The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development (original) (raw)
Abstract
MicroRNAs (miRNAs) are endogenous 21–24-nt RNAs that can down-regulate gene expression by pairing to the messages of protein-coding genes to specify mRNA cleavage or repression of productive translation. They act within the RNA-induced silencing complex (RISC), which in animals contains a member of the Argonaute family of proteins. In the present study, we show that Arabidopsis ago1 mutants have increased accumulation of mRNAs known to be targeted for cleavage by miRNAs. In hypomorphic ago1 alleles, this compromised miRNA function occurs without a substantial change in miRNA accumulation, whereas in null alleles it is accompanied by a drop in some of the miRNAs. Therefore, AGO1 acts within the Arabidopsis miRNA pathway, probably within the miRNA-programmed RISC, such that the absence of AGO1 destabilizes some of the miRNAs. We also show that targeting of AGO1 mRNA by miR168 is needed for proper plant development, illustrating the importance of feedback control by this miRNA. Transgenic plants expressing a mutant AGO1 mRNA with decreased complementarity to miR168 overaccumulate AGO1 mRNA and exhibit developmental defects partially overlapping with those of dcl1, hen1, and hyl1 mutants showing a decrease in miRNA accumulation. miRNA targets overaccumulate in miR168-resistant plants, suggesting that a large excess of AGO1 protein interferes with the function of RISC or sequesters miRNAs or other RISC components. Developmental defects induced by a miR168-resistant AGO1 mRNA can be rescued by a compensatory miRNA that is complementary to the mutant AGO1 mRNA, proving the regulatory relationship between miR168 and its target and opening the way for engineering artificial miRNAs in plants.
Keywords: AGO1, miRNA, negative feedback regulation, posttranscriptional gene silencing, RNAi
Noncoding small RNAs (20–24 nt long) have recently been recognized as important regulators in both plants and animals, acting in the control of invading nucleic acids or in the regulation of gene expression (Hannon 2002; Zamore 2002; Ambros 2003; Bartel 2004). Small RNAs that derive from long double-stranded RNAs are called siRNAs. siRNAs can direct the cellular immune system to silence exogenous invading nucleic acids (viruses, transgenes) by RNA cleavage through phenomena called posttranscriptional gene silencing (PTGS) in plants or RNA interference (RNAi) in animals (Hannon 2002; Zamore 2002). siRNAs can also trigger changes in the chromatin state that silence endogenous invading and/or repeated elements (transposons, centromers) at the transcriptional level (Finnegan and Matzke 2003). Endogenous small RNAs that are processed from stem–loop precursor RNAs and regulate genes that are distinct from the genes from which they derive are called miRNAs (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001; Bartel 2004). Such RNAs have been cloned from worm, fly, fish, mouse, human, and plant (Bartel 2004). Many of them are conserved between species, indicating an evolutionarily conserved role in gene regulation.
In animals, miRNAs are derived from long primary transcripts (pri-miRNAs) that are cleaved by the nuclear-localized RNase III Drosha into 70-nt precursors (pre-miRNAs; Lee et al. 2003). These 70-nt precursors are actively exported out of the nucleus by Ran-GTP and the export receptor Exportin-5 (Yi et al. 2003; Bohnsack et al. 2004; Lund et al. 2004). Pre-miRNAs are subsequently cleaved by another RNAse III with an RNA helicase domain called Dicer to generate short-lived double-stranded miRNA intermediates (Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001; Lee et al. 2003). One strand of these duplex RNAs accumulates as the mature miRNA and down-regulates gene expression by pairing to the messages of protein-coding genes to specify repression of productive translation or mRNA cleavage (Lee et al. 1993; Wightman et al. 1993; Olsen and Ambros 1999; Bartel 2004; Yekta et al. 2004). The miRNAs act within a ribonucleoprotein complex that is similar if not identical to the RNA-induced silencing complex (RISC; Hutvagner and Zamore 2002; Mourelatos et al. 2002; Doench et al. 2003; Zeng et al. 2003). The RISC was first described as the ribonucleoprotein complex that mediates RNAi (Hammond et al. 2001). When purified from either fly or human cells, it contains a member of the ARGONAUTE protein family that is considered a core component of the complex (Hammond et al. 2001; Caudy et al. 2002, 2003; Hutvagner and Zamore 2002; Ishizuka et al. 2002; Martinez et al. 2002; Mourelatos et al. 2002). ARGONAUTE proteins (also known as PPD proteins because they carry PAZ and PIWI domains) have a β-barrel core that together with a side appendage weakly bind to single-stranded RNAs at least 5 nt in length and also to double-stranded RNA (Lingel et al. 2003; Song et al. 2003; Yan et al. 2003), suggesting that ARGONAUTE proteins could be directly associated with siRNAs/miRNAs before and after they recognize their mRNA targets. RISC-associated proteins also include the nuclease Tudor-SN, the putative RNA-binding VIG protein and the Fragile X-related protein (Hammond et al. 2001; Caudy et al. 2002, 2003; Hutvagner and Zamore 2002; Ishizuka et al. 2002; Martinez et al. 2002).
In plants, most knowledge of miRNA biogenesis comes from the analysis of mutants originally isolated during developmental screens, which turned out to be partially defective in the accumulation of miRNAs. Plant mutants that are totally defective for miRNA accumulation have not been reported yet, and it is highly probable that plants totally depleted in miRNAs are inviable. Indeed, mutants that lack the nuclear DICER-LIKE1 (DCL1) protein are embryo-lethal, indicating that DCL1 is required for plant viability, at least during reproduction and/or at early stages of development (Schauer et al. 2002). Partial loss-of-function dcl1 mutants exhibiting point mutations in the RNA helicase domain (dcl1-7, dcl1-8) or truncation of the second dsRNA-binding domain (dcl1-9) are viable but show developmental defects including sterility (Schauer et al. 2002). Accompanying these defects are greatly reduced miRNA levels, suggesting the crucial role of miRNAs during plant development and reproduction (Park et al. 2002; Reinhart et al. 2002). The proper accumulation of miRNAs also depends on the activity of two other nuclear proteins: HEN1 and HYL1 (Park et al. 2002; Boutet et al. 2003; Han et al. 2004; Vazquez et al. 2004). _hen1_- and _hyl1_-null alleles show reduced miRNA levels and developmental defects that overlap with that of partial loss-of-function dcl1 mutants. However, in contrast to _dcl1_-null alleles, _hen1_- and _hyl1_-null alleles are viable. This suggests either that HEN1 and HYL1 do not have essential functions in the miRNA pathway or that other genes encode proteins with partially redundant functions.
