Investigation of the Evolutionary Development of the Genus Bifidobacterium by Comparative Genomics (original) (raw)
Abstract
The Bifidobacterium genus currently encompasses 48 recognized taxa, which have been isolated from different ecosystems. However, the current phylogeny of bifidobacteria is hampered by the relative paucity of genotypic data. Here, we reassessed the taxonomy of this bacterial genus using genome-based approaches, which demonstrated that the previous taxonomic view of bifidobacteria contained several inconsistencies. In particular, high levels of genetic relatedness were shown to exist between particular Bifidobacterium taxa which would not justify their status as separate species. The results presented are here based on average nucleotide identity analysis involving the genome sequences for each type strain of the 48 bifidobacterial taxa, as well as phylogenetic comparative analysis of the predicted core genome of the Bifidobacterium genus. The results of this study demonstrate that the availability of complete genome sequences allows the reconstruction of a more robust bifidobacterial phylogeny than that obtained from a single gene-based sequence comparison, thus discouraging the assignment of a new or separate bifidobacterial taxon without such a genome-based validation.
INTRODUCTION
Bifidobacteria represent a group of microorganisms that are often dominant inhabitants of the mammalian gut during its suckling phase (for a review, see references 1 to 3). The ecological distribution of bifidobacteria spans a wide range of hosts including not only mammals but also birds and social insects (4). Currently, the genus Bifidobacterium encompasses 48 taxa, representing 39 species and 9 subspecies (2, 5–8). The DNA-DNA reassociation method has been considered the cornerstone of (bifido)bacterial taxonomy (5). However, in addition to problems associated with the reproducibility of this methodology, DNA reassociation values do not represent actual sequence identity or gene content differences since DNA heteroduplexes will form only between strands that show at least 80% sequence complementarity (9). Another, universally accepted method used in modern bacterial taxonomy is the comparison of 16S rRNA gene sequences (5). The limitation of this approach is that there are known cases where two taxa have high 16S rRNA gene sequence identity but low DNA-DNA hybridization (DDH) values (10, 11).
Recently, bifidobacterial taxonomy has benefited from the use of a multigenic approach involving alternative molecular markers to 16S rRNA genes such as clpC, dnaB, dnaG, dnaJ1, purF, rpoC, and xfp (12). Such a multigenic approach was shown to result in a higher resolution between closely related bifidobacterial taxa and to provide a more robust phylogeny of the Bifidobacterium genus (12). With the availability of complete genome sequences, it has become possible to reconstruct phylogenies on the basis of a much larger sequence data set per species, allowing a more reliable and representative inference of the tree of life. Several approaches to build trees from complete genomes have been introduced (13–22). Recently, the draft genome sequences of all 48 currently recognized bifidobacterial taxa have been decoded, representing the largest genome sequencing project involving bifidobacteria performed thus far (23–26, 74). Here, we inferred phylogeny within the genus Bifidobacterium through the use of genomic-based data, thereby allowing a reassessment of the current status of bifidobacterial taxonomy.
MATERIALS AND METHODS
Bacterial strains.
The genome sequences from 48 type strains or the reference chromosome sequences indicated by the National Center for Biotechnology Information (NCBI) of Bifidobacterium were retrieved from the NCBI public database (Table 1). In this context, we used the Bifidobacterium stercoris DSM 24849 genome, which was recently reclassified as a member of the B. adolescentis species (27), as a separate species in order to validate the new classification through genome-based analyses.
TABLE 1.
Bifidobacterium genome list
| Taxon no. | Bifidobacterium strain | Genome size (nt) | GC content (%) | No. of ORFs | Source of isolation | GenBank accession no. | Reference_a_ |
|---|---|---|---|---|---|---|---|
| 1 | B. actinocoloniiforme DSM 22766 | 1,823,388 | 62.71 | 1,484 | Bumblebee digestive tract | JGYK00000000 | 7 |
| 2 | B. adolescentis ATCC 15703 | 2,089,645 | 59.18 | 1,649 | Intestine of adult | AP009256.1 | 49 |
| 3 | B. angulatum LMG 11039 | 2,003,806 | 59.41 | 1,523 | Human feces | JGYL00000000 | 50 |
| 4 | B. animalis subsp. animalis LMG 10508 | 1,915,007 | 60.47 | 1,527 | Rat feces | JGYM00000000 | 51 |
| 5 | B. animalis subsp. lactis DSM 10140 | 1,938,606 | 60.48 | 1,518 | Fermented milk | CP001606.1 | 52 |
| 6 | B. asteroides LMG 10735 | 2,167,304 | 60.05 | 1,653 | Honeybee hindgut | CP003325.1 | 53 |
| 7 | B. biavatii DSM 23969 | 3,252,147 | 63.10 | 2,557 | Feces of tamarin | JGYN00000000 | 6 |
| 8 | B. bifidum LMG 11041 | 2,208,468 | 62.67 | 1,704 | Brest-feed infant feces | JGYO00000000 | 54 |
| 9 | B. bohemicum DSM 22767 | 2,052,470 | 57.45 | 1,632 | Bumblebee digestive tract | JGYP00000000 | 7 |
| 10 | B. bombi DSM 19703 | 1,895,239 | 56.08 | 1,454 | Bumblebee digestive tract | ATLK00000000 | 8 |
| 11 | B. boum LMG 10736 | 2,171,356 | 59.31 | 1,726 | Bovine rumen | JGYQ00000000 | 55 |
| 12 | B. breve LMG 13208 | 2,263,780 | 58.88 | 1,887 | Infant intestine | JGYR00000000 | 49 |
| 13 | B. callitrichos DSM 23973 | 2,887,313 | 63.52 | 2,364 | Feces of common marmoset | JGYS00000000 | 6 |
| 14 | B. catenulatum LMG 11043 | 2,082,756 | 56.11 | 1,664 | Adult intestine | JGYT00000000 | 56 |
| 15 | B. choerinum LMG 10510 | 2,096,123 | 65.53 | 1,672 | Piglet feces | JGYU00000000 | 55 |
| 16 | B. coryneforme LMG 18911 | 1,755,151 | 60.51 | 1,364 | Honeybee hindgut | CP007287 | 53 |
| 17 | B. crudilactis LMG 23609 | 2,362,816 | 57.72 | 1,883 | Raw cow milk | JHAL00000000 | 57 |
| 18 | B. cuniculi LMG 10738 | 2,531,592 | 64.87 | 2,194 | Rabbit feces | JGYV00000000 | 55 |
| 19 | B. dentium LMG 11045 | 2,636,367 | 58.54 | 2,129 | Oral cavity | CP001750.1 | 56 |
| 20 | B. gallicum LMG 11596 | 2,004,594 | 57.61 | 1,507 | Adult intestine | JGYW00000000 | 58 |
| 21 | B. gallinarum LMG 11586 | 2,160,836 | 64.22 | 1,654 | Chicken cecum | JGYX00000000 | 59 |
| 22 | B. indicum LMG 11587 | 1,734,546 | 60.49 | 1,352 | Insect | CP006018 | 53 |
| 23 | B. kashiwanohense DSM 21854 | 2,307,960 | 56.20 | 1,948 | Infant feces | JGYY00000000 | 60 |
| 24 | B. longum subsp. infantis ATCC 15697 | 2,832,748 | 59.86 | 2,500 | Intestine of infant | AP010889.1 | 49 |
| 25 | B. longum subsp. longum LMG 13197 | 2,384,703 | 60.33 | 1,899 | Adult intestine | JGYZ00000000 | 49 |
| 26 | B. longum subsp. suis LMG 21814 | 2,335,832 | 59.96 | 1,955 | Pig feces | JGZA00000000 | 61 |
| 27 | B. magnum LMG 11591 | 1,822,476 | 58.72 | 1,507 | Rabbit feces | JGZB00000000 | 56 |
| 28 | B. merycicum LMG 11341 | 2,280,236 | 60.33 | 1,741 | Bovine rumen | JGZC00000000 | 62 |
| 29 | B. minimum LMG 11592 | 1,892,860 | 62.73 | 1,590 | Sewage | JGZD00000000 | 63 |
| 30 | B. mongoliense DSM 21395 | 2,170,490 | 62.78 | 1,798 | Fermented mare's milk | JGZE00000000 | 64 |
| 31 | B. moukalabense DSM 27321 | 2,515,335 | 59.87 | 2,046 | Feces of gorilla | AZMV01000000 | 65 |
| 32 | B. pseudocatenulatum LMG 10505 | 2,283,767 | 56.36 | 1,771 | Infant feces | JGZF00000000 | 55 |
| 33 | B. pseudolongum subsp. globosum LMG 11569 | 1,935,255 | 63.39 | 1,574 | Bovine rumen | JGZG00000000 | 66 |
| 34 | B. pseudolongum subsp. pseudolongum LMG 11571 | 1,898,684 | 63.06 | 1,495 | Swine feces | JGZH00000000 | 51 |
| 35 | B. psychraerophilum LMG 21775 | 2,615,078 | 58.75 | 2,122 | Pig cecum | JGZI00000000 | 67 |
| 36 | B. pullorum LMG 21816 | 2,153,559 | 64.22 | 1,691 | Chicken feces | JGZJ00000000 | 68 |
| 37 | B. reuteri DSM 23975 | 2,847,572 | 60.45 | 2,149 | Feces of common marmoset | JGZK00000000 | 6 |
| 38 | B. ruminantium LMG 21811 | 2,249,807 | 59.18 | 1,832 | Bovine rumen | JGZL00000000 | 62 |
