Drugging MYCN through an allosteric transition in Aurora Kinase A (original) (raw)
. Author manuscript; available in PMC: 2015 Sep 8.
Published in final edited form as: Cancer Cell. 2014 Aug 28;26(3):414–427. doi: 10.1016/j.ccr.2014.07.015
Summary
MYC proteins are major drivers of cancer, yet are considered undruggable, as their DNA binding domains are composed of two extended alpha helices with no apparent surfaces for small molecule binding. Proteolytic degradation of MYCN protein is regulated in part by a kinase-independent function of Aurora A. We describe a class of inhibitors that disrupts the native conformation of Aurora A, and drives degradation of MYCN protein across MYCN-driven cancers. Comparison of co-crystal structures with structure-activity relationships across multiple inhibitors and chemotypes, coupled with mechanistic studies and biochemical assays, delineates an Aurora A conformation-specific effect on proteolytic degradation of MYCN, rather than simple nanomolar-level inhibition of Aurora A kinase activity.
Introduction
MYC proteins are considered undruggable, as their DNA binding domains are composed of two extended alpha helices with no apparent surfaces for small molecule binding. MYC also regulates as much as a third of the genome, with overexpression proposed to amplify cell-type specific gene expression rather than modulate a MYC-specific group of genes (Lin et al., 2012; Nie et al., 2012). The transcription of both MYC and MYCN targets may be blocked through bromodomain inhibitors (Delmore et al., 2011; Filippakopoulos et al., 2010; Mertz et al., 2011). Other methods, such as synthetic lethal screens for potential targets, have revealed druggable targets that may act downstream of MYC (Gustafson and Weiss, 2010; Toyoshima et al., 2012). Using an inducible dominant negative MYC protein, others have shown that systemic MYC inhibition is a viable cancer therapeutic strategy (Soucek et al., 2013). However, using current medicinal chemistry, direct and efficient pharmacologic targeting of MYC transcription factors has proven challenging if not impossible (Prochownik and Vogt, 2010).
MYC genes contribute to a wide range of human tumors through overexpression, amplification, translocation, or stabilizing point mutations. The normal concentration of MYC in cells is tightly regulated at the level of protein stability through canonical upstream kinase signaling pathways, including PI3K/mTOR, CDK2, and MAPK. These kinases direct sequential phosphorylation and dephosphorylation of conserved residues in MYC proteins, which target them for ubiquitination and degradation by the proteasome (reviewed in (Gustafson and Weiss, 2010)).
The MYC family member MYCN, named based on its association with MYCN amplification in the childhood tumor neuroblastoma, is stabilized by Aurora A in a kinase-independent fashion involving protein-protein interaction (Otto et al., 2009). Independent of its effects on MYCN, Aurora A is an attractive cancer target, as it regulates entry into mitosis, maturation of centrosomes, cytokinesis, and formation of the bipolar spindle, in part through phosphorylation of key regulators of proliferation and survival such as p53, BRCA1, and Histone H3 (Crosio et al., 2002; Liu et al., 2004; Ouchi, 2004; Scrittori et al., 2001; Zhao et al., 2008). Increased Aurora A expression is a negative prognostic factor in neuroblastoma (Shang et al., 2009), and pre-clinical testing with MLN8237, a specific Aurora A inhibitor, showed significant promise in cell line xenograft experiments (Maris et al., 2010). Furthermore, the co-crystal structure of MLN8054 (the predecessor of MLN8237) with Aurora A shows a partial shift away from the active state of the kinase and treatment of MYCN-expressing neuroblastoma with MLN8237 or MLN8054 modestly decreases MYCN (Brockmann et al., 2013; Dodson et al., 2010). This partial effect on MYCN of these compounds may therefore result from the prolonged inhibition of Aurora A kinase activity or a partial shift in the tertiary structure of Aurora A, which subtly weakens the Aurora A-MYCN complex. Consistent with this modest effect on MYCN, early phase clinical testing of MLN8237 in patients with _MYCN_-amplified neuroblastoma has shown little efficacy, underscoring the need for inhibitors of Aurora A that more potently block MYCN (Mosse et al., 2012).
Results
Initial screen for conformation-disrupting Aurora A inhibitors
We hypothesized that the kinase-independent stabilization of MYCN requires a distinct conformation of Aurora A, and that we could rationally design specific and potent conformation-disrupting (CD) inhibitors that perturb this protein-protein interaction, effecting degradation of MYCN. To identify such CD inhibitors we synthesized a set of compounds with either diaminopyrimidine (VX-680-like) or pyrazolopyrimidine (PP-1-like) scaffolds (Figure 1A) predicted to induce a large structural shift in Aurora A. Derivatives of each of these scaffolds were known to bind to Aurora A. Structural data were available on both scaffolds bound to related kinases, and routes to their synthesis were tractable. To these ATP-competitive cores, we fused biphenyl urea and amide moieties predicted to stabilize distinct conformations of Aurora A (Dietrich et al., 2010; Filomia et al., 2010).
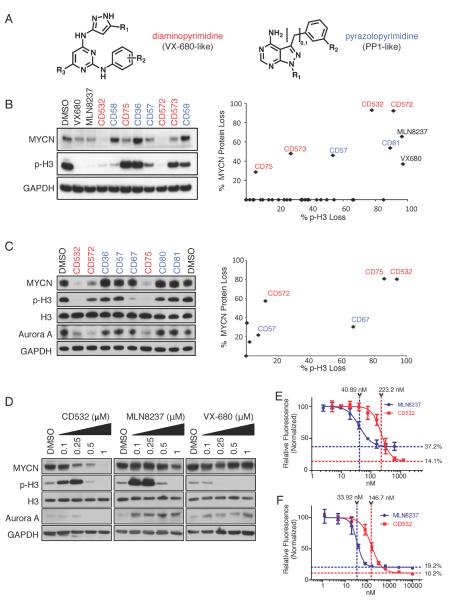
Figure 1.
Screening and characterization of conformation disrupting (CD) Aurora A inhibitor compounds. (A) The diaminopyrimidine (VX-680-like, red text) and the pyrazolopyrimidine (PP1-like, blue text) scaffolds used for generating the initial screening panel of compounds. Cell lines were treated for 24 hr with 1 μM of 32 different compounds. (B, C) Cell extracts of Kelly cells (B) or SK-N-BE(2) cells (C) treated with the indicated compounds at 1 ppfor 24 hr were examined by immunoblot as indicated (left) and the quantification results (right) are expressed as percent of untreated control. (D) Dose response of SK-N-BE(2) cells to increasing concentrations of CD532, MLN8237, and VX-680. (E, F). Dose responses of MLN8237 and CD532 at 72 hr using a cyquant assay in SK-N-BE(2) (E) and Kelly (F) MYCN-amplified neuroblastoma cells. Error bars represent mean +/− SD. See also Figure S1.
To test whether this panel of 32 putative CD inhibitors would destabilize MYCN, we initially treated Kelly _MYCN-_amplified neuroblastoma cells with these compounds and measured MYCN protein by western blot. We also assessed for phosphorylation of Histone H3 (p-H3), a known substrate for Aurora A and Aurora B and a marker for mitosis. Treatment with several members of the screening panel decreased levels of both MYCN and p-H3 (Figures 1B and S1A). In contrast, and as predicted, known inhibitors of Aurora A, VX-680 and MLN8237, blocked Histone H3 phosphorylation at 1 μM yet demonstrated very modest effects on the MYCN protein level. Candidate CD inhibitors were subsequently screened against a second _MYCN_-amplified neuroblastoma cell line, SKN-BE(2) (Figure 1C), substantiating CD532 as our most active lead compound.
CD532 potently inhibits Aurora A, causes loss of MYCN, and is cytotoxic in _MYCN_-amplified neuroblastoma cells
To determine the potency of CD532, we first measured its activity using purified Aurora A protein and revealed it as a potent Aurora A kinase inhibitor with an IC50 of 45 nM (Figure S1B–C). CD532 inhibited Aurora A kinase activity in cells as measure by both p-Aurora A (T288) and p-H3 at short time points to rule out off-target effects (Figure S1D). Treatment of multiple cell lines with CD532, MLN8237, and VX-680 showed dose-dependent loss of MYCN protein with CD532, and little or no response to high concentrations of MLN8237 (Figures 1D and S1E–F).