Two other proteins have been proposed to play a role in the plant miRNA pathway: HASTY (HST) and ARGONAUTE 1 (AGO1). HST encodes a protein that is homologous to Exportin-5, and hst mutants have developmental phenotypes, suggesting that HST participates in the transport of miRNAs or pre-miRNAs from the nucleus to the cytoplasm (Bollman et al. 2003; Yi et al. 2003; Bohnsack et al. 2004; Lund et al. 2004). AGO1 is the founding member of the ARGONAUTE protein family, which comprises 10 members in Arabidopsis (Morel et al. 2002). The first _ago1_-null alleles were isolated in a screen for mutants exhibiting early developmental defects (Bohmert et al. 1998). Additional _ago1_-null or hypomorphic alleles were isolated during a screen for PTGS-deficient mutants (Fagard et al. 2000; Morel et al. 2002). AGO1 was subsequently shown to be specifically required for siRNA accumulation and DNA methylation triggered by sense transgenes (S-PTGS) but not inverted-repeat transgenes (IR-PTGS; Beclin et al. 2002; Boutet et al. 2003). These results prompted the early proposal that AGO1 is not part of RISC but rather is acting upstream from the mRNA degradation step in the S-PTGS pathway, and that S-PTGS and IR-PTGS are two branches of the PTGS pathway that converge toward a common RISC that contains other AGO proteins (Beclin et al. 2002). Indeed, some of the other proteins in the AGO family are associated with specific functions or processes, including AGO4, which is linked to transposon siRNAs as well as DNA and histone methylation (Zilberman et al. 2003), ZIP/AGO7, which controls vegetative phase changes (Hunter et al. 2003), and PNH/ZLL/AGO10, which controls meristem maintenance (Moussian et al. 1998, 2003; Lynn et al. 1999). Alternatively, it is possible that that ago1 mutants are specifically deficient for S-PTGS because AGO1 is part of the S-PTGS RISC but is absent or can be replaced by other family members in the IR-PTGS RISC.
Based on our knowledge of RISC in animals, one or multiple AGO proteins should be a component of the miRNA RISC that mediates cleavage of plant mRNAs. AGO1, ZIP/AGO7, and PNH/ZLL/AGO10 are good candidates because ago1, zip/ago7, and pnh/zll/ago10 mutations affect development. Indeed, it is expected that mutants defective in the miRNA RISC would exhibit developmental defects overlapping those of dcl1, hen1, or hyl1 mutants. AGO1 is a particularly attractive candidate to act in the miRNA RISC owing to the possible feedback regulation of AGO1 mRNA by miR168 (Rhoades et al. 2002). Such feedback regulation would be analogous to that proposed for DCL1 mRNA by miR162 (Xie et al. 2003). Among the 10 plant AGO mRNA homologs, only AGO1 has extensive complementary to miR168 or any of the other known miRNAs, suggesting that AGO1 might be the only member of the ARGONAUTE family that is regulated by an miRNA, just as DCL1, the Dicer family member known to be required for miRNA accumulation, is the only one of the four Dicer homologs known to be an miRNA target. 5′-RACE experiments revealed that AGO1 mRNA fragments that terminate precisely at the predicted site of miR168-directed cleavage accumulate in wild-type plants, showing that miR168 directs the cleavage of AGO1 mRNA (Vazquez et al. 2004). Furthermore, the steady-state level of uncleaved AGO1 mRNA increases in flowers of hen1 mutants that show reduced accumulation of miR168, indicating that miR168-directed cleavage of AGO1 mRNA is important for AGO1 regulation (Vazquez et al. 2004).
In this paper, we present evidence for a role of AGO1 in the miRNA pathway. We propose that AGO1 acts in RISC; however, we cannot rule out that other AGO proteins also participate in the miRNA RISC, together with AGO1 or in cells where AGO1 is not present. We also demonstrate that the feedback regulation of AGO1 mRNA by the miRNA pathway through the action of miR168 is crucial for proper plant development; decreasing the complementarity of AGO1 mRNA with miR168 results in increased accumulation of AGO1 mRNA and to developmental defects. Finally, we show that these defects can be rescued by expressing a compensatory miRNA that is complementary to the mutant AGO1 mRNA, proving the regulatory relationship between miR168 and its target and opening the way for the engineering of artificial miRNAs.
Results
ago1 mutants exhibit elevated steady-state levels of miRNA targets and developmental defects overlapping with those of dcl1, hen1_, and_ hyl1 mutants
We previously reported the identification of an allelic series of ago1 mutants through a screen for PTGS-deficient mutants (Fagard et al. 2000; Morel et al. 2002). Three of these alleles (ago1-22, ago1-23, ago1-24) exhibit a phenotype similar to that of the _ago1-3_-null allele previously identified through a phenotypic screen for developmental mutants (Fig. 1). Three other alleles (ago1-25, ago1-26, ago1-27) exhibit less dramatic developmental defects, although they are deficient for PTGS, suggesting that PTGS is more sensitive to perturbation in AGO1 than is development. Interestingly, ago1 hypomorphic mutants exhibit plant stature, leaf shape, and flower phenotypes partially overlapping those of dcl1, hen1, or hyl1 mutants, which are impaired in the accumulation of miRNAs (Fig. 1), suggesting that AGO1 could play a role in the miRNA pathway.
Figure 1.
ago1, hen1, and hyl1 mutants exhibit overlapping developmental defects. (A) Rosettes of plants grown under short-day conditions. (B) Flowers of plants grown under long-day conditions.
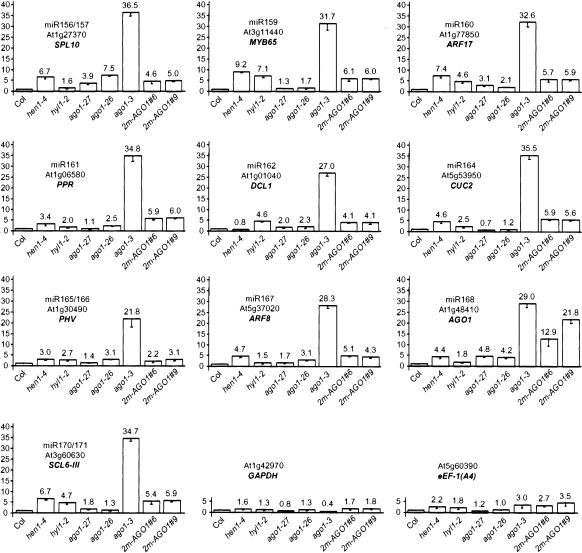
In dcl1, hen1, and hyl1 mutants, the impairment of the miRNA pathway results in increased steady-state levels of uncleaved target mRNAs or decreased steady-state levels of cleavage products (Kasschau et al. 2003; Vazquez et al. 2004), with the exception of AP2 mRNA, for which regulation by miR172 relies on translational repression (Aukerman and Sakai 2003; Chen 2004). To determine if AGO1 also participates in the regulation of endogenous mRNAs by miRNAs, we quantified steady-state levels of 10 mRNAs targeted for cleavage by miRNAs of 10 different families. As controls we quantified the level of GAPDH and eEF-1(A4) mRNA, which are not thought to be targeted by miRNAs. Because it is not possible to extract enough RNA from flowers of _ago1_-null alleles, we analyzed mRNA steady-state levels in rosettes of representative ago1 mutants: ago1-27 (hypomorphic), ago1-26 (intermediate), ago1-3 (null), as well as _hen1-4_- and _hyl1-2_-null alleles (all in the Col ecotype) grown in short days. A limited increase in the accumulation of miRNA targets was observed in hen1, hyl1, and ago1 hypomorphic mutants (Fig. 2), each target responding to a different extent in the different genetic backgrounds as previously reported in leaves or flowers of dcl1, hen1, and hyl1 mutants (Kasschau et al. 2003; Vazquez et al. 2004). A consistent and stronger increase in the accumulation of miRNA targets but not of GAPDH and eEF-1(A4) mRNAs was observed in the _ago1-3_-null allele, which was confirmed on a second set of independent plants (data not shown). The observe increase in ago1-3 should be interpreted with caution. Most of the miRNA targets analyzed are expressed at much higher levels in meristem than in leaves. Because _ago1_-null alleles develop smaller leaves than hen1, hyl1, and ago1 hypomorphic mutants (Fig. 1A), the ratio between meristematic cells and leaf cells in a rosette is probably higher in _ago1_-null alleles, thus introducing a possible bias in the analysis. Nevertheless, the similar increase observed in hen1, hyl1, and ago1 hypomorphic mutants, which all exhibit similar development, indicates that miRNA-directed mRNA cleavage involves AGO1.
