Noninvasive optical inhibition with a red-shifted microbial rhodopsin (original) (raw)
. Author manuscript; available in PMC: 2014 Oct 3.
Published in final edited form as: Nat Neurosci. 2014 Jul 6;17(8):1123–1129. doi: 10.1038/nn.3752
Abstract
Optogenetic inhibition of the electrical activity of neurons enables the causal assessment of their contributions to brain functions. Red light penetrates deeper into tissue than other visible wavelengths. We present a red-shifted cruxhalorhodopsin, Jaws, derived from Haloarcula (Halobacterium) salinarum (strain Shark) and engineered to result in red light–induced photocurrents three times those of earlier silencers. Jaws exhibits robust inhibition of sensory-evoked neural activity in the cortex and results in strong light responses when used in retinas of retinitis pigmentosa model mice. We also demonstrate that Jaws can noninvasively mediate transcranial optical inhibition of neurons deep in the brains of awake mice. The noninvasive optogenetic inhibition opened up by Jaws enables a variety of important neuroscience experiments and offers a powerful general-use chloride pump for basic and applied neuroscience.
Optogenetic inhibition, the use of light-activated ion pumps to enable transient activity suppression of genetically targeted neurons by pulses of light1–3, is valuable for the causal parsing of neural circuit component contributions to brain functions and behaviors. A major limit to the utility of optogenetic inhibition is the addressable quantity of neural tissue. Previous optogenetic hyperpolarizing proton pumps (Arch1, ArchT3, Mac1) and chloride pumps (eNpHR4, eNpHR3.0 (ref. 2)) have successfully inhibited volumes of approximately a cubic millimeter, but many neuroscience questions require the ability to suppress larger tissue volumes. A number of pharmacogenetic, chemical and genetic strategies have been used for this purpose5–7, but it would ideally be possible to address these large brain volumes with the millisecond temporal precision of optogenetic tools.
Another common desire in optogenetic experiments is to minimize invasiveness from inserting optical fibers into the brain, which displaces brain tissue and can lead to side effects such as brain lesion, neural morphology changes, glial inflammation and motility, or compromise of asepsis8–10. Less invasive strategies that do not require an implanted optical device would also increase experimental convenience and enable longer timescale experiments than often feasible with fragile implants. While a number of previous studies using channelrhodopsins have attempted to address this problem11–16, noninvasive optical inhibition has not yet been possible.
To enable noninvasive large-volume optogenetic inhibition, we engineered and characterized Jaws, a spectrally shifted cruxhalorhodopsin derived from the species H. salinarum (strain Shark)17, which mediates strong red light–driven neural inhibition. Jaws is capable of powerful optical hyperpolarization in a variety of neuroscientific contexts: it successfully enabled suppression of visually evoked neural activity in mice, functioned in cone photoreceptors to restore greater light sensitivity in mouse models than possible with previous opsins and enabled the noninvasive transcranial inhibition of neurons in brain structures up to 3 mm deep. This new reagent thus makes a variety of important experiments amenable to optogenetic investigation.
RESULTS
Engineering a red light–sensitive chloride pump
In earlier work, we identified two cruxhalorhodopsins from the haloarcula H. marismortui and H. vallismortis that possessed the most red-shifted action spectra known for any hyperpolarizing opsins1,18. While their low photocurrents made them poor candidates for in vivo use1, their spectra suggested they might be good scaffolds for further engineering. Because red light is less absorbed by hemoglobin than blue, green or yellow wavelengths, we reasoned this red-light sensitivity might render deep brain regions more accessible. We validated this through Monte Carlo modeling and direct measurement in the live mouse brain (Supplementary Fig. 1).
We therefore screened members of the cruxhalorhodopsin class in primary neuronal culture to identify molecules with both red-shifted action spectra and robust photocurrents (Fig. 1a and Supplementary Fig. 2) and identified a hyperpolarizer we named Halo57, the cruxhalorhodopsin from H. salinarum (strain Shark), with less than 60% homology to the Natronomonas pharaonis halorhodopsin (NpHR/halo; Supplementary Fig. 3a). We subsequently engineered Halo57 by identifying K200R and W214F point mutations19,20 (Fig. 1b), which significantly boosted photocurrents without altering its red action spectrum (n = 9 or 10 cells; P = 0.02, ANOVA with Dunnett’s post hoc test), and by appending the KGC21 or ER2 (ref. 22) trafficking sequences from the potassium channel Kir2.1 to result in the final molecules Jaws (KGC + ER2) and Jaws-ER2 (ER2) (Fig. 1c and Supplementary Fig. 3b). In accord with the low sequence conservation, the two homologous point mutations did not enhance the N. pharaonis halorhodopsin (Supplementary Fig. 3a,c,d). Ion-specific solutions confirmed Jaws to be a light-driven chloride pump (Supplementary Fig. 3e,f).
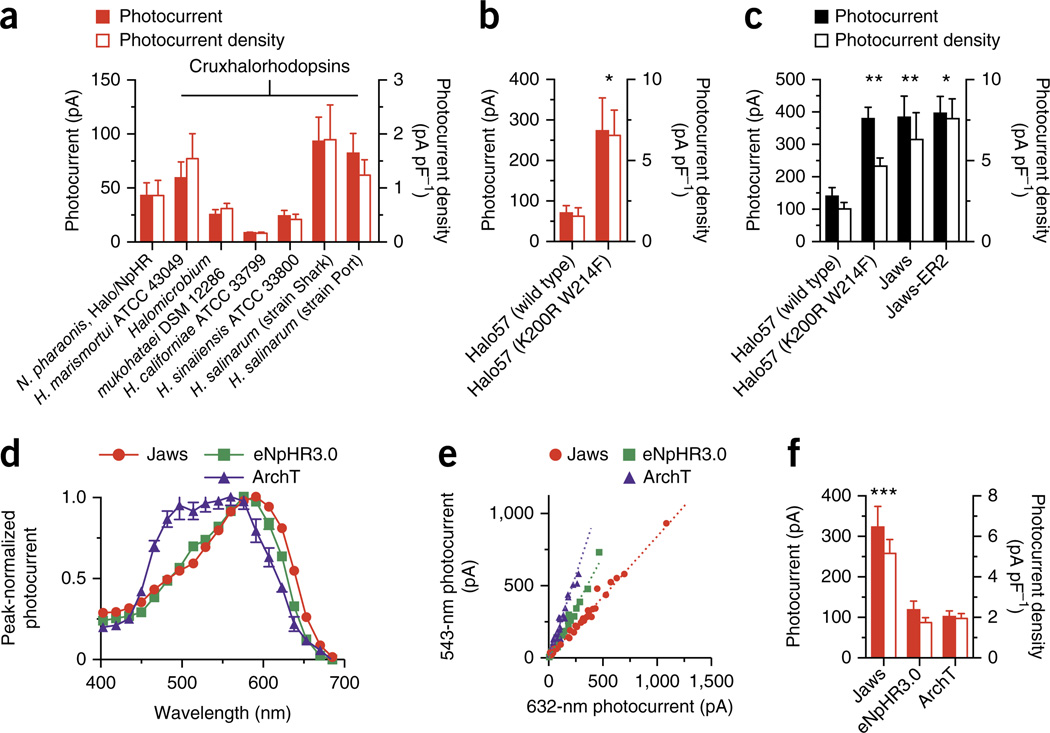
Figure 1.
Engineering and in vitro characterization of Jaws, a red-shifted cruxhalorhodopsin. (a) Red light photocurrents (left _y_-axis, red bars) and photocurrent densities (right _y_-axis, white bars) from cruxhalorhodopsins and the N. pharaonis halorhodopsin in cultured neurons (632 nm, 5 mW/mm2; n = 6 cells for each). ATCC, American Type Culture Collection; DSM, German Collection of Microorganisms and Cell Cultures. (b) The K200R W214F Halo57 mutant (n = 9 cells) demonstrates enhanced photocurrents over wild-type Halo57 in cultured neurons (n = 10 cells; 632 nm, 5 mW/mm2). (c) Photocurrents (black bars) and photocurrent densities (white bars) for wild-type Halo57 (n = 9 cells), Halo57 (K200R W214F) (n = 21 cells), Jaws (n = 11 cells) and Jaws-ER2 in cultured neurons (n = 5 cells; 5 mW/mm2, 543 nm; P < 0.01 for all variants compared to the wild-type Halo57). (d) Action spectra for Jaws (n = 12 cells), ArchT (n = 13 cells) and eNpHR3.0 (n = 15 cells; measured in cultured neurons using equal photon fluxes of ~3.0 × 1021 photons s−1 m−2). (e) Red:green photocurrent ratios for Jaws (n = 33 cells), ArchT (n = 36 cells) and eNpHR3.0 in cultured neurons (n = 29 cells; 5 mW/mm2 at 632 nm or 543 nm). Regression lines are shown for each opsin, indicating distinct spectral shifts. (f) Red-light mediated photocurrents (left _y_-axis, red bars) and photocurrent densities (right _y_-axis, white bars) for Jaws (n = 33 cells), ArchT (n = 36 cells) and eNpHR3.0 in cultured neurons (n = 29 cells; 5 mW/mm2 at 632 nm; P < 0.001 for Jaws compared to ArchT or eNpHR3.0). Data in each panel were measured from 2 batches of neuron culture. Values throughout are mean ± s.e.m. In b, *P = 0.020; in c, from left to right, **P = 0.0017, **P = 0.0047, *P = 0.0183; in f, ***P < 0.001. Unpaired _t_-tests in b, ANOVA in c with Dunnett’s post hoc test using wild-type Halo57 as the reference, ANOVA in f with Newman-Keuls post hoc test. t = 2.567 for b; F = 5.473 for c and 14.02 for f.
In vivo optogenetic inhibition has previously only been possible with blue, green and yellow light, as existing optogenetic inhibitors operate at only 10–30% of their peak capacity at red wavelengths. Because Jaws has a 14-nm red shift relative to the N. pharaonis halorhodopsin (NpHR/halo, eNpHR, eNpHR3.0), we hypothesized that it might be a good candidate for potent red-light neural inhibition (Fig. 1d,e). To further characterize this, we transfected Jaws, eNpHR3.0 or ArchT into primary neuron cultures along with a secondary cytosolic tdTomato plasmid and selected cells for whole-cell patch clamp characterization solely on the basis of the presence of tdTomato (Supplementary Fig. 4), to prevent selection bias for opsin expression levels and more accurately represent the full range of in vivo expression: we found Jaws to have significantly higher red light (632 nm) photocurrents than eNpHR3.0 or ArchT across all tested light powers (Fig. 1f and Supplementary Fig. 4d; n = 21–30 cells from 2 batches of culture; P < 0.01 for 0.1–20 mW/mm2, ANOVA with Newman-Keuls post hoc test). The robust in vitro photocurrents we observed seemed promising, so we next moved to assess Jaws in a variety of in vivo contexts.