MLN8237 is a relatively selective inhibitor of Aurora A with of 1.2 nM and 396.5 nM for Aurora A and Aurora B respectively, while VX-680 is potent against both Aurora A and Aurora B, with IC50s of 0.6 nM and 18 nM respectively (Harrington et al., 2004; Lin et al., 2012; Manfredi et al., 2011; Nie et al., 2012; Otto et al., 2009). Notably, the in vitro (cell line) activity of CD532 against MYCN paralleled its cell-free in vitro IC50 for Aurora A by approximately 10 fold (Figures 1D and S1E–F). By contrast MLN8237 and VX-680 treatment effected little loss of MYCN protein even at doses 100 to 1000 times greater than their IC50s for Aurora A. MLN8237 and VX-680 upregulated or had little effect on Aurora A protein. CD532, in contrast, downregulated Aurora A protein across cell lines at higher concentrations consistent with distinct mechanisms of binding underlying these differential effects. At low concentrations of CD532 and short time points however, loss of MYCN was apparent while levels of Aurora A protein were unaffected. These observations are consistent with degradation of MYCN resulting from CD532 binding, rather than from loss of Aurora A protein.
Histone H3 is a known substrate for both Aurora A and Aurora B. Accordingly, dual inhibition of Aurora A and Aurora B with VX-680 abrogates phosphorylation of Histone H3 at S10. In contrast, MLN8237 caused an initial increase in S10 phosphorylation at lower concentrations, followed by a sharp drop at higher concentrations (Figures 1D and S1E–F). This increase in phosphorylation of Histone H3 in response to MLN8237 has been described previously, and results from Aurora A inhibition with feedback increase in Aurora B activity (Görgün et al., 2010; Wen et al., 2012). CD532 behaves similarly to MLN8237 with regard to Histone H3 phosphorylation, consistent with an Aurora A-selective effect.
We determined the cellular EC50 at 72 hr against two different _MYCN_-amplified neuroblastoma cell lines SK-N-BE(2) and Kelly as 223.2 nM and 146.7 nM, respectively, for CD532 and 40.89 nM and 33.92 nM, respectively, for MLN8237 (Figures 1E and F). These values are directly proportionate to the cell-free IC50 for Aurora A inhibition by CD532 (45 nM) and MLN8237 (4 nM) by ~10 fold. Additionally, the IC50 of CD532 for on-target MYCN knockdown in SK-N-BE(2) cells (~250 nM--Figure 1D) is consistent with the cellular EC50 (223.2 nM--Figure 1E). Notably the maximal cytotoxicity (Emax) for each compound is proportionate to the degree of MYCN knockdown rather than the degree of Aurora A inhibition in _MYCN_-amplified neuroblastoma lines. These data argue for an Aurora A-dependent effect on inhibition of cell growth and a MYCN-dependent effect on loss of viability.
Degradation of MYCN requires phosphorylation and proteasomal degradation of MYCN
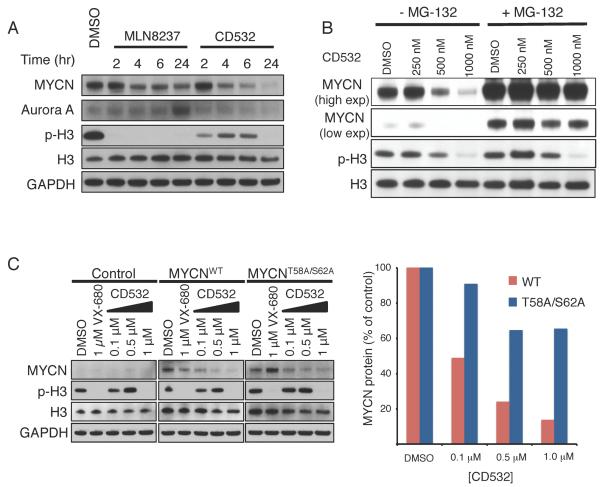
Upon loss of Aurora A scaffolding function by siRNA knockdown, MYCN is degraded through canonical ubiquitination and proteasomal degradation (Otto et al., 2009). As such, we would expect rapid degradation of MYCN protein to occur within hours of dissociation of the MYCN-Aurora A complex. We observed a clear and time-dependent loss of MYCN protein at time points as short as 4 hr of treatment with CD532. In contrast, treatment with MLN8237 although results in a similarly rapid decrease in the MYCN level, the decrease is more modest that does not change over time (Figure 2A). Treatment of MYCN-amplified IMR32 cells with increasing concentrations of CD532 in the presence of the proteasome inhibitor MG-132 shows that MG-132 protected MYCN from degradation but has no effect on inhibition of H3 phosphorylation (Figure 2B).
Figure 2.
Degradation of MYCN requires its phosphorylation and is proteasome-dependent. (A) Immunoblot analysis of the indicated proteins in SK-N-BE(2) cells treated with MLN8237 or CD532 at 1 μM for the indiated durations. (B) Immunoblot analysis of the indicated proteins in IMR-32 cells treated with the indicated concentrations of CD532 for 2 hr in the absence or presence of MG-132 (4 hr). (C) Immunoblot analysis of the indicated proteins in the control SHEP cells and SHEP cells ectopically expressing the indicated MYCN treated with the indicated compounds at indicated concentrations for 24 hr.
MYCN is sequentially phosphorylated at S62 and T58 before it is ubiquitinated and targeted for degradation. However, when bound in a complex with Aurora A, ubiquitinated MYCN is protected from degradation (Gustafson and Weiss, 2010; Otto et al., 2009). To test whether the activity of CD532 is dependent on these phospho-residues, we treated SHEP _MYCN_-non-amplified neuroblastoma cells engineered to express either MYCNWT or a non-phosphorylatable mutant of MYCN (MYCNT58A/S62A) with CD532. CD532 dose-dependently decreased the wild-type MYCN protein but was less effective in degrading MYCNT58A/S62A suggesting that CD532 potentiates loss of MYCN through the canonical phosphorylation and ubiquitination pathway. Notably, even high concentrations of VX-680, which stabilizes Aurora A in the active conformation (Zhao et al., 2008), had little effect on MYCN protein levels in this system (Figure 2C).
CD532 stabilizes a DFG-in, inactive conformation of Aurora A
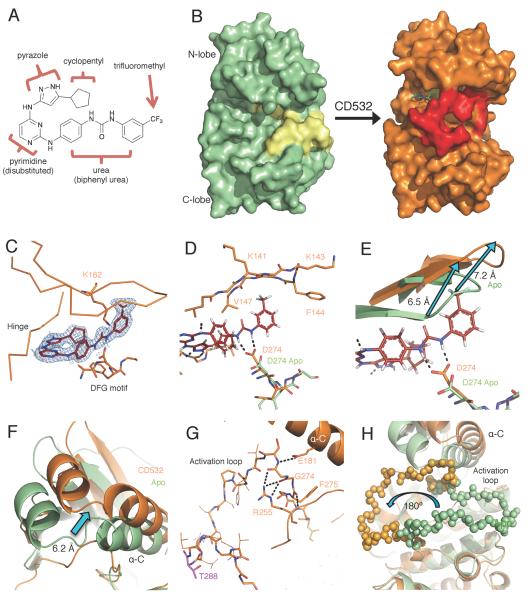
CD532 consists of an aminopyrazole-pyrimidine ATP-mimetic backbone, similar to VX-680, but includes a 3-trifluoromethyl-biphenyl urea as its conformation-disrupting pharmacophore (Figure 3A). To determine how CD532 binding affects the conformation of Aurora A, we determined the crystal structure of the catalytic domain of Aurora A (residues 123–390) both alone (Apo) and bound to CD532, to resolutions of 3.14 Å and 1.85 Å, respectively (Figure 3B and Table S1). While the B-factor of the relatively disordered activation loop in both structures is high, the tracing of the polypeptide backbone was unambiguous. Electron density for CD532 within the active site was well defined (Figure 3C).
Figure 3.
