MgrB Inactivation Is a Common Mechanism of Colistin Resistance in KPC-Producing Klebsiella pneumoniae of Clinical Origin (original) (raw)
Abstract
Klebsiella pneumoniae strains producing KPC-type carbapenemases (KPC-KP) are challenging multidrug-resistant pathogens due to their extensively drug-resistant phenotypes and potential for epidemic dissemination in health care settings. Colistin is a key component of the combination antimicrobial regimens used for treatment of severe KPC-KP infections. We previously reported that insertional inactivation of the mgrB gene, encoding a negative-feedback regulator of the PhoQ-PhoP signaling system, can be responsible for colistin resistance in KPC-KP, due to the resulting upregulation of the Pmr lipopolysaccharide modification system. In this work we investigated the status of the mgrB gene in a collection of 66 colistin-resistant nonreplicate clinical strains of KPC-KP isolated from different hospitals in Italy and Greece. Overall, 35 strains (53%) exhibited alterations of the mgrB gene, including insertions of different types of mobile elements (IS_5_-like, IS_1F_-like, or IS_Kpn14_), nonsilent point mutations, and small intragenic deletions. Four additional strains had a larger deletion of the mgrB locus, while the remaining 27 strains (41%) did not show mgrB alterations. Transcriptional upregulation of the phoQ and pmrK genes (part of the phoPQ and pmrHFIJKLM operon, respectively) was observed in all strains with mgrB alterations. Complementation experiments with a wild-type mgrB gene restored colistin susceptibility and basal expression levels of phoQ and pmrK genes in strains carrying different types of mgrB alterations. The present results suggest that mgrB alteration can be a common mechanism of colistin resistance among KPC-KP in the clinical setting.
INTRODUCTION
The dissemination of carbapenemase-producing Enterobacteriaceae (CPE) is an emerging global problem (1, 2). Among CPE, Klebsiella pneumoniae producing KPC-type carbapenemases (KPC-KP) are likely the most challenging pathogens due to their extensively drug-resistant phenotypes and potential for rapid dissemination in health care settings, with a remarkable impact on morbidity and mortality (2–4).
Polymyxins (polymyxin B and colistin) are among the few antimicrobial agents that retain activity against KPC-KP, being a key component of the combination antimicrobial regimens used for the treatment of severe KPC-KP infections (2, 5). The emergence of polymyxin-resistant KPC-KP, however, has repeatedly been reported (6–9) and is a matter of major concern. In some settings, polymyxin resistance has achieved alarming proportions among KPC-KP, 18.6% among isolates from a Greek hospital in 2007 to 2010 (7) and 22.4% among isolates from the first Italian countrywide survey on carbapenem-resistant Enterobacteriaceae (CRE), carried out in 2011 (9).
We recently showed that inactivation of the mgrB gene, encoding a negative feedback regulator of the PhoQ-PhoP signaling system, is responsible for acquired colistin resistance in KPC-KP by upregulating PhoQ-PhoP, which, in turn, activates the Pmr system responsible for modification of the lipopolysaccharide polymyxin target (10). This resistance mechanism was identified in a colistin-resistant isolate from a patient with bacteremia who had been exposed to colistin treatment, and it was easily reproduced in vitro after exposure to colistin of the susceptible progenitor strain (10). The same resistance mechanism has also been detected in a non-KPC-producing carbapenemase- and colistin-resistant K. pneumoniae isolate from Spain (11).
In this work we investigated the occurrence of mgrB gene alterations as a mechanism of resistance in a large collection of colistin-resistant clinical isolates of KPC-KP collected during the ongoing Greek and Italian KPC-KP epidemics.
MATERIALS AND METHODS
Bacterial strains.
