Cellular Requirements for LARK in the Drosophila Circadian System (original) (raw)
. Author manuscript; available in PMC: 2014 Aug 18.
Published in final edited form as: J Biol Rhythms. 2012 Jun;27(3):183–195. doi: 10.1177/0748730412440667
Abstract
RNA-binding proteins mediate posttranscriptional functions in the circadian systems of multiple species. A conserved RNA recognition motif (RRM) protein encoded by the lark gene is postulated to serve circadian output and molecular oscillator functions in Drosophila and mammals, respectively. In no species, however, has LARK been eliminated, in vivo, to determine the consequences for circadian timing. The present study utilized RNA interference (RNAi) techniques in Drosophila to decrease LARK levels in clock neurons and other cell types in order to evaluate the circadian functions of the protein. Knockdown of LARK in timeless (TIM)– or pigment dispersing factor (PDF)–containing clock cells caused a significant number of flies to exhibit arrhythmic locomotor activity, demonstrating a requirement for the protein in pacemaker cells. There was no obvious effect on PER protein cycling in lark interference (RNAi) flies, but a knockdown within the PDF neurons was associated with increased PDF immunoreactivity at the dorsal termini of the small ventral lateral neuronal (s-LNv) projections, suggesting an effect on neuropeptide release. The expression of lark RNAi in multiple neurosecretory cell populations demonstrated that LARK is required within pacemaker and nonpacemaker cells for the manifestation of normal locomotor activity rhythms. Interestingly, decreased LARK function in the prothoracic gland (PG), a peripheral organ containing a clock required for the circadian control of eclosion, was associated with weak population eclosion rhythms or arrhythmicity.
Keywords: clock output, posttranscriptional, RNA binding, locomotor activity, eclosion
In both prokaryotic and eukaryotic species, circadian rhythms in biochemistry, physiology, and behavior are governed by endogenous cellular clocks. Feedback loops that drive rhythmic changes in gene transcription are important components of the clocks governing circadian behavior (Hardin, 2005; Kadener et al., 2008). However, there is evidence in several species that certain types of circadian oscillators can function in the complete absence of rhythmic clock gene transcription (Lakin-Thomas, 2006; O’Neill et al., 2011; O’Neill and Reddy, 2011). In multicellular organisms, circadian clocks are connected, via biochemical and cellular output pathways, to rhythmic physiological processes, and there is evidence in many species that both transcriptional and posttranscriptional mechanisms function in such output pathways (Jackson et al., 2005; Taghert and Shafer, 2006; Benito et al., 2007; Garbarino-Pico and Green, 2007; Kojima et al., 2010).
Perhaps the best studied clock output factor is Drosophila pigment dispersing factor (PDF), a neuropeptide essential for circadian behavior that is rhythmically released from a ventral subset (the ventral lateral neuron [LNv] cells) of approximately 150 neurons comprising the fly clock neuronal circuit (Renn et al., 1999; Park et al., 2000; Myers et al., 2003; Shafer et al., 2006). PDF acts through a class II G protein–coupled receptor (PDFR) (Hyun et al., 2005; Lear et al., 2005; Mertens et al., 2005) to stimulate intracellular cAMP signaling (Mertens et al., 2005); recent studies indicate that most fly clock neurons respond to PDF and that this coordinates the circadian network (Shafer et al., 2008; Im and Taghert, 2010).
The intracellular pathways regulating neuropeptide release from fly clock neurons have not been well defined, although there is evidence that the CLOCK and CYCLE clock proteins might regulate pdf mRNA expression in the LNv (Park et al., 2000), even though the gene does not exhibit transcriptional rhythms (Park and Hall, 1998). In addition, it is likely that posttranscriptional mechanisms contribute to PDF synthesis, storage, or release, and possible candidate regulatory factors include RNA-binding proteins (RBPs), several of which have been implicated in circadian control in various organisms (Heintzen et al., 1997; McNeil et al., 1998; Morales et al., 2002; Dockendorff et al., 2002; Baggs and Green, 2003; Iliev et al., 2006; Hassidim et al., 2007; Green et al., 2007). Our previous work characterized a Drosophila RBP of the RNA recognition motif (RRM) class called LARK that functions in the control of circadian behavior (Newby and Jackson, 1993; McNeil et al., 1998; Schroeder et al., 2003; Huang et al., 2007). LARK is an evolutionarily conserved RBP (Jackson et al., 1997) that exhibits circadian rhythms in abundance in the nervous systems of both flies and mice (McNeil et al., 1998; Zhang et al., 2000; Kojima et al., 2007). In flies, LARK posttranscriptionally regulates RNA targets thought to function in clock output (Huang et al., 2007), whereas the mouse homolog translationally regulates expression of the PERIOD1 clock protein (Kojima et al., 2007). The fly protein has pan-neuronal distribution and nuclear localization in the nervous system (McNeil et al., 2001). Previous studies show that overexpression of LARK in Drosophila clock neurons causes arrhythmic adult eclosion and locomotor activity (Schroeder et al., 2003; Huang et al., 2009). This phenotype depends on the presence of functional RNA-binding (RRM) domains within LARK, suggesting it is due to misregulated expression of a target RNA (Huang et al., 2007).
Because LARK is essential for development, at least in Drosophila, it has not been possible to generate null mutants for behavioral studies. In this report, we employ RNA interference (RNAi) technology to knock down Drosophila LARK in a cell-specific manner in order to examine the effects of reduced abundance on circadian rhythmicity.
MATERIALS AND METHODS
Construction of lark RNAi Transgenic Flies
Two different types of lark RNAi constructs were generated (Fig. 1A): one (RNAi-SI) contains an inverted repeat that targets the 5′UTR (bp 611–1490 as the sense strand, and bp 837–1234 as the complement). PCR-generated clones containing the 2 sequences were ligated into the pUAST transformation vector to form an inverted repeat. The second RNAi transgene (called RNAi) utilized a sequence from the 3′ end of the lark gene (313 bp that includes 76 bp of coding sequence and the 3′UTR). This segment was cloned as an inverted repeat into the pWIZ _P_-element vector. Both the RNAi-SI and RNAi constructs were sequenced prior to production of transgenic strains using standard procedures. Two independent genomic insertions of the RNAi transgene were obtained: RNAiA and RNAiB.
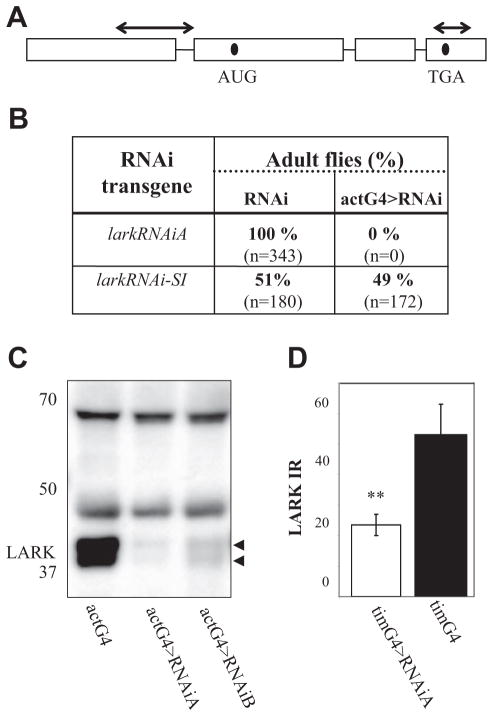
Figure 1.