| 39 | B. saeculare LMG 14934 | 2,263,283 | 63.75 | 1,857 | Rabbit feces | JGZM00000000 | 62 |
| 40 | B. saguini DSM 23967 | 2,787,036 | 56.35 | 2,321 | Feces of tamarin | JGZN00000000 | 6 |
| 41 | B. scardovii LMG 21589 | 3,141,793 | 64.63 | 2,480 | Blood | JGZO00000000 | 69 |
| 42 | B. stellenboschense DSM 23968 | 2,812,864 | 65.34 | 2,202 | Feces of tamarin | JGZP00000000 | 6 |
| 43 | B. stercoris DSM 24849 | 2,304,613 | 59.38 | 1,891 | Adult feces | JGZQ00000000 | 70 |
| 44 | B. subtile LMG 11597 | 2,790,088 | 60.92 | 2,260 | Sewage | JGZR00000000 | 63 |
| 45 | B. thermacidophilum subsp. porcinum LMG 21689 | 2,079,368 | 60.20 | 1,738 | Piglet feces | JGZS00000000 | 71 |
| 46 | B. thermacidophilum subsp. thermacidophilum LMG 21395 | 2,233,072 | 60.38 | 1,823 | Anaerobic digester | JGZT00000000 | 72 |
| 47 | B. thermophilum JCM 1207 | 2,099,496 | 59.91 | 1,700 | Swine feces | JGZV00000000 | 51 |
| 48 | B. tsurumiense JCM 13495 | 2,164,426 | 52.84 | 1,629 | Hamster dental plaque | JGZU00000000 | 73 |
Phylogenetic comparison.
For each genome pair, a value for the average nucleotide identity (ANI) was calculated using the program JSpecies, version 1.2.1 (28). To evaluate relationships between species, two software-based approaches were used to compare the nucleotide sequences, BLAST (29) and MUMmer (30). For each genome pair, the list of DNA maximal unique matches (MUMs) was generated on both strands using the MUMmer software, version 3.0 (30). This software, provided as a supplement package by Deloger et al. (31), was then used to trim the overlapping MUMs and to generate MUM index (MUMi) values for the genome relatedness comparison. The similarity/identity matrix between all 16S and 23S rRNA bifidobacterial gene sequences was calculated with MatGat, version 2.03 (Matrix Global Alignment Tool), using the BLOSUM 50 alignment matrix (32).
Bifidobacterium core gene selection.
For all bifidobacterial genomes used in this study, a core gene calculation was performed using the pan-genomes analysis pipeline (PGAP) (33). The open reading frame (ORF) content of the examined genomes was organized in functional gene clusters using the gene family (GF) method involving comparisons of each protein against all other proteins using BLAST analysis (E value cutoff of 1 × 10−4 and 50% identity across at least 50% of each of the two protein sequences); sequences were then clustered into protein families, named BifCOGs (bifidobacterium-specific clusters of orthologous groups), using a graph theory-based Markov clustering algorithm (MCL) (34). Protein families shared between all genomes, named core BifCOGs, were defined by selecting the families that contained at least one single protein member for each genome. Each set of orthologous proteins constituting core COGs with one member per genome were aligned using MAFFT (35), and phylogenetic trees were constructed using the neighbor-joining method in Clustal W, version 2.1 (36). The supertree was built using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). The values of pairwise distances between the concatenated core genes sequences, the 16S rRNA genes sequences, and the 23S rRNA genes sequences were calculated using MEGA, version 6.06, by using the Kimura two-parameter model.
Positive selection in coding genes.
The Ka/Ks ratio (ratio of the number of nonsynonymous substitutions per nonsynonymous site [_Ka_] to the number of synonymous substitutions per synonymous site [_Ks_]) of selected bifidobacterial gene pairs was evaluated using the KaKs_Calculator software (37).
Bifidobacterial clustering and principal coordinate analysis.
The clustering of combined data collected by in silico analyses (distance matrix, average nucleotide identity, and genome distance index) was performed using the J. Craig Venter Institute's MultiExperiment Viewer (TMeV) software (38). The principal coordinate analysis (PCoA) applied to the data collected was performed using a script implemented in the software suite Quantitative Insights into Microbial Ecology (QIIME) (39).
Nucleotide sequence accession numbers.
Sequences of the newly described bifidobacterial taxa have been deposited in the GenBank under the accession numbers listed in Table 1.
RESULTS AND DISCUSSION
Phylogenetic analyses of the genus Bifidobacterium based on RNA genes.
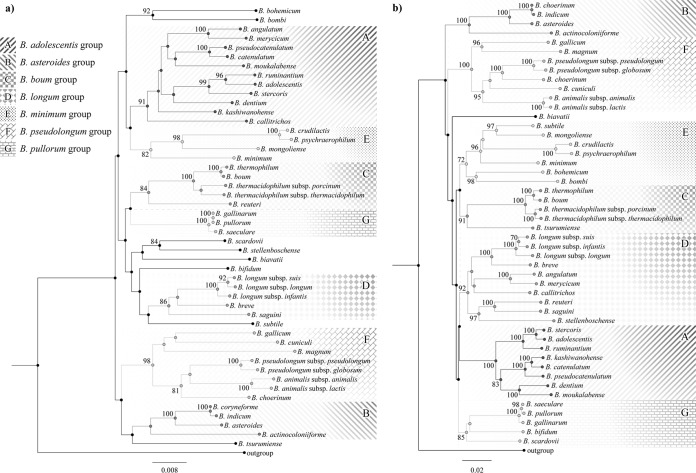
Ribosomal genes such as the 16S rRNA or 23S rRNA genes are considered valuable phylogenetic markers to investigate the evolutionary development of prokaryotes (5). Thus, we decided to investigate the phylogenetic relationships between the 48 currently recognized members of the Bifidobacterium genus using 16S rRNA and 23S rRNA gene sequences. These sequences were retrieved from the genomes of the 48 type strains of each taxon belonging to the genus Bifidobacterium and used to build a phylogenetic tree based on either the 16S rRNA gene sequences or the 23S rRNA gene (Fig. 1). We decided not to concatenate both trees because the 16S rRNA-based tree alone is commonly used to infer microbial phylogeny, and so it was used as a reference condition. As displayed in Fig. 1, the obtained 16S rRNA-based tree is consistent with the six previously recognized phylogenetic groups (i.e., the B. asteroides group, B. pseudolongum group, B. longum group, B. adolescentis group, B. pullorum group, and B. boum group) (12). In the same way, the existence of these six groups was validated by the topology of the 23S rRNA-based tree. However, the 16S and 23S rRNA sequences of some of the recently sequenced bifidobacterial taxa seem to occupy positions that are separate from any of the above-mentioned groups. With the assistance of the obtained rRNA sequence-based trees, it is now possible to expand previously recognized bifidobacterial groups and to identify new ones. The delineation of the bifidobacterial groups based on the rRNA trees is displayed in Fig. 1, with bootstrap values in excess of 70%. The 23S rRNA tree displays more robust and delineated groups than the 16S rRNA tree. In fact, it shows tree-associated bootstrap values greater than 70% for 35 nodes in contrast to the 24 nodes of the 16S rRNA sequence-based tree (Fig. 1) and thus within the rRNA-based trees provides the more reliable phylogenetic image of the genus Bifidobacterium. The B. asteroides, B. longum, B. pullorum, and B. boum groups appear to be conserved in both rRNA trees, while on the other hand the B. pseudolongum and B. longum groups exhibit a number of differences when the 16S- and 23S rRNA-based trees are compared. In addition to the previously recognized groups (12), a novel 16S/23S rRNA-based group was identified which includes Bifidobacterium minimum LMG 11592, Bifidobacterium mongoliense DSM 21395, Bifidobacterium psychraerophilum LMG 21775, and Bifidobacterium crudilactis LMG 23609. Furthermore, we evaluated DNA identity based on the 16S rRNA and 23S rRNA gene sequences (see Table S1 in the supplemental material). Such analyses showed that a set of 19 bifidobacterial pairs (Table 2) have a level of 16S and 23S rRNA gene sequence identity higher than 97%, a value typically considered the cutoff for species assignment (11). Moreover, in the case of the bifidobacterial taxa listed in Table 2, we also considered the possible effect of microheterogeneity that may exist between the various 16S rRNA loci within the same genome; however, the level of 16S rRNA identity was observed to be above 97% in all examined cases (see Table S1). Notably, bifidobacterial pairs that have previously been classified as subspecies have a level of 16S rRNA and 23S rRNA identity above 97%. However, these analyses also reveal a high level of 16S rRNA identity for bifidobacterial taxa that are currently recognized as separate species, for example, as seen for the Bifidobacterium pseudocatenulatum LMG 10505-Bifidobacterium catenulatum LMG 11043 (98.9%), pair, the Bifidobacterium angulatum LMG 11039-Bifidobacterium merycicum LMG 11341 (98.2%) pair, and both the Bifidobacterium asteroides LMG 10735-Bifidobacterium indicum LMG 11587 and B. asteroides LMG 10735-Bifidobacterium coryneforme LMG 18911 pairs (98%).
