairpin RNA (shRNA) lentivirus containing a puromycin resistance cassette. Foxp3 protein expression of puromycin-resistant Treg cells was clearly decreased as a result of USP7 knockdown (Figures 5E and S5E). Accordingly, the protein expression of CD25, a Foxp3 transcriptional target, was also reduced after USP7 knockdown (Figure S5F). Importantly, CD25 expression was unaffected when USP7 was knocked down in Foxp3− Teff cells (Figure S5G), providing support that the effect of USP7 on CD25 expression is mediated through Foxp3. To further validate that USP7 directly reduced Foxp3 protein expression because of enhanced degradation and not indirectly through additional mechanisms, we analyzed the activation status of multiple signaling pathways in the presence or absence of USP7. No significant differences in the downstream targets of TCR and TGF-β signaling (phospho-ERK, phospho-SMAD3, or phospho-NFκB) were observed between control and USP7 knockdown Treg cells (Figure S5H).
Figure 5. USP7 Deubiquitinates Foxp3 in Treg Cells.
(A) HEK 293T cells were transfected with HA-Foxp3 or HA-Foxp3 K22xR and Myc-USP7. Cell lysates were quantified and analyzed by western blotting with anti-HA and anti-HSP90.
(B) Cells were cotransfected with HA-Foxp3 and Myc-USP7, Myc-USP14, or FLAG-UCHL3. Cell lysates were analyzed for Foxp3 expression by western blotting with anti-HA.
(C) Ubiquitin pull-down for HA-Foxp3 or HA-Foxp3 K22xR as described in the Experimental Procedures.
(D) Cells were transfected with USP7 siRNA along with HA-Foxp3. Equalized protein lysates were immunoblotted and analyzed for HA.
(E) USP7 was knocked down in human Treg cells with shRNA lentivirus containing a puromycin resistance cassette. Endogenous USP7 and Foxp3 expression of puromycin-resistant Treg cells were analyzed by flow cytometry.
(F) Primary Treg cells were isolated from Foxp3-Ires-GFP mice and activated with anti-CD3 (5 μg/ml) and anti-CD28 (2 μg/ml) in the presence of 100 U/ml IL-2 for 24 hr followed by either heat shock treatment for 30 min at 42°C or replacement of fresh media with either LPS (1 mg/ml) or IL-6 (20 ng/ml) and cultured for an additional 24 hr before being harvested for the western blots and probed with the indicated antibodies.
(G) Primary Treg cells were isolated from Foxp3-Ires-GFP mice and treated for 1 hr with IL-6 (50 ng/ml). Foxp3-USP7 association was visualized with an in situ proximity ligation assay. Punctate staining (green) indicates a Foxp3-USP7 interaction as detected by the assay, and DAPI was used to visualize the nuclei (blue). All results are representative for at least three independent experiments. SC, scrambled; SP, smart pool; IP, immunoprecipitation; WB, western blot.
See also Figure S5.
Foxp3 is acetylated at lysines 31, 249, 251, 263, and 267 (Kwon et al., 2012; Song et al., 2012). Because we have previously reported that acetylation can stabilize Foxp3 by preventing ubiquitination, we wished to determine whether the expression of the described mutants could still be influenced by USP7 (van Loosdregt et al., 2010). HEK 293T cells were transfected with Foxp3 K3R (lysines 31, 263, and 267), K4R (lysines 249, 251, 263, and 267), or K5R (lysines 249, 251, 263, 267, and 393) in the presence or absence of USP7, and Foxp3 expression was determined. Cotransfection of USP7 increased the expression of all the mutants, demonstrating that the expression of these specific mutants can still be regulated by USP7 (Figure S5I).
Next, we sought to determine which signals could regulate USP7 expression and, thus, Foxp3 stability. Primary Treg cells from Foxp3-GFP mice were activated and stimulated with lipopolysaccharide (LPS), heat shock, or IL-6 and cultured for 24 hr. USP7 expression was reduced by these treatments, and, accordingly, Foxp3 protein expression was also reduced in these samples (Figure 5F). To analyze whether IL-6 could also disrupt association of Foxp3 and USP7, we treated freshly isolated Foxp3-GFP Treg cells with IL-6 for 1 hr, and a PLA was performed. As depicted in Figure 5G, short-term IL-6 treatment disrupted the association between Foxp3 and USP7. Taken together, these data demonstrate that USP7 activity in Treg cells stabilizes Foxp3, a process that can be disrupted by inflammatory stimuli.