Potential for optogenetic visual therapeutics
Optogenetic neural hyperpolarization is being explored not only as a tool for basic neuroscience, but also as a prototype therapeutic. As one example, retinitis pigmentosa is a visual disorder that results first in night blindness and then in overall blindness as a result of photoreceptor degeneration23. A potential therapy for patients with cone photoreceptor atrophy would be to resensitize the cone cells to light by genetically expressing light-activated hyperpolarizing ion pumps in the cone photoreceptors, which in their healthy state hyperpolarize in response to optical stimulation.
When expressed in retinal cones of retinitis pigmentosa mouse models, eNpHR has previously been shown to be capable of transducing spikes in downstream retinal ganglion cells and mediating visually guided behaviors. However, the resultant spiking rates are limited to less than 200 Hz, substantially less than the full wild-type range of 15–450 Hz (ref. 24), and thus potentially limit fine visual perception in future clinical use. Additionally, while eNpHR can be activated using light powers that are safe for human use24, more light-sensitive hyperpolarizers would require less light for stimulation and thus provide a greater margin of efficacy and safety given the inevitable unknowns that might crop up in potential future trials in humans. Thus, better neural hyperpolarizers remain of great interest for potential clinical use in the human eye.
We decided to first compare Jaws’s performance against those of other known hyperpolarizers by injecting adeno-associated virus (AAV), serotype 8 (AAV8), encoding various light-driven proton or chloride pumps into the cone photoreceptors of Pde6brd1 mice, also called fast retinal degeneration mice24,25 (Fig. 2a and Supplementary Fig. 5b), which are blind by postnatal day (P) 28. We conducted extracellular retinal ganglion cell recordings (Fig. 2b) 4–6 weeks after injection (P64–75) and found peak wavelength photostimulation of Jaws-expressing photoreceptors to induce significantly more retinal ganglion spiking, indicated by mean spiking frequencies greater than those for eNpHR, ArchT, Mac or the wild-type Halo57 (Fig. 2c and Supplementary Fig. 5a). Jaws also enabled a broader peak spike frequency distribution than other opsins (P < 0.05 for Jaws versus all other opsins, Kolmogorov-Smirnov test), with a mean frequency of 67.8 ± 7.0 Hz with eNpHR versus 189.5 ± 15.9 Hz with Jaws (values throughout are mean ± s.e.m. unless otherwise stated), increasing the spiking bandwidth by threefold (Fig. 2d). We additionally observed Jaws to mediate the highest ganglion spiking rates over various irradiances of 600 nm, 550 nm and 470 nm light (Fig. 2e and Supplementary Fig. 5c,d), indicating its high light sensitivity, which alongside Jaws’s spectral peak at 600 nm may confer benefits because of the relative safety of 600-nm light26. From this comparison of light-sensitive hyperpolarizers, we conclude that Jaws may represent an optogenetic reagent with molecular properties better suited than previous opsins for therapeutic cone reactivation for a subset of patients with retinitis pigmentosa.
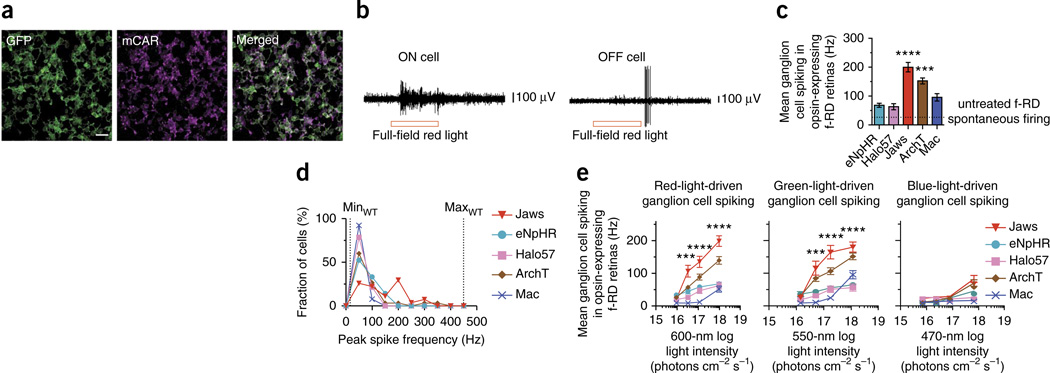
Figure 2.
Jaws-mediated light responses in mouse retinitis pigmentosa retinas. (a) Confocal fluorescence images from Jaws-GFP expressing retina with the fast form of retinal degeneration (f-RD), stained for GFP (green) and mouse cone arrestin (mCAR) (magenta), 4 weeks after injection with AAV8-mCAR-Jaws-GFP virus. Scale bar, 20 µm. (b) Raw traces recorded from ON and OFF retinal ganglion cells in retinas expressing Jaws in cone photoreceptors, optically stimulated with 9.6 × 1017 photons cm−2 s−1 at 600 nm for 1 s. (c) Comparison of mean spiking in ganglion cells downstream of neurons expressing eNpHR (n = 21 cells from 1 mouse), Halo57 (n = 14 cells from 1 mouse), ArchT (n = 30 cells from 1 mouse), Mac (n = 13 cells from 2 mice) or Jaws (n = 27 cells from 1 mouse) at opsin peak wavelength sensitivity. Light intensity was 1.2 × 1018 photons cm−2 s−1 for green light (ArchT, Mac) and 9.6 × 1017 photons cm−2 s−1 for red light (Jaws, eNpHR, Halo57). Dotted line indicates baseline f-RD firing per ref. 49. (d) Population distribution of retinal ganglion cell peak firing rates at opsin peak wavelength sensitivity. Light intensities as indicated in c; dotted lines indicate the minimum (minWT) and maximum (maxWT) of the wild-type dynamic range24. (e) Retinal ganglion spike rate versus red, green and blue irradiances, measured in ganglion cells downstream of opsin-expressing cones in f-RD retina. Values are means ± s.e.m.; n values for c–e are as indicated in c. In c–e, ***P < 0.001, *P < 0.0001. ANOVAs in c,e with Dunnett’s post hoc test using eNpHR as the reference; Kolmogorov-Smirnov test in d. F = 22.13 for c and 37.08, 32.53 and 10.43 for e. In c, ****P < 0.0001 for Jaws, ***P < 0.001 for ArchT, P = 0.9973 for Halo57 and P = 0.4929 for Mac. In d, as compared to Jaws, P < 0.0001 for eNpHR, P < 0.0001 for Halo57, P = 0.0375 for ArchT and P = 0.0008 for Mac. In eP = 0.4679, P = 0.001, P < 0.0001, P < 0.0001 for green and P = 0.2703, P = 0.001, P < 0.0001 and P < 0.0001 for red, from lowest to highest irradiance.
In vivo suppression of visually evoked cortical activity
We next evaluated the performance of Jaws in a common neuroscientific experimental context: the suppression of stimulus-evoked neural activity. Given the proximity of yellow light to all existing inhibitory opsins’ peak excitation wavelengths1,2, we decided, for this specific experiment, to directly compare Jaws’s in vivo performance in the mouse brain against that of other inhibitors, using yellow light. Although all previous studies characterizing optogenetic hyperpolarizers have focused on inhibiting spontaneous neural activity1–4, many neuroscientific questions focus on stimulus-evoked or event-associated neural activity. The ability to suppress the activity of a given cell type responding to behaviorally relevant inputs is therefore critical.
To assess Jaws’s potential as a general-purpose inhibitor of stimulus-evoked neural activity, we injected a Cre recombinase–dependent AAV, serotype 5 (AAV5-FLEX), expressing eNpHR, eNpHR3.0, Arch, ArchT or Jaws into the primary visual cortex of Emx1-cre transgenic mice and delivered visual stimuli while delivering yellow light with an optetrode (593 nm; 35 mW/mm2 out of a 200 µm fiber tip). Inhibition of visually evoked neural activity was strong (Fig. 3a,b), with an 86 ± 3% reduction of visually evoked activity in Jaws-expressing cortex (Fig. 3c; from 4 mice, n = 27 units with a significant reduction in firing; n = 3 units showed no change and n = 3 units showed a significant increase in firing) over a range of evoked firing rates (1.8–12.5 Hz; Supplementary Fig. 6a), and consistent across a range of input contrast strengths (Fig. 3d). This comparison of neural inhibitors thus reveals that Jaws is capable of mediating excellent inhibition of evoked neural activity in a biologically meaningful context.
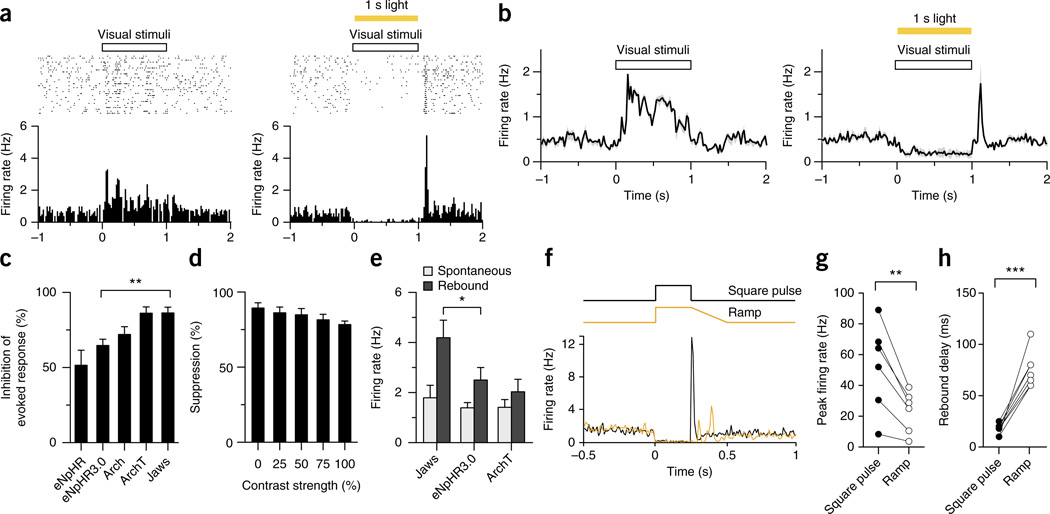
Figure 3.