Generation of lark RNAi transgenes and confirmation of their effectiveness. (A) Schematic representation of the lark gene. Arrows above the diagram indicate the regions targeted for knockdown by the RNAi-SI (5′) or RNAiA transgenes. (B) Ubiquitous knockdown of LARK using the RNAiA transgene results in lethality at pupal stages. The actG4 driver was used to drive ubiquitous expression of RNAi-SI or RNAi. The embedded table shows the percentage of eclosed LARK knockdown flies and sibling controls. The experiment was carried out at 25 °C. Four independent RNAi-SI and 2 independent RNAi strains (A, B) were tested with similar results. The RNAiA transgene was most effective and was used for subsequent experiments. (C) Ubiquitous knockdown of LARK using the RNAi transgene results in reduced LARK levels. Protein lysates from larvae expressing either of 2 independent LARK RNAi transgenes, actG4>RNAiA and actG4>RNAiB, were probed with affinity-purified LARK antibody (McNeil et al., 1998). Similar results were obtained in 2 independent experiments. Note that 2 LARK bands are always observed by Western blotting, and these are thought to represent distinct isoforms of the protein (McNeil et al., 1998). (D) An approximately 2- to 3-fold reduction in LARK-IR was observed in PDF cells, using tim-uasG4 for a clock cell knockdown of the protein. Histograms show quantification of LARK-IR within the large PDF (l-LNv) cells of tim-uasG4>RNAiA (n = 8) or control tim-uasG4 (n = 12) brains. Control RNAiA and tim-uasG4 brains were similar. The y-axis shows arbitrary units of fluorescence intensity. Error bars represent SEM. **Different from control at p < 0.01 (Student t test).
Fly Stocks and Growth Conditions
Various Gal4 and UAS strains were obtained from the Indiana University Stock Center or individual investigators (see Acknowledgments). Strains were maintained and crosses carried out using a standard culture medium (Newby and Jackson, 1991). Crosses were reared in a cycle consisting of 12 hours of light and 12 hours of dark (LD 12:12) with constant humidity (~60%) at 23 °C to 29 °C, depending on the experiment. Standard meiotic recombination was used to generate a third chromosome containing the lark1 mutation, a null allele, and the RNAiA transgene. This chromosome is designated RNAiA, lark1 in the text.
Locomotor Activity Assays
Locomotor activity was monitored at 30 °C or 25 °C unless otherwise indicated. Flies were collected under CO2 anesthesia, aged for 3 to 4 days, and then placed in Trikinetics activity monitors (Waltham, MA). They were entrained to LD 12:12 for 4 days, followed by transfer to constant darkness for approximately 2 weeks. Activity data were analyzed for periodicity using the MatLab-based fly toolbox package (Levine et al., 2002).
Eclosion Assays
Crosses of 30 females and 20 males were reared at 29 °C with entrainment to LD 12:12 for at least 5 days prior to the experiment. Newly emerged flies were counted by hand at the same temperature at 2-hour intervals under LD 12:12 or constant dark (DD) conditions (in red light using a 7.5-W bulb and a Kodak GBX-2 filter [Rochester, NY]). Assays of eclosion in phmN1G4>lark RNAi and control populations were performed at 20 °C in DD, following entrainment to LD 12:12 for 3 days, using Trikinetics automated eclosion monitors. Data were analyzed using the Maximum Entropy Spectral Analysis (MESA) function of the fly toolbox software package (Levine et al., 2002).
Immunostaining and Western Blotting
Standard laboratory immunostaining procedures were employed with whole mounts of the fly brain (Ng et al., 2011). Primary antibody dilutions were the following: rabbit anti-PER (1:300 preabsorbed against per0 embryos; from R. Stanewsky), rabbit anti-PDF (1:20,000; from K. Rao), guinea pig anti-PAP (1:2000; from P. Taghert), mouse anti-PDF (1:100; from the University of Iowa Hybridoma Bank), and rabbit anti-LARK (1:2000; from G. McNeil). The following secondary antibodies, all from Molecular Probes (Eugene, OR), were used at a 1:1000 dilution: goat anti-rabbit, Alexa-488 or Cy3 conjugated; goat anti-guinea pig, Alexa-488 or Cy3 conjugated; and donkey anti-mouse, Alexa-488 conjugated. Confocal images were acquired using a Leica TCS SP2 confocal microscope (Wetzlar, Germany). Image intensities (PER and PDF) were assessed using ImageJ 1.33 (National Institutes of Health, Bethesda, MD). Statistical significance of PER and PDF immunoreactive signal intensities was determined using either an ANOVA or t test (InStat, GraphPad Software, La Jolla, CA).
Standard procedures were used to prepare and analyze larval protein extracts (McNeil et al., 2001). Blocking of blots was performed using 5% skim milk in Tris-buffered saline, pH 8.0, containing 0.1% Tween-20 (TBST). They were incubated overnight with anti-LARK antibody (1:15,000) diluted in blocking solution, washed with TBST (4 × 20 minutes), and then incubated with HRP-tagged donkey anti-rabbit antibody (1:3000 in TBST) prior to chemiluminescence detection (GE Healthcare, Little Chalfont, UK). Protein band intensities were quantified using Kodak 1D Image Analysis Software and normalized against the intensity of a nonspecific band or MAP kinase (detected using an antibody dilution of 1:25,000; Sigma, St. Louis, MO).
RESULTS
Ubiquitous Expression of lark RNAi Causes Lethality
We employed the Gal4/UAS binary expression system and RNAi transgenes to study the functions of LARK in specific cell populations. As lark RNAi transgenes were not available from public resources at the time these experiments were initiated, we generated 2 different types for our studies (Fig. 1A): one called RNAi-SI, and another called RNAi (see Materials and Methods). Two independent insertions of the second transgene were named RNAiA and RNAiB.
To test the efficacy of our several different RNAi transgenes, ubiquitous knockdowns of LARK were performed using the act5C-Gal4 (actG4) driver, which is expressed in all cell types. We predicted that effective RNAi would result in a substantial decrease in LARK levels and perhaps mimic the lark null phenotype that is associated with pupal lethality. Indeed, _actG4_-driven expression of either RNAiA or RNAiB (i.e., actG4>RNAi) was associated with pupal lethality in pilot crosses (no RNAi-expressing adults observed) (Fig. 1B), although a few escaper flies were observed using the B transgene. In contrast, flies expressing the RNAi-SI transgene had normal viability perhaps because it is less effective than the other RNAi transgenes.
Expression of lark RNAi Decreases LARK Abundance
The analysis of larval protein lysates, using an anti-LARK antibody (Zhang et al., 2000), demonstrated _RNAiA_- and _RNAiB_-mediated knockdowns of LARK and indicated that the knockdown is more extreme with RNAiA (>5-fold reduced with RNAiA v. ~3-fold with RNAiB) (Fig. 1C). Immunostaining of adult brains indicated that expression of the RNAiA transgene in clock cells, using tim-uasG4, resulted in decreased amounts of LARK in the PDF neurons (~2- to 3-fold reduced) (Fig. 1D).