CD532 stabilizes an inactive, DFG-in conformation of Aurora A. (A) The structure of CD532 with key components highlighted. (B) Surface representations of Aurora A Apo (green, activation loop in yellow) and of Aurora A bound to CD532 (orange, activation loop in red). (C) CD532 (red sticks) in ATP binding pocket, overlaid with electron density before ligand fitting (blue mesh). (D) Interactions between CD532 (red), the DFG motif (D274) and β1/β2 (K141-V147) (E) Displacement of glycine rich loop in drug-bound structure (orange) as compared to Apo (green) due to drug binding. (F) Displacement of α-C helix of N-terminal domain allows for (G) a network of polar contacts between E181, R255, and DFG motif. (H) Stabilization of inactive orientation of the activation loop (activation loop in balls). Structural comparisons are all C-terminal alignments. See also Movie S1 and Table S1.
The ATP-binding hinge region of the Aurora A active site makes polar contacts with the aminopyrazole portion of CD532, consistent with our choice of ATP-mimetic scaffold. The catalytic D274 achieves polar contacts with the urea moiety of CD532 to stabilize the biphenyl urea in its orientation towards the N-terminal β1 and β2 strands forming part of the ATP binding pocket (Figures 3C–D). The polar contacts between the urea moiety and CD532 allow for a ~7 Å displacement of the β1 and β2 strands in the N-terminal domain, via steric clash with the trifluoromethylphenyl moiety of CD532 (Figure 3E). These β1 and β2 strands form part of a β-sheet that is the core of the relatively rigid N-terminal domain. Thus displacement of these strands by CD532 disrupts the conformation of Aurora A (Apo), rotating and shifting the N-terminal domain by 6.2 Angstroms, relative to the C-terminal domain (Figure 3F, Movie S1).
The highly conserved HRD kinase regulatory sequence is located at the lip of the active site. Coordination between this HRD arginine and a phospho-threonine in the activation loop (R255 and T288 respectively, in the case of Aurora A) orients the HRD catalytic aspartic acid to be primed for catalysis. By this mechanism, the catalytic activity of HRD-containing kinases can be regulated through phosphorylation of their activation loop. In the presence of CD532 R255 and T288 are displaced by a considerable distance (Figure 3G). In fact, CD532-bound Aurora sequesters R255 in a manner that displaces the catalytic HRD aspartic acid from its catalytically functional orientation, disengaging HRD regulation and stabilizing the kinase in a catalytically inactive conformation.
Indeed, the displaced α-C helix and R255 together trap the most N-terminal portion of the activation loop in a network of hydrogen bonds (Figure 3G). This interaction positions the activation loop backbone in a manner that stabilizes the entire activation loop in its inactive orientation, flipped 180° relative to its active state (Figure 3H). Thus, CD532 stabilizes Aurora A in a distinct conformation, associated with a 6.2 Å shift in the position of the N-terminal domain relative to the C-terminal domain, a disengaged state of the regulatory HRD motif, and a 180° flip in the activation loop.
Degradation of MYCN requires conformation-specific inhibition of Aurora A
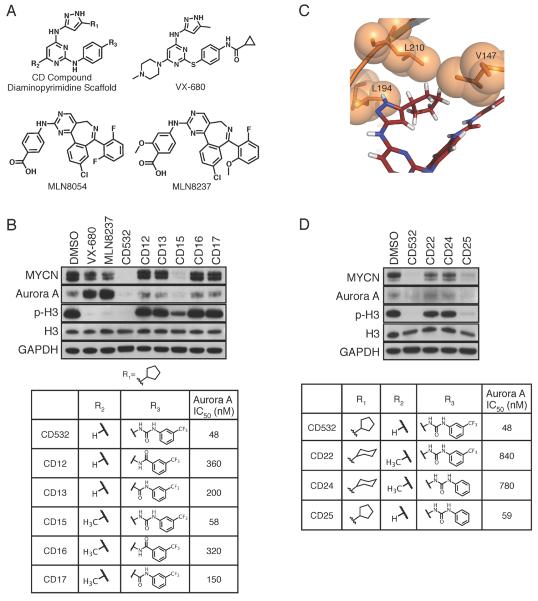
Although both VX-680 and CD532 bind to the ATP-binding `hinge' of Aurora A in an identical manner through their aminopyrazole-pyrimidine core, each contains distinct chemical components that produce highly divergent effects on MYCN in cells (Figure 3A and 4A). Our crystallographic data suggest that several chemical moieties of CD532 were critical for its ability to destabilize MYCN. As expected, altering the urea moiety of CD532 decreased biochemical potency against Aurora A, as well as efficacy against MYCN in neuroblastoma cell lines (Figure 4B). Our structural data also show that the 6-position of the pyrimidine backbone is oriented towards solvent, and addition of a methyl group to this position (CD15) maintained both cell-free potency and efficacy against MYCN (Figure 4B and S2). These data are consistent with degradation of MYCN occurring as a consequence of on-target Aurora A Kinase conformation-disrupting activity of CD532.
Figure 4.
Structure-activity relationships in inhibiting Aurora A and reducing MYCN. (A) Chemical structures of CD compound diaminopyrimidine scaffold, VX-680, MLN8054, and MLN8237. (B) Immunoblot analysis of the indicated proteins in SK-N-BE(2) cells treated with 1 μM of the indicated compound for 24 hr (Top) and the structures of CD compounds and their IC50s in inhibiting Aurora A kinase activity (bottom). (C) Hydrophobic packing of cyclopentyl of CD532 between V147, L194, and gatekeeper L210 of Aurora A (D) Immunoblot analysis of the indidated proteins in SK-N-BE(2) cells treated with 1 μM of the indicated compound for 24 hr (Top) and the structures of CD compounds and their IC50s in inhibiting Aurora A kinase activity (bottom). See also Figure S2.
The cyclopentyl moiety of CD532 packs neatly in a hydrophobic pocket made by V147, L194, and the leucine gatekeeper (L210) (Figure 4C). Thus our crystallographic data suggests that an additional methylene and adoption of the resulting six-membered ring into a chair conformation would preclude binding to Aurora A without abrogating binding to other kinases with a less bulky gatekeeper. Indeed, compounds CD22 and CD24 lost both potency against Aurora A and efficacy against MYCN (Figure 4D and S2).
The sterically bulky trifluoromethyl interacts with and displaces the β1 and β2 strands, which stabilizes a global conformational change in Aurora A that is unable to protect MYCN from degradation (Figure 3E). We hypothesized that replacement of this group with a hydrogen would decrease the magnitude of the N-terminal displacement of Aurora A without altering binding affinity. Indeed, CD25 retained potency against Aurora A activity, demonstrated both biochemically and by loss of Histone H3 phosphorylation, but was less effective than CD532 in driving MYCN loss, suggesting that the magnitude of the N-terminal shift of Aurora A contributes to MYCN destabilization (Figure 4D).
CD532 blocks S-phase entry
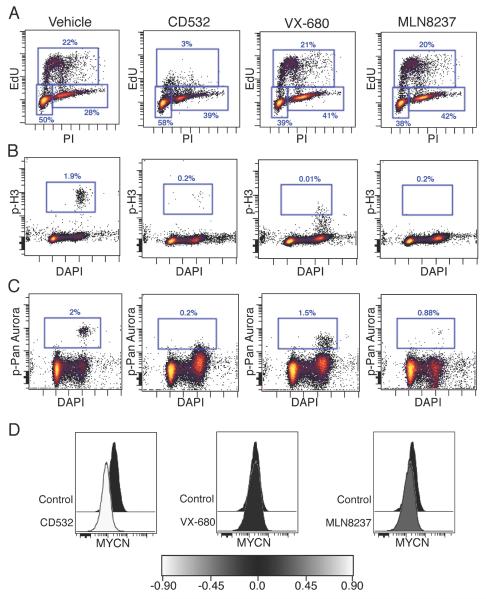
Both Aurora A and MYCN are critical to different phases of the cell cycle, and the functional consequences of Aurora A kinase inhibition and MYCN loss are distinct. Inhibition of Aurora A blocks mitosis, causing a G2/M arrest (Manfredi et al., 2011). In contrast, MYC family proteins drive S-phase entry. Knockdown of MYCN protein blocks entry into S-phase causing a subsequent G0/G1 arrest (Gogolin et al., 2013). To compare functional differences between conventional Aurora A kinase inhibition (MLN8237 or VX-680) with conformation disrupting Aurora A kinase inhibition, we treated MYCN amplified neuroblastoma cells and measured cell cycle by flow cytometry. As expected, treatment with MLN8237 or VX-680 resulted in G2/M arrest (Figures 5A and S3), consistent with inhibition of Aurora A kinase without a significant inhibition of MYCN. By contrast, CD532 resulted in potent loss of S-phase entry even after only 4 or 6 hr of treatment, a result expected in response to inhibition of MYCN. This loss of S-phase was concomitant with loss of p-Histone H3 (Figures 5A and B), loss of p-pan-Aurora (Figure 5C), and with loss of MYCN protein (Figure 5D). Aurora kinase inhibitors all caused loss of phospho-pan-Aurora, detectable in a small fraction of cells by flow cytometry (Figure 5D). All aurora kinase inhibitors caused loss of phospho-pan-Aurora but only CD532 also caused a loss of S-phase and MYCN (Figures 5).