FIG 1.
Phylogenetic trees of the Bifidobacterium genus. (a) The 16S rRNA gene-based tree of the 48 bifidobacterial taxa currently recognized. (b) The 23S rRNA gene-based tree of the same 48 bifidobacterial taxa as shown in panel a. For each tree, bootstrap values higher than 70 are marked near the respective node, and phylogenetic clusters are identified by shading as indicated on the figure. With the exception of a small number of species, seven conserved clusters (designated groups A to G) are present in each tree although the groups are arranged in different positions.
TABLE 2.
Bifidobacterial pairs with both 16S and 23S identities higher than 97%
| Bifidobacterial pair | % identity | ANI (%) | MUMi | |
|---|---|---|---|---|
| 16S rRNA gene | 23S rRNA gene | |||
| B. indicum LMG 11587-B. coryneforme LMG 18911 | 100 | 99.7 | 98.1 | 0.12 |
| B. adolescentis ATCC 15703-B. stercoris DSM 24849 | 98.1 | 98.6 | 98.1 | 0.26 |
| B. pullorum LMG 21816-B. saeculare LMG 14934 | 99.7 | 99.6 | 97.3 | 0.27 |
| B. pullorum LMG 21816-B. gallinarum LMG 11586 | 99.9 | 99.3 | 97.0 | 0.26 |
| B. saeculare LMG 14934-B. gallinarum LMG 11586 | 99.7 | 99.4 | 97.0 | 0.29 |
| B. catenulatum LMG 11043-B. kashiwanohense DSM 21854 | 98.9 | 98.1 | 96.9 | 0.33 |
| B. longum subsp. suis LMG 21814-B. longum subsp. longum LMG 13197 | 99.7 | 98 | 96.6 | 0.35 |
| B. longum subsp. infantis ATCC 15697-B. longum subsp. suis LMG 21814 | 99.2 | 98.6 | 96.2 | 0.44 |
| B. animalis subsp. animalis LMG 10508-B. animalis subsp. lactis DSM 10140 | 98.8 | 98.6 | 96.0 | 0.28 |
| B. thermacidophilum subsp. thermacidophilum LMG 21395-B. thermacidophilum subsp. porcinum LMG 21689 | 98.7 | 97.5 | 95.9 | 0.29 |
| B. longum subsp. infantis ATCC 15697-B. longum subsp. longum LMG 13197 | 99.3 | 97.7 | 95.5 | 0.47 |
| B. thermophilum JCM 1207-B. boum LMG 10736 | 99.7 | 97.6 | 94.9 | 0.37 |
| B. pseudolongum subsp. pseudolongum LMG 11571-B. pseudolongum subsp. globosum LMG 11569 | 99.3 | 98.1 | 93.9 | 0.40 |
| B. thermophilum JCM 1207-B. thermacidophilum subsp. porcinum LMG 21689 | 98.3 | 98.9 | 90.4 | 0.71 |
| B. thermophilum JCM 1207-B. thermacidophilum subsp. thermacidophilum LMG 21395 | 98.8 | 97.5 | 90.2 | 0.72 |
| B. adolescentis ATCC 15703-B. ruminantium LMG 21811 | 98.5 | 97.4 | 89.6 | 0.71 |
| B. boum LMG 10736-B. thermacidophilum subsp. thermacidophilum LMG 21395 | 98.6 | 98.7 | 88.7 | 0.79 |
| B. boum LMG 10736-B. thermacidophilum subsp. porcinum LMG 21689 | 98 | 97.7 | 88.7 | 0.78 |
| B. psychraerophilum LMG 21775-B. crudilactis LMG 23609 | 99.5 | 97.8 | 85.9 | 0.92 |
Phylogenomic analyses of members of the genus Bifidobacterium.
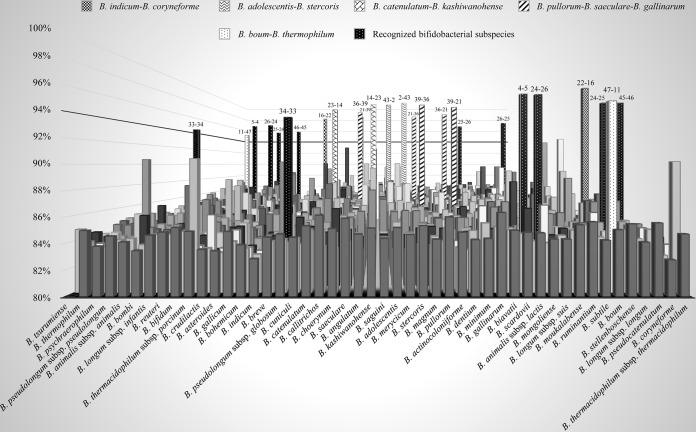
The availability of the 48 bifidobacterial genome sequences allowed us to obtain insights into the various levels of genetic similarities between them. As described in Table S2 in the supplemental material, the average nucleotide identity (ANI) (Ks) (40) between the examined members of the Bifidobacterium genus ranged between 81.3% and 98.1%. In this context, the Bifidobacterium adolescentis ATCC 15703-Bifidobacterium stercoris DSM 24849, B. indicum LMG 11587-B. coryneforme LMG 18911, and B. catenulatum LMG 11043-Bifidobacterium kashiwanohense DSM 21854 genome pairs and the Bifidobacterium pullorum LMG 21816-Bifidobacterium saeculare LMG 14934-Bifidobacterium gallinarum LMG 11586 set have ANI values above 95% (Table 2). According to the DNA-DNA hybridization (DDH) rules, two strains belong to the same species when they possess a DNA-DNA similarity above 70% (41). However, in the era of bacterial genomics, the ANI approach based on draft genome sequences for prokaryotic species definition has been proposed as a powerful alternative to DDH (28). Thus, the observed high ANI values between these bifidobacterial taxa would not support their classification as distinct species. However, it is worth mentioning that our in silico analyses are based on genome sequences that are still fragmented in a varying number of contigs (ranging from 1 to 56 contigs), having been assembled from a sequence coverage ranging from 45- to 281-fold. Nevertheless, a 50-fold sequence coverage of prokaryotic genomes, as obtained from next-generation sequencing (NGS) platforms, is predicted to represent more than 99% of the complete genome sequence (42), which implies an adequate reliability of the in silico analyses of ANI. Furthermore, it has been shown that for highly related genomes (i.e., ANI values of >94%), draft genome sequences that reach at least 50% of genome coverage for both strains result in ANI values that have been recognized as adequate for the calculation of nucleotide identity (28). Notably, our analysis demonstrated that comparing the genomes of Bifidobacterium thermophilum JCM 1207 and Bifidobacterium boum LMG 10736 generated an ANI value of 94.9%, which is very close to the generally accepted threshold for species recognition (see Table S2 in the supplemental material) (43). Furthermore, a second bifidobacterial pair, Bifidobacterium pseudolongum subsp. pseudolongum LMG 11571 and Bifidobacterium pseudolongum subsp. globosum LMG 11569, has an ANI value of 93.9%, which is below the threshold value obtained for genomes of bacteria that belong to different species. This therefore indicates that these taxa should be considered separate species rather than subspecies. On the other hand, other currently recognized bifidobacterial subspecies, i.e., the Bifidobacterium animalis subsp. animalis LMG 10508-Bifidobacterium animalis subsp. lactis DSM 10140 pair, the Bifidobacterium thermacidophilum subsp. thermacidophilum LMG 21395-Bifidobacterium thermacidophilum subsp. porcinum LMG 21689 pair, and the Bifidobacterium longum subsp. longum LMG 13197-Bifidobacterium longum subsp. infantis ATCC 15697-Bifidobacterium longum subsp. suis LMG 21814 set, were shown to be associated with ANI values ranging from ≥95% to ≤97%, thus confirming the current bifidobacterial subspecies assignment for these taxa (Fig. 2).