USP7-Mediated Foxp3 Deubiquitination Improves Treg Cell Functionality
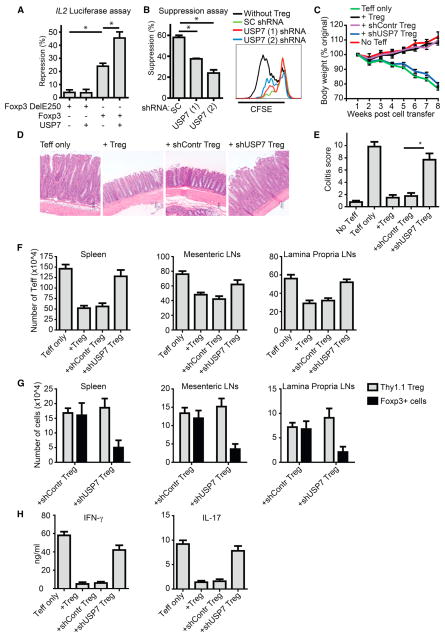
Given that we observed a clear effect of USP7 on Foxp3 protein stability, we subsequently analyzed the effect of USP7 on Foxp3 transcriptional activity. Promoter activity of the canonical Foxp3 transcriptional target IL2 was evaluated with an IL2 promoter luciferase reporter. Foxp3 expression resulted in a clear repression of IL2 promoter activity, whereas the well-characterized IPEX mutant Foxp3 Del250 was inactive (Figure 6A). Although cotransfection of Foxp3 Del250 with USP7 alone did not influence IL2 reporter activity, USP7 significantly increased IL2 promoter repression mediated by WT Foxp3. To determine whether USP7 can specifically modulate Treg-cell-mediated suppression, we performed a USP7 knockdown with two distinct shRNAs, and Treg cell functionality was addressed with an in vitro suppression assay (Figures 6B and S6A). Both USP7 shRNAs significantly abrogated Treg-cell-mediated suppression in comparison to the scrambled shRNA control without affecting apoptosis (Figure S6B). Accordingly, IL-2 expression was also increased in Treg cells upon USP7 knockdown (Figure S6C).
Figure 6. USP7 Modulates Treg Cell Function In Vitro and In Vivo.
(A) IL-2 promoter luciferase activity was analyzed in HEK 293T cells by cotransfecting NFAT with Foxp3 Del250 or WT Foxp3 with USP7. Repression of IL2 reporter activity is depicted, and all values were normalized for cotransfected Renilla.
(B) USP7 knockdown in human Treg cells was performed with two different USP7 shRNAs (#1 or #2), and scrambled (SC) shRNA was used as control was used as control. Analysis of Treg-cell-mediated suppression was performed with a standard suppression assay.
(C) Colitis was induced by intravenous coinjection of 1 ×106 CD4+CD25−CD62Lhigh T cells and 2 × 105 CD4+CD25high Treg cells per _Rag2_−/− mouse. Changes in body weight over time were expressed as a percentage of the original weight. Data are represented as the mean + SEM of eight mice in each group.
(D) Histological examination of the colon at 8 weeks after cell transfer.
(E) Histological scores were determined at 8 weeks after cell transfer as described in the Experimental Procedures.
(F) Spleen, mesenteric lymph node and lamina propria lymph node cells were isolated from the mice at 8 weeks after adoptive T cell transfer, and the number of Teff cells was determined by flow cytometry.
(G) Cell numbers of injected Thy1.1+ Treg cells and Foxp3+ cells were determined by flow cytometry.
(H) IFN-γ and IL-17 production by CD4+ lamina propria lymphocytes was analysed by ELISA. Data are represented as mean + SEM. *p < 0.05.
See also Figure S6.