Figure 5.
CD532 inhibits Aurora A kinase activity and downregulates MYCN. (A–D) SKN-BE(2) cells were treated for 6 hr with the indicated compounds at 1 μM and EdU was added 1 hr prior to harvest to measure cell cycle by EdU incorporation and propidium iodide (PI) staining (A), p-H3 (B), pan-Aurora (A, B, and C isoforms) phosphorylation (C), and MYCN protein (D) by flow cytometry. See also Figure S3.
CD532 is a MYC-directed therapy
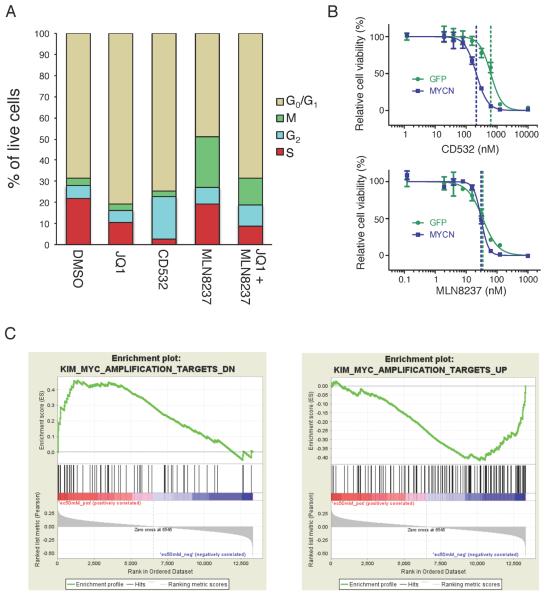
CD532 has the dual effect of blocking Aurora A kinase activity and driving degradation of MYCN. To further characterize the effects of CD532 on the cell cycle, we compared it with the bromodomain inhibitor JQ1, which has been shown to block the transcriptional activity of MYCN and the transcription of MYCN itself in neuroblastoma (Puissant et al., 2013). Treatment of MYCN amplified neuroblastoma cells with JQ1 for 24 hr resulted in downregulation of MYCN, blockade of S-phase entry, and accumulation of cells in G0/G1 (Figures 6A, S4A and S4B). Treatment with CD532 for 4 hr resulted in a rapid and potent loss of S-phase (consistent with the rapid and potent loss of MYCN protein) and accumulation in both G0/G1 and G2, consistent with a mixed Aurora A and MYCN effect. Treatment with MLN8237 for 4 hr resulted in a modest downregulation of MYCN and accumulation of cells in G2 and M phase, which has been described previously (Manfredi et al., 2011). When combining treatments with JQ1 for 24 hr and MLN8237 for 4 hr, an additive loss of S-phase and accumulation in G2/M was observed, similar to CD532.
Figure 6.
CD532 acts as a MYCN inhibitor in cell lines. (A) Quantification of cell cycle of SK-N-BE(2) cells treated with CD532 (1 μM, 4 hr), MLN8237 (0.1 μM, 4 hr), JQ1 (2 μM, 24 hr), or MLN8237 (0.1 μM, 4 hr) in combination with JQ1 (2 μM, 24 hr). (B) Viability of SHEP cells transduced with MYCN or GFP after 72 hr of treatment with CD532 (top) or MLN8237 (bottom). Error bars represent mean +/− SD. (C) Gene set enrichment analysis of 87 cancer cell lines against CD532 dose response showing positive correlation between MYC genes down and EC50 (left) and negative correlation between MYC genes up and EC50 (right). See also Figure S4 and Table S2.
That the cell cycle and viability activity of CD532 but not MLN8237 is related to degradation of MYCN suggests that expression of MYCN might confer sensitivity to CD532. We therefore determined the cellular EC50 for these compounds against both GFP-and MYCN-transduced SH-EP neuroblastoma cells, which express little to no MYCN. Transduction of MYCN conferred sensitivity to CD532 but not to MLN8237 (Figure 6B). In addition, CD532-driven loss of S-phase in these cells could be rescued by the stabilizing MYCNT58A/S26A mutant (Figures S4C and D). These data suggest that the efficacy of CD532 is due primarily to loss of MYCN, whereas that of MLN8237 is due primarily to inhibition of Aurora A.
To determine whether MYCN might serve as a biomarker of sensitivity to CD532, we screened a panel 169 distinct tumor-derived and genetically characterized cell lines, including 93 lines for which the information of MYCN copy number was available, and 87 lines for which mRNA expression data were available (Garnett et al., 2012). CD532 showed activity in most cell lines, with EC50s in the nanomolar range, consistent with our results in neuroblastoma (Table S2). Sensitivity to CD532 correlated with expression of MYCN/MYC mRNA in neuroblastoma cells (Fig S4E). _MYCN_-amplified cell lines were significantly more susceptible to CD532 than non-amplified lines (p=0.0010). In validation of this analysis, MYCN amplified lines were significantly more susceptible to JQ1 than non-amplified lines (p=0.0069), whereas MYCN amplified and non-amplified lines showed similar sensitivity to VX-680 (p=0.618; Figure S4F–H). Gene-set enrichment analysis revealed that susceptibility to CD532 correlated with a MYC signature, i.e. lowest EC50 in cells with highest expression of MYC targets and highest EC50 in cells with downregulated MYC targets (Figure 6C). These data support a broad potential for CD inhibitors of Aurora A against tumors in addition to neuroblastoma, and suggest a role for Aurora A CD inhibitors in both MYC- and MYCN-driven diseases.
CD532 reduces MYCN and is effective in vivo
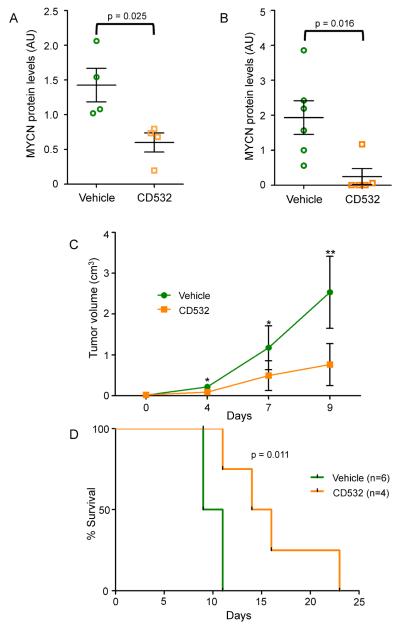
While CD532 is a compound in development not yet optimized for in-vivo pharmacokinetics, its efficacy in cell culture was substantial enough to warrant testing in vivo. Studies in mice revealed a serum half-life of ~1.5 hr, providing for an AUC0-24 of 27 μM*h when delivered at 20 mg/kg (Fig S5A). This is in contrast to the clinically developed MLN8237, which has an AUC0-24 of 78.4 μM*h when delivered at the same dose (Carol et al., 2011). Nonetheless, treatment of _MYCN_-amplified neuroblastoma xenografts with CD532 led to decreased levels of MYCN protein (Figures 7A and S5B), demonstrating that CD532 can block MYCN protein in vivo.
Figure 7.
In vivo activities of CD532. (A) MYCN protein levels in MYCN-amplified SMS-KCN xenografts tumors from mice treated for 2 days with 60 mg/kg CD532. (B–D) MYCN protein levels in tumors (B), tumor burden (C), and survival (D) of mice with subcutaneous SHH-subtype medulloblastoma treated with vehicle (n=6) or 25 mg/kg CD532 (n=4) twice weekly for up to three weeks *p<0.05, **p<0.005; two-tailed student's t-test for 7A–C, bars are mean +/−SEM; log rank test for 7D, AU = arbitrary units. See also Figure S5.