Figure 2.
ago1, hen1, and hyl1 mutants have increased steady-state levels of miRNA targets. RNA extracted from rosettes of isogenic wild-type or mutant siblings deriving from heterozygote parents and of untransformed plants or 2m-AGO1 transformants was quantified for the indicated mRNA by real-time quantitative PCR using primers surrounding the cleavage site. GAPDH and eEF-1(A4) were used as nontarget controls. Quantifications were normalized to that of ACTIN2, then to the value of the wild-type plants or wild-type siblings, which was arbitrarily fixed to 1.
miRNA accumulation in ago1 mutants
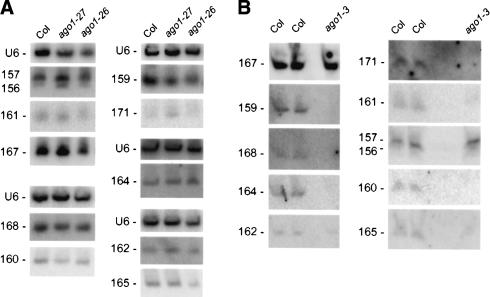
The increased accumulation of target mRNAs observed in dcl1, hen1, and hyl1 mutants is likely caused by reduced miRNA accumulation and subsequent reduced cleavage efficiency (Kasschau et al. 2003; Vazquez et al. 2004). To examine at which step ago1 mutants are impaired in the miRNA pathway, we analyzed miRNA accumulation in rosettes of wild-type plants and ago1 mutants using the same plant material used for the RT–qPCR experiments. We observed that miRNAs are present in ago1-27 and ago1-26 at levels more or less similar to those observed in wild-type plants (Fig. 3A), suggesting that AGO1 acts downstream from DCL1, HEN1, and HYL1 in the miRNA pathway to promote cleavage of target mRNAs. In the _ago1-3_-null allele, only miR156/157 and miR167 accumulated to a level similar to that of wild-type plants (Fig. 3B). For the eight other miRNAs examined, accumulation in ago1-3 was reduced, in some cases to below the level of detection. We suggest that AGO1 is important for the stabilization of miRNAs, although we cannot rule out a role in miRNA production. It seems likely that Arabidopsis AGO1 functions similarly to Drosophila AGO2 or human eIF2C (Hammond et al. 2001; Mourelatos et al. 2002), which associate with miRNAs in the RISC, which, in turn, mediates the posttranscriptional regulation of target messages. Other possibilities exist, including the idea that AGO1 might even act before miRNA processing, as suggested by the recent observation that the precursor of miR165 is mislocalized in ago1 mutants (Kidner and Martienssen 2004).
Figure 3.
miRNA accumulation in ago1 mutants. miRNA accumulation was determined by RNA gel blot analysis using 30 μg (A) or 10μg (B) of the same RNA used for RT–qPCR analyses. Blots were successively hybridized to different probes complementary to miRNAs. (A) miRNA accumulation in the ago1-26 and ago1-27 hypomorphic alleles. (B) miRNA accumulation in the _ago1-3_-null allele.
Plants impaired in miR168-directed cleavage of AGO1 exhibit developmental defects
The results presented in Figure 2 show that the steady-state level of uncleaved AGO1 mRNA is increased in hen1, hyl1, and ago1 mutants, strongly supporting the idea that AGO1 mRNA undergoes a negative feedback regulation by the miRNA pathway through the action of miR168. If the negative feedback regulation of AGO1 mRNA by the miRNA pathway is essential to maintain a proper regulation of plant development by miRNAs, plants impaired in this feedback regulation but expressing functional AGO1, DCL1, HEN1, and HYL1 proteins should exhibit developmental defects. To test this hypothesis, we introduced silent mutations in the AGO1 gene to decrease the complementarity between AGO1 mRNA and miR168 without changing the AGO1 protein sequence, a strategy that has already proven to be successful to specifically reduce miRNA-directed cleavage of MYB, TCP, and REV mRNAs (Emery et al. 2003; Palatnik et al. 2003). In a first step, the wild-type AGO1 gene was subcloned as an 8-kb fragment carrying the entire transcribed region plus 1.5 kb upstream of the transcription start and 0.5 kb downstream from the polyadenylation signal. Introduction of this construct (WT-AGO1) into the ago1-27 hypomorphic allele or the _ago1-1_-null allele restored a wild-type phenotype in ∼80% of the transformants, indicating that the WT-AGO1 construct contains all the upstream and downstream regulatory elements required for wild-type function. Introduction of WT-AGO1 into wild-type plants had no effect on plant development in 94 out of 131 transformants analyzed. The remaining 37 transformants looked normal at early stages of development but progressively exhibited the characteristics of ago1 hypomorphic alleles, including late flowering, serrated leaves, fused flowers, and limited fertility (data not shown), suggesting that the WT-AGO1 construct had triggered late cosuppression of the endogenous AGO1 gene. To test this hypothesis, we introduced the WT-AGO1 construct into the cosuppression-deficient sgs2 mutant (Mourrain et al. 2000). None of the 130 transformants analyzed showed an ago1 hypomorphic phenotype, thus confirming that the phenotype observed in transformed wild-type plants resulted from cosuppression of the endogenous AGO1 gene.