Pan-Neuronal Knockdown of LARK Decreases Robustness of the Locomotor Activity Rhythm
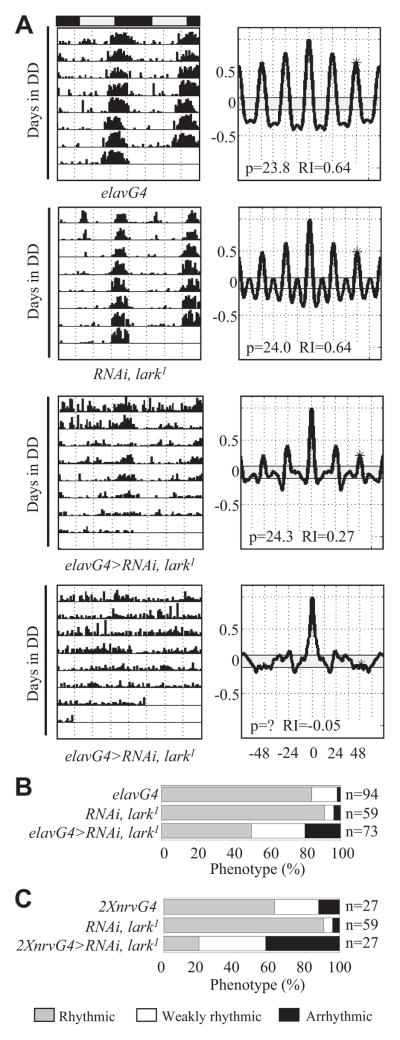
We initially examined adult locomotor activity in flies with a pan-neuronal knockdown of LARK using the elavG4 driver (elavG4>RNAiA). Data were analyzed for changes in the patterning of activity, period length, and robustness of rhythmicity using the Rhythmicity Index (RI) (Levine et al., 2002); experimental and control flies were categorized as rhythmic (RI ≥ 0.3), weakly rhythmic (RI = 0.1–0.3), or arrhythmic (RI ≤ 0.1 and obvious aperiodic activity). All behavioral results were verified in 2 or more independent experiments.
When flies were reared and assayed at 25 °C, there were no significant differences observed between elavG4>RNAiA and control flies (data not shown). However, when reared at 29 °C and assayed at 29 °C to 30 °C, 21% of elavG4>RNAiA flies were arrhythmic, and 16% were weakly rhythmic, whereas only 0% to 2% of control flies were arrhythmic (Fig. 2A and 2B; see Table 1 for statistics for this and all other genotypes). We attribute the effects on rhythmicity at higher temperatures to increased Gal4 activity and/or a more efficacious RNAi process (Fortier and Belote, 2000), although we have not examined LARK knockdown at the 2 different temperatures.
Figure 2.
A neuron-specific knockdown of LARK alters the locomotor activity rhythm. Pan-neuronal knockdown of LARK was carried out in flies heterozygous for lark1 using either the elavG4 or nrvG4 driver to express RNAiA. Unless otherwise indicated, in this and all subsequent activity experiments, flies were reared at 29 °C, entrained to LD 12:12 for 4 days at 30 °C, and then tested for locomotor activity at the same temperature. (A) Representative actograms and correlograms for control elavG4, control RNAiA, lark1 (flies carrying RNAiA in a lark1/+ background), weakly rhythmic elavG4>RNAiA, lark1, and arrhythmic elavG4>RNAiA, lark1 flies. (B) Categorization of flies based on the robustness of activity rhythms: rhythmic (gray), weakly rhythmic (white), and arrhythmic (black). (C) Effects of LARK knockdown using 2 copies (2×) of nrvG4 driving expression of RNAiA in lark1 heterozygotes.
Table 1.
Locomotor activity rhythm phenotypes resulting from decreased LARK function.
| GAL4 Type | N | R, % | WR, % | AR, % | RI ± SEM | τ ± SEM |
|---|---|---|---|---|---|---|
| elavG4>RNAiA | 57 | 63 | 16 | 21 | 0.39 ± 0.02 | 23.8 ± 0.06 |
| elavG4>RNAiA, lark1 | 73 | 50 | 30 | 20 | 0.26 ± 0.02a | 23.5 ± 0.08 |
| elavG4 | 94 | 84 | 14 | 2 | 0.39 ± 0.03 | 23.8 ± 0.08 |
| nrvG4; nrvG4>RNAiA | 20 | 45 | 35 | 20 | 0.19 ± 0.03b | 24.0 ± 0.10 |
| nrvG4; nrvG4>RNAiA, lark1 | 27 | 20 | 38 | 42 | 0.19 ± 0.04b | 23.7 ± 0.06 |
| nrvG4; nrvG4 | 27 | 63 | 25 | 12 | 0.37 ± 0.02 | 23.5 ± 0.05 |
| tim-uasG4>RNAiA | 21 | 57 | 25 | 18 | 0.30 ± 0.03c | 23.9 ± 0.13 |
| tim-uasG4>RNAiA (replicate) | 42 | 55 | 51 | 24 | 0.31 ± 0.03c | 23.9 ± 0.12 |
| tim-uasG4>RNAiA, lark1 | 32 | 20 | 33 | 29 | 0.18 ± 0.02d | 23.7 ± 0.10 |
| tim-uasG4>RNAiA/RNAiA, lark1 | 41 | 20 | 29 | 51 | 0.15 ± 0.03 | 24.1 ± 0.07 |
| tim-uasG4>RNAiA/RNAiA | 43 | 21 | 60 | 19 | 0.20 ± 0.03 | 24.0 ± 0.05 |
| tim-uasG4 | 23 | 91 | 4.5 | 4.5 | 0.39 ± 0.03 | 23.8 ± 0.07 |
| RNAiA | 21 | 86 | 14 | 0 | 0.45 ± 0.02 | 23.7 ± 0.09 |
| RNAiA, lark1 | 27 | 70 | 26 | 4 | 0.34 ± 0.02 | 23.5 ± 0.07 |
| RNAiA/RNAiA, lark1 | 14 | 36 | 50 | 14 | 0.20 ± 0.03 | 23.7 ± 0.34 |
| RNAiA/RNAiA | 28 | 61 | 36 | 4 | 0.29 ± 0.02 | 23.7 ± 0.08 |
| pdfG4>RNAiA | 59 | 95 | 5 | 0 | 0.54 ± 0.04 | 23.8 ± 0.02 |
| pdfG4>RNAiA, lark1 | 21 | 86 | 14 | 0 | 0.52 ± 0.03 | 24.0 ± 0.05 |
| pdfG4>RNAiA/RNAiA, lark1 | 38 | 78 | 21 | 0 | 0.36 ± 0.03 | 23.1 ± 0.07 |
| pdfG4>dcr2; RNAiA/RNAiA, lark1 | 113 | 12 | 51 | 37 | 0.18 ± 0.01 | 23.5 ± 0.05 |
| pdfG4>dcr2 | 31 | 97 | 0 | 3 | 0.46 ± 0.02 | 23.9 ± 0.05 |
| RNAiA | 23 | 96 | 4 | 0 | 0.56 ± 0.04 | 23.8 ± 0.10 |
| pdfG4 | 46 | 96 | 4 | 0 | 0.56 ± 0.03 | 24.1 ± 0.06 |
| RNAiA, lark1 | 27 | 93 | 4 | 4 | 0.63 ± 0.03 | 23.7 ± 0.05 |
| c929G4>RNAiA | 27 | 48 | 33 | 19 | 0.24 ± 0.02e | 23.3 ± 0.08 |
| c929G4>RNAiA, lark1 | 58 | 50 | 21 | 29 | 0.23 ± 0.06 | 24.0 ± 0.05 |
| pdfGal80; c929>RNAiA, lark1 | 48 | 52 | 17 | 31 | 0.38 ± 0.02f | 23.4 ± 0.05 |