In addition to neuroblastoma, MYCN prominently drives other cancer types including medulloblastoma (Swartling et al., 2010). The sonic hedgehog (SHH) subtype of medulloblastoma shows high expression of MYCN, as SHH signaling promotes both expression and post-transcriptional stabilization of MYCN (Kenney et al., 2003; Thomas et al., 2009). In order to test activity in medulloblastoma in vivo, we treated a MYCN-expressing SHH-subtype medulloblastoma allograft derived from Ptch+/−;_p53_−/−mice (Kim et al., 2013; Romer et al., 2004). CD532 at 25 mg/kg twice per week substantially reduced MYCN levels, reduced tumor burden and extended survival in these mice (Figures 7B–D and S5C). Notably, mice tolerated this dosing regimen without obvious short or long-term toxicity or weight loss.
Disruption of the MYCN-Aurora A complex depends on the magnitude of conformational change in Aurora A
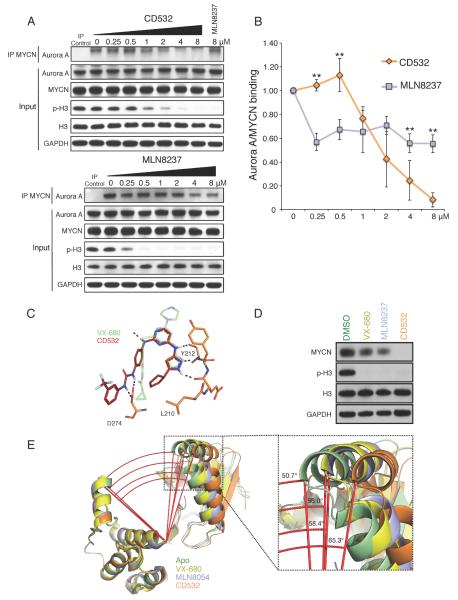
Despite its potency against Aurora A kinase activity and modest effect on the conformation of Aurora A (Dodson et al., 2010), MLN8237 subtly decreased MYCN protein levels compared to CD532 (Figures 1D, 2A, S1E–F). To test how the degree of conformational shift in Aurora A affects binding of MYCN and Aurora A, we measured the MYCN-Aurora A interaction in _MYCN_-amplified neuroblastoma cells treated with increasing concentrations of CD532 or MLN8237. CD532 inhibited histone H3 phosphorylation at concentrations 10-fold higher than MLN8237, consistent with their respective biochemical IC50s and cellular EC50 (Figure 8A). However, CD532 caused a dose-dependent and complete dissociation of the MYCN-Aurora A complex at 2 hr whereas MLN8237 only modestly disrupted this interaction (Figures 8A, B). This dissociation did not occur with VX-680 treatment (data not shown). The effect of CD532 on the MYCN-Aurora A interaction was specific in that it did not affect the MYCN-MAX binding (Figures S6A–B). Notably, disruption of the MYCN-Aurora A complex by CD532 occurred at doses comparable to those required to block p-H3, consistent with conformation change of MYCN as a consequence of CD532 binding. This is in contrast with MLN8237, which showed only partial disruption of the complex upon maximal Aurora A inhibition (Figure 8A). Thus MLN8237, a more potent Aurora A binder, only modestly decreased the interaction of Aurora A with MYCN. By comparison, CD532 binds Aurora A with lower affinity but has a dramatic effect on Aurora A binding to MYCN (Figure 8B).
Figure 8.
MYCN loss and dissociation of the MYCN-Aurora A complex track with the degree of conformational change in Aurora A. (A) Representative immunoblot analysis of immunoprecipitation (IP) of MYCN and total cell lysate (Input) from MYCN-amplified IMR32 cells treated for 4 hr with MG-132 and 2 hr with increasing concentrations of CD532 or MLN8237. (B) Quantification of Aurora A and MYCN binding from triplicate experiments (*p < 0.05, two-tailed student's t-test, bars are mean +/−SD). (C) Comparison of binding modes of VX-680 and CD532. (D) Immunoblot of MYCN protein after 24 hr treatment of SK-N-BE(2) cells with VX-680, MLN8237, and CD532. See also Figure S6. (E) Angle between α-carbons of T333, E308, and A172 of Aurora A Apo (PDB: 4J8N, green), Aurora A with VX-680 (PDB: 3E5A, yellow), Aurora A with MLN8054 (PDB: 2WTV, purple), and Aurora A with CD532 (PDB: 4J8M, orange).
As intended through use of the diaminopyrimidine scaffold for screening, CD532 binds to Aurora A at the hinge region via a pyrazole moiety in a manner similar to VX-680 (Figure 8C), yet interacts with other parts of the Aurora A binding pocket to confer distinct biological effects (loss of MYCN, decreased viability, and loss of S-phase), biophysical effects (shift in tertiary structure), and biochemical effects (disruption of the Aurora A/MYCN complex). Data in Figure 8D demonstrate that VX680, MLN8237 and CD532 show increasing activity in driving destabilization of MYCN protein in MYCN amplified cell lines. Comparing the published structures of Aurora A bound to VX-680 and to MLN8054 with our structure of Aurora A bound to CD532 demonstrates a progressive disruption of the conformation of Aurora A (Figure 8E). Thus the ability of VX680, MLN8237 and CD532 to progressively displace the α-C helix in Aurora (a structural measure which tracks directly with MYCN proteolysis) illustrates how a starting scaffold can be modified to effect divergent biochemical and biological activities.
Discussion
Earlier studies of Aurora kinases clarified a central role for Aurora A in mitosis and transformation. Inhibitors of Aurora A have therefore been developed as therapeutics, and are currently being tested across a range of cancers. Aurora A shares significant structural and sequence similarity with Aurora B, although these proteins have distinct mitotic functions and distinct subcellular localizations. These differences in both function and localization are attributed in part to the association of each kinase with a unique group of cofactor proteins (reviewed in (Carmena et al., 2009)).
Here we describe a class of compounds that were initially designed to bind Aurora A in a type II fashion, defined by the DFG-out orientation of D274, as a strategy for disrupting the conformation of this kinase. Thus it was surprising to observe that CD532 binds Aurora A as DFG-in, yet still induces a conformational disruption not achieved by nonselective tool inhibitors which induce a DFG-out conformation in Aurora A in vitro (Martin et al., 2012). Comparing CD532-bound Aurora A to the Apo structure shows the activation loop in the inactive orientation, accompanied by a shift in the entire N-terminal domain. Although the activation loop flip is consistent with an inactive conformation of Aurora A, the urea moiety of CD532 locks the DFG motif in the active “DFG-in” orientation. This concurrence of features from otherwise distinct states of the kinase is achieved through a steric clash of the trifluoromethylphenyl moiety of CD532 with Aurora's N-terminal β1 and β2 strands, displacing the N-terminal lobe of Aurora A and allowing a unique network of hydrogen bonds to stabilize the activation loop in an inactive orientation.
Our structural data also suggest a mechanism through which an inhibitor can stabilize the inactive conformation of a kinase. Previously described inhibitors that stabilize kinases in their inactive conformation displaced the aspartic acid of the catalytic DFG motif, with a concomitant crankshaft-like 180° rotation of the DFG backbone. In contrast, CD532 induces this inactive conformation through interaction with the β1 and β2 strands of the N-terminal domain, without reorienting the DFG motif. Our structure thus reveals an “uncoupling” of the DFG-flip from the inactive state of a kinase. Whether such uncoupling plays a role in the physiological state of the kinase, perhaps as part of its regulation, or only occurs in the presence of specific pharmacological entities remains to be determined.
Can these specific associations be exploited to identify inhibitors of Aurora kinases that also disrupt interactions with cofactor proteins? The resulting conformation of CD532--bound Aurora A blocks both kinase-dependent and kinase-independent functions of Aurora A. CD532 inhibits Aurora A at low nanomolar concentrations and, in parallel, effects proteolytic degradation of MYCN. Importantly, we were unable to uncouple kinase inhibition and MYCN proteolysis through structural modification of CD532, consistent with disruption of Aurora A's scaffold as a result of bulky pharmacophores that extend from an ATP-competitive core.