FIG 2.
Graphical representation in three-dimensional columns of the average nucleotide identity (ANI) matrix shown in Table S4 in the supplemental material, based on the analysis of the 48 bifidobacterial type strain genome sequences. On the x axis are located the 48 bifidobacterial taxa to generate a matrix; the y axis reflects the ANI percentages such that each column represents the ANI observed between a given bifidobacterial species pair. All bifidobacterial pairs that show an ANI value of >94% are highlighted according to the legend at the top of the figure. The paired numbers above the columns correspond to the numbering identifying the organisms described in Table 1.
Bifidobacterial genome similarities.
In order to validate the results achieved with ANI analysis, we employed another bioinformatics approach. This approach, based on DNA maximal unique matches (MUM), called MUMi (MUM index) (31), was used to determine the genomic distance for all considered bifidobacterial pairs. MUMi values ranged from 0.11 (between B. indicum LMG 11587 and B. coryneforme LMG 18911) to 0.99 (between Bifidobacterium tsurumiense JCM 13495 and Bifidobacterium asteroides LMG 10735) (see Table S3 in the supplemental material). According to what had previously been demonstrated, i.e., that a MUMi value of 0.33 ± 0.03 corresponds to an ANI value of 95% ± 0.5% (30), the MUMi assignments for the bifidobacterial B. indicum LMG 11587-B. coryneforme LMG 18911 (0.12) pair, the B. adolescentis ATCC 15703-B. stercoris DSM 24849 (0.26) pair, the B. pullorum LMG 21816-B. saeculare LMG 14934-B. gallinarum LMG 11586 (0.27) set, and the B. catenulatum LMG 11043-B. kashiwanohense DSM 21854 (0.33) pair clearly confirmed a very high level of genetic relatedness, which does not support their classification into separate species (Table 2). Furthermore, the MUMi value for the bifidobacterial pair B. thermophilum JCM 1207 and B. boum LMG 10736 (0.37) confirms the ANI value that is close to the recognized threshold of 95%, thus raising questions around the validity of their previous assignment into two separate species. In contrast, the MUMi value for the subspecies pair B. pseudolongum subsp. pseudolongum LMG 11571 and B. pseudolongum subsp. globosum LMG 11569 (0.4) confirms the results of the ANI analysis; i.e., both support limited nucleotide identity between the genomes of these particular taxa.
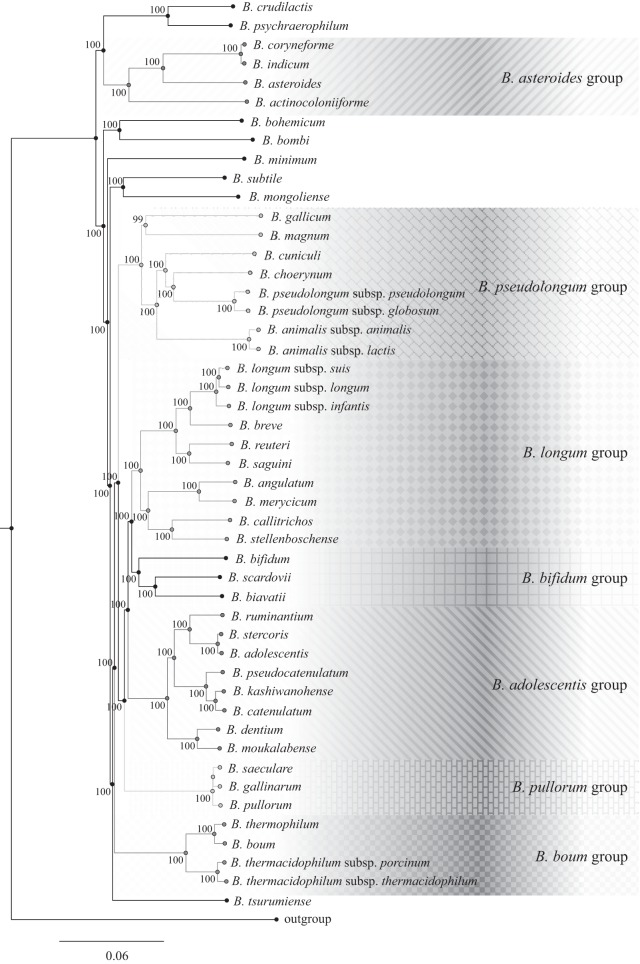
The Bifidobacterium supertree.
The availability of genome sequences for each of the 48 currently recognized members of the genus Bifidobacterium also allows a more robust reconstruction of the phylogeny of members of this genus. A comparative study was undertaken to determine putative orthology between the 48 bifidobacterial genome sequences, which resulted in the identification of 18,435 BifCOGs (bifidobacterium-specific clusters of orthologous genes). Analysis of the predicted BifCOGs allowed the identification of 534 COGs that were shared among all these genomes, representing the core of bifidobacterial genome coding sequences (core BifCOGs). This core BifCOG collection represents, at the time of writing, the most updated core genome sequences of the genus Bifidobacterium. Conserved genes, which represented paralogs within bifidobacterial genomes, were not taken into account further. A concatenated protein sequence that represents the protein products of the thus identified 411 core genes was used to build a Bifidobacterium supertree (Fig. 3). This Bifidobacterium supertree analysis produced an evolutionary positioning of all 48 bifidobacterial taxa of the genus Bifidobacterium by placing the type strains of B. adolescentis ATCC 15703 and B. stercoris DSM 24849 in the same cluster. In a similar way, the concatenated sequences of the type strains of B. indicum LMG 11587 and B. coryneforme LMG 18911 are located on the same branch of the tree as well as those of the B. pullorum LMG 21816-B. saeculare LMG 14934-B. gallinarum LMG 11586 set and the B. catenulatum LMG 11043-B. kashiwanohense DSM 21854 pair (Fig. 3). Furthermore, all validated bifidobacterial subspecies cluster in the same branches of the core gene-based tree, as well as the B. thermophilum JCM 1207-B. boum LMG 10736 pair. This finding indicates the absence of any substantial amino acid sequence differences between the individual core proteins of these strains.
FIG 3.
Supertree of the Bifidobacterium genus based on the concatenation of the 411 core BifCOG amino acid sequences identified by the PGAP analysis (core BifCOGs that include bifidobacterial paralogs have been excluded in this analysis). Bootstrap values higher than 70 are marked near the respective nodes showing a robust phylogenetic reconstruction, and the seven phylogenetic clusters are highlighted by patterned shading.
Comparison between bifidobacterial supertree and ribosomal gene trees.
The bifidobacterial core genome-based tree allowed the identification of seven phylogenetic clusters, named B. longum, B. adolescentis, B. pseudolongum, B. boum, B. asteroides, B. pullorum, and B. bifidum, which largely overlap the phylogenetic rRNA-based groups so far recognized within the genus Bifidobacterium (see above). In this context, we identified a novel phylogenetic cluster named B. bifidum, consisting of Bifidobacterium bifidum LMG 11041, Bifidobacterium scardovii LMG 21589, and Bifidobacterium biavatii DSM 23969 (Fig. 3).