To determine whether USP7 activity could also modulate Treg-cell-mediated suppression in vivo, we once again used a mouse colitis model. Here, naïve CD4+CD25−CD62Lhigh T cells were cotransferred with CD4+CD25high USP7 knockdown Treg cells into _Rag2_−/− mice. In contrast to WT or control knockdown Treg cells, USP7 knockdown Treg cells could not suppress the development of disease based on both body weight loss and histological analysis of the colon (Figures 6C, 6D, and 6E). Equivalently, Teff cell numbers in the spleen, mesenteric, and lamina propria lymph nodes were increased in mice that received shUSP7 Treg cells in comparison to controls (Figure 6F). In addition, the numbers of Thy1.1 Treg cells were similar in both the control and USP7 knockdown group, demonstrating that USP7 knockdown did not alter Treg cell survival or homing to lymph nodes (Figures 6G and S6D). Importantly, the percentage of Foxp3+ cells within the Thy1.1 population was decreased in mice that received USP7 knockdown Treg cells, indicating that knockdown of USP7 resulted in decreased Foxp3 expression, which was in line with our in vitro observations. Accordingly, IFN-γ and IL-17 production by CD4+ lamina propria lymphocytes was increased in mice that received USP7 knockdown Treg cells (Figure 6H). Collectively, these data demonstrate that USP7 can regulate Treg-cell-mediated suppression both in vitro and in vivo.
DISCUSSION
The transcription factor Foxp3 is crucial for Treg cell development and function and, therefore, is critical in controlling immune responses (Khattri et al., 2003; Fontenot et al., 2003; Hori et al., 2003; Williams and Rudensky, 2007). Because Foxp3 protein expression is directly associated with the suppressive capacity of Treg cells, many studies have focused on the transcriptional regulation of Foxp3 expression (Wan and Flavell, 2007; Kim and Leonard, 2007; Tone et al., 2008). Here, we show that Foxp3 protein expression can also be regulated by the deubiquitinase USP7. USP7 expression was found to be upregulated in Treg cells and to interact with and deubiquitinate Foxp3, thereby increasing Foxp3 protein amounts. USP7 knockdown in Treg cells resulted in decreased Foxp3 protein expression and impaired Treg-cell-mediated suppression both in vitro and in vivo. These data provide insights into the molecular mechanisms regulating Foxp3 protein expression and, therefore, Treg cell functionality.
We also demonstrate that stimulation with IL-6, heat shock, or LPS reduces USP7 protein amounts and, thus, Foxp3 expression. Our data are in accordance with a recent study by Yang et al. (2012) that reports that, in colon tumors, IL-6 can inhibit USP7 expression in a STAT3-dependent manner. Furthermore, we show that the association of USP7 and Foxp3 is disrupted upon short-term IL-6 stimulation. These data indicate that the effects of IL-6 stimulation on Foxp3 protein expression can be both rapid through the disruption of USP7-Foxp3 association and prolonged by the reduction of USP7 expression.
Although it was initially thought that Foxp3 expression was unique for Treg cells, several studies have reported that Foxp3 can also be expressed in activated T cells. The stimulation of human CD4 cells in vitro with anti-CD3 and anti-CD28 or phorbol 12-meristate 13-acetate and Ca2+ ionophore resulted in the transient expression of Foxp3, although protein amounts were lower in comparison to nTreg cells (Gavin et al., 2006; Allan et al., 2005). Analysis of these T cells transiently expressing Foxp3 revealed that these cells cannot suppress cytokine production or proliferation of cocultured T cells (Gavin et al., 2006; Allan et al., 2005). In contrast, TCR-stimulated CD4+CD25− cells expressing high and stable Foxp3 amounts develop suppressive capacity, whereas the ablation of Foxp3 in mature Treg cells in mice with a loxP-flanked Foxp3 allele resulted in a loss of Treg-cell-mediated suppression and the production of IL-2 and Th1 cytokines (Williams and Rudensky, 2007; Wang et al., 2007; Gavin et al., 2006). Here, we demonstrate that the turnover of Foxp3 in primary Treg cells is relatively high. Given that Foxp3 transcription rates are normally equal to its degradation rates, the total pool of Foxp3 protein is stable. This high turnover allows for a rapid regulation of Foxp3 protein expression by the modulation of Foxp3 polyubiquitination and degradation. These data demonstrate that stable Foxp3 expression is a necessity for the suppressive phenotype of Treg cells, suggesting that the stabilization of Foxp3 by USP7 may provide an essential step in the conversion of activated T cells into iTreg cells. In agreement with this hypothesis, we observed a more than 10-fold increase of USP7 mRNA in Treg cells induced from a naïve T cell pool in comparison to other Th subsets.