The difference in the kinetics of complex dissociation between CD532 and MLN8237 coupled with their respective IC50s and crystallographic information provide insight into the biophysical basis for disruption of the Aurora A-MYCN interaction. While MLN8237 is a potent inhibitor of Aurora A, it only modestly disrupts the conformation of Aurora A. In contrast, CD532 is a weaker inhibitor of Aurora A, however saturating doses lead to complete dissociation of the complex. Taken together with structural data, these observations suggest that the equilibrium of dissociation of the MYCN-Aurora A complex is dependent upon the degree of conformational disruption of Aurora A.
Several other inhibitors of Aurora kinase are in clinical development, all of which act as mitotic poisons much like current cytotoxic chemotherapy agents. Our functional data show that CD532 acts more as a potent MYCN inhibitor rather than a conventional Aurora A inhibitor in neuroblastoma, and has potential to act as a MYC inhibitor in other cell types. While the pharmacokinetic properties of CD532 have not been optimized, CD532 could effect loss of MYCN protein in neuroblastoma xenografts as well as reduce tumor burden and improve survival in a mouse model of medulloblastoma, providing motivation for additional medicinal chemistry and optimization of this family of compounds for clinical use.
Neuroblastoma is the most common extracranial solid tumor of childhood and MYCN amplification is the best-described genetic lesion marking high-risk, chemotherapy resistant disease. Targeted expression of MYCN drives neuroblastoma in mice and zebrafish (Weiss et al., 1997; Zhu et al., 2012). We have previously finessed destabilization of MYCN through blockade of PI3K/mTOR (Chanthery et al., 2012; Chesler et al., 2006) and have shown in vivo efficacy through an alternative approach to block MYCN and its transcriptional targets using BRD4-based bromodomain inhibitors (Puissant et al., 2013). Here we propose a third strategy to block MYCN in cancer. These three interventions, at distinct nodes in the same oncogenic pathway, present a unique opportunity for combinatorial, targeted therapeutics to block emergent resistance, while maximizing the blockade of MYCN in neuroblastoma and potentially in other _MYCN_- and _MYC_-driven cancers.
Allostery is most generally defined as a phenomenon whereby a perturbation by an effector at one site of the molecule leads to a functional change at another through alteration of shape and/or dynamics (Nussinov and Tsai, 2013). There are several recent examples of allosteric inhibitors for the treatment of cancer including arsenic trioxide, an anti-leukemic, that binds to zinc fingers within the PML-RARAα fusion protein of acute promyelocytic leukemia to induce a conformational change favoring oligomerization and eventual degradation (Zhang et al., 2010) and biculutamide that binds to the androgen receptor to block androgen receptor mediated transcription in prostate cancer (Osguthorpe and Hagler, 2011). Enzymes, including but not exclusive to kinases like Aurora A, may have important non-enzymatic activities including scaffolding, regulation, and localization of other proteins. As such, many molecular interactions necessary for cellular function and carcinogenesis are not targetable directly with small molecules either because they have no amenable binding pocket (as with MYC proteins) or because their affinity for natural substrate is too high (as with many GTPases such as RAS). By contrast, orthosteric targeting of small molecules to enzymes like kinases has become relatively trivial. Here we refer to an ATP-mimetic ligand that binds the active site of Aurora A to alter its kinase-independent stabilization of MYCN, but also, obligately, its kinase activity. We propose that such an inhibitor be referred to as “amphosteric”, denoting an inhibitor that is simultaneously both orthosteric (inhibiting kinase activity) and allosteric (disrupting protein-protein interactions). Thus, CD532 represents the prototype of a class of amphosteric inhibitors that induce an allosteric change to disrupt non-enzymatic functions of enzymes. As these amphosteric effects are neglected in most current inhibitor screening, development of small molecule screens for other amphosteric inhibitors has the potential to target other undruggable oncoprotein targets.
Experimental Procedures
Cell culture, inhibitors, and western blotting
Neuroblastoma tumor cell lines were obtained from the University of California San Francisco Cell Culture Facility (Kelly, SK-N-BE2, and SH-EP). SMS-KCN, SHEP MYCNwt and MYCNT58A/S62A cells were obtained from Martin Eilers lab. All cells were grown in RPMI with 10% FBS. Neuroblastoma cells were harvested and lysed with Cell Signaling Lysis buffer + 1% SDS, sonicated and supernatants boiled in LDS sample buffer (Invitrogen). Western blots were performed as described previously (Chanthery et al., 2012), with primary antibodies to MYCN (ab24193, Abcam), Histone H3, P-Histone H3 (S10), Aurora A (Cell signaling), and GAPDH (Millipore). Western blot quantitation performed with ImageJ software. VX-680 (S1048) and MLN8237 (S1133) were obtained from Selleck chemicals.
Flow cytometry and viability
Neuroblastoma cells were treated for the indicated time, trypsinized, washed, stained with Dylight 800 at 0.3 μg/mL (Pierce, 46421), fixed with 1.5% PFA, and permeabilized with 100% methanol. Cells were then stained with antibodies against p-MPM2 (Millipore, 16–155), p-pan-Aurora (Cell Signaling, 2914), MYCN (Thermo, PA5-17403), rabbit IgG (Invitrogen, A10542), or mouse IgG (BioLegend, 405307). Cells were stained with DAPI at 0.3 μg/mL (Invitrogen, D21490) and analyzed on the BD LSR II flow cytometer. For cell cycle analysis, cells were stimulated with EdU for 2 hours prior to harvest, then probed using the Click-iT EdU Flow Cytometry Assay Kit (Invitrogen, C10424). Cells were stained with propidium iodide (BD, 556547) and analyzed on the BD FACSCalibur flow cytometer. Data was gated using Cytobank. For viability studies, neuroblastoma cells were plated in 96-well plates at a density of 1,000 cells/well for SHEP or 4,000 cells/well for Kelly or SK-N-BE2 cells, then incubated with indicated concentrations of drug for 72 hours at 37°C. Plates were frozen at −80°C to induce cell lysis. CyQUANT reagent mixture (Invitrogen, C7026) was added to thawed plates, then fluorescence was measured. Alternatively, resazurin (Sigma-Aldrich, R7017) was added directly to wells following drug treatment then incubated for 4 hours at 37°C prior to measuring fluorescence. Data was analyzed using GraphPad Prism software.
Pulldowns
Cells were pretreated with MG-with drug (CD532, MLN8237, or VX-680) for 2 hours before lysis with TNN lysis buffer in the presence of protease inhibitor (Sigma-Aldrich, P8849). Pulldowns were performed with anti-MYCN antibody (Santa Cruz, SC-53993) and Protein G sepharose beads (Sigma-Aldrich, P3296). Immunoblots were performed as described above.
Chemical synthesis
Starting materials were purchased from Sigma-Aldrich or Alfa Aesar. Unless otherwise noted, reactions were performed in dry, argon-charged, glass roundbottom flasks and monitored by thin layer chromatography (TLC) or liquid chromatography-mass spectrometry (LCMS). Compounds were characterized by LCMS and nuclear magnetic resonance (NMR) spectroscopy. LCMS retention times (RT) are reported in minutes based on a gradient of 5–95% ACN/H2O from t=0.1–1.9 min. NMR shifts (δ) are reported in ppm as singlets (s), doublets (d), quartets (q), quintets (quin), or multiplets (m). High-performance liquid chromatography (HPLC) was conducted using a Waters 2545 binary gradient module, Waters 2767 sample manager, and Waters 2998 photodiode array detector running MassLynx v4.1. Flash/silica gel chromatography was performed on an AnaLogix Intelliflash using SuperFlash Si50 columns (Agilent). Synthetic procedures can be found in supplemental information.
Expression and purification of Aurora A Kinase
Purification and expression of Aurora A was performed as described previously (Martin et al., 2012), with the following modifications. Aurora A (residues 123–390, T287D) was cloned into a pET28a plasmid providing fusion with a PreScission Protease-cleavable hexahistidine tag. The protein was overexpressed in BL-21(DE3) cells at 18° C. Digestion with PreScission protease was performed overnight at 4° C in a 10 kDa molecular weight cut off (MWCO) dialysis cartridge (Thermo Scientific, Inc) with dialysis buffer containing 50 mM MES (pH 6.5), 300 mM NaCl, and 1 mM DTT, followed by 4 hours of dialysis with buffer containing 50 mM MES (pH 6.5) and 1 mM DTT before loading onto ion exchange column. Pooled fractions were concentrated to 5 mg/mL (Amicon Ultra 10 kD MWCO, Millipore) and loaded onto a HiLoad Prep Grade Superdex 200 column (GE Healthcare) equilibrated with 50 mM HEPES (pH 7.4) and 1 mM DTT to yield monomeric enzyme for use in both kinase assays and crystallization.