A comparison between the supertree and the single-marker tree based on 16S rRNA gene sequences involving the same set of bifidobacterial strains was also performed. Such analysis showed that 24 nodes and 45 (to 46) total nodes were supported by bootstrap values greater than 70 for the 16S rRNA gene-based tree and the core gene-based tree, respectively (Fig. 1). The discriminatory power of the tree based on the concatenated core genes, as based on the analysis of pairwise distances, is significantly higher than that observed for the 16S rRNA gene-based tree: the mean pairwise distance of the concatenated core gene-based protein sequences was determined to be 0.25, versus a mean 16S rRNA gene sequence-based distance of 0.051 (see Table S4 in the supplemental material). In addition, the increase in sequence length as well as the use of protein-based sequences allowed a considerable increase in tree robustness. In this context, the concatenation showed an increase in deep-node bootstrap values. For example, the conserved cluster B. adolescentis, which is supported by a bootstrap value of 29 for the single 16S rRNA tree, is supported by a bootstrap value of 100 in the concatenated tree. Data concatenation therefore provides a powerful means of increasing the robustness of the final tree (Fig. 3). The observed substantial increase in bootstrap values demonstrates that the phylogenetic tree calculated from the concatenation of the bifidobacterial core proteins identified here as alternative molecular markers to the 16S rRNA gene may considerably improve the phylogenetic relevance. In a similar way, a comparison with the phylogenetic tree based on 23S rRNA gene sequences was also performed, showing lower bootstrap and pairwise distance values (0.108) than the core protein-based tree (Fig. 1) (see Table S4). However, it is interesting that the distribution of the bifidobacterial 23S rRNA clusters largely reflects the bifidobacterial core protein clusters, showing more affinity to the core protein tree than the 16S rRNA gene-based tree. As displayed in Fig. 3, a comparison with the 23S rRNA gene-based tree highlights differences in two clusters represented in Fig. 1 as the B. pullorum group and B. minimum group. In both cases, we noticed a different distribution of the bifidobacterial taxa, which might be the consequence of a high level of genetic adaptation to a specific ecological niche. The bifidobacterial supertree displays in this case two separate clusters (designated the B. pullorum and B. bifidum clusters) instead of the extended B. pullorum group, as exhibited by the 23S rRNA gene-based tree, which may reflect speciation in accordance with their ecological origins (rabbit and chicken feces for B. pullorum LMG 21816, B. saeculare LMG 14934, and B. gallinarum LMG 11586 and human-derived samples for B. bifidum LMG 11041 and B. scardovii LMG 21589) (Table 1). Furthermore, the B. minimum group identified in the 23S rRNA gene-based tree is displayed as separated branches in the core protein-based tree. A large part of bifidobacterial taxa that did not fall in any core protein cluster are represented by Bifidobacterium species isolated from milk and sewage, except for Bifidobacterium bohemicum DSM 22767 and Bifidobacterium bombi DSM 19703, which represent isolates from the digestive tract of a bumblebee. Therefore, it is possible that the observed inconsistencies between the supertree and the rRNA-based trees are linked to genetic adaptations of these bifidobacterial taxa to specific niches.
Thus, based on the higher level of robustness (higher number of nodes with bootstrap values greater than 70) as well as the level of pairwise distance, we can argue that the bifidobacterial core genome-based tree provides a more reliable phylogenetic image than the 23S rRNA gene-based- or 16S rRNA gene-based tree.
Bifidobacterium genus evolution rate.
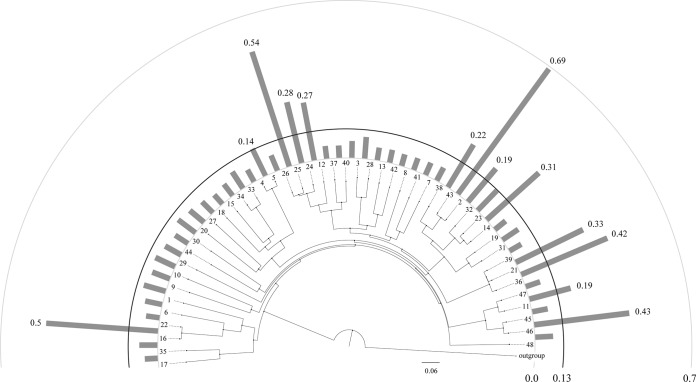
The strength of purifying selection acting on a given Bifidobacterium species can be estimated by evaluating synonymous substitution (Ks) and nonsynonymous substitution (Ka) from coding sequence alignments of the orthologous genes shared by bifidobacterial pairs. Such calculated substitution rates are of substantial significance in reconstructing phylogeny and understanding evolutionary dynamics of protein-coding sequences across closely related yet diverged species (44). The Ka/Ks ratio (ratio of the number of nonsynonymous substitutions per nonsynonymous site to the number of synonymous substitutions per synonymous site) for every bifidobacterial pair was evaluated using KaKs_Calculator (45). In this study, we considered all orthologous genes between bifidobacterial pairs that are not assigned to the Bifidobacterium core gene pool. Generally, between members harboring two distinct species, the Ka value is much less than the Ks value (i.e., Ka/Ks < 1) because a mutation that changes a protein is much less probable than one which is silent (46). The average _Ka /Ks_ values for all gene pairs obtained for every bifidobacterial genome pair are shown in Fig. 4, and these values indicate purifying selection for the validated separated species pair. Notably, the _B. indicum_ LMG 11587-_B. coryneforme_ LMG 18911 pair, the _B. adolescentis_ ATCC 15703-_B. stercoris_ DSM 24849 pair, the _B. pullorum_ LMG 21816-_B. saeculare_ LMG 14934-_B. gallinarum_ LMG 11586 set, and the _B. catenulatum_ LMG 11043-_B. kashiwanohense_ DSM 21854 and _B. thermophilum_ JCM 1207-_B. boum_ LMG 10736 pairs have average _Ka /Ks_ distances of >0.1, revealing a higher average of Ka substitutions than other bifidobacterial pairs. Furthermore, the subspecies pair B. pseudolongum subsp. pseudolongum LMG 11571 and B. pseudolongum subsp. globosum LMG 11569 shows an average Ka /Ks distance of <0.1, which suggests a purifying selection similar to the other separated species pair. In contrast, two exceptions are represented by _B. longum_ subsp. _infantis_ ATCC 15697 and _B. adolescentis_ ATCC 15703 that have an average _Ka/Ks_ distance of >0.2 with the bifidobacterial strains Bifidobacterium breve LMG 13208 and Bifidobacterium ruminantium LMG 21811, respectively (Fig. 4). With the exception of the B. longum subsp. infantis ATCC 15697 and B. adolescentis ATCC 15703, all bifidobacterial pairs that belong to the same species have clearly different Ka/Ks rates than the remaining recognized species.
FIG 4.
Average Ka/Ks ratio between bifidobacterial pairs represented by a fan-shaped chart. The diagram was built on the bifidobacterial core gene supertree presented in Fig. 2, and the numbers placed at the supertree leaves represent the related bifidobacterial taxa presented in Table 1. The columns indicate the average Ka/Ks ratios of the flanking bifidobacterial taxa. For bifidobacterial taxa pairs having high Ka/Ks ratios, the corresponding values are indicated above the columns.
Evaluation of the bifidobacterial taxon relatedness based on a polyphasic approach.