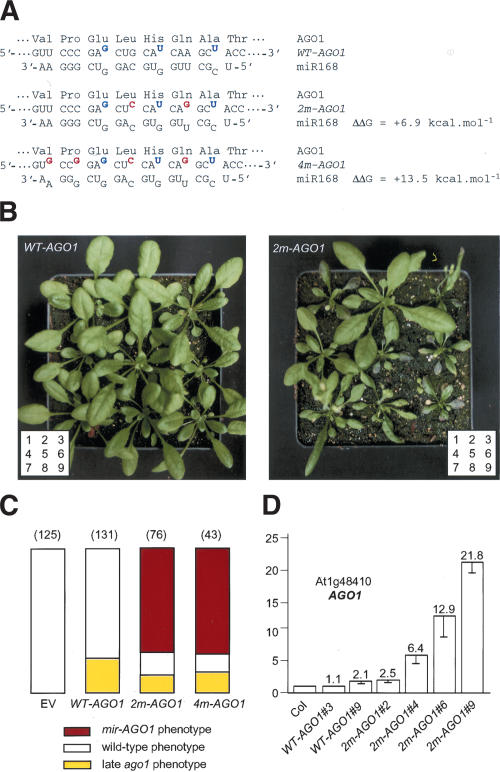
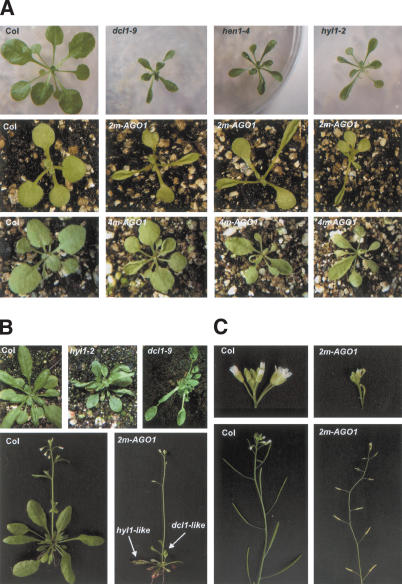
The wild-type AGO1 gene encodes an mRNA that naturally contains three mismatches with miR168 (Rhoades et al. 2002). Two mutant constructs (2m-AGO1 and 4m-AGO1) were made by introducing two or four silent mutations into the WT-AGO1 construct, thus adding two or four mismatches with miR168 that increase the predicted free energy of the miRNA/target duplex by 6.9 kcal/mole and 13.5 kcal/mole, respectively (Fig. 4A). When introduced into wild-type plants, the 2m-AGO1 and 4m-AGO1 constructs yielded 13%–18% of transformants with a cosuppressed phenotype similar to the 28% of cosuppressed transformants observed with the WT-AGO1 construct (Fig. 4C). In addition, the 2m-AGO1 and 4m-AGO1 constructs yielded 63%–68% of transformants exhibiting developmental defects (referred to as the miR-resistant AGO1 or mir-AGO1 phenotype) that were not observed in plants carrying the WT-AGO1 construct (Fig. 4B,C). When introduced into the cosuppression-deficient mutant sgs2, the 4m-AGO1 construct yielded 91% of transformants with the mir-AGO1 phenotype and no transformants with the cosupppression phenotype (data not shown). In young seedlings, the mir-AGO1 phenotype was mostly characterized by the emergence of curled leaves resembling those of hen1 and hyl1 mutants (Fig. 5A). Additional developmental defects were also observed, including abnormal cotyledons. As plants grew, developmental defects became more variable from plant to plant and from one leaf to another (Fig. 5A). Adult transformants exhibited a shorter stature, asymmetric rosette leaf formation, twisted or spoon-shaped leaves resembling those observed in hyl1 and dcl1 mutants, respectively, a disorganized phyllotaxy, and an accelerated senescence of leaves exemplified by photobleaching spots on a highly anthocyaned background (Figs. 4B, 5B). In the most affected transformants, the shoot apical meristem aborted and the plants died before flowering. In less severely affected transformants, a short stem developed bearing degenerate flowers that were mostly sterile (Fig. 5C). The rest of the transformants produced seeds, the amount of which inversely correlated with the severity of the developmental defects observed in the vegetative phase.
Figure 4.
Silent mutations in the miR168 complementary site of the AGO1 mRNA induce developmental defects. (A) The WT-AGO1 mRNA naturally contains three mismatches with miR168 (in blue), including a G:U wobble pair. Silent mutations in 2m-AGO1 and 4m-AGO1 constructs introduce two and four additional mismatches (in red), reducing complementarity with miR168. ΔΔ_G_ was calculated using mfold. (B) Representative sets of transformants carrying the WT-AGO1 or 2m-AGO1 construct. (C) Proportion of transformants showing a wild-type phenotype (open bar), an ago1 phenotype caused by late cosuppression (yellow bar), or an mir-AGO1 phenotype caused by AGO1 overexpression (red bar). Plants were transformed with either an empty vector (EV) or the WT-AGO1, 2m-AGO1, or 4m-AGO1 constructs. The number of transformants analyzed is indicated in parentheses. (D) AGO1 mRNA accumulation determined by real-time quantitative PCR in untransformed plants (Col) or plants transformed with the WT-AGO1 or 2m-AGO1 constructs. Quantifications were normalized to that of ACTIN2. The value in Col was arbitrarily fixed to 1. Numbers (#) correspond to the plants shown in B.
Figure 5.
Developmental defects in 2m-AGO1 transformants. (A, top row) Wild-type plant (Col) and dcl1, hen1, and hyl1 mutants. (Middle row) Wild-type plant (Col) and 2m-AGO1 transformants exhibiting curled leaves resembling those of hen1 and hyl1 mutants, at 10 d. Transformants with aberrant cotyledons were occasionally observed (right). (Bottom row) Wild-type plant (Col) and 4m-AGO1 transformants exhibiting a variety of developmental defects, including asymmetric rosette leaf formation and curled or twisted leaves, at 20 d. (B, top row) Wild-type plant (Col) and dcl1 and hyl1 mutants. (Bottom row) Adult wild-type plant (Col) and a representative 2m-AGO1 transformant exhibiting spoon-shaped or twisted anthocyaned leaves resembling those of dcl1 and hyl1 mutants. (C, top row) Inflorescence of a wild-type plant (Col) and of a representative 2m-AGO1 transformant. (Bottom row) Stems and siliques (seed pots) of the same plants. The wild-type Col plant is fertile, whereas the 2m-AGO1 transformant is sterile with aborted siliques.
In the progeny of fertile transformants exhibiting a weak phenotype, a 3:1 ratio of abnormal/wild-type plants was observed that is consistent with the expected dominant character of constructs triggering ectopic gene expression. This result also indicates that developmental changes induced by 2m-AGO1 or 4m-AGO1 constructs are reversible by segregation of the constructs at meiosis and do not induce inherited imprints, consistent with the posttranscriptional level of deregulation induced by the constructs. Transformants exhibiting more severe phenotypes produced small siliques containing either no seeds or very few seeds. In the progeny of such transformants, the ratio of abnormal/wild-type plants was less than 3:1 and in the case of the most affected fertile transformant declined to 1:5. This deficit in abnormal plants strongly suggests that expression of the 2m-AGO1 or 4m-AGO1 construct might compromise gamete or embryo viability. This hypothesis is consistent with the lower number of transformants generated with the 2m-AGO1 construct compared with the WT-AGO1 construct and the even lower number obtained with the 4m-AGO1 construct (Fig. 4C). Because the T-DNA stably integrates into mature female gametes during the floral dipping procedure used to transform Arabidopsis (Bechtold et al. 2000, 2003), it is likely that only the transformed embryos that do not express the 2m-AGO1 or 4m-AGO1 construct at high levels can survive and develop into seeds.