In vivo studies
For pharmacokinetic studies, CD532 was formulated at 20 mg/ml in 7.5% DMSO and 92.5% PEG300. LC-MS/MS detection of CD532 was performed using a Waters 2545 binary gradient module, Waters 2767 sample manager, and Waters 2998 photodiode array detector running MassLynx v4.1. For neuroblastoma studies, NOD scid gamma mice (Jackson Laboratory) were implanted with 106 SMS-KCN cells in growth media with 50% Geltrex (Invitrogen) into renal capsule. At 21 days post implantation, tumors were palpable and treated for two days with 60 mg/kg CD532 before harvesting and flash freezing for analysis. For medulloblastoma studies, homozygous nu/nu mice (Simonsen) with flank subcutaneous allografts (106 cells implanted per mouse) of SHH-subtype MYCN-expressing medulloblastoma were started on treatment once tumors reached 25 mm3 in volume (~14 days). Mice were treated with vehicle (5% DMSO in PEG300) or CD532 (25 mg/kg, formulated at 7.5 mg/ml) twice per week, delivered by intraperitoneal injection. Mice were euthanized once maximum tumor length reached 2.0 cm. Difference in tumor burden was evaluated by two-tailed student's T-test, and difference in survival by log-rank test. All experiments on live vertebrates or higher invertebrates were performed in accordance with relevant institutional and national guidelines and approved by the UCSF Animal Care and Use Committee (IACUC).
Crystallization and data collection
After gel separation, purified fractions of Aurora A were pooled and concentrated in the presence of drug to a final concentration of 20 mg/ml Aurora A and 1 mM drug. All crystallization reagents were obtained from Hampton Research (Aliso Viejo, CA). Crystals were generated by hanging drop vapor diffusion at room temperature using a 1:1 mixture of protein solution and well solution. For Aurora A apo, well solution consisted of 10% Tacsimate (pH 7.0) and 20% PEG 3350. For Aurora A with CD532, well solution consisted of 0.2 M magnesium acetate tetrahydrate, 0.1 M sodium cacodylate trihydrate and 20% w/v PEG8,000 at pH 6.0. Crystals did not grow in the Apo conditions in the presence of drug, or in the drug conditions in the absence of compound. CD532-bound and apo crystals were cryoprotected with well solution supplemented with 10% and 25% ethylene glycol, respectively, and stored in liquid nitrogen. Diffraction data were recorded on Beamline 8.2.2 at the Lawrence-Berkeley Advanced Light Source at a temperature of 100 K and wavelength of 1.0088 nm. Data were indexed using HKL2000 (HKL Research, Inc). The drug-bound crystals belong to the C2221 space group with one monomer in the asymmetric unit, and apo crystals belong to the P31 space group with four monomers in the asymmetric unit. Molecular replacement and refinement were performed using Phaser-MR and phenix.refine in PHENIX (Adams et al., 2010), model building was performed using Coot (Emsley et al., 2010), and figures were drawn using MacPYMOL 1.5.0 (Schrodinger, LLC). RCSB validation reports are shown in Supplemental Information.
Supplementary Material
01
02
03
Highlights.
- A class of inhibitors disrupts the conformation of Aurora A and destabilizes MYCN.
- CD532 potently inhibits the activities of Aurora A and MYCN.
- CD532 blocks MYCN in vitro and in vivo, across tumor types.
Significance.
MYC genes contribute to a range of cancers including neuroblastoma and medulloblastoma, where amplification confers a poor prognosis. MYC also regulates as much as a third of the genome, with overexpression proposed to amplify cell-type specific gene expression. Despite their prominence in cancer, MYC proteins are considered undruggable. We synthesize and characterize a class of conformation-disrupting inhibitors of Aurora A that destabilize interactions between Aurora A and MYCN. These small molecules represent candidate agents to target MYC and MYCN-driven cancers, as well as prototypes for inhibitors that induce an allosteric change to block non-enzymatic functions of kinases.
Acknowledgments
We thank Ulf Peters, Arvin Dar, and Chris Waddling for technical assistance, Yoon-Jae Cho for medulloblastoma cells; Alex Warkentin, Mike Lopez, Greg Hamilton, Qi Wen Fan, Miller Huang, and other members of the Shokat and Weiss labs for helpful discussion and critical review. This research was supported by: NIH K08NS079485 (WCG); Alex's Lemonade Stand (WCG, KKM, and WAW); Frank A. Campini Foundation (WCG, KKM); P01CA081403 (WAW, KS, KKM, RS); F30CA174154 (JGM); CureSearch Grand Challenge Award (WAW); NIH LINCS grant 1U54HG006097-01 (CB); R01CA102321 (WAW); R01CA159859 (WAW); R01CA148699 (WAW); P30CA82103 (WAW); Katie Dougherty Foundation (WAW, KKM); Howard Hughes Medical Institute (KMS); The Samuel Waxman Cancer Research Foundation (KMS, WAW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession number Atomic coordinates and structure factors for CD532-bound and Apo Aurora A have been deposited in the PDB as 4J8M and 4J8N, respectively.
Procedures for chemical synthesis, gene set enrichment analysis, and in vitro kinase assays are described in supplemental information.
Author Contributions W.C.G., J.G.M., and E.A.N. designed, performed and analyzed experiments. E.C. performed and analyzed experiments. J.G.M. and N.T.H. performed chemical synthesis. J.G.M. carried out crystallographic and biochemical studies. J.C. performed bioinformatics analysis. E.F.S. designed and performed flow cytometry experiments. K.K.M., K.M.S. and W.A.W. designed and analyzed experiments. C.B. designed and performed cell line screens. M.E. and R.S. provided reagents. W.C.G., J.G.M., and W.A.W. wrote the manuscript. All authors edited the manuscript. W.C.G. and J.G.M. share equal authorship.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann M, Poon E, Berry T, Carstensen A, Deubzer HE, Rycak L, Jamin Y, Thway K, Robinson SP, Roels F, et al. Small Molecule Inhibitors of Aurora-A Induce Proteasomal Degradation of N-Myc in Childhood Neuroblastoma. Cancer Cell. 2013 doi: 10.1016/j.ccr.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Ruchaud S, Earnshaw WC. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 2009;21:796–805. doi: 10.1016/j.ceb.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol H, Boehm I, Reynolds CP, Kang MH, Maris JM, Morton CL, Gorlick R, Kolb EA, Keir ST, Wu J, et al. Efficacy and pharmacokinetic/pharmacodynamic evaluation of the Aurora kinase A inhibitor MLN8237 against preclinical models of pediatric cancer. Cancer Chemotherapy and Pharmacology. 2011;68:1291–1304. doi: 10.1007/s00280-011-1618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanthery YH, Gustafson WC, Itsara M, Persson A, Hackett CS, Grimmer M, Charron E, Yakovenko S, Kim G, Matthay KK, et al. Paracrine signaling through MYCN enhances tumor-vascular interactions in neuroblastoma. Sci Transl Med. 2012;4:115ra3. doi: 10.1126/scitranslmed.3002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler L, Schlieve C, Goldenberg DD, Kenney A, Kim G, McMillan A, Matthay KK, Rowitch D, Weiss WA. Inhibition of phosphatidylinositol 3-kinase destabilizes Mycn protein and blocks malignant progression in neuroblastoma. Cancer Res. 2006;66:8139–8146. doi: 10.1158/0008-5472.CAN-05-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis CD, Sassone-Corsi P. Mitotic Phosphorylation of Histone H3: Spatio-Temporal Regulation by Mammalian Aurora Kinases. Mol Cell Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET Bromodomain Inhibition as a Therapeutic Strategy to Target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich J, Hulme C, Hurley LH. The design, synthesis, and evaluation of 8 hybrid DFG-out allosteric kinase inhibitors: a structural analysis of the binding interactions of Gleevec, Nexavar, and BIRB-796. 2010;18:5738–5748. doi: 10.1016/j.bmc.2010.05.063. [DOI] [PubMed] [Google Scholar]