| c929G4 | 26 | 88 | 13 | 0 | 0.38 ± 0.02 | 23.8 ± 0.07 |
| pdfGal80; RNAiA, lark1 | 51 | 98 | 2 | 0 | 0.47 ± 0.02 | 23.4 ± 0.03 |
| mai179G4>RNAiA | 15 | 40 | 47 | 13 | 0.30 ± 0.03g | 23.1 ± 0.06 |
| mai179G4>RNAiA, lark1 | 49 | 41 | 35 | 24 | 0.17 ± 0.03h | 23.5 ± 0.12 |
| pdfGal80; mai179>RNAiA, lark1 | 34 | 56 | 21 | 24 | 0.42 ± 0.02f | 23.3 ± 0.05 |
| mai179G4 | 15 | 81 | 19 | 0 | 0.38 ± 0.03 | 23.5 ± 0.09 |
| RNAiA | 15 | 75 | 19 | 6 | 0.36 ± 0.04 | 23.3 ± 0.05 |
| RNAiA, lark1 | 16 | 81 | 19 | 0 | 0.36 ± 0.03 | 23.2 ± 0.06 |
| phmN1G4>RNAiA | 26 | 88 | 8 | 4 | 0.23 ± 0.02 | 24.1 ± 0.10 |
| phmN1G4 | 23 | 91 | 4 | 4 | 0.26 ± 0.03 | 23.5 ± 0.12 |
| RNAi | 29 | 97 | 0 | 3 | 0.57 ± 0.02 | 24.1 ± 0.04 |
| RNAiA, lark1 | 16 | 100 | 0 | 0 | 0.53 ± 0.02 | 23.8 ± 0.04 |
We examined the locomotor activity of flies carrying another pan-neuronal driver, NirvanaGal4 (nrvG4). Similar to results using elavG4, these nrvG4; nrvG4>RNAiA flies, which carried 2 copies of the driver, exhibited less robust rhythmicity when assayed at 30 °C (Table 1 and Fig. 2C). Indeed, when flies of a similar genotype were heterozygous for lark1, 42% of them were arrhythmic, whereas controls for this genotype (nrvG4; nrvG4 and RNAiA, lark1) were mostly rhythmic (only 4%–12% arrhythmicity) (Fig. 2C). A more severe phenotype when flies were heterozygous for lark1 suggests specificity for the observed RNAi-mediated knockdown (compare similar genotypes with or without lark1 in Figs. 2 and 3 and Table 1).
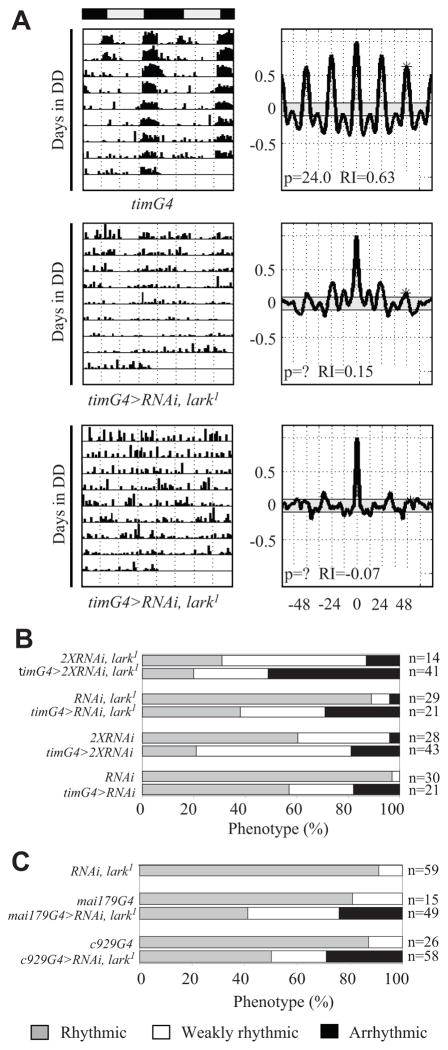
Figure 3.
Knockdown of LARK using tim-uasG4 results in weak rhythms or arrhythmic behavior. (A) Representative actograms and correlograms for tim-uasG4>RNAiA, lark1 and control flies (top panels). The middle panels show a weakly rhythmic fly, whereas the bottom panels show an arrhythmic fly. (B) Percentages of rhythmic, weakly rhythmic, or arrhythmic flies in various tim-uasG4 knockdown and control genotypes. (C) Behavior with knockdown of LARK using c929G4 or mai179G4, which drive expression in clock and other neurosecretory cells. Activity was examined at 30 °C.
Knockdown of LARK in Clock Neurons Causes Arrhythmicity
A group of approximately 150 clock neurons comprises the pacemaker driving behavioral rhythmicity (Nitabach and Taghert, 2008). To determine whether LARK is required within clock neurons, we expressed the RNAiA transgene in all such neurons (alone or in a lark1 background) using either the perG4 or the tim-uasG4 driver. When flies were behaviorally assayed at 29 °C, we observed altered rhythmicity in tim-uasG4>RNAiA flies (Fig. 3A and 3B and Table 1). The phenotype was more severe in tim-uasG4>RNAiA/RNAiA flies, which carry 2 copies of the RNAi transgene: only 21% were strongly rhythmic versus 57% for tim-uasG4>RNAiA flies. We note that little or no effect was seen using the per-Gal4 or Cry-G4 drivers (data not shown) perhaps because these are weaker drivers compared to tim-uasG4, which is expressed at high levels due to the presence of UAS elements in the construct that provide a positive feedback of Gal4 to the tim promoter (Blau and Young, 1999). To determine if further reductions in LARK resulted in a more extreme phenotype, we assayed tim-uasG4>RNAiA/RNAiA, lark1 flies (2 copies of RNAiA with lark1). That genotype exhibited an extreme phenotype (51% arrhythmic and 29% weakly rhythmic) compared to other RNAi-expressing flies or control genotypes (RNAiA/RNAiA, lark1) (Fig. 3B and Table 1). Similarly, more extreme phenotypes were observed in flies carrying recombinant RNAiB, lark1 or both the RNAiA and RNAiB transgenes, relative to RNAiB or either transgene alone, respectively (data not shown). Flies carrying 2 copies of tim-uasG4 were not analyzed because such flies were arrhythmic even without the RNAi transgene, presumably due to a genomic insertion effect (data not shown). Altogether, our results demonstrate a dosage dependency for the observed effects and verify specificity of the RNAi transgenes. Although tim-uasG4 drives expression in both neurons and glia, LARK cannot be detected in glial cells (McNeil et al., 2001; Huang et al., 2009); thus, we interpret our results to indicate a requirement for the RBP within clock neurons.