Using MS, we identified five distinct lysine residues that were ubiquitinated (K249, K251, K263, K267, and K393). Although a mutant of Foxp3 lacking all lysine residues (Foxp3 K22xR) was not ubiquitinated, the addition of the five identified lysines in the Foxp3 K22xR background restored Foxp3 ubiquitination. The addition of K249, K251 and K393, or K263 and K267 was also sufficient to restore Foxp3 ubiquitination to WT levels, suggesting a level of promiscuity in which lysine residues are ubiquitinated. Furthermore, generating an N-terminal ubiquitin-Foxp3 fusion protein dramatically increased Foxp3 polyubiquitination, demonstrating that the specific location of Foxp3 polyubiquitinated lysine residues is apparently not critical. The promiscuity of ubiquitinated lysine residues is a commonly observed phenomenon and has been described for a variety of proteins (Lee et al., 2004; Lu et al., 2007; Huang et al., 2006). For example, with the use of MS, the DUB UCH-L1 was also found to be ubiquitinated on at least four lysine residues (Meray and Lansbury, 2007). The mutation of all four lysine residues did not result in reduced ubiquitination levels, only a UCH-L1 protein in which all 16 lysine residues were mutated showed severely reduced ubiquitination levels. In addition, the mutation of single ubiquitinated lysine residues may result in ubiquitination of secondary lysine residues that are not normally ubiquitinated, as has been observed in TCRξ chain and Gpa1 (Hou et al., 1994; Marotti et al., 2002). This promiscuity potentially allows the possibility for multiple mechanisms to regulate Foxp3 protein expression.
Interestingly, USP7 polymorphisms have been associated with multiple autoimmune diseases. In a genome-wide association study, 14,000 cases of seven common diseases were compared to 3,000 controls (Wellcome Trust Case Control Consortium, 2007). Numerous single-nucleotide polymorphisms in the USP7 gene were highly significantly correlated with the autoimmune diseases, including Crohn’s disease, insulin-dependent diabetes mellitus, and rheumatoid arthritis. The precise effect of these polymorphisms on USP7 function remains unclear. Because we demonstrate that USP7 increases Treg-cell-mediated suppression in multiple models, it is tempting to speculate that these USP7 polymorphisms may inhibit USP7 function, resulting in reduced Foxp3 expression, abrogating Treg cell functionality, and, thus, leading to autoimmunity.
Treg cells are found in increased numbers both within and in close proximity to solid tumors. They have been shown to inhibit tumor-specific T cell immunity and contribute to the growth of human tumors in vivo (Yang et al., 2006; Curiel et al., 2004). The depletion of CD4+CD25+ cells in vivo augments the generation of tumor-specific T cells and promotes the rejection of tumors derived from myeloma, sarcoma, and melanoma (Turk et al., 2004; Onizuka et al., 1999). These data suggest that the inhibition of Treg cell numbers and function could also provide a potent antitumor therapy. Compounds inhibiting USP7 could potentially be clinically relevant when used to treat diseases, given that we have demonstrated that the inhibition of USP7 results in decreased Foxp3 protein expression, preventing Treg-cell-mediated suppression.
We propose a molecular mechanism regulating the efficiency of Treg-cell-mediated immune modulation. We demonstrate that Foxp3 protein expression can be tightly regulated by polyubiquitination-mediated proteosomal degradation. The modulation of USP7 activity may provide a novel molecular therapy aimed to control (inappropriate) immune responses.