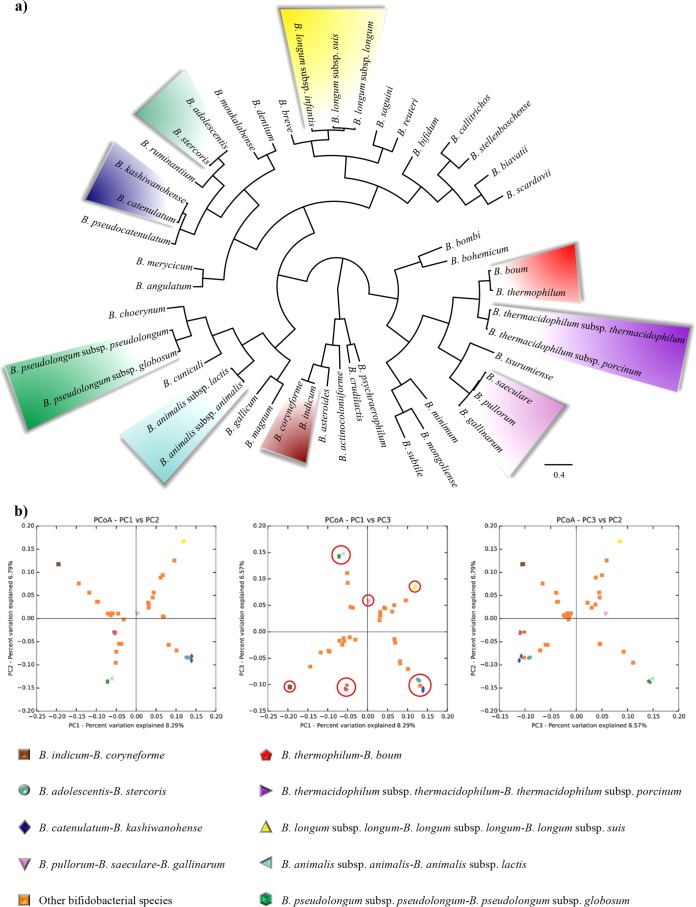
In order to reliably infer phylogeny within the genus Bifidobacterium, we decided to implement a consensus bifidobacterial clustering approach based on previously described analyses. Such a consensus clustering method includes the phylogenetic data presented above in the form of distance matrices, as well as the level of nucleotide identity between genomes and the genome distance index detected between the 48 bifidobacterial taxa. As displayed in Fig. 5, all bifidobacterial pairs that we propose here as being members of the same species fell in the same cluster, showing a substantiated correlation between the analyses described above. Furthermore, in order to visualize the correlation between these collected data, a principal coordinate analysis (PCoA) was performed (Fig. 5), which highlights the very close genetic relatedness of the B. indicum LMG 11587-B. coryneforme LMG 18911, B. adolescentis ATCC 15703-B. stercoris DSM 24849, B. catenulatum LMG 11043-B. kashiwanohense DSM 21854, and B. thermophilum JCM 1207-B. boum LMG 10736 pairs and the set of B. pullorum LMG 21816, B. saeculare LMG 14934, and B. gallinarum LMG 11586, as well as all bifidobacterial subspecies pairs. Thus, these observations suggest that a serious revision of the taxonomy of the genus Bifidobacterium needs to be considered.
FIG 5.
Consensus clustering based on the inclusion of results from the phylogenetic distance matrix, the level of nucleotide identity between genomes, and the genome distance index detected between all 48 bifidobacterial taxa. (a) Polar cluster where all bifidobacterial taxa proposed here as members of the same species fall in the same cluster, highlighted by different colors. (b) Principal coordinate analysis (PCoA) of the same data set used to generate the polar cluster. In the central PCoA image, recognized bifidobacterial subspecies and related species discussed in the main text are circled in red to separate them from the remaining bifidobacterial species, which are represented by orange squares.
Conclusions.
Genome sequencing has revolutionized the microbial taxonomy by providing large genetic data sets that are particularly useful for bacterial phylogeny analyses. Thanks to genome sequencing efforts involving all currently recognized bifidobacterial species, we were able to identify a set of 411 genes representing the core bifidobacterial genome sequences that act as alternative molecular markers to the 16S rRNA gene sequences. The multisequence approach applied here is in line with recommendations of the ad hoc committee for reevaluation of the definition of bacterial species (11). This approach has been used successfully also for the delineation of other bacterial genera such as Lactobacillus and Streptococcus (47, 48). The concatenation of these 411 proteins provides a reliable phylogenetic tree that is discriminatory and robust compared to the 16S rRNA gene-based tree.
In addition, investigation of ANI values based on bifidobacterial genomes identified inconsistencies in the current taxonomy of bifidobacteria. In fact, the high ANI values obtained from genome pairs representing the type strains of B. adolescentis ATCC 15703 and B. stercoris DSM 24849, B. indicum LMG 11587 and B. coryneforme LMG 18911, and B. catenulatum LMG 11043 and B. kashiwanohense DSM 21854, and from the set of B. pullorum LMG 21816, B. saeculare LMG 14934, and B. gallinarum LMG 11586 did not support the assignment of the bacteria as different species. Such a high level of genetic relatedness between the above-mentioned bifidobacterial taxa was also confirmed by the investigation of MUMi and genome-to-genome distance calculator (GGDC) values. Furthermore, a lower ANI value detected between the type strains of B. pseudolongum subsp. pseudolongum LMG 11571 and B. pseudolongum subsp. globosum LMG 11569 did not support the assignment of subspecies due to an ANI value lower than 94%, as supported by MUMi and GGDC analysis. Moreover, as shown in many different analyses carried out, borderline bifidobacterial pairs emerge, represented by B. boum LMG 10736 and B. thermophilum JCM 1207. Thus, altogether our analyses clearly suggest that the taxonomy of the genus Bifidobacterium should be redefined with the recognition of only 34, as opposed to the currently 39, described species, while we furthermore propose to consider B. pseudolongum subsp. globosum LMG 11569 and B. pseudolongum subsp. pseudolongum LMG 11571 to be two separate species. Furthermore, in contrast to what is currently known, we have demonstrated the existence of seven phylogenetic group within the genus Bifidobacterium instead of six (15), including the new B. bifidum phylogenetic cluster encompassing bifidobacterial species isolated from human. Even though this study revised the evolutionary development represented by the current taxonomy of the genus Bifidobacterium, it should also affect approaches followed in the future for species assignment within this genus. In fact, it is desirable that bifidobacterial taxonomy in the near future give more consideration to multigene or whole-genomic features of one hypothetical new taxon rather than focus on a single molecular marker, as in the current practice.
Supplementary Material
Supplemental material
ACKNOWLEDGMENTS
We thank GenProbio s.r.l. for financial support of the Laboratory of Probiogenomics. This work was financially supported by a FEMS Jensen Award to F.T. and by a Ph.D. fellowship (Spinner 2013, Regione Emilia Romagna) to S.D. D.V.S. and F.T. are members of The Alimentary Pharmabiotic Centre, and D.V.S. is also a member of the Alimentary Glycoscience Research Cluster; both are funded by Science Foundation Ireland through the Irish Government's National Development Plan (grant numbers 07/CE/B1368, SFI/12/RC/2273, and 08/SRC/B1393).