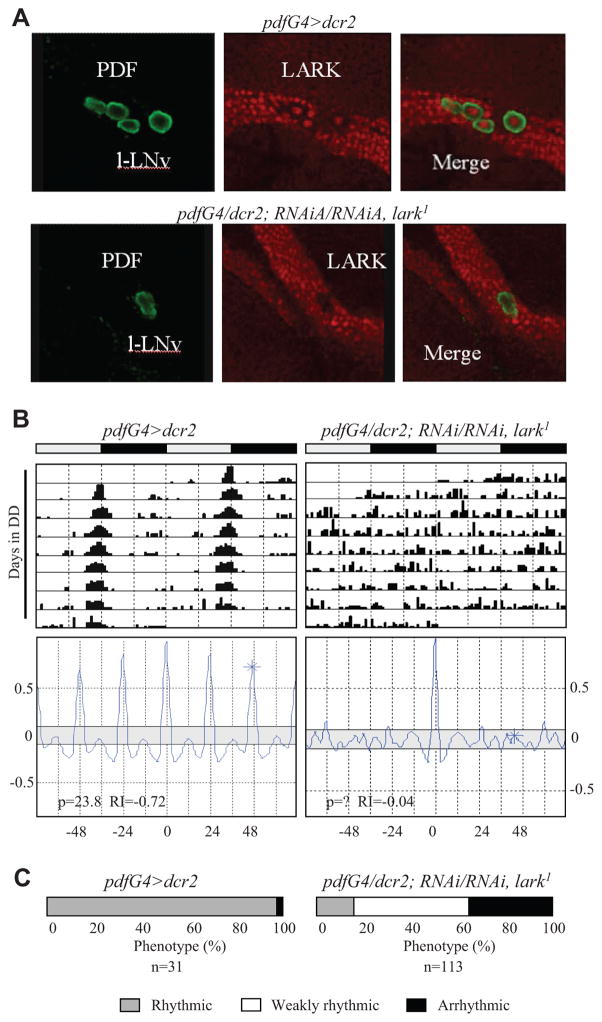
Among the approximately 150 clock neurons, there are 16 PDF-containing LNv, which serve as pacemakers for the regulation of free-running rhythms in constant dark conditions (Nitabach and Taghert, 2008). To refine the spatial requirement for LARK, we selectively knocked down the RBP in PDF neurons using the pdfG4 driver. Surprisingly, there was little or no effect on the percentage of rhythmicity even in pdfG4>RNAiA/RNAiA, lark1 flies, which carry multiple copies of the RNAi transgene (Table 1). However, upon examination of LARK immunoreactivity in the RNAi flies, we did not detect a knockdown of the protein (Suppl. Fig. S1B). Thus, we included a UAS-dicer2 (dcr2) transgene in the background of RNAi flies (pdfG4>dcr2; RNAiA/RNAiA, lark1 in Table 1), which is known to enhance RNAi effects (Dietzl et al., 2007). In flies expressing dcr2, there was an obvious knockdown of LARK in the PDF neurons compared to controls lacking the pdfG4 driver (Fig. 4A and Suppl. Fig. S1). Importantly, there was no obvious effect on cell viability or neuronal projections with knockdown of LARK in the PDF cells (data not shown). As shown in Figure 4B and 4C, pdfG4>dcr2; RNAiA/RNAiA, lark1 flies exhibited altered behavior: only 12% of the flies were strongly rhythmic, and 37% were arrhythmic, whereas 97% of controls (run at the same time) were strongly rhythmic. Correspondingly, the average RI value for the knockdown population was only 0.18 versus 0.46 for the pdfG4>dcr2 controls (Table 1). We conclude that LARK is required within PDF neurons for robust rhythmicity.
Figure 4.
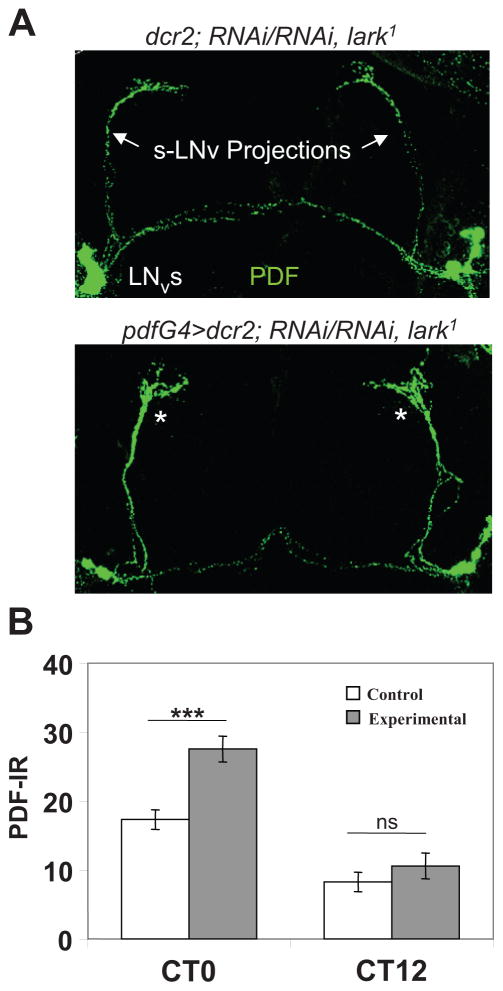
A strong knockdown of LARK in PDF neurons causes arrhythmicity. (A) Dicer (dcr2) expression greatly increases effectiveness of knockdown of LARK in the PDF neurons. High-magnification optical sections for control (upper panels) and LARK (lower panels) knockdown brains. Note the “hole” where LARK nuclear staining ought to be in the PDF cells of knockdown brains (lower right panel). Only 2 large PDF cells (l-LNv), in close proximity, are shown for the LARK knockdown in this optical section, but l-LNv cells in other optical sections also exhibited a severe knockdown of the protein; similarly, there was decreased LARK staining in the s-LNv cells. Green = PDF; red = nuclear LARK. (B) Actograms and correlograms for control (left) and knockdown (right) flies. (C) Percentages of rhythmic, weakly rhythmic, and arrhythmic flies for control and knockdown populations. Flies employed in these experiments were reared and tested at 30 °C.
We examined 2 additional Gal4 drivers, c929 and mai179, that express in the PDF neurons as well as other neurosecretory cells. The c929 driver expresses in approximately 100 peptidergic neurons, including the PDF-containing l-LNv cells (the large PDF neurons) (Taghert et al., 2001). It has been shown (Grima et al., 2004) that mai179 is expressed in adult clock neurons including some of the l-LNv, most of the LNd (which are PDF negative), and all s-LNv (4/5 being PDF positive). Interestingly, knockdown of LARK using c929G4 or mai179G4 produced similar results, with 50% or fewer of the G4>RNAiA flies possessing robust rhythms (Fig. 3C and Table 1). As expected, the percentage of such flies showing arrhythmic behavior increased when the knockdown was performed in a lark1 heterozygous background (Table 1).
With regard to the clock neuronal population, the overlap of the c929 and mai179 expression patterns is restricted to the l-LNv cells (the only clock neurons in which c929 expresses). Thus, either the l-LNv or a nonclock population of neurons requires LARK for normal behavioral rhythms. In contrast to pdfG4, however, we did not observe a significant knockdown of LARK in the PDF neurons using either the mai179 or c929 driver (Suppl. Fig. S2). In fact, there seemed to be an increase in LARK amounts in the l-LNv cells of c929G4>RNAiA flies. These results suggest a requirement for LARK in non-PDF cell types in which c929G4 or mai179G4 is expressed. To confirm such a requirement, we examined pdfGal80; c929G4>RNAiA, lark1 and pdf-Gal80; mai179G4>RNAiA, lark1 flies, which express the lark knockdown transgene in PDF neurons but also express an inhibitor of Gal4 (Gal80) in the same cells (effectively turning off Gal4 activity in PDF cells but allowing RNAi effects in other neurosecretory cells). These populations were arrhythmic to a similar extent as c929G4>RNAiA, lark1 and mai179G4>RNAiA, lark1 flies (Table 1), indicating that there is a neuronal requirement for LARK outside of the PDF population.