EXPERIMENTAL PROCEDURES
Antibodies, DNA Constructs, and Reagents
The following antibodies were used: mouse anti-Foxp3 clone PCH101 for fluorescence-activated cell sorting analysis (eBioscience), mouse anti-FLAG M2 from Sigma-Aldrich, mouse anti-HA clone 12CA5 from Santa Cruz Biotechnology, and mouse anti-tubulin (Sigma-Aldrich), and anti-Myc monoclonal Ab were made with a hybridoma cell line. Foxp3 was cloned from MIGR1-Foxp3 (which was kindly provided by S. Sakaguchi) (Hori et al., 2003) into pMT2 containing a HA-tag-generating pMT2-HA-Foxp3. pMT2-FLAG-Foxp3, pMT2-FLAG-Foxp3ΔE250, and pRSV-NFATC/A have been described previously (van Loosdregt et al., 2013). HA-Ubi and HA-UbiK7xR were described previously (van der Horst et al., 2006). Foxp3 lysine mutants were obtained by site-directed mutagenesis. Myc-USP7 has been described previously (Meulmeester et al., 2005). pcDNA3, CHX, and MG132 were purchased from Sigma-Aldrich. The HA-DUB probe and DUB inhibitor have been described previously (Borodovsky et al., 2002; Colombo et al., 2010; Patent WO, 2007).
Luciferase Assays
Luciferase reporter assays were performed as previously described (van Loosdregt et al., 2011).
Confocal Imaging
Localization Studies
Localization experiments were performed as previously described (van Loosdregt et al., 2010).
Proximity Ligation Assay
PLA detection was performed with a Duolink II kit (Olink Bioscience) as previously described (van Loosdregt et al., 2011).
Pulse Chase Analysis
Transfected HEK 293T cells were washed and depleted from methionine and cysteine for 30 min in depletion medium (Dulbecco’s modified Eagle’s medium [DMEM] without Met and Cys, Invitrogen), pulsed for 17 min with depletion medium containing 35S Met + Cys (100 μCi/ml, PerkinElmer), washed, and subsequently chased in chase medium (DMEM with Met and Cys + 100 μM methionine). Cells were washed in stop buffer (PBS + 100 μM N-ethylmaleimide) and lysed in lysis buffer (1% Triton, 1 mM EDTA, and Halt Protease Inhibitor Cocktail [1:100] in PBS). Lysates were incubated with Anti-FLAG M2 Affinity Gel-coupled beads (Sigma-Aldrich) for 2 hr at 4°C. Beads were washed three times, boiled once in sample buffer, and subjected to gel electrophoresis. Radioactive labeled FLAG-Foxp3 was analysed with phosphorscreen and a Storm 860 phosphorimager (GE Healthcare).
TUBEs Ubiquitination Assay
Ubiqutination of endogenously expressed protein was determined with Agarose-TUBE 2 according to the manufacturer’s protocol (LifeSensors).
Isolation of Active DUBs with the HA-DUB Probe
HA immunoprecipitation of the HA-DUB probe was performed as described previously (Borodovsky et al., 2002).
Immunoprecipitation
Immunoprecipitation experiments were performed as described previously (van Loosdregt et al., 2010).
In Vitro Suppression Assay
Human CD4+CD25highCD127low Treg cells (top 2%) were sorted and cocultured with CFSE-labeled PBMCs (1:5 ratio) in anti-CD3 (clone OKT3)-coated 96-well plates. Cells were cultured for 4 days in RPMI medium supplemented with 10% fetal calf serum (FCS), 100 units/ml penicillin, 100 μg/ml streptomycin, and 5 × 10−5 M 2-mercaptoethanol. Proliferation of CD4+ and CD8+ was determined by measuring CFSE dilution with a FACSCANTO flow cytometer (BD Biosciences).
Generation of iTreg Cells
CD4+CD25− cells were isolated from cord blood by magnetic-activated cell sorting and cultured in RPMI 1640 supplemented 10% FCS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 5 × 10−5 M 2-mercaptoethanol. Foxp3 expression was induced by culturing the cells for 5 days in combination with anti-CD3 anti-CD28 Dynabeads, 300 IU IL-2, and 10 nM TGF-β.
Ubiquitin Pull-Down Assay
Cells transfected with both His-tagged ubiquitin and HA-tagged Foxp3 were treated with 20 μM MG132 and 10 μM DUBi for 3 hr. Cells were lysed in a pH 8 urea buffer (8 M urea, 100 mM Na2HPO4, 10 mM TRIS [pH 8.0], 0.2% TX-100, 10 mM imidazole, and 1 mM N-ethylmaleimide) and tumbled with Ni-NTA beads for 2 hr at room temperature. The beads were washed twice in pH 8 urea buffer, twice in pH 6.3 urea buffer (8 M urea, 100 mM Na2HPO4, 10 mM TRIS [pH 6.3], 0.2% TX-100, and 10 mM imidazole), and once in a wash buffer (20 mM TRIS [pH 8.0], 100 mM NaCl, 20% glycerol, 1 mM dithiothreitol, and 10 mM imidazole). Beads were boiled in sample buffer for 5 min and separated on SDS-PAGE, and Foxp3-specific polyubiquitination was determined by western blot analysis with anti-HA.