Footnotes
Published ahead of print 8 August 2014
REFERENCES
- 1.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71:495–548. 10.1128/MMBR.00005-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ventura M, Turroni F, Lugli GA, van Sinderen D. 2014. Bifidobacteria and humans: our special friends, from ecological to genomic perspectives. J. Sci. Food Agric. 94:163–168. 10.1002/jsfa.6356 [DOI] [PubMed] [Google Scholar]
- 3.Turroni F, Ventura M, Butto LF, Duranti S, O'Toole PW, O'Connell Motherway M, van Sinderen D. 2014. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell. Mol. Life Sci. 71:183–203. 10.1007/s00018-013-1318-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turroni F, Ribbera A, Foroni E, van Sinderen D, Ventura M. 2008. Human gut microbiota and bifidobacteria: from composition to functionality. Antonie Van Leeuwenhoek 94:35–50. 10.1007/s10482-008-9232-4 [DOI] [PubMed] [Google Scholar]
- 5.Bergey DH, Goodfellow M, Whitman WB, Parte AC. 2012. The Actinobacteria. Bergey's manual of systematic bacteriology, 2nd ed, vol 5 Springer, New York, NY [Google Scholar]
- 6.Endo A, Futagawa-Endo Y, Schumann P, Pukall R, Dicks LM. 2012. Bifidobacterium reuteri sp. nov., Bifidobacterium callitrichos sp. nov., Bifidobacterium saguini sp. nov., Bifidobacterium stellenboschense sp. nov. and Bifidobacterium biavatii sp. nov. isolated from faeces of common marmoset (Callithrix jacchus) and red-handed tamarin (Saguinus midas). Syst. Appl. Microbiol. 35:92–97. 10.1016/j.syapm.2011.11.006 [DOI] [PubMed] [Google Scholar]
- 7.Killer J, Kopecny J, Mrazek J, Koppova I, Havlik J, Benada O, Kott T. 2011. Bifidobacterium actinocoloniiforme sp. nov. and Bifidobacterium bohemicum sp. nov., from the bumblebee digestive tract. Int. J. Syst. Evol. Microbiol. 61:1315–1321. 10.1099/ijs.0.022525-0 [DOI] [PubMed] [Google Scholar]
- 8.Killer J, Kopecny J, Mrazek J, Rada V, Benada O, Koppova I, Havlik J, Straka J. 2009. Bifidobacterium bombi sp. nov., from the bumblebee digestive tract. Int. J. Syst. Evol. Microbiol. 59:2020–2024. 10.1099/ijs.0.002915-0 [DOI] [PubMed] [Google Scholar]
- 9.Rossello-Mora R, Amann R. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39–67. 10.1111/j.1574-6976.2001.tb00571.x [DOI] [PubMed] [Google Scholar]
- 10.Fox GE, Wisotzkey JD, Jurtshuk P., Jr 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166–170. 10.1099/00207713-42-1-166 [DOI] [PubMed] [Google Scholar]
- 11.Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kampfer P, Maiden MC, Nesme X, Rossello-Mora R, Swings J, Truper HG, Vauterin L, Ward AC, Whitman WB. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043–1047. 10.1099/ijs.0.02360-0 [DOI] [PubMed] [Google Scholar]
- 12.Ventura M, Canchaya C, Del Casale A, Dellaglio F, Neviani E, Fitzgerald GF, van Sinderen D. 2006. Analysis of bifidobacterial evolution using a multilocus approach. Int. J. Syst. Evol. Microbiol. 56:2783–2792. 10.1099/ijs.0.64233-0 [DOI] [PubMed] [Google Scholar]
- 13.Brown JR, Douady CJ, Italia MJ, Marshall WE, Stanhope MJ. 2001. Universal trees based on large combined protein sequence data sets. Nat. Genet. 28:281–285. 10.1038/90129 [DOI] [PubMed] [Google Scholar]
- 14.Daubin V, Gouy M, Perriere G. 2002. A phylogenomic approach to bacterial phylogeny: evidence of a core of genes sharing a common history. Genome Res. 12:1080–1090. 10.1101/gr.187002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitz-Gibbon ST, House CH. 1999. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 27:4218–4222. 10.1093/nar/27.21.4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grishin NV, Wolf YI, Koonin EV. 2000. From complete genomes to measures of substitution rate variability within and between proteins. Genome Res. 10:991–1000. 10.1101/gr.10.7.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huson DH, Steel M. 2004. Phylogenetic trees based on gene content. Bioinformatics 20:2044–2049. 10.1093/bioinformatics/bth198 [DOI] [PubMed] [Google Scholar]
- 18.Qi J, Wang B, Hao BI. 2004. Whole proteome prokaryote phylogeny without sequence alignment: a K-string composition approach. J. Mol. Evol. 58:1–11. 10.1007/s00239-003-2493-7 [DOI] [PubMed] [Google Scholar]
- 19.Snel B, Bork P, Huynen MA. 1999. Genome phylogeny based on gene content. Nat. Genet. 21:108–110. 10.1038/5052 [DOI] [PubMed] [Google Scholar]
- 20.Tekaia F, Lazcano A, Dujon B. 1999. The genomic tree as revealed from whole proteome comparisons. Genome Res. 9:550–557 [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf YI, RIB. Grishin NV, Tatusov RL, Koonin EV. 2001. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol. Biol. 1:8. 10.1186/1471-2148-1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang S, Doolittle RF, Bourne PE. 2005. Phylogeny determined by protein domain content. Proc. Natl. Acad. Sci. U. S. A. 102:373–378. 10.1073/pnas.0408810102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ventura M, Turroni F, Zomer A, Foroni E, Giubellini V, Bottacini F, Canchaya C, Claesson MJ, He F, Mantzourani M, Mulas L, Ferrarini A, Gao B, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Gupta RS, Zhang Z, Beighton D, Fitzgerald GF, O'Toole PW, van Sinderen D. 2009. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 5:e1000785. 10.1371/journal.pgen.1000785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. U. S. A. 107:19514–19519. 10.1073/pnas.1011100107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bottacini F, Milani C, Turroni F, Sanchez B, Foroni E, Duranti S, Serafini F, Viappiani A, Strati F, Ferrarini A, Delledonne M, Henrissat B, Coutinho P, Fitzgerald GF, Margolles A, van Sinderen D, Ventura M. 2012. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One 7:e44229. 10.1371/journal.pone.0044229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lugli GA, Duranti S, Milani C, Turroni F, Viappiani A, Mangifesta M, van Sinderen D, Ventura M. 2014. The genome sequence of Bifidobacterium moukalabense DSM 27321 highlights the close phylogenetic relatedness with the Bifidobacterium dentium taxon. Genome Announc. 2(1):e00048-14. 10.1128/genomeA.00048-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Killer J, Sedlacek I, Rada V, Havlik J, Kopecny J. 2013. Reclassification of Bifidobacterium stercoris Kim et al. 2010 as a later heterotypic synonym of Bifidobacterium adolescentis. Int. J. Syst. Evol. Microbiol. 63:4350–4353. 10.1099/ijs.0.054957-0 [DOI] [PubMed] [Google Scholar]
- 28.Richter M, Rossello-Mora R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106:19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 30.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deloger M, El Karoui M, Petit MA. 2009. A genomic distance based on MUM indicates discontinuity between most bacterial species and genera. J. Bacteriol. 191:91–99. 10.1128/JB.01202-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campanella JJ, Bitincka L, Smalley J. 2003. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4:29. 10.1186/1471-2105-4-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. 2012. PGAP: pan-genomes analysis pipeline. Bioinformatics 28:416–418. 10.1093/bioinformatics/btr655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30:1575–1584. 10.1093/nar/30.7.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z, Li J, Zhao XQ, Wang J, Wong GK, Yu J. 2006. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4:259–263. 10.1016/S1672-0229(07)60007-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378 [DOI] [PubMed] [Google Scholar]
- 39.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 102:2567–2572. 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wayne LG. 1988. International Committee on Systematic Bacteriology: announcement of the report of the ad hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 268:433–434 [DOI] [PubMed] [Google Scholar]