PDF Immunoreactivity (PDF-IR) Is Altered by LARK Knockdown
We examined PER cycling in the adult brains of tim-uasG4>RNAiA/RNAiA, lark1 and pdfG4>dcr2; RNAiA/RNAiA, lark1 flies to determine if the observed arrhythmicity was caused by altered molecular oscillator function. However, both genotypes exhibited neuronal PER abundance and nuclear entry rhythms on DD day 2 that were indistinguishable from those observed in control brains (Suppl. Fig. S3; data not shown). We next examined PDF-IR in pdfG4>dcr2; RNAiA/RNAiA, lark1 knockdown brains based on our previous finding that increased LARK expression within PDF cells, which also causes arrhythmicity, resulted in decreased neuropeptide signaling in LNv projections (Schroeder et al., 2003). Whereas PDF-IR in the LNv cell bodies of knockdown flies was similar to that of controls (Suppl. Fig. S3B), immunosignals for the peptide were increased in the dorsal projections of the s-LNv (Fig. 5), an effect opposite to the decrease observed in LARK-overexpressing flies. Although PDF-IR continued to cycle, as expected, in knockdown brains, with a high at CT0 and a low at CT12, the signal at the high point was increased relative to controls in 4 independent experiments. On average, it was approximately 55% stronger at the termini of the s-LNv projections than that observed in control brains (Fig. 5B). To determine if peak PDF-IR was shifted in RNAi flies relative to controls, we examined signals at CT21, CT0, and CT3. As shown in Supplementary Figure S4, RNAi flies exhibited a significant increase in PDF-IR at CT21 and CT0 relative to controls, but signals were similar in the 2 genotypes by CT3 (see Discussion). As decreased PDF signals in the s-LNv projections at the beginning of the day are thought to represent neuropeptide release (Park et al., 2000), which drives rhythmic locomotor activity, our results suggest that LARK knockdown flies may be defective in this process.
Figure 5.
PDF-IR is abnormally high in s-LNv projections at CT0 with _pdfG4_-driven LARK knockdown. (A) Representative brain hemispheres showing PDF-IR in control and experimental (pdfG4>dcr2; RNAiA/RNAiA, lark1) brains showing abnormally high signals in the s-LNv dorsal projections. (B) Quantification of PDF-IR signals in the 2 types of brains on DD day 2. ***Difference from the control: p < 0.001 (ANOVA with Tukey-Kramer multiple comparisons).
LARK Is Required in the Prothoracic Gland for Normal Rhythmicity in Adult Eclosion
LARK was discovered in a _P-_element screen for mutants with defects in the daily timing of population eclosion (Newby and Jackson, 1993). The lark1 allele is lethal, but we found in those earlier studies that lark1/+ heterozygotes emerged earlier in the cycle than did control populations, indicating a role for LARK in the temporal gating of eclosion. We note that this early eclosion phenotype has not been reproducibly observed in more recent studies (using stocks maintained for many generations in the laboratory). Nevertheless, other studies suggest that LARK functions in the clock regulation of eclosion (Schroeder et al., 2003; Park et al., 2003).
To determine the effects of LARK knockdown on the timing of eclosion, we examined eclosion profiles for several different Gal4>RNAi and control populations. In experiments performed at 29 °C, LARK knockdowns with the elav, crustacean cardioactive peptide (ccap), eclosion hormone (eh), c929, pdf, or tim-uasG4 drivers did not cause significant arrhythmicity in LD or DD, although we observed elevated night-time eclosion with _tim-uasG4_–driven lark RNAi (data not shown), reminiscent of that previously reported for lark/+ heterozygotes (Newby and Jackson, 1993).
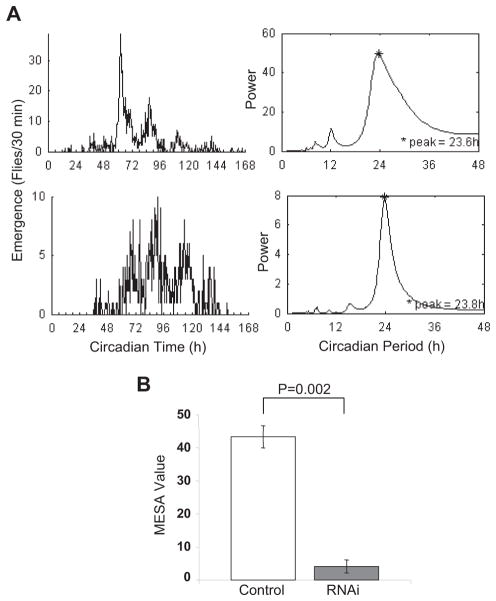
The tim gene is expressed in the Drosophila nervous system and peripheral tissues including the prothoracic gland (PG), which synthesizes the insect molting hormone ecdysone. Furthermore, it is known that a PER/TIM-based oscillator exists in the PG (Emery et al., 1997), and it is thought to be important for the clock control of eclosion (Myers et al., 2003). We previously showed that LARK can be detected in the PG (Zhang et al., 2000). Thus, it is possible that the observed effects on the eclosion rhythm with tim-uasG4 result from decreased LARK in this tissue. To test this idea, we employed 2 different PG drivers, mai60G4 and phmN1G4, to restrict knockdown of LARK to the PG. Both drivers have been described as PG specific in pupae (Myers et al., 2003; Mirth et al., 2005). In particular, phmN1G4 reflects expression of the phantom gene, which is involved in steroidogenesis and is exclusively expressed in the PG during postembryonic stages. Whereas mai60G4>RNAiA populations showed normal daily eclosion profiles (data not shown), phmN1G4>RNAi populations, expressing any 1 of 3 different lark RNAi transgenes (RNAiA and 2 others from the Japanese National Institute of Genetics), exhibited a less robust eclosion rhythm in DD. However, there was variability among experiments, which might reflect a hypomorphic effect of the LARK knockdown. With phmN1G4>RNAiA populations, for example, eclosion profiles were arrhythmic in 5 experiments, weakly rhythmic in 5 others, and rhythmic in 4 experiments (as assessed using the MESA statistic; see Materials and Methods). In contrast, control populations (n = 3 experiments) uniformly showed robust rhythms. Inclusion of dcr2 in the genetic background seemed to reduce variability. In 3 experiments, phmN1G4>dcr2; RNAiA populations were weakly rhythmic and significantly different from controls, which were robustly rhythmic (Fig. 6A and 6B; note scale differences between control and RNAi MESA plots in panel A). Thus, we conclude that LARK is required within the PG for robust free-running rhythms of population eclosion. We note that adult locomotor activity rhythms were completely normal in phmN1G4>RNAiA flies (Table 1), consistent with degeneration of the PG after metamorphosis.
Figure 6.
LARK is required in the PG for normal free-running eclosion rhythms. (A) Plots of adult eclosion and MESA statistics for phmN1G4>dcr2/RNAiA (lower) and control phmN1G4>dcr2 (upper) populations (note different scales for MESA plots). Eclosion was assayed at 20 °C using an automated (Trikinetics) monitor (see Materials and Methods). (B) Average MESA values for phmG4>dcr2/RNAiA and control phmN1G4>dcr2 populations (n = 3 experiments for both genotypes).