Plants expressing a miR168-resistant AGO1 mRNA exhibit elevated steady-state levels of miRNA targets and developmental defects similar to those of dcl1, hen1_, and_ hyl1
The accumulation of uncleaved AGO1 mRNA was determined by RT–qPCR in rosettes of representative transformants (Fig. 4D). No difference or a slight increase of 2.1-fold was observed between wild-type plants and transformants carrying the WT-AGO1 construct (WT-AGO1 #3 and #9). A limited increase (2.5-fold and 6.4-fold) was observed in 2m-AGO1 transformants that did not exhibit a strong phenotype (2m-AGO1 #2 and #4). A stronger increase (12.9-fold and 21.7-fold) was observed in 2m-AGO1 transformants exhibiting a strong phenotype (2m-AGO1 #6 and #9). These results are consistent with RT–qPCR analyses showing that AGO1 mRNA strongly overaccumulates in the ago1-3 mutant, in which miRNA-mediated cleavage is generally impaired (Fig. 2), and confirm that miR168 regulates the AGO1 mRNA level through cleavage. They also indicate that plants can tolerate a limited increase in the amount of AGO1 mRNA accumulation without dramatically affecting development, whereas a strong increase in the amount of AGO1 mRNA triggers dramatic developmental defects.
Because 2m-AGO1 and 4m-AGO1 transformants exhibited spoon-shaped, curled or twisted leaves resembling those of dcl1, hen1, and hyl1 mutants (Fig. 5A,B), we analyzed the steady-state level of mRNAs targeted for cleavage by miRNAs in rosettes of two 2m-AGO1 transformants exhibiting a strong phenotype (2m-AGO1 #6 and #9). For every miRNA target examined, the level in 2m-AGO1 transformants was slightly higher than that in wild-type plants (Fig. 2), suggesting that an increase in the amount of AGO1 perturbates the miRNA pathway. Different hypotheses can be proposed to explain this phenomenon. It is possible that the excess of AGO1 protein interferes with the formation or the functioning of RISC by displacing other AGO proteins. Alternatively, the excess of free AGO1 protein could independently titrate miRNAs and other RISC components into separate incomplete complexes. Such a sequestration of miRNAs and/or other RISC components by free AGO1 protein could therefore mimic the effect of dcl1, hen1, or hyl1 mutations that reduce the accumulation of miRNAs, thus leading to some of the same developmental defects.
Phenotypic rescue of 4m-AGO1 plants with a compensatory 4m-MIR168 transgene
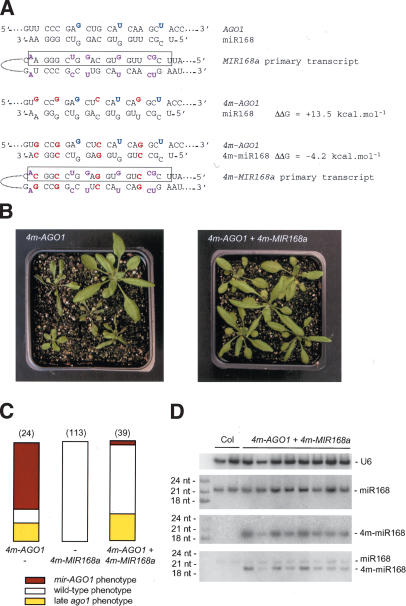
To prove that the developmental abnormalities observed in 4m-AGO1 transformants result from the absence of regulation of 4m-AGO1 mRNA by miR168, we mutagenized the MIR168a gene to introduce compensatory mutations that would allow the production of an miRNA that can pair with the mutant AGO1 mRNA transcribed from the 4m-AGO1 construct. miR168 is processed from an imperfect hairpin in which 15 of 21 miRNA residues are predicted to take part in Watson-Crick pairs involving residues on the other arm of the hairpin (Fig. 6A). When we altered the AGO1 gene to introduce mutations that decrease the complementarity with miR168, we chose nucleotides that kept the amino acid sequence unchanged and also corresponded to paired nucleotides in the miRNA precursor. Therefore, compensatory mutations could be introduced on both sides of the stem of the dsRNA precursor encoded by the MIR168a gene to generate a new miRNA precursor with the same predicted folding geometry as the original precursor (Fig. 6A). We started with MIR168a gene because of the existence of a corresponding EST starting 20 bp upstream of the putative fold-back stem–loop and ending 180 bp downstream, thus indicating that this gene is transcribed. The wild-type MIR168a gene was subcloned as a 2.4 kb-fragment with 1.4 kb upstream of and 1.0 kb downstream from the miR168 sequence so as to include regulatory sequences. The miR168 and miR168* sequences were successively mutagenized to generate the 4m-MIR168a construct (Fig. 6A). The predicted free energy of the 4m-miR168/4m-AGO1 duplex was slightly more favorable than that of the wild-type miR168/wt-AGO1 duplex (by 4.2 kcal/mole), suggesting that the newly generated miRNA would cleave the mutant 4m-AGO1 mRNA at least as well as the wild-type miRNA cleaves the wild-type AGO1 mRNA (Fig. 6A). Wild-type plants were transformed with either 4m-AGO1 or 4m-MIR168a or cotransformed with both constructs (each carried on a different vector with unique selectable markers). The number of transformants carrying the two constructs was low because the frequency of double transformation by independent bacteria is low. Nevertheless, it was higher than that observed with the 4m-AGO1 construct alone, suggesting that the deleterious effect of the 4m-AGO1 construct is abolished in the presence of the 4m-MIR168a construct. Indeed, we observed that only one out of 39 transformants carrying both 4m-AGO1 and 4m-MIR168a constructs displayed the mir-AGO1 phenotype, whereas 17 out of 24 transformants carrying the 4m-AGO1 construct alone displayed this phenotype (Fig. 6B,C). To confirm the efficiency of the rescue, we transformed a larger number of wild-type plants with the two constructs. Only two out of 77 additional double transformants displayed the mir-AGO1 phenotype (data not shown). No phenotype was observed in plants transformed with the 4m-MIR168 construct alone (Fig. 6C). Double transformants that carry both 4m-AGO1 and 4m-MIR168a constructs and exhibit a wild-type phenotype accumulated 4m-miR168, whereas no signal was visible in untransformed plants (Fig. 6D), indicating that the probe does not cross-hybridize with the endogenous miR168 and that the signal corresponds to bona fide 4m-miR168. Rehybridization of the blot with a probe complementary to miR168 revealed that the level of accumulation of miR168 was not affected by the expression of 4m-miR168. Interestingly, 4m-miR168 migrated faster than miR168, probably by 1 nt. These results show that the 4m-MIR168a construct is functional and expresses sufficient 4m-miR168 to rescue the phenotype conferred by the 4m-AGO1 construct. However, the observation that 4m-miR168 is shorter than miR168 by 1 nt indicates that subtle changes in the duplex induced by changes in the primary sequence of the miRNA transcript might affect the boundaries of the cleavage.
Figure 6.