Jaws-mediated inhibition of evoked responses in visual cortex. (a) Representative rasters of visually evoked responses (top left) and Jaws-mediated inhibition of visually evoked responses (top right), and post-stimulus time histograms for visually evoked (bottom left) and Jaws-inhibited visually evoked (bottom right) responses, of a representative neuron (n = 27) in the visual cortex of an anesthetized Emx1-cre mouse, as measured by extracellular tetrode recording (AAV5-FLEX virus; 35 mW/mm2 at 593 nm using a 200-µm fiber). (b) Population average of five simultaneously recorded neurons showing visually evoked responses (left) and Jaws-mediated inhibition of visually evoked responses (right). (c) Inhibition of visually evoked neural responses for eNpHR (n = 8 units from 2 mice), eNpHR3.0 (n = 32 units from 4 mice), Arch (n = 18 units from 3 mice), ArchT (n = 21 units from 3 mice) and Jaws (n = 27 units from 4 mice; P < 0.01 for Jaws versus eNpHR3.0). (d) Jaws-mediated inhibition of visually evoked responses (n = 14 units from 2 mice) for different visual input stimulus strengths. (e) Comparison of spontaneous and immediately post-illumination firing rates for Jaws, eNpHR3.0 and ArchT (n = 32 units from 4 mice for eNpHR3.0, n = 21 units from 3 mice for ArchT, n = 5 units for Jaws from 2 mice; P < 0.05 for Jaws versus eNpHR3.0). (f) Post-stimulus time histogram for a standard, step light pulse (black line) versus ramped illumination (yellow line), for a spontaneously firing visual cortex neuron. (g,h) Comparison of peak firing rates (g) and rebound delay rates (h) for step versus ramped illumination (n = 16 units from 3 mice). Values throughout are mean ± s.e.m. For c,e,g,h, *P < 0.05, **P < 0.01, ***P < 0.001. ANOVAs with Newman-Keuls post hoc tests in c,e; paired _t_-tests in g,hF = 7.379 for c, 2.826 for e; t = 4.485 for g and 9.078 for h.
A rebound burst of action potentials is common after illumination of cells expressing halorhodopsins18,27–30 or archaerhodopsins18,27,31, which respectively pump chloride inward and protons outward. A variety of possible mechanisms have been proposed, including hyperpolarization-activated _I_h currents29,30,32 or changes in chloride reversal potential due to intracellular chloride accumulation33. We similarly observed post-illumination rebound in Jaws-, eNpHR3.0- and ArchT-transduced neurons, and Jaws produced greater effects than eNpHR3.0 or ArchT (Fig. 3e; n = 8–32 units; P < 0.05, ANOVA with Newman-Keuls post hoc test). We attempted to ameliorate this by gradually ramping down illumination over a duration of 200–1,000 ms and found that the mean rebound instantaneous firing rate dropped significantly, from 52 ± 12 Hz to 23 ± 6 Hz (n = 16 units; P = 0.0065, paired _t_-test), suggesting that sculpting of light pulses may enable rebound reduction. Notably, the rebound delay also significantly lengthened, from 18 ± 6 ms to 76 ± 6 ms (n = 16 units; P < 0.001, paired _t_-test) and became more temporally dispersed (Fig. 3f–h and Supplementary Fig. 6b). Like all previous inhibitors, Jaws does yield post-illumination rebound, which likely has implications for careful experiment design comparable to those for earlier inhibitors such as eNpHR3.0. Further attention to light-pulse shape may be merited as a way to reduce rebound for all optogenetic inhibitors.
Red-light performance of Jaws
We next assessed Jaws’s red-light properties via whole-cell patch-clamp recordings in acute cortical slices, as well as extracellular recordings in awake, head-fixed mice. We injected AAV8 encoding Jaws under the Ca2+/calmodulin-dependent protein kinase (Camk2a) promoter or a truncated human synapsin (SYN1) promoter, hereafter referred to as hSyn16, or Jaws-ER2 under the hSyn promoter, into motor cortex and observed robust red light photocurrents, hyperpolarization and neural inhibition (Fig. 4a–c and Supplementary Fig. 7a,c), with ~95% reduction of neural activity achievable in neurons illuminated by a 200-µm fiber (130 mW/mm2 fiber tip irradiance at 635 nm; Supplementary Fig. 8). Jaws’s kinetic properties were comparable to those of other optogenetic inhibitors (Supplementary Fig. 4e and 7c), and its expression did not alter basal cell properties in cultured cortical neurons (Supplementary Fig. 4g), or in cortical or dentate granule neurons in acute brain slice (Supplementary Fig. 7d and 9).
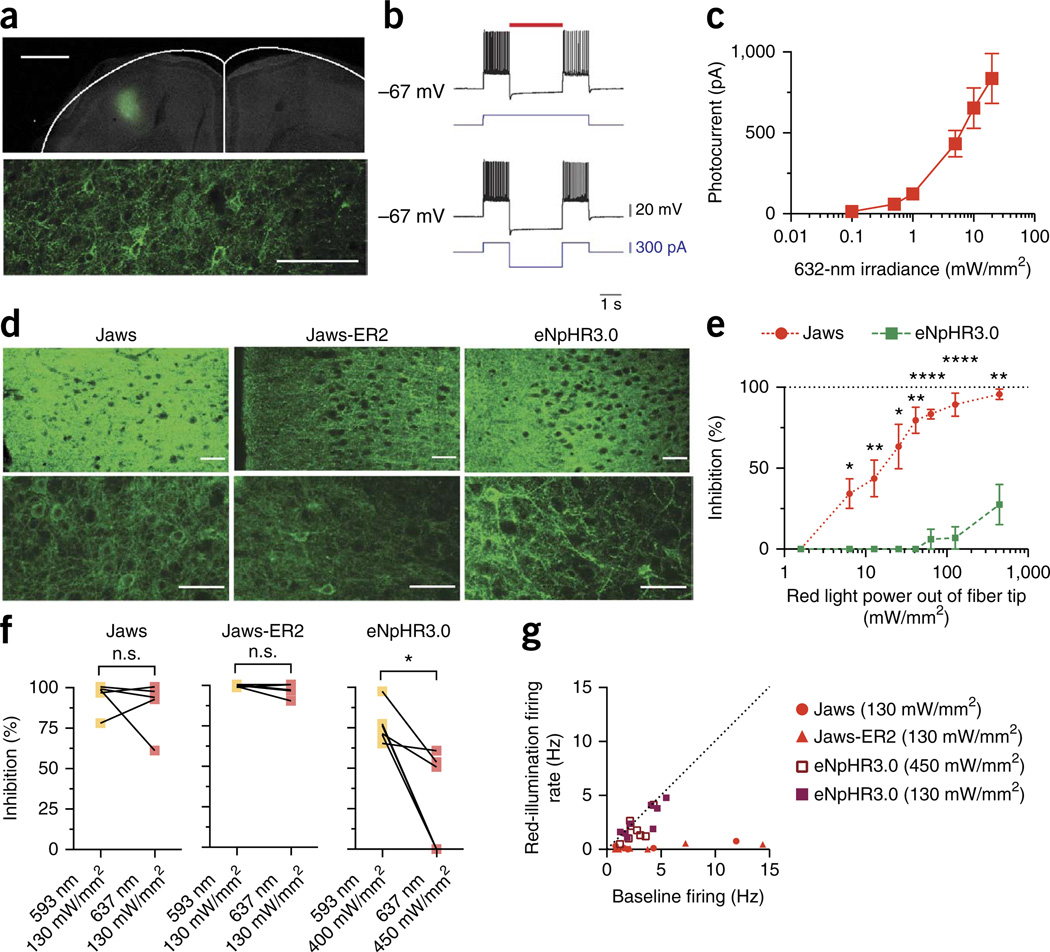
Figure 4.
Jaws-mediated red light inhibition in rodent cortex. (a) Confocal fluorescence images from Jaws-GFP–expressing motor cortex, 6 weeks after injection. Outline indicates brain boundary50. Scale bar, 1 mm (top) or 100 µm (bottom). (b) Representative current-clamp recording of Jaws-expressing neuron undergoing optically evoked (top, 632 nm, 5 mW/mm2) or electrically evoked (bottom) hyperpolarization in acute cortical slice 6 weeks after injection. Red bar indicates optical illumination; blue trace indicates electrical current injection. (c) Quantification of Jaws photocurrents as a function of red light irradiance in acute slice (n = 16 cells from 4 mice). (d) Confocal fluorescence images of Jaws-GFP-, Jaws-GFP-ER2- or eNpHR3.0-expressing motor cortex. Scale bars, 100 µm. (e) Side-by-side comparison of red light–driven inhibition of spontaneous neural activity in motor cortex (n = 6 units from 2 mice for each opsin; light powers were measured at the tip of a 200-µm fiber; 637 nm). (f) Red-light inhibition was equivalent to yellow-light inhibition for Jaws (n = 5 units from 2 mice; P = 0.4685) and Jaws-ER2 (n = 6 units from 2 mice; P = 0.1952), but substantially less potent for eNpHR3.0 (n = 6 units from 2 mice; P = 0.0121). (g) Red light efficaciously inhibited neurons expressing Jaws (n = 5 units from 2 mice) and Jaws-ER2 (n = 6 units from 2 mice) but not eNpHR3.0 (n = 8 units from 2 mice) over a range of firing rates. Values throughout are mean ± s.e.m. In e,f, n.s., not significant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Unpaired _t_-test in e, paired _t_-test in f. From left to right, t = 0.2902, 3.264, 3.494, 2.422, 5.422, 11.4, 9.275 and 4.338 for e and 0.6205, 3.843 and 1.496 for f. In eP = 0.031, 0.0068, 0.0452, 0.0016, <0.0001, <0.0001 and 0.0025 from lowest to highest irradiance. In fP = 0.4685 for Jaws, P = 0.1952 for Jaws-ER2 and *P = 0.0121 for eNpHR3.0.
Having established Jaws as a potent red-light-drivable inhibitor, we next assessed its in vivo performance compared to that of eNpHR3.0. We did titer-matched injections of 2 × 109 viral particles of AAV8-hSyn-Jaws, AAV8-hSyn-Jaws-ER2 or AAV8-hSyn-eNpHR3.0 into mouse motor cortex (Fig. 4d) and performed awake head-fixed recordings to characterize each hyperpolarizer over different red (637-nm) light powers (Fig. 4e). Yellow light (593 nm) was used as a positive control to identify opsin-expressing neurons, owing to its proximity to both Jaws’s and eNpHR3.0’s spectral peaks, and we conducted paired recordings of individual opsin-expressing neurons to assess their performance under different wavelengths (Fig. 4f ). As expected, both Jaws variants robustly inhibited spontaneous neural activity when illuminated with either red (88.8 ± 15.9% inhibition for Jaws, 97.2 ± 3.9% inhibition for Jaws-ER2) or yellow wavelengths (94.2 ± 9.3% inhibition for Jaws, 99.0 ± 1.6% inhibition for Jaws-ER2), while eNpHR3.0’s red-light inhibition dropped significantly to only 27.5 ± 30.3% relative to its peak yellow-light performance of 76.1 ± 11.1% inhibition (Fig. 4f; n = 5 or 6 units, P = 0.0121), and we found these red-light results to hold over a range of firing rates (Fig. 4g; n = 5–8 units for each opsin).