- Dodson CA, Kosmopoulou M, Richards MW, Atrash B, Bavetsias V, Blagg J, Bayliss R. Crystal structure of an Aurora-A mutant that mimics Aurora-B bound to MLN8054: insights into selectivity and drug design. Biochem J. 2010;427:19–28. doi: 10.1042/BJ20091530. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomia F, De Rienzo F, Menziani MC. Insights into MAPK p38alpha DFG flip mechanism by accelerated molecular dynamics. 2010;18:6805–6812. doi: 10.1016/j.bmc.2010.07.047. [DOI] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogolin S, Ehemann V, Becker G, Brueckner LM, Dreidax D, Bannert S, Nolte I, Savelyeva L, Bell E, Westermann F. CDK4 inhibition restores G(1)-S arrest in MYCN-amplified neuroblastoma cells in the context of doxorubicin-induced DNA damage. Cell Cycle. 2013;12:1091–1104. doi: 10.4161/cc.24091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görgün G, Calabrese E, Hideshima T, Ecsedy J, Perrone G, Mani M, Ikeda H, Bianchi G, Hu Y, Cirstea D, et al. A novel Aurora-A kinase inhibitor MLN8237 induces cytotoxicity and cell-cycle arrest in multiple myeloma. Blood. 2010;115:5202–5213. doi: 10.1182/blood-2009-12-259523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson WC, Weiss WA. Myc proteins as therapeutic targets. Oncogene. 2010;29:1249–1259. doi: 10.1038/onc.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- Kim J, Aftab BT, Tang JY, Kim D, Lee AH, Rezaee M, Kim J, Chen B, King EM, Borodovsky A, et al. Itraconazole and arsenic trioxide inhibit Hedgehog pathway activation and tumor growth associated with acquired resistance to smoothened antagonists. Cancer Cell. 2013;23:23–34. doi: 10.1016/j.ccr.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional Amplification in Tumor Cells with Elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kaneko S, Yang L, Feldman RI, Nicosia SV, Chen J, Cheng JQ. Aurora-A abrogation of p53 DNA binding and transactivation activity by phosphorylation of serine 215. 2004;279:52175–52182. doi: 10.1074/jbc.M406802200. [DOI] [PubMed] [Google Scholar]
- Manfredi MG, Ecsedy JA, Chakravarty A, Silverman L, Zhang M, Hoar KM, Stroud SG, Chen W, Shinde V, Huck JJ, et al. Characterization of Alisertib (MLN8237), an investigational small-molecule inhibitor of aurora A kinase using novel in vivo pharmacodynamic assays. Clin Cancer Res. 2011;17:7614–7624. doi: 10.1158/1078-0432.CCR-11-1536. [DOI] [PubMed] [Google Scholar]
- Maris JM, Morton CL, Gorlick R, Kolb EA, Lock R, Carol H, Keir ST, Reynolds CP, Kang MH, Wu J, et al. Initial testing of the aurora kinase A inhibitor MLN8237 by the Pediatric Preclinical Testing Program (PPTP) Pediatr Blood Cancer. 2010;55:26–34. doi: 10.1002/pbc.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MP, Zhu J-Y, Lawrence HR, Pireddu R, Luo Y, Alam R, Ozcan S, Sebti SM, Lawrence NJ, Schönbrunn E. A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A. ACS Chem Biol. 2012 doi: 10.1021/cb200508b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ. Targeting MYC dependence in cancer by inhibiting BET bromodomains. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosse YP, Lipsitz E, Fox E, Teachey DT, Maris JM, Weigel B, Adamson PC, Ingle MA, Ahern CH, Blaney SM. Pediatric Phase I Trial and Pharmacokinetic Study of MLN8237, an Investigational Oral Selective Small-Molecule Inhibitor of Aurora Kinase A: A Children's Oncology Group Phase I Consortium Study. Clinical Cancer Research. 2012;18:6058–6064. doi: 10.1158/1078-0432.CCR-11-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc Is a Universal Amplifier of Expressed Genes in Lymphocytes and Embryonic Stem Cells. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R, Tsai C-J. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- Osguthorpe DJ, Hagler AT. Mechanism of Androgen Receptor Antagonism by Bicalutamide in the Treatment of Prostate Cancer. Biochemistry. 2011;50:4105–4113. doi: 10.1021/bi102059z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto T, Horn S, Brockmann M, Eilers U, Schüttrumpf L, Popov N, Kenney AM, Schulte JH, Beijersbergen R, Christiansen H, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Ouchi M. BRCA1 Phosphorylation by Aurora-A in the Regulation of G2 to M Transition. 2004;279:19643–19648. doi: 10.1074/jbc.M311780200. [DOI] [PubMed] [Google Scholar]
- Prochownik EV, Vogt PK. Therapeutic Targeting of Myc. 2010;1:650–659. doi: 10.1177/1947601910377494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, Nekritz EA, Zeid R, Gustafson WC, Greninger P, et al. Targeting MYCN: A Good BET for Improving Neuroblastoma Therapy? Cancer Discovery. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer JT, Kimura H, Magdaleno S, Sasai K, Fuller C, Baines H, Connelly M, Stewart CF, Gould S, Rubin LL, et al. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1+/−p53−/− mice. Cancer Cell. 2004;6:229–240. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Scrittori L, Hans F, Angelov D, Charra M, Prigent C, Dimitrov S. pEg2 aurora-A kinase, histone H3 phosphorylation, and chromosome assembly in Xenopus egg extract. 2001;276:30002–30010. doi: 10.1074/jbc.M102701200. [DOI] [PubMed] [Google Scholar]
- Shang X, Burlingame SM, Okcu MF, Ge N, Russell HV, Egler RA, David RD, Vasudevan SA, Yang J, Nuchtern JG. Aurora A is a negative prognostic factor and a new therapeutic target in human neuroblastoma. Molecular Cancer Therapeutics. 2009;8:2461–2469. doi: 10.1158/1535-7163.MCT-08-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, Swigart LB, Evan GI. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27:504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartling FJ, Grimmer MR, Hackett CS, Northcott PA, Fan Q-W, Goldenberg DD, Lau J, Masic S, Nguyen K, Yakovenko S, et al. Pleiotropic role for MYCN in medulloblastoma. Genes Dev. 2010;24:1059–1072. doi: 10.1101/gad.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas WD, Chen J, Gao YR, Cheung B, Koach J, Sekyere E, Norris MD, Haber M, Ellis T, Wainwright B, et al. Patched1 deletion increases N-Myc protein stability as a mechanism of medulloblastoma initiation and progression. Oncogene. 2009;28:1605–1615. doi: 10.1038/onc.2009.3. [DOI] [PubMed] [Google Scholar]
- Toyoshima M, Howie HL, Imakura M, Walsh RM, Annis JE, Chang AN, Frazier J, Chau BN, Loboda A, Linsley PS, et al. Functional genomics identifies therapeutic targets for MYC-driven cancer. 2012 doi: 10.1073/pnas.1121119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss WA, Weiss WA, Aldape K, Aldape K, Mohapatra G, Mohapatra G, Feuerstein BG, Feuerstein BG, Bishop JM, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. Embo J. 1997;16:2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Q, Goldenson B, Silver SJ, Schenone M, Dancik V, Huang Z, Wang L-Z, Lewis TA, An WF, Li X, et al. Identification of regulators of polyploidization presents therapeutic targets for treatment of AMKL. Cell. 2012;150:575–589. doi: 10.1016/j.cell.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XW, Yan XJ, Zhou ZR, Yang FF, Wu ZY, Sun HB, Liang WX, Song AX, Lallemand-Breitenbach V, Jeanne M, et al. Arsenic Trioxide Controls the Fate of the PML-RAR Oncoprotein by Directly Binding PML. Science. 2010;328:240–243. doi: 10.1126/science.1183424. [DOI] [PubMed] [Google Scholar]
- Zhao B, Smallwood A, Yang J, Koretke K, Nurse K, Calamari A, Kirkpatrick RB, Lai Z. Modulation of kinase-inhibitor interactions by auxiliary protein binding: crystallography studies on Aurora A interactions with VX-680 and with TPX2. Protein Sci. 2008;17:1791–1797. doi: 10.1110/ps.036590.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Lee J-S, Guo F, Shin J, Perez-Atayde AR, Kutok JL, Rodig SJ, Neuberg DS, Helman D, Feng H, et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell. 2012;21:362–373. doi: 10.1016/j.ccr.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03