Compensatory mutations in the MIR168a gene rescue developmental defects induced by silent mutations in the miR168 complementary site of the AGO1 mRNA. (A) The MIR168a gene encodes a primary transcript that is partially paired (unpaired nucleotides are purple). The miRNA is boxed. Compensatory mutations in the 4m-MIR168a transgene (red) conserved the structure of the primary transcript and restored pairing with the 4m-AGO1 mRNA. Original mismatches (blue) were kept. ΔΔ_G_ was calculated using mfold. (B) Representative sets of transformants carrying the 4m-AGO1 construct alone or the 4m-AGO1 and 4m-MIR168a constructs together. (C) Proportion of transformants showing a wild-type phenotype (open bar), an ago1 phenotype caused by late cosuppression (yellow bar), or a mir-AGO1 phenotype caused by AGO1 overexpression (red bar). Plants were transformed with the 4m-AGO1 or 4m-MIR168a constructs or both. The number of transformants analyzed is indicated in parentheses. (D) Accumulation of the compensatory miRNA (m4-miR168) in double transformants carrying the 4m-AGO1 and 4m-MIR168a constructs. RNA gel blot analysis was performed using 20 μg of total RNA extracted from two nontransformed plants (Col) and eight independent double transformants. The blot was hybridized with a probe complementary to 4m-miR168, stripped, rehybridized with a probe complementary to miR168, stripped, and finally rehybridized with the two probes simultaneously.
Discussion
In this paper, we show that Arabidopsis ago1 mutants that exhibit a range of developmental defects (Fig. 1) also exhibit increased accumulation of mRNAs targeted by miRNAs (Fig. 2). miRNA accumulation is not affected in ago1 hypomorpohic alleles that retain a PAZ domain but is reduced in _ago1_-null alleles (Fig. 3), supporting a role for AGO1 in the miRNA pathway. AGO1 could participate in the distribution of miRNAs or miRNA precursor transcripts as recently suggested by Kidner and Martienssen (2004) or could function in RISC, similar to eIF2C2 in human, AGO2 in Drosophila, or QDE-2 in Neurospora (Hammond et al. 2001; Catalanotto et al. 2002; Hutvagner and Zamore 2002; Martinez et al. 2002; Mourelatos et al. 2002). However, it is unlikely that AGO1 is the only AGO protein that associates with miRNAs in RISC. Indeed, because _dcl1_-null alleles lacking the enzyme that processes miRNA precursors are not viable, it is expected that plants lacking a functional RISC are not viable either. However, _ago1_-null alleles are viable, although they exhibit dramatic developmental defects (Fig. 1). This viability is likely caused by the function of other AGO protein(s), in particular PINHEAD/ZWILLE, which is 75% similar to AGO1 and has a pattern of expression overlapping with that of AGO1. Indeed, Lynn et al. (1999) reported that plants homozygous for a _pnh_-null mutation and heterozygous for an _ago1_-null mutation exhibit developmental defects stronger than those of single homozygous pnh mutants, whereas plants homozygous for an _ago1_-null mutation and heterozygous for a _pnh_-null mutation exhibit developmental defects stronger than those of single homozygous ago1 mutants, clearly indicating the redundant functions. In addition, plants homozygous for both _ago1_-null and _pnh_-null mutations are embryo-lethal, pointing out the crucial roles of these two proteins in plant development (Lynn et al. 1999). Interestingly, ago1 hypomorphic mutants strongly resemble transgenic plants expressing the viral suppressor of PTGS HC-Pro (Kasschau et al. 2003). The molecular phenotypes are also similar. Both ago1 mutants and HC-Pro-expressing plants have accumulation of miRNA targets and in both miRNAs are still present and accumulate to similar or higher levels compared with wild-type plants (Fig. 3A; Kasschau et al. 2003). The overlapping physical and molecular phenotypes of ago1 mutants and HC-Pro transgenic plants suggest that HC-Pro could alter the miRNA pathway by interfering with the action of AGO1.
We find that decreasing the complementarity of AGO1 mRNA with miR168 increases the level of AGO1 mRNA (Fig. 4) and has dramatic consequences on plant development and reproduction (Fig. 5). This observation, together with our finding that AGO1 is needed for proper miRNA function, supports the previous speculation that AGO1 mRNA is subjected to negative feedback regulation through the action of miR168. In this scenario, if the amount of AGO1 activity in a wild-type cell decreases below a critical level, the efficiency of miR168-mediated cleavage of AGO1 mRNA would also decrease, allowing more AGO1 mRNA to be translated into AGO1 protein and restoring activity to the initial level. Of course, the miR162 regulation of DCL1 mRNA (Xie et al. 2003) could also come into play, evoking more complex scenarios. For example, lowered AGO1 activity would decrease the amount of miR162-directed DCL1 mRNA cleavage, which could increase the amount of miRNAs produced, potentially increasing the amount of AGO1 mRNA cleavage and further lowering AGO1 activity. Thus, the outcome of a change in AGO1 activity could depend on many factors, including which of the components, DCL1, AGO1, miR162, and miR168, are limiting or in excess at the time of the perturbation. Another consideration is that a large increase in AGO1 mRNA, and presumably AGO1 protein, results in an apparent decrease in RISC activity (Fig. 2, 2m-AGO1 plants). Whether such a large increase is attainable in a wild-type plant, or whether marginal increases in AGO1 are instead held in check by miR168 feedback repression, is not known. Clearly, in transgenic plants expressing an miR168-resistant AGO1 mRNA, the system cannot return to the equilibrium because the mutant AGO1 mRNA is insensitive to feedback regulation by miR168.
The expression of a mutant miRNA that is able to pair with the mutant AGO1 mRNA can restore the feedback regulation and rescue the developmental defects induced by the mutant AGO1 mRNA (Fig. 6). This phenotypic rescue proves the regulatory relationship between miR168 and its target. It also demonstrates the feasibility of engineering artificial miRNAs in plants. These have potential as tools for targeted silencing of a single gene or a gene family, as well as utility for exploring facets of miRNA maturation and function.
Materials and methods
Plant material and growth conditions
The ago1-1, ago1-3, ago1-25, ago1-26, ago1-27, dcl1-9, hen1-4, and hyl1-2 mutants have been described previously (Bohmert et al. 1998; Morel et al. 2002; Boutet et al. 2003; Vazquez et al. 2004). Plants were grown under cool-white light in long days (16 h of light, 8 h of dark) at 23°C or short days (8 h of light, 16 h of dark) at 17°C.
RNA analysis
Total RNA was extracted as described previously (Vazquez et al. 2004), separated by denaturing 15% polyacrylamide gel electrophoresis, and blotted to a nylon membrane (Genescreen Plus; PerkinElmer Inc.) as described (Lau et al. 2001). MicroRNA probes were prepared by end-labeling antisense oligonucleotides using T4 polynucleotide kinase (New England Biolabs). Blots were rehybridized with a probe complementary to U6.