Noninvasive optogenetic inhibition
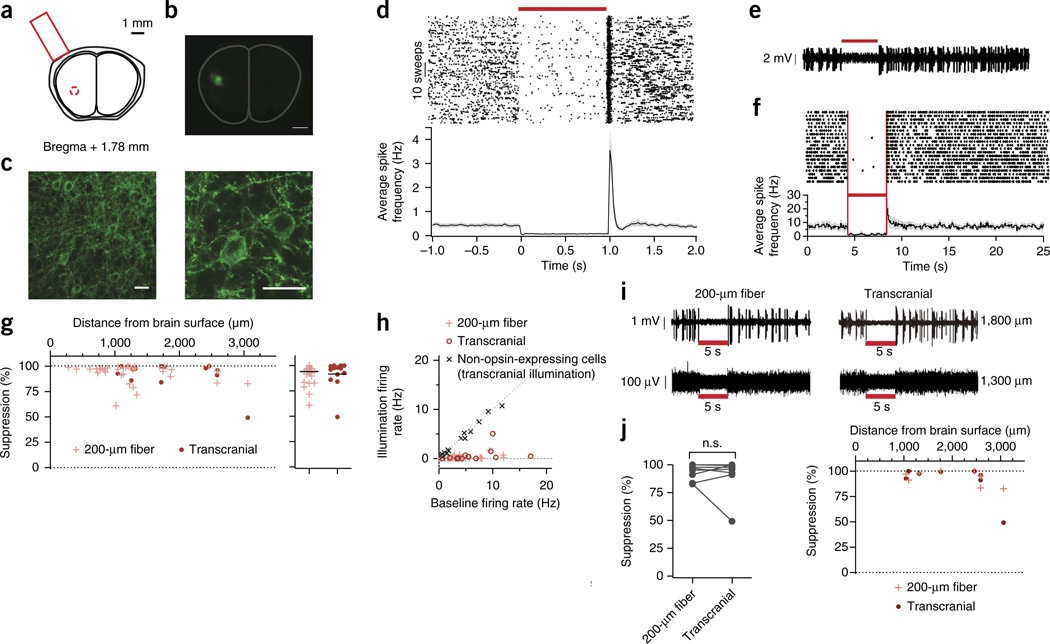
Having demonstrated Jaws’s efficacy as a red-light-drivable reagent in a variety of contexts, we next sought to determine its potency as a noninvasive and long-distance neural inhibitor. Using red illumination (Fig. 5a), we found we could transdurally inhibit neurons in the medial prefrontal cortex of a mouse, with a 86 ± 13% decrease (mean ± s.d.) in firing for neurons showing suppression (n = 66 units across 2 anesthetized mice; Fig. 5d). Using a similar strategy, we successfully inhibited neurons in the awake mouse cortex by delivering red light transcranially through the intact skull (635 nm, 10 mW/mm2 out of a 1,500 µm fiber tip; Fig. 5a,b) to regions 1–3 mm below the brain’s surface (Fig. 5c,e,f). Transcranial inhibition in the motor, somatosensory, insular and piriform cortices was efficient, with a 92 ± 14% decrease in firing for neurons showing suppression (mean ± s.d.; from 7 mice, n = 13 units; n = 9 units showed no change and n = 3 units showed an increase in activity; Fig. 5f).
Figure 5.
Noninvasive red-light inhibition of neural activity. (a) Schematic of noninvasive red light delivery through the intact skull. (b,c) Fluorescence images from mouse insular cortex (b; scale bar, 1 mm) and motor cortex (c; scale bars, 25 µm) 6 weeks after injection with AAV8-hSyn-Jaws virus. Outline indicates brain boundary50. (d) Raster plot (top) and population average (bottom) of transdural inhibition of neurons in the medial prefrontal cortex of anesthetized mice, showing light-induced suppression 6–8 weeks after injection with AAV8-hSyn-Jaws virus (n = 26 units from 2 mice; red bar, 1 s pulse, 25 mW/mm2 fiber tip irradiance, 635 nm). Red bars denote red light illumination. Vertical red lines in f also demarcate the illumination period. (e) Representative glass-pipette extracellular recording (n = 13) of a transcranially illuminated cortical neuron 2,700 µm below the brain surface in an awake mouse (red bar, 10 mW/mm2 fiber tip irradiance, 635 nm; 4 weeks after injection with AAV8-hSyn-Jaws virus). (f) Spike rasters taken from a representative neuron (top) and population average (bottom; n = 13 units from 6 mice) of instantaneous firing rate in neurons from awake mice undergoing transcranial light-induced suppression 4–8 weeks after injection with AAV8-hSyn-Jaws virus (5 s pulse, 10 mW/mm2 635 nm fiber tip irradiance; black line, mean; gray lines, mean ± s.e.m.). (g) Percentage reduction in neural activity for neurons recorded at different depths in awake mouse brain, 6–8 weeks after injection with AAV8-hSyn-Jaws virus. Red light delivered via traditional 200-µm fiber (n = 37 units from 11 mice) or transcranially (n = 13 units from 6 mice). (h) Firing rate averaged over the illumination period versus baseline firing rate for neurons recorded in awake mouse brain (n = 37 units from 11 mice for 200-µm-fiber illumination; n = 13 units from 6 mice for transcranial illumination; n = 14 units from 5 mice for transcranial illumination of non-opsin-expressing neurons). (i) Representative extracellular recordings of neurons 1,800 µm (top) and 1,300 µm (bottom) below the surface of the awake brain undergoing illumination via a 200-µm fiber (red bar, ~130 mW/mm2, 635 nm) 500 µm above the electrode tip (left) or transcranially (right) through the intact skull (~10 mW/mm2; 635 nm), recorded 8 weeks after injection with AAV8-hSyn-Jaws virus. (j) Transcranial and 200-µm-fiber illumination were equally efficacious (left) in inhibiting neurons (n = 8 units from 6 mice; P = 0.5894; n.s., not significant) over depths of up to 3 mm (right). Values are means ± s.e.m. Paired _t_-test in j t = 0.5655.
The efficacy of transcranial optogenetic inhibition was similar across several millimeters of the awake mouse brain (Fig. 5g) and over a variety of baseline firing rates (Fig. 5h). To directly address the possibility that opsin-expressing apical dendrites near the brain’s surface were being hyperpolarized, resulting in somatic or downstream network inhibition, we next conducted paired recordings using a standard acutely inserted 200-µm fiber as a positive control for transcranial illumination outside the mouse’s intact skull. Illumination from a 200-µm fiber has been shown to fall off rapidly within a few hundred microns of the fiber tip34,35, and the likelihood of optogenetic inhibition has been similarly demonstrated to fall off over a similar distance31. Our paired recordings revealed similar degrees of suppression over a range of 1–3 mm below the brain’s surface (Fig. 5i,j). From this, we conclude Jaws is capable of noninvasive optogenetic inhibition comparable to the current standard of invasive optical delivery.
DISCUSSION
We here report Jaws, a red light sensitive opsin with the most red-shifted spectrum of any optogenetic inhibitor known to us. Jaws enabled efficacious transcranial inhibition of neural activity in awake mouse brain in response to red light, with a similar efficacy to standard invasive light delivery with a 200-µm optical fiber. Additionally, Jaws enabled significantly more inhibition of stimulus-evoked neural activity than possible with previous halorhodopsins. Finally, it restored photosensory responses in retinitis pigmentosa retinas in a fashion achieving greater spike rates than previously achievable and with a more naturalistic frequency range, which may be important for retinal neural coding. Thus, Jaws has demonstrated utility across a wide variety of key neuroscience applications, both basic science and preclinical. We also demonstrate that light pulse sculpting can ameliorate the amplitude of the post-inhibition rebound that has been widely reported for optogenetic inhibitors.
It has been previously well established that optical penetrance of mammalian tissues in the red to near-infrared (600–1,000 nm) wavelengths is better than that of bluer wavelengths, as a result of substantially less absorption from hemoglobin, myoglobin and lipid. Despite this better red light penetrance, however, myelination may still affect optical propagation in neural tissue and should therefore be taken into account given the experimental system in question36,37. The relative inexpensiveness and stability of red light sources may additionally be of use to experimental investigators, as well as the ability to avoid potentially confounding experimental visual artifacts, since mouse visual pigments peak at approximately 380 nm (ultraviolet cones) and 500 nm (green cones), but mouse vision above 600 nm is poor38,39.
As others have reported, we observed a small number of neurons across multiple in vivo experiments that increased firing during optical illumination delivered either transcranially or from a 200-µm fiber, most likely as a result of network inhibition from upstream neurons3,40,41. It is not possible to noninvasively illuminate only deep volumes with our experimental setup without also targeting surface and intermediate layers. We therefore cannot explicitly rule out the possibility that the observed noninvasive results were the result of indirect network activity or apical dendritic illumination: a caveat for any in vivo investigation of noninvasive optogenetics. However, the robust photocurrents we observed both in vitro and in acute slices over a range of irradiances are consistent with and corroborate the functional inhibition we observed in vivo in awake mice. We therefore believe our paired recordings comparing transcranial illumination with local 200-µm fiber illumination to be highly suggestive of direct inhibition.
In scenarios where brain integrity is mandatory, noninvasive optogenetics may be invaluable in helping to prevent the many known issues associated with implants8–10. The efficacy and simplicity of transcranial inhibition could be helpful for chronic optogenetic experiments involving long-term imaging42,43, longitudinal monitoring of disease progression44, or developmental studies, during which developmental changes in brain structures might preclude the chronic implantation of a fiber at a given target45. In principle, the need for brain surgery could be eliminated altogether by using transcranial illumination with a Jaws-expressing transgenic mouse strain27. Neural inhibition in animals with large brains, such as the rhesus macaque, in which perturbation of behavior with optogenetic stimulation or inhibition of neurons has recently been demonstrated46,47, may particularly benefit from the use of Jaws. Finally, with increasing interest in the potential for therapeutic optogenetics24,48, the opsin described here may prove useful in the context of the many prototype therapies being explored.
ONLINE METHODS
Animal procedures
All procedures were in accordance with the National Institute for Laboratory Animal Research Guide for the Care and Use of Laboratory Animals and approved by the Boston University Institute Institutional Care and Use Committee (for extracellular red light experiments conducted transcranially or with a 200 um fiber), the Massachusetts Institute of Technology Committee on Animal Care (for all in vitro experiments, slice electrophysiology and light propagation measurements) or the Yale University Institute Institutional Care and Use Committee (for inhibition of evoked response and transdural red light experiments). The Swiss Veterinary Office approved all animal experiments and procedures for viral delivery of opto-genes and multi-electrode array recordings (for the retina experiments). All mice in this study were housed 3–5 per cage, maintained with a 12-h light-dark cycle, and had no previous experimental history.
Plasmid construction and site directed mutagenesis
Opsin genes were mammalian codon–optimized and synthesized (Genscript). Point mutants were generated using the QuikChange kit (Stratagene) on the opsin-GFP fusion cassette in the pEGFP-N3 backbone (Invitrogen). Cultured neuron experiments were carried out by subcloning all genes into a lentiviral backbone containing the Camk2a promoter and with a C-terminal GFP fusion. AAV vectors were constructed by subcloning opsin-GFP cassettes into AAV vectors behind the Camk2a, CAG or hSyn truncated human synapsin (SYN1) promoters. For cone photoreceptor delivery, opsin-GFP cassettes were subcloned into pAAV2-mCAR-EGFP, replacing the EGFP gene24.