To determine why phmN1G4, and not mai60G4, was associated with altered eclosion rhythms, we reexamined the expression patterns of both drivers, using UAS-GFP as a reporter. Surprisingly, we did not detect fluorescence in the PG of mai60G4>GFP pharate adults, although intense fluorescence was observed in the salivary glands of these animals (Suppl. Fig. S5). The previous study reporting the mai60G4 expression pattern utilized a UAS-lacZ transgene and β-galactosidase staining to visualize expression of the driver (Myers et al., 2003), and the increased sensitivity associated with this assay may explain the discrepancy between our results and that study. In contrast, phmN1G4>GFP pharate adults exhibited an intense fluorescence specifically in the PG (Suppl. Fig. S5). Considered with the lark RNAi results using this driver, we conclude that LARK is required in the PG for normal eclosion rhythms.
DISCUSSION
LARK Is Required in Clock Neurons for Robust Activity Rhythms
This report documents a requirement for LARK in circadian pacemaker neurons. Behavioral arrhythmicity was observed with knockdown of the RBP in either PDF- or TIM-containing neurons, although RNAi flies appeared to entrain normally to LD 12:12. With knockdown of LARK in PDF cells, there was a corresponding effect on PDF-IR; immunoreactive signaling was significantly increased in the termini of the s-LNv projections at the time of peak neuropeptide release. A decrease in PDF-IR in the projections of the s-LNv has previously been interpreted as neuropeptide release (Park et al., 2000); thus, it is possible that increased immunoreactive signals with LARK knockdown reflect an inhibition of peptide release. Examination of PDF-IR at several different circadian times showed that signaling is significantly higher in RNAi versus control flies at both CT21 and CT0, but similar in the 2 genotypes by CT3, that is, not significantly delayed. Given the arrhythmic phenotype observed in many lark knockdown flies, we favor the idea that there is decreased PDF release in such individuals, perhaps with a subsequent intracellular degradation of the peptide (because the immunosignal is normal by CT3). That all flies with decreased LARK function are not arrhythmic (i.e., the phenotype is not completely penetrant) is presumably due to the inherently “hypomorphic” nature of RNAi-mediated knockdowns. Together with previous results showing that LARK overexpression is associated with decreased PDF-IR within s-LNv projections (Schroeder et al., 2003), our findings suggest that LARK may regulate release of the neuropeptide. In this regard, it is of interest that putative target RNAs have been identified for LARK (Huang et al., 2007), and many of them encode proteins predicted to function in synaptic transmission (Huang et al., 2007).
LARK Is Required in Nonclock Cells for Normal Activity Rhythms
We observed arrhythmicity with LARK knockdown by mai179G4 or c929G4, drivers that express broadly in neurosecretory cells of the adult brain including neurons of the clock circuitry. Arrhythmicity was also observed with these drivers even when flies carried pdfGal80, which is predicted to specifically inhibit RNAi expression in the PDF neurons. Thus, LARK is required in PDF and non-PDF neurosecretory cells for normal circadian behavior. Perhaps the RBP regulates a posttranscriptional event within neurons downstream of the clock circuitry to mediate clock output. A requirement for LARK in other neurosecretory cells also suggests that the RBP may generally regulate neuropeptide secretion.
LARK Expression within the PG Is Required for Robust Adult Eclosion Rhythms
Although the lark1 mutant was isolated on the basis of an eclosion rhythm phenotype (Newby and Jackson, 1993), it has never been determined which tissues require the RBP for normal daily eclosion rhythms. A complication with this determination is the ubiquitous expression of LARK and its vital role during oogenesis and zygotic development. The present study indicates that LARK knockdown with either of 2 different Gal4 drivers (tim and phmN1) results in abnormal population eclosion rhythms, although effects are more prominent using phmN1. Importantly, phmN1 is exclusively expressed in the PG of developing adults, indicating a requirement for LARK within that tissue. Differences in phenotypic severity for the 2 drivers may reflect the strength of expression within the PG or modification of the phenotype by expression within the nervous system (tim).
Previous studies have shown that a PER-based oscillator exists in the PG and that this peripheral oscillator may interact with the neuronal pacemaker to regulate the timing of eclosion (Emery et al., 1997; Myers et al., 2003). The only defined function of the PG is the synthesis of ecdysteroid (Riddiford, 1993), and the hormone is known to be constitutively released from that tissue. Thus, our results, surprisingly, suggest that LARK may have a role in ecdysteroid biosynthesis perhaps by post-transcriptional regulation of a factor required for this process. Synthesis of the hormone is known to be rhythmic in the PG of certain insects (Vafopoulou and Steel, 1991), but it is unknown whether such a rhythm exists in Drosophila. Furthermore, it has not been determined whether LARK oscillates in abundance within the PG, although the RBP shows circadian changes in the nervous system (McNeil et al., 1998). It will be of interest to examine the hypothesis that LARK participates in the interactions between central and peripheral oscillators (Myers et al., 2003) that govern the daily timing of eclosion events.
Supplementary Material
Acknowledgments
We thank all members of the Jackson laboratory for help with experiments, in particular Dr. Michelle Tangredi for help with scoring of PDF immunoreactive signals and Dr. Yanmei Huang who did the immunostaining shown in Figure 4A to demonstrate LARK knockdown using the pdfG4 driver. We also thank R. Carthew for the pWIZ vector; J. Belote for helpful discussions about fly RNAi transgenes; R. Stanewsky, P. Taghert, G. McNeil, and K. Rao for antibodies; the Indiana University (Bloomington) Drosophila Stock Center and J. Hall, J. Blau, R. Allada, J. Park, F. Rouyer, and M. Rosbash for fly stocks; the CNR Imaging Core and Alenka Lovy-Wheeler for help with confocal microscopy; and FlyBase for access to Drosophila genetics and genomics information. This work was supported by NIH R01 HL59873 and NS065900 to F.R.J. and a center grant from NINDS (NIH P30 NS047243) to F.R.J. F.S.N. was supported by training grant NIH T32 HD049341. C.M. was supported by FONDECYT grant 3090019; J.E. was supported by NIH R21 NS053833 and a Millennium “Centro Interdisciplinario de Neurociencia” grant.