T Cell Differentiation
We cultured 100,000 sorted CD4+CD27+CD45RO− naïve T cells derived from human PBMCs in 96-well plates coated with 0.3 μg/ml anti-CD3 and 2 μg/ml soluble anti-CD28 for 4 days. For skewing towards different T cells subsets, the cultures were supplemented with the following cytokines and antibodies: Th1, IL-12 (200 U/ml) + anti-iL-4 (10 μg/ml); Th2, IL-4 (1000 U/ml) + anti-iFNγ (5 μg/ml); Th17, TGFβ (10 ng/ml) + IL-6 (20 ng/ml) + IL1β (10 ng/ml) + IL-23 (100 ng/ml); Treg, IL-2 (300 U/ml) + TGFβ (10 ng/ml).
Induced Colitis Mouse Model for DUBi-Treated Treg Cells
_Rag1_−/− mice were kept in the animal facility of the Utrecht University under specific pathogen-free conditions. The experiments were approved by the Animal Experiment Committee of the Faculty of Veterinary Medicine (Utrecht University). Immunodeficient _Rag1_−/− mice were injected with 4 × 105 CD4+CD45RBhigh cells in order to induce colitis. After 21 days, 2 × 105 Treg cells isolated from Foxp3-GFP mice were pretreated with 10 μM DUBi for 1 hr, washed, and intravenously injected. The mice were sacrificed 3 weeks later, and tissue slides of the colon were prepared. Scoring was performed as described previously (Zaiss et al., 2013).
Induced Colitis Mouse Model for USP7 KD Treg Cells
Naïve CD4+CD25−CD62Lhigh T cells were isolated from BALB/c and intravenously injected into BALB/c _Rag2_−/− immunodeficient recipients (1 × 106 per mouse). CD4+CD25+ Treg cells or siRL- or siUSP7-transduced Treg cells (2 × 105) isolated from Thy1.1 BALB/c were intravenously coinjected where indicated. After 8 weeks, mice were sacrificed, and tissue slides of the colon were prepared and scored as described.
Statistical Analysis
Statistical analysis was performed with the Mann-Whitney test (GraphPad Prism). p < 0.05 was considered statistically significant.
Supplementary Material
Suppl Data
Acknowledgments
The authors like to thank M. Klein, S. Vervoort, I. Kuper, and A. Veenstra for technical assistance and S. Sakaguchi (Institute for Physical and Chemical Research, Yokohama, Japan) and B.M.T. Burgering (University Medical Center Utrecht) for providing several constructs. H.O. was supported by a VIDI grant from the Netherlands Foundation for Scientific Research. M.M.M. is supported by the Dutch Cancer Society (2006-3508), Utrecht University (High Potential Program), and the European Research Council (starting grant 242958). F.P.’s research is partially supported by grants from the National Institutes of Health (R01AI099300), the Melanoma Research Alliance, a Stewart Trust Award, and a Kelly’s Dream Award. J.v.L. and V.F. were supported by a grant from the Dutch Rheumatism Foundation. H.O. is a cofounder of UbiQ B.V. and a member of its scientific advisory board.