- 42.Desai A, Marwah VS, Yadav A, Jha V, Dhaygude K, Bangar U, Kulkarni V, Jere A. 2013. Identification of optimum sequencing depth especially for de novo genome assembly of small genomes using next generation sequencing data. PLoS One 8:e60204. 10.1371/journal.pone.0060204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konstantinidis KT, Ramette A, Tiedje JM. 2006. The bacterial species definition in the genomic era. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1929–1940. 10.1098/rstb.2006.1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fay JC, Wu CI. 2003. Sequence divergence, functional constraint, and selection in protein evolution. Annu. Rev. Genomics Hum. Genet. 4:213–235. 10.1146/annurev.genom.4.020303.162528 [DOI] [PubMed] [Google Scholar]
- 45.Zhang Z, Yu J. 2006. Evaluation of six methods for estimating synonymous and nonsynonymous substitution rates. Genomics Proteomics Bioinformatics 4:173–181. 10.1016/S1672-0229(06)60030-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hurst LD. 2002. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18:486. 10.1016/S0168-9525(02)02722-1 [DOI] [PubMed] [Google Scholar]
- 47.Claesson MJ, van Sinderen D, O'Toole PW. 2008. Lactobacillus phylogenomics—towards a reclassification of the genus. Int. J. Syst. Evol. Microbiol. 58:2945–2954. 10.1099/ijs.0.65848-0 [DOI] [PubMed] [Google Scholar]
- 48.Anisimova M, Bielawski J, Dunn K, Yang Z. 2007. Phylogenomic analysis of natural selection pressure in Streptococcus genomes. BMC Evol. Biol. 7:154. 10.1186/1471-2148-7-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reuter G. 1963. Comparative studies on the bifidus flora in the feces of infants and adults. With a contribution to classification and nomenclature of bifidus strains. Zentralbl. Bakteriol. Orig. 191:486–507 (In German.) [PubMed] [Google Scholar]
- 50.Scardovi V, Sgorbati B. 1974. Electrophoretic types of transaldolase, transketolase, and other enzymes in bifidobacteria. Antonie Van Leeuwenhoek 40:427–440. 10.1007/BF00399355 [DOI] [PubMed] [Google Scholar]
- 51.Mitsuoka T. 1969. Comparative studies on bifidobacteria isolated from the alimentary tract of man and animals (including descriptions of Bifidobacterium thermophilum nov. spec. and Bifidobacterium pseudolongum nov. spec.). Zentralbl. Bakteriol. Orig. 210:52–64 (In German.) [PubMed] [Google Scholar]
- 52.Masco L, Ventura M, Zink R, Huys G, Swings J. 2004. Polyphasic taxonomic analysis of Bifidobacterium animalis and Bifidobacterium lactis reveals relatedness at the subspecies level: reclassification of Bifidobacterium animalis as Bifidobacterium animalis subsp. animalis subsp. nov. and Bifidobacterium lactis as Bifidobacterium animalis subsp. lactis subsp. nov. Int. J. Syst. Evol. Microbiol. 54:1137–1143. 10.1099/ijs.0.03011-0 [DOI] [PubMed] [Google Scholar]
- 53.Scardovi V, Trovatelli LD. 1969. New species of bifid bacteria from Apis mellifica L. and Apis indica F. A contribution to the taxonomy and biochemistry of the genus Bifidobacterium. Zentralbl. Bakteriol. Parasitenkd. Infektionskr. Hyg. 123:64–88 [PubMed] [Google Scholar]
- 54.Tissier H. 1900. Recherchers sur la flora intestinale normale et pathologique du nourisson. University of Paris, Paris, France [Google Scholar]
- 55.Scardovi V, Trovatelli LD, Biavati B, Zani G. 1979. Bifidobacterium cuniculi, Bifidobacterium choerinum, Bifidobacterium boum, and Bifidobacterium pseudocatenulatum: four new species and their deoxyribonucleic and homology relationships. Int. J. Syst. Evol. Bacteriol. 29:291–311. 10.1099/00207713-29-4-291 [DOI] [Google Scholar]
- 56.Scardovi V, Crociani F. 1974. Bifidobacterium catenulatum, Bifidobacterium dentium, and Bifidobacterium angulatum: three new species and their deoxyribonucleic and homology relationships. Int. J. Syst. Evol. Bacteriol. 24:6–20. 10.1099/00207713-24-1-6 [DOI] [Google Scholar]
- 57.Delcenserie V, Gavini F, Beerens H, Tresse O, Franssen C, Daube G. 2007. Description of a new species, Bifidobacterium crudilactis sp. nov., isolated from raw milk and raw milk cheeses. Syst. Appl. Microbiol. 30:381–389. 10.1016/j.syapm.2007.01.004 [DOI] [PubMed] [Google Scholar]
- 58.Lauer E. 1990. Bifidobacterium gallicum sp. nov. isolated from human feces. Int. J. Syst. Bacteriol. 40:100–102. 10.1099/00207713-40-1-100 [DOI] [PubMed] [Google Scholar]
- 59.Watabe J, Benno Y, Mitsuoka T. 1983. Bifidobacterium gallinarum sp. nov., a new species isolated from the ceca of chickens. Int. J. Syst. Evol. Microbiol. 33:127–132 [Google Scholar]
- 60.Morita H, Nakano A, Onoda H, Toh H, Oshima K, Takami H, Murakami M, Fukuda S, Takizawa T, Kuwahara T, Ohno H, Tanabe S, Hattori M. 2011. Bifidobacterium kashiwanohense sp. nov., isolated from healthy infant faeces. Int. J. Syst. Evol. Microbiol. 61:2610–2615. 10.1099/ijs.0.024521-0 [DOI] [PubMed] [Google Scholar]
- 61.Matteuzzi D, Crociani F, Zani G, Trovatelli LD. 1971. Bifidobacterium suis n. sp.: a new species of the genus Bifidobacterium isolated from pig feces. Z. Allg. Mikrobiol. 11:387–395. 10.1002/jobm.3630110504 [DOI] [PubMed] [Google Scholar]
- 62.Biavati B, Mattarelli P. 1991. Bifidobacterium ruminantium sp. nov. and Bifidobacterium merycicum sp. nov. from the rumens of cattle. Int. J. Syst. Bacteriol. 41:163–168. 10.1099/00207713-41-1-163 [DOI] [PubMed] [Google Scholar]
- 63.Scardovi V, Trovatelli LD. 1974. Bifidobacterium animalis (Mitsuoka) comb. nov. and minimum and subtile groups of new Bifidobacteria found in sewage. Int. J. Syst. Bacteriol. 24:21–28. 10.1099/00207713-24-1-21 [DOI] [Google Scholar]
- 64.Watanabe K, Makino H, Sasamoto M, Kudo Y, Fujimoto J, Demberel S. 2009. Bifidobacterium mongoliense sp. nov., from airag, a traditional fermented mare's milk product from Mongolia. Int. J. Syst. Evol. Microbiol. 59:1535–1540. 10.1099/ijs.0.006247-0 [DOI] [PubMed] [Google Scholar]
- 65.Tsuchida S, Takahashi S, Mbehang Nguema PP, Fujita S, Kitahara M, Yamagiwa J, Ngomanda A, Ohkuma M, Ushida K. 2014. Bifidobacterium moukalabense sp. nov. isolated from the faeces of wild west lowland gorilla (Gorilla gorilla gorilla) in Gabon. Int. J. Syst. Evol. Microbiol. 64:449–455. 10.1099/ijs.0.055186-0 [DOI] [PubMed] [Google Scholar]
- 66.Scardovi V, Trovatelli LD, Crociani F, Sgorbati B. 1969. Bifid bacteria in bovine rumen. New species of the genus Bifidobacterium: B. globosum n.sp. and B. ruminale n.sp. Arch. Mikrobiol. 68:278–294 [PubMed] [Google Scholar]
- 67.Simpson PJ, Ross RP, Fitzgerald GF, Stanton C. 2004. Bifidobacterium psychraerophilum sp. nov. and Aeriscardovia aeriphila gen. nov., sp. nov., isolated from a porcine caecum. Int. J. Syst. Evol. Microbiol. 54:401–406. 10.1099/ijs.0.02667-0 [DOI] [PubMed] [Google Scholar]
- 68.Trovatelli LD, Crociani F, Pedinotti M, Scardovi V. 1974. Bifidobacterium pullorum sp. nov.: a new species isolated from chicken feces and a related group of bifidobacteria isolated from rabbit feces. Arch. Microbiol. 98:187–198. 10.1007/BF00425281 [DOI] [PubMed] [Google Scholar]
- 69.Hoyles L, Inganas E, Falsen E, Drancourt M, Weiss N, McCartney AL, Collins MD. 2002. Bifidobacterium scardovii sp. nov., from human sources. Int. J. Syst. Evol. Microbiol. 52:995–999. 10.1099/ijs.0.02065-0 [DOI] [PubMed] [Google Scholar]
- 70.Kim MS, Roh SW, Bae JW. 2010. Bifidobacterium stercoris sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 60:2823–2827. 10.1099/ijs.0.019943-0 [DOI] [PubMed] [Google Scholar]
- 71.Zhu L, Li W, Dong X. 2003. Species identification of genus Bifidobacterium based on partial HSP60 gene sequences and proposal of Bifidobacterium thermacidophilum subsp. porcinum subsp. nov. Int. J. Syst. Evol. Microbiol. 53:1619–1623. 10.1099/ijs.0.02617-0 [DOI] [PubMed] [Google Scholar]
- 72.Dong X, Xin Y, Jian W, Liu X, Ling D. 2000. Bifidobacterium thermacidophilum sp. nov., isolated from an anaerobic digester. Int. J. Syst. Evol. Microbiol. 50:119–125. 10.1099/00207713-50-1-119 [DOI] [PubMed] [Google Scholar]
- 73.Okamoto M, Benno Y, Leung KP, Maeda N. 2008. Bifidobacterium tsurumiense sp. nov., from hamster dental plaque. Int. J. Syst. Evol. Microbiol. 58:144–148. 10.1099/ijs.0.65296-0 [DOI] [PubMed] [Google Scholar]
- 74.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M, Sanchez B, Viappiani A, Mancabelli L, Taminiau B, Delcenserie V, Barrangou R, Margolles A, van Sinderen D, Ventura M. 2014. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl. Environ. Microbiol. 80:6290–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material