Footnotes
CONFLICT OF INTEREST STATEMENT
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- Baggs JE, Green CB. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol. 2003;13:189–198. doi: 10.1016/s0960-9822(03)00014-9. [DOI] [PubMed] [Google Scholar]
- Benito J, Zheng H, Hardin PE. PDP1 epsilon functions downstream of the circadian oscillator to mediate behavioral rhythms. J Neurosci. 2007;27:2539–2547. doi: 10.1523/JNEUROSCI.4870-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dockendorff TC, Su HS, Mcbride SMJ, Yang ZH, Choi CH, Siwicki KK, Sehgal A, Jongens TA. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron. 2002;34:973–984. doi: 10.1016/s0896-6273(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Emery IF, Noveral JM, Jamison CF, Siwicki KK. Rhythms of Drosophila period gene expression in culture. Proc Natl Acad Sci U S A. 1997;94:4092–4096. doi: 10.1073/pnas.94.8.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier E, Belote JM. Temperature-dependent gene silencing by an expressed inverted repeat in Drosophila. Genesis. 2000;26:240–244. doi: 10.1002/(sici)1526-968x(200004)26:4<240::aid-gene40>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Garbarino-Pico E, Green CB. Posttranscriptional regulation of mammalian circadian clock output. Cold Spring Harb Symp Quant Biol. 2007;72:145–156. doi: 10.1101/sqb.2007.72.022. [DOI] [PubMed] [Google Scholar]
- Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, Lourim D, Keller SR, Besharse JC. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci U S A. 2007;104:9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Hassidim M, Yakir E, Fradkin D, Hilman D, Kron I, Keren N, Harir Y, Yerushalmi S, Green RM. Mutations in chloroplast RNA binding provide evidence for the involvement of the chloroplast in the regulation of the circadian clock in Arabidopsis. Plant J. 2007;51:551–562. doi: 10.1111/j.1365-313X.2007.03160.x. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Nater M, Apel K, Staiger D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1997;94:8515–8520. doi: 10.1073/pnas.94.16.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Genova G, Roberts M, Jackson FR. The LARK RNA-binding protein selectively regulates the circadian eclosion rhythm by controlling E74 protein expression. PLoS ONE. 2007;2:e1107. doi: 10.1371/journal.pone.0001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Howlett E, Stern M, Jackson FR. Altered LARK expression perturbs development and physiology of the Drosophila PDF clock neurons. Mol Cell Neurosci. 2009;41:196–205. doi: 10.1016/j.mcn.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang JK, Shin J, Lee J, Jeon K, Hwang S, et al. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Iliev D, Voytsekh O, Schmidt EM, Fiedler M, Nykytenko A, Mittag M. A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol. 2006;142:797–806. doi: 10.1104/pp.106.085944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SH, Taghert PH. PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J Comp Neurol. 2010;518:1925–1945. doi: 10.1002/cne.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson FR, Banfi S, Guffanti A, Rossi E. A novel zinc finger-containing RNA-binding protein conserved from fruitflies to humans. Genomics. 1997;41:444–452. doi: 10.1006/geno.1997.4704. [DOI] [PubMed] [Google Scholar]
- Jackson FR, Genova GK, Huang Y, Kleyner Y, Suh J, Roberts MA, Sundram V, Akten B. Genetic and biochemical strategies for identifying Drosophila genes that function in circadian control. In: Young MW, editor. Circadian Rhythms. New York: Elsevier; 2005. pp. 663–682. [DOI] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Schoer R, Rosbash M. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 2008;6:e119. doi: 10.1371/journal.pbio.0060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Gatfield D, Esau CC, Green CB. MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS One. 2010;5:e11264. doi: 10.1371/journal.pone.0011264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Matsumoto K, Hirose M, Shimada M, Nagano M, Shigeyoshi Y, Hoshino S, Ui-Tei K, Saigo K, Green CB, et al. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci U S A. 2007;104:1859–1864. doi: 10.1073/pnas.0607567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakin-Thomas PL. Transcriptional feedback oscillators: maybe, maybe not. J Biol Rhythms. 2006;21:83–92. doi: 10.1177/0748730405286102. [DOI] [PubMed] [Google Scholar]
- Lear BC, Merrill CE, Lin JM, Schroeder A, Zhang L, Allada R. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron. 2005;48:221–227. doi: 10.1016/j.neuron.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3:1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil GP, Schroeder AJ, Roberts MA, Jackson FR. Genetic analysis of functional domains within the Drosophila LARK RNA-binding protein. Genetics. 2001;159:229–240. doi: 10.1093/genetics/159.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil GP, Zhang XL, Genova G, Jackson FR. A molecular rhythm mediating circadian clock output in Drosophila. Neuron. 1998;20:297–303. doi: 10.1016/s0896-6273(00)80457-2. [DOI] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Mirth C, Truman JW, Riddiford LM. The role of the prothoracic gland in determining critical weight for metamorphosis in Drosophila melanogaster. Curr Biol. 2005;15:1796–1807. doi: 10.1016/j.cub.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Morales J, Hiesinger PR, Schroeder AJ, Kume K, Verstreken P, Jackson FR, Nelson DL, Hassan BA. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron. 2002;34:961–972. doi: 10.1016/s0896-6273(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Myers EM, Yu JJ, Sehgal A. Circadian control of eclosion: interaction between a central and peripheral clock in Drosophila melanogaster. Curr Biol. 2003;13:526–533. doi: 10.1016/s0960-9822(03)00167-2. [DOI] [PubMed] [Google Scholar]
- Newby LM, Jackson FR. Drosophila ebony mutants have altered circadian activity rhythms but normal eclosion rhythms. J Neurogenet. 1991;7:85–101. doi: 10.3109/01677069109066213. [DOI] [PubMed] [Google Scholar]
- Newby LM, Jackson FR. A new biological rhythm mutant of Drosophila melanogaster that identifies a gene with an essential embryonic function. Genetics. 1993;135:1077–1090. doi: 10.1093/genetics/135.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng FS, Tangredi MM, Jackson FR. Glial cells physiologically modulate clock neurons and circadian behavior in a calcium-dependent manner. Curr Biol. 2011;21:625–634. doi: 10.1016/j.cub.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–R93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Hall JC. Isolation and chronobiological analysis of a neuropeptide pigment-dispersing factor gene in Drosophila melanogaster. J Biol Rhythms. 1998;13:219–227. doi: 10.1177/074873098129000066. [DOI] [PubMed] [Google Scholar]
- Park JH, Helfrich-Förster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Schroeder AJ, Helfrich-Forster C, Jackson FR, Ewer J. Targeted ablation of CCAP neuropeptide-containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development. 2003;130:2645–2656. doi: 10.1242/dev.00503. [DOI] [PubMed] [Google Scholar]
- Renn SCP, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Riddiford LM. Hormones and Drosophila development. In: Bate M, Arias AM, editors. The Development of Drosophila melanogaster. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1993. pp. 899–939. [Google Scholar]
- Schroeder AJ, Genova K, Roberts MA, Kleyner Y, Suh J, Jackson FR. Cell-specific expression of the lark RNA-binding protein in Drosophila results in morphological and circadian behavioral phenotypes. J Neurogenet. 2003;17:139–169. [PubMed] [Google Scholar]
- Shafer OT, Helfrich-Forster C, Renn SCP, Taghert PH. Reevaluation of Drosophila melanogaster’s neuronal circadian pacemakers reveals new neuronal classes. J Comp Neurol. 2006;498:180–193. doi: 10.1002/cne.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert PH, Hewes RS, Park JH, O’Brien MA, Han M, Peck ME. Multiple amidated neuropeptides are required for normal circadian locomotor rhythms in Drosophila. J Neurosci. 2001;21:6673–6686. doi: 10.1523/JNEUROSCI.21-17-06673.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert PH, Shafer OT. Mechanisms of clock output in the Drosophila circadian pacemaker system. J Biol Rhythms. 2006;21:445–457. doi: 10.1177/0748730406293910. [DOI] [PubMed] [Google Scholar]
- Vafopoulou X, Steel CGH. Circadian regulation of synthesis of ecdysteroids by prothoracic glands of the insect Rhodnius prolixus: evidence of a dual oscillator system. Gen Comp Endocrinol. 1991;83:27–34. doi: 10.1016/0016-6480(91)90102-c. [DOI] [PubMed] [Google Scholar]
- Zhang X, McNeil GP, Hilderbrand-Chae MJ, Franklin TM, Shroeder AJ, Jackson FR. Circadian regulation of the LARK RNA-binding protein within identifiable neurosecretory cells. J Neurobiol. 2000;45:14–29. doi: 10.1002/1097-4695(200010)45:1<14::aid-neu2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.