All constructs were verified by sequencing, and codon-optimized sequences were submitted to GenBank (accession codes KM000925, KM000926, KM000927 and KM000928) and made available at http://syntheticneurobiology.org/protocols/.
In vitro culture, transfection and imaging
Hippocampal neuron cultures were prepared from postnatal day 0 or 1 Swiss Webster mice (Taconic), plated at a density of 16,000–20,000 per glass coverslip coated with Matrigel (BD Biosciences)51, and transfected at 3–5 d in vitro (DIV) using calcium phosphate (Invitrogen)1,3. GFP fluorescence was used to identify transfected neurons for the cruxhalorhodopsin screen and HEK293FT characterizations; all other experiments were conducted by cotransfecting with an independent tdTomato plasmid, in which case neurons were picked based solely on tdTomato fluorescence.
Whole cell patch clamp recordings were made using a Multiclamp 700B, a Digidata 1440 and a PC running pClamp (Molecular Devices). Neurons were recorded 14–24 DIV, bathed in room temperature Tyrode’s solution containing 125 mM NaCl, 2 mM KCl, 3 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 30 mM glucose, 0.01 mM NBQX and 0.01 mM GABAzine (Sigma) at pH 7.3 (NaOH-adjusted) and osmolarity 300 mOsm (sucrose-adjusted). HEK cells were bathed in an identical Tyrode’s bath solution lacking GABAzine and NBQX. No all-_trans_-retinal was supplemented for any recordings. 3–9 MΩ borosilicate glass pipettes (Warner Instruments) were pulled with a P-97 micropipette puller (Sutter Instruments) and filled with a solution containing (in mM) 125 potassium gluconate, 8 NaCl, 0.1 CaCl2, 0.6 MgCl2, 1 EGTA, 10 HEPES, 4 Mg-ATP and 0.4 Na-GTP at pH 7.3 (KOH-adjusted) and osmolarity 298 mOsm (sucrose-adjusted).
Ion selectivity tests were carried out in chloride-free recording solution containing (in mM) 125 sodium gluconate, 2 potassium gluconate, 3 CaSO4, 1 MgSO4, 10 HEPES, 30 glucose, at pH 7.3 (NaOH-adjusted) and 305–310 mOsm (sucrose-adjusted), and using intracellular solution containing (in mM) 125 potassium gluconate, 8 sodium gluconate, 0.1 CaSO4, 0.6 MgSO4, 1 EGTA, 10 HEPES, 4 Mg-ATP, 0.4 Na-GTP, pH 7.3 (KOH-adjusted), 295–300 mOsm (sucrose-adjusted).
No action spectrum data were gathered from cells voltage-clamped at −65 mV with a leak current greater than −200 pA or with a resting membrane potential more positive than −45 mV in current-clamp mode. Access resistance was 5–30 MΩ. Resting membrane potential was ~−65 mV for neurons and ~−30 mV for HEK 293FT cells in current-clamp recording. All parameters were monitored throughout recording.
All in vitro action spectra, photocurrent and voltage data were taken with a Leica DMI6000B microscope. Action spectra were taken with a monochromator (Till Photonics Polychrome V, 15 nm bandwidth centered around each value). Spectra for a given cell were taken by averaging red-to-UV (685 to 387 nm) and UV-to-red (387 to 685 nm) spectra, to eliminate history dependence. Photon fluxes for all wavelengths were ~3.0 × 1021 photons/s/m2 (0.756 mW/mm2 at 670 nm). Normalized action spectra were obtained by dividing the averaged photocurrent data by the highest observed data point.
Trafficking in vitro photocurrents were taken with a DG-4 optical switch with 300 W xenon lamp (Sutter Instruments), delivered with a 575 ± 25 nm bandpass filter (Chroma). All other in vitro photocurrents were measured using 470 nm, 530 nm or 625 nm LEDs (Thorlabs). LED spectra were bandpass filtered with the following (Semrock): 530 nm LED with 543 nm ± 11 nm filter or 625 nm LED with 632 nm ± 11 nm. All light powers were measured out of the objective lens with a PM200B photodetector (Thorlabs). Data were analyzed using Clampfit (Molecular Devices) and Matlab (MathWorks, Inc.).
AAV preparation
The AAV particles used for cone photoreceptor targeting were produced in the laboratory of B.R.52, with titers between 5.2 × 1011 and 6.8 × 1012 GC/ml. All other AAV constructs were produced by the University of North Carolina Chapel Hill Vector Core at a titer of ~6 × 1012 c.f.u./ml. All viral dilutions were carried out in phosphate-buffered saline (PBS; Life Technologies).
In vivo rodent electrophysiology
All light powers were measured and reported at the fiber tip (rather than calculated some distance away); for example, we measured 4 mW emitted from a 200-µm fiber, which we calculated and reported as 127 mW/mm2 density out of the fiber tip.
Injections for glass-pipette electrophysiology were made under isoflurane anesthesia and buprenorphine analgesia, and 1 µl AAV was injected through a craniotomy made in the mouse skull into the motor cortex (1.78 mm anterior, 1.5 mm lateral and 1.75 mm deep, relative to bregma) or the piriform, insular or somatosensory/motor cortices (1.78 mm anterior, 2.0 mm lateral and, respectively, 4.0, 3.0 and 2.0 mm deep, relative to bregma) of female C57BL6 mice 5–9 weeks old. Comparisons with eNpHR3.0 and Jaws were carried out by injecting 2 × 109 viral particles in 1 µl of an AAV-PBS mixture into motor cortex.
All viruses were injected at a rate of 0.15 µl/min through a 34-gauge injection needle, after which the needle was allowed to rest at the injection site for 10 min to allow viral diffusion. The craniotomy was marked with Examix NDS (GC America), headplates were affixed to the skull with skull screws (JL Morris), and the craniotomy and headplate were covered with dental cement (C&B Metabond).
Expression in Jaws-targeted populations of neurons in the prefrontal cortex was achieved by intracranial injection of AAV8-hSyn-Jaws in P60–120 male C57BL/6 mice. Opsins were expressed in excitatory neurons in primary visual cortex by injecting AAV5-CAG-FLEX viruses into Emx1-cre or Pvalb-cre (ref. 53) mice. Mice were anesthetized with 1.5% isoflurane and virus was injected at 0.1µl/min; mice were given 4–5 weeks for recovery.
Extracellular recordings to measure inhibition of spontaneous neural activity were made in the cortex of headfixed awake mice 1–2 months after virus injection, using 3–10 MΩ saline-filled glass microelectrodes containing silver/silver chloride electrodes1,3. Signals were amplified with a Multiclamp 700B and digitized with a Digidata 1440, using pClamp software (Molecular Devices). A 635-nm, 200-mW laser (Shanghai Laser Optics and Century) was coupled to a 200-µm-diameter optical fiber. An optical fiber was attached to the recording glass electrode, with the tip of the fiber 600 µm laterally from and 500 ± 50 µm above the tip of the electrode, and guided into the brain with a Sutter manipulator. Transcranial recordings were carried out with a 635-nm red laser coupled to a 1,500-µm-diameter optical fiber, and the recording electrode was also attached to a 200-µm-diameter fiber, coupled to a different 635-nm laser, as described above. Comparisons between Jaws and eNpHR3.0 were carried out using a 593-nm yellow laser (Shanghai Laser Optics and Century) and a 637-nm red laser (Coherent Lasers), which were both coupled into the same 200-µm-diameter optical fiber. All lasers were controlled via Digidata-generated TTL pulses. Light powers were measured with an integrating sphere (S142C, Thorlabs) and PM200B photodetector (Thorlabs).
In vivo whole cell patching was conducted in the cortex of anesthetized mice with an autopatcher54. A 200-µm fiber was coupled to a 635-nm red laser, and attached to a patch electrode as described for extracellular glass pipette recordings. Data were acquired and analyzed using pClamp software (Molecular Devices).
Extracellular recordings to measure suppression of evoked neural activity were conducted in mice anesthetized with 0.2–0.4% isoflurane and 0.8 mg/kg fentanyl. Extracellular recordings were performed with moveable arrays of tetrodes (Thomas Recording) with a 250-µm interelectrode distance. Data acquisition at 40 kHz and online spike identification by waveform analysis used Cheetah data acquisition software (Neuralynx). An optical fiber (200 or 1,500 µm) was placed on the dura next to the electrode array and connected to either a 593-nm or a 635-nm laser. To activate Jaws, 1-s pulses of light were given at 0.1 Hz at varying light intensities (10, 25, 50 or 75 mW/mm2). Visually evoked responses were driven by randomized sequences of drifting gratings at 16 orientations and a spatial frequency of 0.05 cycles/degree on a background of mean luminance. The size of the visual stimulus was optimized for the receptive fields of the recorded population. Response measurements for each neuron were taken at the optimal orientation as identified by post hoc analysis.
Data were analyzed using Matlab (MathWorks)1,3. Briefly, spikes were detected and sorted offline using Offline Sorter (Plexon) or waveform template matching and principal component analysis discrimination (KlustaKwik and custom-written Igor software, J.A.C.55). Only clusters with clear refractory periods in the autocorrelogram, indicating a single-unit source, were used for further analysis. As a further precaution against over-representation of the same unit, cross-correlograms were run for every pair of identified units in a set. Clusters that were not easily separable were discarded. Light-modulated units were identified by performing a paired _t_-test between the 1- or 5-s illumination period against the baseline firing period of the same time duration immediately before illumination, thresholding at P < 0.05. The degree of suppression by Jaws activation was calculated for each cell by dividing the mean firing rate during the light stimulus by the mean baseline firing rate during the same time duration before light stimulation onset. Rebound firing rates were calculated in a window immediately after the cessation of the light stimulus. The width of the window was determined as the duration for which firing rates were elevated >3 s.d. of the mean spontaneous firing rate.
Light onset latency to inhibition was calculated by sweeping a 20-ms sliding window to identify the earliest 20-ms period deviating from the baseline firing rate, as assessed by performing a paired _t_-test for the firing rate during each window versus during the baseline period, averaged across all trials for each neuron. The time for after-light suppression to recover back to baseline was calculated by taking the median of all trials, owing to a nonparametric distribution.