Real-time RT–PCR
RNA was extracted from mutant and wild-type siblings segregating from heterozygote parents grown in short days for 4 mo. Poly(dT) cDNAs were made by using the Invitrogen cDNA first-strand synthesis system. Quantifications were performed on a Bio-Rad IQcycler apparatus with the Quantitech SYBR green kit (QIAGEN) upon recommendations of the manufacturer. PCR was carried out in 96-well optical reaction plates heated for 10 min to 95°C to activate hot start Taq DNA polymerase, followed by 50 cycles of denaturation for 30 sec at 95°C and annealing-extension for 45 sec at 60°C. Target quantifications were performed with specific primer pairs designed for each side of the cleavage site by using Beacon Designer from Biosoft. Primers used for At1g27370/SPL10, At3g11440/MYB65, At1g77850/ARF17, At1g06580/PPR, At5g53950/CUC2, At5g37020/ARF8, At1g48410/AGO1, At3g60630/SCL6-III, and At3g18780/ACTIN2 have been described (Vazquez et al. 2004). The primers used to quantify additional mRNAs are At1g01040/DCL1, 5′-GATCCATTCCTAAGCGAAGTTTCAGAG-3′ and 5′-GCCC GAGCAACATAAAGATCCATAG-3′; At1g30490/PHV, 5′-AG ACCTTGGCGGAGTTCCTTTG-3′ and 5′-GTTGCGTGAAA CAGCTACGATACC-3′; At1g42970/GAPDH, 5′-TCTTTCCC TGCTCAATGCTCCTC-3′ and 5′-TTTCGCCACTGTCTCTC CTCTAAC-3′; At5g60390/eEF-1(A4), 5′-CTGGAGGTTTTGAG GCTGGTAT-3′ and 5′-CCAAGGGTGAAAGCAAGAAGA-3′.
For each cDNA synthesis, quantifications were made in triplicate. For each quantification, conditions were, as recommended, 1 ≥ E ≥ 0.85 and r2 ≥ 0.985, where E is the PCR efficiency and r2 corresponds to the correlation coefficient obtained with the standard curve. For each quantification, a melt curve was realized at the end of the amplification experiment by steps of 0.5°C from 55°C to 95°C, to ensure that quantification was not caused by primer self-amplification but by a pure and common PCR product. Results were normalized to that of ACTIN2, then to the value of the isogenic wild-type sibling. For each mutant analyzed, results were considered as acceptable if the variation between the wild-type sibling and a true wild-type plant was <15%.
Molecular cloning and plant transformation
A KpnI–SalI fragment carrying the 3′-half of the AGO1 gene was subcloned from BAC F11A17 (position 12600–18030) into the binary vector pBin+. The 5′-half of the AGO1 gene was amplified by PCR from BAC F11A17 using the following pair of primers: 5′-CTCGACTCTCGAGGTAGTATTAATTAACGAGTT CTAAGTTCTTCTTCCGTTATGAG-3′ and 5′-GGTTCTGGT ACCTGGGTAGGACTCACCTCAGACAGTGTAGGCTGAG AAGACACCGC-3′, cut with XhoI and KpnI (at positions 10,050 and 12,600 on BAC F11A17) and cloned into the Bluescript vector pKS+. The 5′-primer introduced a PacI site downstream from the XhoI site so that the 5′-half of the AGO1 gene can be mobilized as a PacI–KpnI fragment and cloned into the pBin+ vector containing the 3′-half of the gene to reconstitute a complete AGO1 gene (WT-AGO1). Silent mutations were introduced into the miR168 complementary site using the Quick Change Site-Directed Mutagenesis Kit (Stratagene) and the following pair of primers: 5′-CCACCGCAGAGACAATCAGTG CCGGAGCTCCATCAGGCTACCTCACCTACTTATCAAG CG-3′ and 5′-CGCTTGATAAGTAGGTGAGGTAGCCTGAT GGAGCTCCGGCACTGATTGTCTCTGCGGTGG-3′. The 4m-AGO1 construct resulted from the perfect replacement of the wild-type sequence by the primer sequence, whereas the 2m-AGO1 construct resulted from partial replacement of the wild-type sequence by the primer sequence. The wild-type and mutagenized PacI–KpnI fragments were entirely sequenced to ensure that no other mutations have been introduced and transferred from pKS+ into the pBin+ vector containing the 3′-half of the gene to reconstitute a complete AGO1 gene.
The MIR168a gene was subcloned from BAC T5K18 into the Bluescript vector pKS+ as a PstI–ClaI fragment (position 55287–57718 on T5K18). Compensatory mutations that restore complementarity to the 4m-AGO1 mRNA were introduced into the MIR168a gene using the Quick Change Site-Directed Mutagenesis Kit (Stratagene). The miR168 sequence was first mutageneized using the following pair of primers: 5′-CACCA TCGGGCTCGGATTCGCCTGGTGGAGGTCCGGCACCAA TTCGGCTGACACAGCC-3′ and 5′-GGCTGTGTCAGCCGA ATTGGTGCCGGACCTCCACCAGGCGAATCCGAGCCC GATGGTG-3′. The miR168* sequence was subsequently mutageneized using the following pair of primers: 5′-TTGGTTT GTGAGCAGGGATTGGAGCCGGCCTTCCATCAGCTGAA TCGGATCCTCGAGGTGTA-3′ and 5′-TACACCTCGAGGA TCCGATTCAGCTGATGGAAGGCCGGCTCCAATCCCTG CTCACAAACCAA-3′. The mutagenized PstI–ClaI fragment was sequenced to ensure that no other mutations have been introduced and then was transferred from pKS+ into the pCambia1200 binary vector.
The WT-AGO1, 2m-AGO1, and 4m-AGO1 constructs (in pBin+) and the 4m-MIR168a construct (in pCambia1200) were transferred from Escherichia coli to Agrobacterium tumefaciens by triparental mating. Arabidopsis plants were transformed by the flower-dipping method (Bechtold and Pelletier 1998; Clough and Bent 1998). Transformants were selected by sowing seeds onto a medium supplemented with kanamycin (pBin+) or hygromycin (pCambia1200).
Acknowledgments
We thank Allison Mallory for contributing in RNA gel blot analyses, Nelson Lau for computer assistance, Taline Elmayan for help and advice, and other members of the Bartel and Vaucheret laboratories for stimulating discussions. This work was partly supported by a grant from the French Minister of Research (ACI number 0220580) to P.C. and H.V., a PhD fellowship from the French Minister of Research to F.V., and an NIH grant to D.P.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1201404.
Corresponding authors.