Footnotes
References
- Allan SE, Passerini L, Bacchetta R, Crellin N, Dai M, Orban PC, Ziegler SF, Roncarolo MG, Levings MK. The role of 2 FOXP3 isoforms in the generation of human CD4+ Tregs. J Clin Invest. 2005;115:3276–3284. doi: 10.1172/JCI24685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Schwartz AL. Ubiquitin-mediated degradation of cellular proteins in health and disease. Hepatology. 2002;35:3–6. doi: 10.1053/jhep.2002.30316. [DOI] [PubMed] [Google Scholar]
- Colombo M, Vallese S, Peretto I, Jacq X, Rain JC, Colland F, Guedat P. Synthesis and biological evaluation of 9-oxo-9H-indeno [1,2-b]pyrazine-2,3-dicarbonitrile analogues as potential inhibitors of deubiquitinating enzymes. ChemMedChem. 2010;5:552–558. doi: 10.1002/cmdc.200900409. [DOI] [PubMed] [Google Scholar]
- Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte JH, Zelenay S, Bergman ML, Martins AC, Demengeot J. Natural Treg cells spontaneously differentiate into pathogenic helper cells in lymphopenic conditions. Eur J Immunol. 2009;39:948–955. doi: 10.1002/eji.200839196. [DOI] [PubMed] [Google Scholar]
- Eldridge AG, O’Brien T. Therapeutic strategies within the ubiquitin proteasome system. Cell Death Differ. 2010;17:4–13. doi: 10.1038/cdd.2009.82. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Gavin MA, Torgerson TR, Houston E, DeRoos P, Ho WY, Stray-Pedersen A, Ocheltree EL, Greenberg PD, Ochs HD, Rudensky AY. Single-cell analysis of normal and FOXP3-mutant human T cells: FOXP3 expression without regulatory T cell development. Proc Natl Acad Sci USA. 2006;103:6659–6664. doi: 10.1073/pnas.0509484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, Rudensky AY. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- Hou D, Cenciarelli C, Jensen JP, Nguygen HB, Weissman AM. Activation-dependent ubiquitination of a T cell antigen receptor subunit on multiple intracellular lysines. J Biol Chem. 1994;269:14244–14247. [PubMed] [Google Scholar]
- Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204:1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu N, Mariotti-Ferrandiz ME, Wang Y, Malissen B, Waldmann H, Hori S. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc Natl Acad Sci USA. 2009;106:1903–1908. doi: 10.1073/pnas.0811556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HS, Lim HW, Wu J, Schnölzer M, Verdin E, Ott M. Three novel acetylation sites in the Foxp3 transcription factor regulate the suppressive activity of regulatory T cells. J Immunol. 2012;188:2712–2721. doi: 10.4049/jimmunol.1100903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Hong US, Lee TH, Yoon SK, Yoon JB. Mass spectrometric analysis of tumor necrosis factor receptor-associated factor 1 ubiquitination mediated by cellular inhibitor of apoptosis 2. Proteomics. 2004;4:3376–3382. doi: 10.1002/pmic.200401000. [DOI] [PubMed] [Google Scholar]
- Lu Y, Amleh A, Sun J, Jin X, McCullough SD, Baer R, Ren D, Li R, Hu Y. Ubiquitination and proteasome-mediated degradation of BRCA1 and BARD1 during steroidogenesis in human ovarian granulosa cells. Mol Endocrinol. 2007;21:651–663. doi: 10.1210/me.2006-0188. [DOI] [PubMed] [Google Scholar]
- Marotti LA, Jr, Newitt R, Wang Y, Aebersold R, Dohlman HG. Direct identification of a G protein ubiquitination site by mass spectrometry. Biochemistry. 2002;41:5067–5074. doi: 10.1021/bi015940q. [DOI] [PubMed] [Google Scholar]
- Meray RK, Lansbury PT., Jr Reversible monoubiquitination regulates the Parkinson disease-associated ubiquitin hydrolase UCH-L1. J Biol Chem. 2007;282:10567–10575. doi: 10.1074/jbc.M611153200. [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Miyao T, Floess S, Setoguchi R, Luche H, Fehling HJ, Waldmann H, Huehn J, Hori S. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 2012;36:262–275. doi: 10.1016/j.immuni.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Oldenhove G, Bouladoux N, Wohlfert EA, Hall JA, Chou D, Dos Santos L, O’Brien S, Blank R, Lamb E, Natarajan S, et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 2009;31:772–786. doi: 10.1016/j.immuni.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onizuka S, Tawara I, Shimizu J, Sakaguchi S, Fujita T, Nakayama E. Tumor rejection by in vivo administration of anti-CD25 (interleukin-2 receptor alpha) monoclonal antibody. Cancer Res. 1999;59:3128–3133. [PubMed] [Google Scholar]