Ex vivo slice electrophysiology
For whole-cell patch clamp recordings in motor cortex, coronal 300-µm brain slices were prepared 4 or 6 weeks after viral injection of AAV8-hSyn-Jaws. Male C57BL6 mice were deeply anesthetized with sodium pentobarbital (200 mg/kg), then transcardially perfused with 15–20 ml of ice-cold modified artificial cerebrospinal fluid (ACSF; composition in mM: NaCl 87, KCl 2.5, NaH2PO4 1.3, MgCl2 7, NaHCO3 25, sucrose 75, ascorbate 5, CaCl2 0.5, in ddH2O; osmolarity 320–330 mOsm, pH 7.30–7.40) saturated with carbogen gas (95% oxygen, 5% carbon dioxide). The brain was rapidly removed from the cranial cavity and then sectioned using a vibrating-blade microtome (VT1000S, Leica). Slices were allowed to recover for at least 90 min in a holding chamber containing ACSF (composition in mM: NaCl 126, KCl 2.5, NaH2PO4 1.25, MgCl2 1, NaHCO3 26, glucose 10, CaCl2 2.4, in ddH2O; osmolarity 299–301 mOsm; pH 7.35–7.45) saturated with carbogen gas at 32°C before being transferred to the recording chamber for electrophysiology. Once in the recording chamber, slices were continuously perfused at a rate of 2 ml/min with fully oxygenated ACSF at 32 °C with added picrotoxin (100 µM), NBQX (20 µM; Sigma) and AP5 (50 µM; Tocris) to block fast synaptic transmission in the slice.
For whole-cell patch clamp recordings of dentate granule cells, four male C57BL6 mice 9–10 weeks old were injected with AAV8-Camk2a-Jaws under deep isoflurane anesthesia. 0.5 µl AAV was delivered twice at two locations into the hippocampus (3.2 mm posterior, 3.1 mm lateral and, respectively, 2.3 and 2.7 mm deep, relative to bregma) at 100 µl/min with a 34-gauge needle attached to a Hamilton 5-µl syringe, and the needle was left in place for 3 min to allow viral diffusion. Four weeks after injection, mice were killed by decapitation and horizontal 300 µm brain sections were prepared on a Leica VT1200 S vibratome. Sucrose-based ACSF (composition in mM: sucrose 75, NaCl 67, NaHCO3 26, glucose 25, KCl 2.5, NaH2PO4 1.25, CaCl2 0.5, MgCl2 7; pH 7.4, osmolarity 305– 310 mOsm) was used for cutting (at 4 °C) and subsequent storage of slices (32°C for 20–30 min, then maintenance at room temperature, 23.0–23.5°C). After > 1 h incubation, slices were transferred to a submerged recording chamber continuously perfused at 2 ml/min with ACSF (in mM: NaCl 119, NaHCO3 1.24, glucose 10, KCl 2.5, NaH2PO4 1.24, CaCl2 2.5, MgCl2 1.3, pH 7.4, 295 mOsm) maintained at room temperature. Jaws-infected cells were identified under 460–480 nm blue light, whereas visual guidance of the patch electrode was assisted by infrared differential interference contrast (DIC) microscopy (Olympus BX51).
Electrodes for cortical recordings were pulled from thin-walled borosilicate glass capillary tubing using a P-97 puller (Sutter Instruments) and had resistances of 4–6 MΩ when filled with internal solution (composition in mM: potassium gluconate 125, NaCl 10, HEPES 20, Mg-ATP 3, Na-GTP 0.4 and 0.5% biocytin, in ddH20; osmolarity 289 mOsm; pH 7.31). Capacitance, series resistance and input resistance were frequently measured throughout recording to monitor cell health. Cells were visualized through a 40× water-immersion objective on an upright microscope (Scientifica) equipped with infrared DIC optics and a Q-imaging Retiga Exi camera (Q Imaging).
Tip electrode resistance for dentate granule cell recordings was 4.6–7.4 MΩ in ACSF. Patch electrode solution consisted of (in mM) potassium gluconate 122.5, KCl 12.5, KOH-HEPES 10, KOH-EGTA 0.2, Mg-ATP 2, Na3-GTP 0.3, NaCl 8 (pH 7.35, mOsm 296), and 0.2–0.4 mg/ml biocytin was added immediately before use. 10-pA step hyperpolarization/depolarization square current pulses were used to determine the input-output relationship and 300 pA ramp depolarization was used for AP generation. Uninfected dentate granule cells recorded from the contralateral dentate gyrus of virus-injected mice or uninjected animals served as controls. None of these cells responded to 625-nm red light. Uncompensated series resistance was typically 14–25 MΩ, and cells with resting membrane potential more positive than −50mV were discarded (n = 3).
Optical activation was delivered to motor cortex using 500-ms pulses from 590-nm or 625-nm LEDs (Thorlabs), which were additionally filtered with bandpass filters (Semrock): 590-nm LED with 590-nm ± 10-nm filter or 625 nm LED with 632-nm ± 11-nm filter, and to dentate granule cells using 1 s 625-nm red light (M625L3-C1, ThorLabs, 68 mW/mm2). All light powers were measured out of the objective lens with a PM200B photometer (Thorlabs). Illumination spot sizes were measured by photobleaching an in-focus microscope slide coated with Alexa 488 dye for 10 min under full-intensity illumination, then imaging with a micrometer calibration slide to determine the photobleached radius.
All recordings were made using a Multiclamp 700B amplifier and Clampex 10 or 10.4 software (Molecular Devices, CA, USA). Cortical recordings were low-pass filtered at 1 Hz and digitized at 10 kHz using a Digidata 1550 (Molecular Devices, CA, USA); dentate granule cell recordings were digitized with a Digidata 1440. All data were analyzed using Clampfit (Molecular Devices) and Matlab (MathWorks).
Retinal multi-electrode array recordings
Rodent information is outlined in Supplementary table 1. Experiments were conducted as previously described24. Briefly, for viral injections to the retina, animals were anesthetized using 3% isoflurane. A small incision was made with a sharp 30-gauge needle in the sclera near the lens and 2 µl AAV particles were injected slowly into the subretinal space using a blunt 5-µl Hamilton syringe held in a micromanipulator.
To record spike trains from retinal ganglion cells, isolated mouse retinas were placed on a flat MEA60 200 Pt GND array (30-µm-diameter microelectrodes spaced 200 µm apart) (Ayanda Biosystems or Multi Channel Systems). The retina was continuously superfused in oxygenated Ringer’s solution (110 mM NaCl, 2.5 mM KCl, 1.0 mM CaCl2, 1.6 mM MgCl2, 22 mM NaHCO3 and 10 mM d-glucose (pH 7.4 with 95% O2/5% CO2)) at 36 °C during experiments. Signals were recorded (MEA1060-2-BC, Multi-Channel Systems) and filtered between 500 Hz (low cut-off) and 3,500 Hz (high cut-off). Action potentials were extracted with a threshold of greater than 4 times the s.d. of the recorded trace (Matlab, MathWorks).
Histology
We performed histology on n = 6 mice injected with AAV8-hSyn-Jaws, n = 2 mice injected with AAV8-hSyn-eNpHR3.0 and n = 2 mice injected with AAV8-hSyn-Jaws-ER2, representative images from each of which are shown in Figures 4 and 5. We examined n = 7 opsin-expressing retinas, and the images shown in Figure 2 and Supplementary Figure 5 are representative of the whole population.
Mice were terminally anesthetized with isoflurane, then perfused through the left cardiac ventricle with 4% paraformaldehyde in PBS. The brain was removed and sectioned into 40 µm coronal sections on a cryostat and subsequently mounted with Vectashield HardSet (Vector Labs). Acute brain slices were fixed in 4% paraformaldehyde overnight at 4 °C after recording and then washed in PBS. The slices were blocked for 1 h at room temperature in PBS containing 3% normal donkey serum (NDS) and 0.3% Triton, followed by incubation with Alexa 405-conjugated streptavidin (1:1,000; Life Technologies) in PBS containing 3% NDS and 0.3% Triton, for 2 h at room temperature, to reveal biocytin labeling. Brain slices were subsequently washed in PBS, mounted on glass slides and coverslipped with PVA-DABCO (Sigma).
Cryosectioned slides were visualized and imaged with a Zeiss LSM 510 confocal microscope using 20× and 63× objective lenses, or a Nikon TI-E microscope using a 20× objective lens. Acute slices were imaged using a Olympus FV1000 confocal laser scanning microscope through a 10×, 0.40 numerical aperture objective or a 40×, 1.30 numerical aperture oil-immersion objective using Fluoview software (Olympus). Images were subsequently processed in Adobe Photoshop CS6 (Adobe Systems).
Retinal immunostaining was conducted as previously described24. Briefly, retinas were isolated and fixed in 4% paraformaldehyde in PBS for 30 min at room temperature, and washed in PBS overnight at 4 °C. Retinal whole mounts were incubated in 30% sucrose treated with three freeze-thaw cycles, after which all steps were performed at room temperature. Retinas were incubated in a blocking solution (10% normal donkey serum (NDS, Chemicon), 1% bovine serum albumin (BSA) and 0.5% Triton X-100 in PBS, pH 7.4) for 1 h. Primary and secondary antibody applications were done in 3% NDS, 1% BSA, 0.02% sodium azide and 0.5% Triton X-100 in PBS.
Primary antibodies for GFP (rat-GFP, 1:500, Nacalai/Brunschwig 04404-84) and mCAR (rabbit-cone-arrestin, 1:200, Millipore AB15282) were applied for 3 d. After washing the retina three times for 10 min in PBS, the retina was incubated with cyanine dye-conjugated secondary antibodies (1:200; donkey anti-rat–Cy5, Jackson Labs 712–175-153, or donkey anti-rabbit–Cy3, Jackson Labs 711-165-152) and 10 µg/ml DAPI (Roche Diagnostics) for 2 h. After another three 10-min washes in PBS, the retina was mounted on a glass slide with ProLong Gold antifade reagent (Invitrogen). Confocal images of antibody-stained retinas were taken using a Axio Imager Z2 equipped with a LSM 700 scanning head.
In vivo light propagation measurements and Monte Carlo modeling
Isotropic light measurement probes were constructed by gluing a 300-µm-diameter spherical ruby ball lens to the end of a 400-µm-diameter multimode optical fiber with transparent, UV-cured adhesive. Emitted fluorescence was measured using an HR2000 CCD spectrometer (Ocean Optics) and recorded using SpectraSuite (Ocean Optics). Prior to testing, a dark spectrum measurement was taken and a nonlinearity correction was applied using default coefficients. The probe was calibrated in water (n = 1.33) with collimated light.
532- or 635-nm lasers (Shanghai Laser Optics Company) were connected to an optical shutter via a 200-µm-diameter multimode fiber, and the shutter was FC-coupled to a 1.5-mm-diameter plastic optical fiber. Five male C57BL/6J mice were used for green light measurements and five different C57BL/6J mice were used for red light measurements. All measurements were taken under pentobarbital analgesia (50 mg/kg, i.p.). Of the 10 mice × 3 depths × 5 powers, one mouse was not analyzed because the 3-mm-depth files were inadvertently not saved owing to manual save process clunkiness; 6 other files were also inadvertently not saved, but at least 3 power levels were measured for every depth and mouse other than the one noted above.