References
- Ambros V. 2003. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell 113**:** 673–676. [DOI] [PubMed] [Google Scholar]
- Aukerman M.J. and Sakai, H. 2003. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15**:** 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116**:** 281–297. [DOI] [PubMed] [Google Scholar]
- Bechtold N. and Pelletier, G. 1998. In planta _Agrobacterium_-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82**:** 259–266. [DOI] [PubMed] [Google Scholar]
- Bechtold N., Jaudeau, B., Jolivet, S., Maba, B., Vezon, D., Voisin, R., and Pelletier, G. 2000. The maternal chromosome set is the target of the T-DNA in the in planta transformation of Arabidopsis thaliana. Genetics 155**:** 1875–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N., Jolivet, S., Voisin, R., and Pelletier, G. 2003. The endosperm and the embryo of Arabidopsis thaliana are independently transformed through infiltration by Agrobacterium tumefaciens. Transgenic Res. 12**:** 509–517. [DOI] [PubMed] [Google Scholar]
- Beclin C., Boutet, S., Waterhouse, P., and Vaucheret, H. 2002. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12**:** 684–688. [DOI] [PubMed] [Google Scholar]
- Bohmert K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., and Benning, C. 1998. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17**:** 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack M.T., Czaplinski, K., and Gorlich, D. 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10**:** 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollman K.M., Aukerman, M.J., Park, M.Y., Hunter, C., Berardini, T.Z., and Poethig, R.S. 2003. HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130**:** 1493–1504. [DOI] [PubMed] [Google Scholar]
- Boutet S., Vazquez, F., Liu, J., Beclin, C., Fagard, M., Gratias, A., Morel, J.B., Crete, P., Chen, X., and Vaucheret, H. 2003. Arabidopsis HEN1: A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13**:** 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C., Azzalin, G., Macino, G., and Cogoni, C. 2002. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes & Dev. 16**:** 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy A.A., Myers, M., Hannon, G.J., and Hammond, S.M. 2002. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes & Dev. 16**:** 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy A.A., Ketting, R.F., Hammond, S.M., Denli, A.M., Bathoorn, A.M., Tops, B.B., Silva, J.M., Myers, M.M., Hannon, G.J., and Plasterk, R.H. 2003. A micrococcal nuclease homologue in RNAi effector complexes. Nature 425**:** 411–414. [DOI] [PubMed] [Google Scholar]
- Chen X. 2004. A MicroRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303**:** 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J. and Bent, A.F. 1998. Floral dip: A simplified method for _Agrobacterium_-mediated transformation of Arabidopsis thaliana. Plant J. 16**:** 735–743. [DOI] [PubMed] [Google Scholar]
- Doench J.G., Petersen, C.P., and Sharp, P.A. 2003. siRNAs can function as miRNAs. Genes & Dev. 17**:** 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. 2003. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13**:** 1768–1774. [DOI] [PubMed] [Google Scholar]
- Fagard M., Boutet, S., Morel, J.B., Bellini, C., and Vaucheret, H. 2000. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. 97**:** 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan E.J. and Matzke, M.A. 2003. The small RNA world. J. Cell Sci. 116**:** 4689–4693. [DOI] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106**:** 23–34. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293**:** 1146–1150. [DOI] [PubMed] [Google Scholar]
- Han M.H., Goud, S., Song, L., and Fedoroff, N. 2004. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. 101**:** 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G.J. 2002. RNA interference. Nature 418**:** 244–251. [DOI] [PubMed] [Google Scholar]
- Hunter C., Sun, H., and Poethig, R.S. 2003. The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13**:** 1734–1739. [DOI] [PubMed] [Google Scholar]
- Hutvagner G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297**:** 2056–2060. [DOI] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293**:** 834–838. [DOI] [PubMed] [Google Scholar]
- Ishizuka A., Siomi, M.C., and Siomi, H. 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes & Dev. 16**:** 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev. Cell 4**:** 205–217. [DOI] [PubMed] [Google Scholar]
- Ketting R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15**:** 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner C.A. and Martienssen, R.A. 2004. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428**:** 81–84. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294**:** 853–858. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294**:** 858–862. [DOI] [PubMed] [Google Scholar]
- Lee R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294**:** 862–864. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75**:** 843–854. [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425**:** 415–419. [DOI] [PubMed] [Google Scholar]
- Lingel A., Simon, B., Izaurralde, E., and Sattler, M. 2003. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426**:** 465–469. [DOI] [PubMed] [Google Scholar]
- Lund E., Guttinger, S., Calado, A., Dahlberg, J.E., and Kutay, U. 2004. Nuclear export of microRNA precursors. Science 303**:** 95–98. [DOI] [PubMed] [Google Scholar]
- Lynn K., Fernandez, A., Aida, M., Sedbrook, J., Tasaka, M., Masson, P., and Barton, M.K. 1999. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126**:** 469–481. [DOI] [PubMed] [Google Scholar]
- Martinez J., Patkaniowska, A., Urlaub, H., Luhrmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110**:** 563. [DOI] [PubMed] [Google Scholar]
- Morel J.B., Godon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., Proux, F., and Vaucheret, H. 2002. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in posttranscriptional gene silencing and virus resistance. Plant Cell 14**:** 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. 2002. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 16**:** 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.B., Jouette, J., Lacombe, A.M., Nikic, S., Picault, N., et al. 2000. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101**:** 533–542. [DOI] [PubMed] [Google Scholar]
- Moussian B., Schoof, H., Haecker, A., Jurgens, G., and Laux, T. 1998. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 17**:** 1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B., Haecker, A., and Laux, T. 2003. ZWILLE buffers meristem stability in Arabidopsis thaliana. Dev. Genes Evol. 213**:** 534–540. [DOI] [PubMed] [Google Scholar]
- Olsen P.H. and Ambros, V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216**:** 671–680. [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425**:** 257–263. [DOI] [PubMed] [Google Scholar]
- Park W., Li, J., Song, R., Messing, J., and Chen, X. 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12**:** 1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. 2002. MicroRNAs in plants. Genes & Dev. 16**:** 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. 2002. Prediction of plant microRNA targets. Cell 110**:** 513–520. [DOI] [PubMed] [Google Scholar]
- Schauer S.E., Jacobsen, S.E., Meinke, D.W., and Ray, A. 2002. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 7**:** 487–491. [DOI] [PubMed] [Google Scholar]
- Song J.J., Liu, J., Tolia, N.H., Schneiderman, J., Smith, S.K., Martienssen, R.A., Hannon, G.J., and Joshua-Tor, L. 2003. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 10**:** 1026–1032. [DOI] [PubMed] [Google Scholar]
- Vazquez F., Gasciolli, V., Crete, P., and Vaucheret, H. 2004. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14**:** 346–351. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha, I., and Ruvkun, G. 1993. Posttranscriptional regulation of the heterochromic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75**:** 855–862. [DOI] [PubMed] [Google Scholar]
- Xie Z., Kasschau, K.D., and Carrington, J.C. 2003. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13**:** 784–789. [DOI] [PubMed] [Google Scholar]
- Yan K.S., Yan, S., Farooq, A., Han, A., Zeng, L., and Zhou, M.M. 2003. Structure and conserved RNA binding of the PAZ domain. Nature 426**:** 468–474. [DOI] [PubMed] [Google Scholar]
- Yekta S., Shih, I.-H., and Bartel, D.P. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304**:** 594–596. [DOI] [PubMed] [Google Scholar]
- Yi R., Qin, Y., Macara, I.G., and Cullen, B.R. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 17**:** 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D. 2002. Ancient pathways programmed by small RNAs. Science 296**:** 1265–1269. [DOI] [PubMed] [Google Scholar]
- Zeng Y., Yi, R., and Cullen, B.R. 2003. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. 100**:** 9779–9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Cao, X., and Jacobsen, S.E. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299**:** 716–719. [DOI] [PubMed] [Google Scholar]