- Ouyang W, Beckett O, Ma Q, Paik JH, DePinho RA, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. 2010;11:618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- Passerini L, Allan SE, Battaglia M, Di Nunzio S, Alstad AN, Levings MK, Roncarolo MG, Bacchetta R. STAT5-signaling cytokines regulate the expression of FOXP3 in CD4+CD25+ regulatory T cells and CD4+CD25− effector T cells. Int Immunol. 2008;20:421–431. doi: 10.1093/intimm/dxn002. [DOI] [PubMed] [Google Scholar]
- 2007/017758 A2. Patent WO. 2007b Jun 14;
- Polansky JK, Schreiber L, Thelemann C, Ludwig L, Krüger M, Baumgrass R, Cording S, Floess S, Hamann A, Huehn J. Methylation matters: binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J Mol Med. 2010;88:1029–1040. doi: 10.1007/s00109-010-0642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powrie F, Leach MW, Mauze S, Menon S, Caddle LB, Coffman RL. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity. 1994;1:553–562. doi: 10.1016/1074-7613(94)90045-0. [DOI] [PubMed] [Google Scholar]
- Rubtsov YP, Niec RE, Josefowicz S, Li L, Darce J, Mathis D, Benoist C, Rudensky AY. Stability of the regulatory T cell lineage in vivo. Science. 2010;329:1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Li B, Xiao Y, Chen C, Wang Q, Liu Y, Berezov A, Xu C, Gao Y, Li Z, et al. Structural and biological features of FOXP3 dimerization relevant to regulatory T cell function. Cell Rep. 2012;1:665–675. doi: 10.1016/j.celrep.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- Turk MJ, Guevara-Patiño JA, Rizzuto GA, Engelhorn ME, Sakaguchi S, Houghton AN. Concomitant tumor immunity to a poorly immunogenic melanoma is prevented by regulatory T cells. J Exp Med. 2004;200:771–782. doi: 10.1084/jem.20041130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- van Loosdregt J, Vercoulen Y, Guichelaar T, Gent YY, Beekman JM, van Beekum O, Brenkman AB, Hijnen DJ, Mutis T, Kalkhoven E, et al. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood. 2010;115:965–974. doi: 10.1182/blood-2009-02-207118. [DOI] [PubMed] [Google Scholar]
- van Loosdregt J, Brunen D, Fleskens V, Pals CE, Lam EW, Coffer PJ. Rapid temporal control of Foxp3 protein degradation by sirtuin-1. PLoS ONE. 2011;6:e19047. doi: 10.1371/journal.pone.0019047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loosdregt J, Fleskens V, Tiemessen MM, Mokry M, van Boxtel R, Meerding J, Pals CE, Kurek D, Baert MR, Delemarre EM, et al. Canonical Wnt Signaling Negatively Modulates Regulatory T Cell Function. Immunity. 2013;39:298–310. doi: 10.1016/j.immuni.2013.07.019. this issue. [DOI] [PubMed] [Google Scholar]
- Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–770. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- Wang J, Ioan-Facsinay A, van der Voort EI, Huizinga TW, Toes RE. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur J Immunol. 2007;37:129–138. doi: 10.1002/eji.200636435. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- Yang ZZ, Novak AJ, Ziesmer SC, Witzig TE, Ansell SM. Attenuation of CD8(+) T-cell function by CD4(+)CD25(+) regulatory T cells in B-cell non-Hodgkin’s lymphoma. Cancer Res. 2006;66:10145–10152. doi: 10.1158/0008-5472.CAN-06-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, Shah B, Chang SH, Schluns KS, Watowich SS, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Huo S, Shan Y, Liu H, Xu Y, Yao K, Li X, Zhang X. STAT3 repressed USP7 expression is crucial for colon cancer development. FEBS Lett. 2012;586:3013–3017. doi: 10.1016/j.febslet.2012.06.025. [DOI] [PubMed] [Google Scholar]
- Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, Laurence A, Robinson GW, Shevach EM, Moriggl R, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss DM, van Loosdregt J, Gorlani A, Bekker CP, Gröne A, Sibilia M, van Bergen en Henegouwen PM, Roovers RC, Coffer PJ, Sijts AJ. Amphiregulin enhances regulatory T cell-suppressive function via the epidermal growth factor receptor. Immunity. 2013;38:275–284. doi: 10.1016/j.immuni.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl Data