In Matlab, we performed Monte Carlo simulations of light scattering and absorption in the brain from light emitted from the end of an optical fiber by dividing a cube of gray matter into a 100 × 100 × 100 grid of voxels, each voxel 50 µm × 50 µm × 50 µm in dimension, using previously published algorithms56,57. To achieve accurate simulations of light propagation close to the optical fiber, before the orientation of photon trajectories is randomized by multiple scattering events, we used an anisotropic scattering model with either Henyey-Greenstein or Gegenbauer kernel phase functions, as indicated below. We interpolated data from ref. 58 to obtain scattering coefficients for gray matter and white matter at wavelengths of 532 and 635 nm (below). Because ref. 58 used samples washed of blood, the effects of blood on optical properties of these tissues would not be reflected in their measurements; to accurately simulate in vivo conditions, the effects of blood scattering must be added. We took the brain’s blood volume fraction to be 4% (ref. 59) and the red cell volume fraction of blood (hematocrit) to be 45% (ref. 60). Thus, excluding large blood vessels, the brain contains dilute blood that has about a 1.8% hematocrit. In this red blood cell concentration regime, the anisotropy coefficient is constant and the scattering and absorbing coefficients are directly proportional to hematocrit61. To calculate the total scattering coefficient, we added the scattering coefficient due to brain tissue alone to the scattering coefficient due to the blood in the tissue. To calculate the total absorption coefficient, we likewise added the absorption coefficient due to brain tissue alone to the absorption coefficient due to the blood in the tissue. To calculate the anisotropy coefficient, we took the weighted average anisotropy coefficients of the tissue and blood, weighted by the relative likelihood of each of the two components of causing a scattering event. We chose the Gegenbauer kernel for the phase function when blood was present, as blood is a large component of the scattering and has been shown to be modeled well using that phase function61. With no blood present, as was assumed for the bone, we used the Henyey-Greenstein phase function as in ref. 58. Optical properties of bone at 532 nm and 633 nm were interpolated from data in refs. 62,63.
We used simplified geometries of gray matter and skull bone to illustrate the effects of tissue variations in several experimental locations. In each case, the tissue properties were set on a per-voxel basis, with no corrections for specular reflections or refraction at boundaries between tissue types.
We launched 2 × 106 packets of photons in a fiberlike radiation pattern through fibers modeled on the experimentally used fiber—1.5 mm diameter, 0.5 NA—and modeled their propagation into the brain on the basis of the algorithm of ref. 64. In essence, whenever a photon packet entered a voxel, our program probabilistically calculated the forecasted traveling distance before the next scattering event. If that traveling distance took the photon packet out of the starting voxel, then the packet would be partially absorbed appropriately for the distance it traveled within the voxel and the voxel’s absorption coefficient. The process would then restart upon entry of the photon packet into the new voxel. If the distance traveled before scattering was less than the distance to the edge of the voxel, then the packet would be partially absorbed appropriately for the distance it traveled within the starting voxel, and a new direction of packet propagation would be randomly chosen according to the phase function. Using this model, we generated Supplementary Figure 1a,b, which shows the contours at which the light fluence falls off to various percentages of the light intensity emitted by the fiber.
The coefficients we used were as follows:
Gray matter at 532 nm with 4% v/v blood: absorption coefficient (mm−1) 0.942, scattering coefficient (mm−1) 23.3, anisotropy factor 0.949, using the Gegenbauer kernel phase function.
Gray matter at 633 nm with 4% blood: absorption coefficient (mm−1) 0.071, scattering coefficient (mm−1) 20, anisotropy factor 0.95, using the Gegenbauer kernel phase function.
Bone at 532 nm with no blood: absorption coefficient (mm−1) 0.105, scattering coefficient (mm−1) 34, anisotropy factor 0.93, using the Henyey-Greenstein phase function.
Bone at 633 nm with no blood: absorption coefficient (mm−1) 0.06, scattering coefficient (mm−1) 33, anisotropy factor 0.93, using the Henyey-Greenstein phase function.
White matter at 532 nm with 4% blood: absorption coefficient (mm−1) 1.0, scattering coefficient (mm−1) 55.3, anisotropy factor 0.862, using the Gegenbauer kernel phase function.
White matter at 633 nm with 4% blood: absorption coefficient (mm−1) 0.134, scattering coefficient (mm−1) 53.8, anisotropy factor 0.87, using the Gegenbauer kernel phase function.
To measure skull thickness for Supplementary Figure 1a, male C57BL/6 mice were transcardially perfused with 4% formaldehyde in PBS, and the skulls were removed and imaged with a Nikon XTH160 X-ray micro-CT system. Images were generated at an X-ray voltage of 45 kV and a current of 7.5 W. Sixteen frames per projection were acquired with an integration time of 100 ms, with a total acquisition time of 42 min per skull. Acquired images were reconstructed with CT Pro 3D (Nikon Metrology) and visualized with VGStudio 2.0 (Volume Graphics). Coordinates were stereotactically defined relative to bregma and skull thicknesses determined by measuring _z_-direction differences at a given coordinate.
Statistical analysis
No statistical methods were used to predetermine sample sizes, but sample sizes in this paper were similar to those in previous papers from our group1,3,65,66 and were chosen to reflect sample sizes that might be experienced by end-users. No blinding was carried out, with the exception of cotransfected opsin-GFP and tdTomato data sets, as the chief goal of the paper was to demonstrate the capabilities of a new technology. No randomization was carried out. All replicates were biological.
Statistical analyses were performed using JMP Pro 10 (SAS Software) and Prism 5 (GraphPad). All data were tested for normality with a Shapiro-Wilk test. Two-sample comparisons were characterized with a two-tailed _t_-test, and multiway comparisons for a single variable were characterized with an ANOVA followed by a Newman-Keuls multiple comparison post-test between pairs. Paired tests were conducted with a two-tailed paired sample _t_-test. A Kolmogorov-Smirnov test was used to assess differences in spiking frequency distributions because of its sensitivity to both location and shape of cumulative distribution functions.
Supplementary Material
Supplementary Methods
01
ACKNOWLEDGMENTS
We thank J. Juettner for help making AAV, and Y.K. Cho, D. Schmidt, F. Chen, A. Beyeler, J.M. Zhuo and R.E. Kohman for advice and discussion. A.S.C. acknowledges the Janet and Sheldon Razin ‘59 Fellowship of the Massachusetts Institute of Technology (MIT) McGovern Institute. E.S.B. acknowledges Jerry and Marge Burnett, the US Defense Advanced Research Projects Agency Living Foundries Program HR0011-12-C-0068, Harvard/MIT Joint Grants Program in Basic Neuroscience, Human Frontiers Science Program, Institution of Engineering and Technology A F Harvey Prize, MIT McGovern Institute and McGovern Institute Neurotechnology (MINT) Program, MIT Media Lab, New York Stem Cell Foundation-Robertson Investigator Award, US National Institutes of Health (NIH) Director’s New Innovator award 1DP2OD002002, NIH EUREKA award 1R01NS075421, NIH grants 1R01DA029639, 1RC1MH088182 and 1R01NS067199, US National Science Foundation (NSF) CAREER award CBET 1053233 and NSF grants EFRI0835878 and DMS0848804, the Skolkovo Institute of Science and Technology, a Society for Neuroscience Research Award for Innovation in Neuroscience (RAIN) and the Wallace H. Coulter Foundation. M.L.M. acknowledges funding from NSF DGE 1122492. J.A.C. acknowledges funding from the Whitehall Foundation, the Klingenstein Foundation, the Swebelius Family Trust, the Simons Foundation, an Alfred P. Sloan Fellowship, a NARSAD Young Investigator Award, a Smith Family Award for Excellence in Biomedical Research, NIH R00 EY018407, NIH R01 EY022951 and NIH R01 MH102365. V.B. acknowledges Human Frontier Science Program, Swiss National Science Foundation and Volkswagen Foundation fellowships. B.R. acknowledges the Gebert-Ruf Foundation, SNSF, European Research Council, and European Union SEEBETTER, TREATRUSH, OPTONEURO and 3×3D Imaging grants. X.H. acknowledges funding from an NIH Director’s New Innovator Award (1DP2NS082126), the NINDS (1R01NS087950, 1R21NS078660, 1R01NS081716), NIMH (5R00MH085944), Pew Foundation, Alfred P. Sloan Foundation, Michael J. Fox Foundation, and Brain and Research Foundation. Y.L. acknowledges funding from NIH RO1 MH091220-01. B.Y.C. acknowledges funding from US Defense Advanced Research Projects Agency Living Foundries, the US National Science Foundation Biophotonics and the Brain Research Foundation. K.M.T. acknowledges funding from the Whitehall Foundation, Klingenstein Foundation, JPB Foundation, PIIF Funding, R01-MH102441-01 (NIMH) and DP2-OD-017366-01. G.A.C.M. was supported by the Simons Center for the Social Brain.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
A.S.C. and E.S.B. coordinated all experiments and data analysis. A.S.C. designed and developed Jaws and cloned all constructs. A.S.C. performed in vivo glass pipette extracellular recordings and in vitro electrophysiology. M.L.M. and J.A.C. performed in vivo tetrode extracellular recordings. V.B. performed in vivo multielectrode array recordings. A.S.C., G.A.C.M., A.T.S. and A.Y. performed slice electrophysiology. A.S.C., M.L.M., G.A.C.M., A.T.S., J.A.C., V.B. and M.O. performed in vivo viral injections. A.S.C., M.L.M., V.B., G.A.C.M., A.T.S., S.B.R. and M.O. performed histological processing and fluorescence imaging. S.B.K. and C.R.F. designed or performed autopatch experiments. A.S.C. and N.C.K. performed transfections, cell culture and in vitro viral infections. M.A.H. conducted Monte Carlo modeling. L.C.A. carried out light propagation measurements. R.C.B. and B.D.A. carried out X-ray scans to measure mouse skull thicknesses for the Monte Carlo model. A.S.C., M.L.M., V.B., G.A.C.M., A.T.S., B.Y.C., X.H., J.A.C., B.R. and E.S.B. contributed to study design and data interpretation. J.A.C., B.R., K.M.T., Y.L. and E.S.B. supervised all aspects of the work. A.S.C. and E.S.B. wrote the paper with contributions from the other authors.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
A Supplementary Methods Checklist is available.
References
- 1.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han X, et al. A high-light sensitivity optical neural silencer: development and application to optogenetic control of non-human primate cortex. Front. Syst. Neurosci. 2011;5:18. doi: 10.3389/fnsys.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36:129–139. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ibañez-Tallon I, et al. Tethering naturally occurring peptide toxins for cell-autonomous modulation of ion channels and receptors in vivo. Neuron. 2004;43:305–311. doi: 10.1016/j.neuron.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to ft the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl. Acad. Sci. USA. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kramer RH, Mourot A, Adesnik H. Optogenetic pharmacology for control of native neuronal signaling proteins. Nat. Neurosci. 2013;16:816–823. doi: 10.1038/nn.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polikov VS, Tresco PA, Reichert WM. Response of brain tissue to chronically implanted neural electrodes. J. Neurosci. Methods. 2005;148:1–18. doi: 10.1016/j.jneumeth.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo . Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 10.Xu HT, Pan F, Yang G, Gan WB. Choice of cranial window type for in vivo imaging affects dendritic spine turnover in the cortex. Nat. Neurosci. 2007;10:549–551. doi: 10.1038/nn1883. [DOI] [PubMed] [Google Scholar]
- 11.Drew PJ, et al. Chronic optical access through a polished and reinforced thinned skull. Nat. Methods. 2010;7:981–984. doi: 10.1038/nmeth.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber D, et al. Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice. Nature. 2008;451:61–64. doi: 10.1038/nature06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott NA, Murphy TH. Hemodynamic responses evoked by neuronal stimulation via channelrhodopsin-2 can be independent of intracortical glutamatergic synaptic transmission. PLoS ONE. 2012;7:e29859. doi: 10.1371/journal.pone.0029859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hira R, et al. Transcranial optogenetic stimulation for functional mapping of the motor cortex. J. Neurosci. Methods. 2009;179:258–263. doi: 10.1016/j.jneumeth.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Li X, et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. USA. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat. Neurosci. 2013;16:1499–1508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otomo J, Tomioka H, Sasabe H. Bacterial rhodopsins of newly isolated halobacteria. J. Gen. Microbiol. 1992;138:1027–1037. doi: 10.1099/00221287-138-11-2389. [DOI] [PubMed] [Google Scholar]
- 18.Mattis J, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat. Methods. 2012;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hackett NR, Stern LJ, Chao BH, Kronis KA, Khorana HG. Structure-function studies on bacteriorhodopsin. V. Effects of amino acid substitutions in the putative helix F. J. Biol. Chem. 1987;262:9277–9284. [PubMed] [Google Scholar]
- 20.Rüdiger M, Oesterhelt D. Specifc arginine and threonine residues control anion binding and transport in the light-driven chloride pump halorhodopsin. EMBO J. 1997;16:3813–3821. doi: 10.1093/emboj/16.13.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma D, et al. Role of ER export signals in controlling surface potassium channel numbers. Science. 2001;291:316–319. doi: 10.1126/science.291.5502.316. [DOI] [PubMed] [Google Scholar]
- 22.Hofherr A, Fakler B, Klocker N. Selective Golgi export of Kir2.1 controls the stoichiometry of functional Kir2.x channel heteromers. J. Cell Sci. 2005;118:1935–1943. doi: 10.1242/jcs.02322. [DOI] [PubMed] [Google Scholar]
- 23.Sung CH, Chuang JZ. The cell biology of vision. J. Cell Biol. 2010;190:953–963. doi: 10.1083/jcb.201006020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Busskamp V, et al. Genetic reactivation of cone photoreceptors restores visual responses in retinitis pigmentosa. Science. 2010;329:413–417. doi: 10.1126/science.1190897. [DOI] [PubMed] [Google Scholar]
- 25.Farber DB, Flannery JG, Bowes-Rickman C. The rd mouse story: seventy years of research on an animal model of inherited retinal degeneration. Prog. Retin. Eye Res. 1994;13:31–64. [Google Scholar]
- 26.Busskamp V, Roska B. Optogenetic approaches to restoring visual function in retinitis pigmentosa. Curr. Opin. Neurobiol. 2011;21:942–946. doi: 10.1016/j.conb.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Madisen L, et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat. Neurosci. 2012;15:793–802. doi: 10.1038/nn.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arrenberg AB, Del Bene F, Baier H. Optical control of zebrafsh behavior with halorhodopsin. Proc. Natl. Acad. Sci. USA. 2009;106:17968–17973. doi: 10.1073/pnas.0906252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsunematsu T, et al. Acute optogenetic silencing of orexin/hypocretin neurons induces slow-wave sleep in mice. J. Neurosci. 2011;31:10529–10539. doi: 10.1523/JNEUROSCI.0784-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tønnesen J, Sorensen AT, Deisseroth K, Lundberg C, Kokaia M. Optogenetic control of epileptiform activity. Proc. Natl. Acad. Sci. USA. 2009;106:12162–12167. doi: 10.1073/pnas.0901915106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Znamenskiy P, Zador AM. Corticostriatal neurons in auditory cortex drive decisions during auditory discrimination. Nature. 2013;497:482–485. doi: 10.1038/nature12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardin JA. Dissecting local circuits in vivo: integrated optogenetic and electrophysiology approaches for exploring inhibitory regulation of cortical activity. J. Physiol. Paris. 2012;106:104–111. doi: 10.1016/j.jphysparis.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raimondo JV, Kay L, Ellender TJ, Akerman CJ. Optogenetic silencing strategies differ in their effects on inhibitory synaptic transmission. Nat. Neurosci. 2012;15:1102–1104. doi: 10.1038/nn.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tye KM, et al. Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 2011;471:358–362. doi: 10.1038/nature09820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Al-Juboori SI, et al. Light scattering properties vary across different regions of the adult mouse brain. PLoS ONE. 2013;8:e67626. doi: 10.1371/journal.pone.0067626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giller CA, et al. Validation of a near-infrared probe for detection of thin intracranial white matter structures. J. Neurosurg. 2003;98:1299–1306. doi: 10.3171/jns.2003.98.6.1299. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs GH, Williams GA, Cahill H, Nathans J. Emergence of novel color vision in mice engineered to express a human cone photopigment. Science. 2007;315:1723–1725. doi: 10.1126/science.1138838. [DOI] [PubMed] [Google Scholar]
- 39.Naarendorp F, et al. Dark light, rod saturation, and the absolute and incremental sensitivity of mouse cone vision. J. Neurosci. 2010;30:12495–12507. doi: 10.1523/JNEUROSCI.2186-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anikeeva P, et al. Optetrode: a multichannel readout for optogenetic control in freely moving mice. Nat. Neurosci. 2012;15:163–170. doi: 10.1038/nn.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S-H, et al. Activation of specifc interneurons improves V1 feature selectivity and visual perception. Nature. 2012;488:379–383. doi: 10.1038/nature11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grutzendler J, Kasthuri N, Gan WB. Long-term dendritic spine stability in the adult cortex. Nature. 2002;420:812–816. doi: 10.1038/nature01276. [DOI] [PubMed] [Google Scholar]
- 43.Yoder EJ, Kleinfeld D. Cortical imaging through the intact mouse skull using two-photon excitation laser scanning microscopy. Microsc. Res. Tech. 2002;56:304–305. doi: 10.1002/jemt.10002. [DOI] [PubMed] [Google Scholar]
- 44.Berényi A, Belluscio M, Mao D, Buzsaki G. Closed-loop control of epilepsy by transcranial electrical stimulation. Science. 2012;337:735–737. doi: 10.1126/science.1223154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trachtenberg JT, et al. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- 46.Jazayeri M, Lindbloom-Brown Z, Horwitz GD. Saccadic eye movements evoked by optogenetic activation of primate V1. Nat. Neurosci. 2012;15:1368–1370. doi: 10.1038/nn.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohayon S, Grimaldi P, Schweers N, Tsao DY. Saccade modulation by optical and electrical stimulation in the macaque frontal eye feld. J. Neurosci. 2013;33:16684–16697. doi: 10.1523/JNEUROSCI.2675-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye H, Daoud-El Baba M, Peng RW, Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565–1568. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- 49.Menzler J, Zeck G. Network oscillations in rod-degenerated mouse retinas. J. Neurosci. 2011;31:2280–2291. doi: 10.1523/JNEUROSCI.4238-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. compact 2nd edn. Amsterdam and Boston: Elsevier Academic; 2004. [Google Scholar]
- 51.Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat. Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grieger JC, Choi VW, Samulski RJ. Production and characterization of adeno-associated viral vectors. Nat. Protoc. 2006;1:1412–1428. doi: 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
- 53.Hippenmeyer S, et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kodandaramaiah SB, Franzesi GT, Chow BY, Boyden ES, Forest CR. Automated whole-cell patch-clamp electrophysiology of neurons in vivo . Nat. Methods. 2012;9:585–587. doi: 10.1038/nmeth.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cardin JA, et al. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–667. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Binzoni T, Leung TS, Gandjbakhche AH, Rufenacht D, Delpy DT. The use of the Henyey-Greenstein phase function in Monte Carlo simulations in biomedical optics. Phys. Med. Biol. 2006;51:N313–N322. doi: 10.1088/0031-9155/51/17/N04. [DOI] [PubMed] [Google Scholar]
- 57.Wang L, Jacques SL, Zheng L. MCML–Monte Carlo modeling of light transport in multi-layered tissues. Comput. Methods Programs Biomed. 1995;47:131–146. doi: 10.1016/0169-2607(95)01640-f. [DOI] [PubMed] [Google Scholar]
- 58.Yaroslavsky AN, et al. Optical properties of selected native and coagulated human brain tissues in vitro in the visible and near infrared spectral range. Phys. Med. Biol. 2002;47:2059–2073. doi: 10.1088/0031-9155/47/12/305. [DOI] [PubMed] [Google Scholar]
- 59.Hatazawa J, et al. Regional cerebral blood flow, blood volume, oxygen extraction fraction, and oxygen utilization rate in normal volunteers measured by the autoradiographic technique and the single breath inhalation method. Ann. Nucl. Med. 1995;9:15–21. doi: 10.1007/BF03165003. [DOI] [PubMed] [Google Scholar]
- 60.Kreiss P, Bettan M, Crouzet J, Scherman D. Erythropoietin secretion and physiological effect in mouse after intramuscular plasmid DNA electrotransfer. J. Gene Med. 1999;1:245–250. doi: 10.1002/(SICI)1521-2254(199907/08)1:4<245::AID-JGM49>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 61.Roggan A, Friebel M, Dorschel K, Hahn A, Muller G. Optical properties of circulating human blood in the wavelength range 400–2500 nm. J. Biomed. Opt. 1999;4:36–46. doi: 10.1117/1.429919. [DOI] [PubMed] [Google Scholar]
- 62.Bashkatov AN, Genina EA, Kochubey VI, Tuchin VV. Optical properties of human cranial bone in the spectral range from 800 to 2000 nm — art. no. 616310. Saratov Fall Meeting 2005: Optical Technologies in Biophysics and Medicine VII. 2006;6163:16310. [Google Scholar]
- 63.Ugryumova N, Matcher SJ, Attenburrow DP. Measurement of bone mineral density via light scattering. Phys. Med. Biol. 2004;49:469–483. doi: 10.1088/0031-9155/49/3/009. [DOI] [PubMed] [Google Scholar]
- 64.Tsubota T, Ohashi Y, Tamura K, Sato A, Miyashita Y. Optogenetic manipulation of cerebellar Purkinje cell activity in vivo. PLoS ONE. 2011;6:e22400. doi: 10.1371/journal.pone.0022400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 66.Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PLoS ONE. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
01