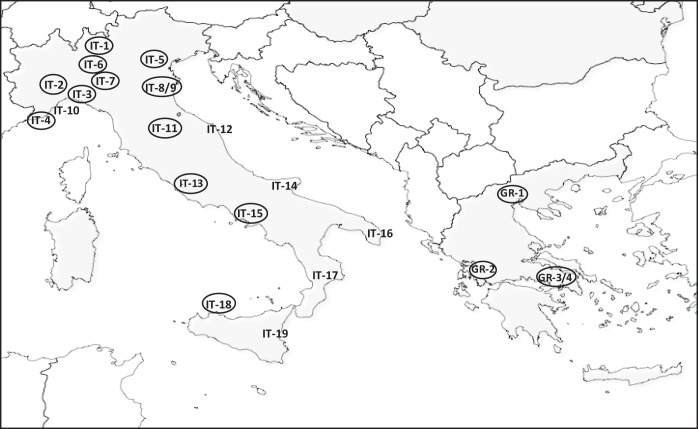
The strains studied in this work included 66 nonreplicate colistin-resistant KPC-KP clinical isolates obtained during the period from 2010 to 2012 from 23 centers (19 in Italy and 4 in Greece) (Fig. 1 and Table 1). Some of these isolates (n = 41) were from the first Italian nationwide survey on carbapenem-resistant Enterobacteriaceae, carried out in mid-2011 (9). The remaining isolates (n = 25) were collected outside that survey by participants in the COLGRIT Study Group. Identification of the isolates was always confirmed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Vitek MS; bioMérieux, Marcy l'Etoile, France). The presence of the _bla_KPC gene was confirmed by PCR, and the _bla_KPC gene variant was determined by direct amplicon sequencing, as previously described (9).
FIG 1.
Geographical distribution of the Greek and Italian centers from which COL-R strains were originated. Centers where strains carrying mgrB lesions were detected are circled. The number of each center corresponds to the location as follows: IT-1, Lecco; IT-2, Turin; IT-3, Genoa; IT-4, Pietra Ligure; IT-5, Verona; IT-6, Milan; IT-7, Pavia; IT-8-9, Modena; IT-10, San Remo; IT-11, Florence; IT-12, Ancona; IT-13, Rome; IT-14, Foggia; IT-15, Naples; IT-16, Lecce; IT-17, Cosenza; IT-18, Palermo; IT-19, Catania; GR-1, Salonika; GR-2, Agrinio; GR-3-4, Athens.
TABLE 1.
Characteristics of the colistin-resistant K. pneumoniae strains investigated in this work
| Strain | Yr | Center_b_ | Source_c_ | ST | KPC type | Colistin MIC (μg/ml) | Additional resistance trait(s)d | mgrB status_e_ |
|---|---|---|---|---|---|---|---|---|
| KP6303 | 2010 | IT-1 | Blood | 258 | KPC-2 | 16 | SXT, TGC | Insertional inactivation, IS_5_-like element at nt 75 (FW) |
| KPENA | 2010 | IT-2 | CVC | 258 | KPC-2 | 4 | SXT, TGC | WT |
| KP04C83_a_ | 2011 | IT-2 | AF | 512 | KPC-3 | 32 | SXT, TGC | Δ18/27 (frameshift and premature termination) |
| KP04C93_a_ | 2011 | IT-2 | Blood | 512 | KPC-3 | 32 | GEN, SXT, TGC | Δ18/27 (frameshift and premature termination) |
| KP04C62 | 2011 | IT-2 | Blood | 512 | KPC-3 | 32 | SXT | Insertional inactivation, IS_5_-like element at nt 126 (FW) |
| KP06C01_a_ | 2011 | IT-3 | Urine | 512 | KPC-3 | 16 | SXT | g109a (G37S) |
| KP06C19_a_ | 2011 | IT-3 | Blood | 512 | KPC-3 | 32 | g109a (G37S) | |
| KP06C02 | 2011 | IT-3 | Urine | 258 | KPC-2 | 16 | SXT | WT |
| KP06C05 | 2011 | IT-3 | Urine | 258 | KPC-3 | 32 | TGC | g109a (G37S) |
| KP06C16_a_ | 2011 | IT-3 | Urine | 258 | KPC-3 | 16 | SXT, TGC | g109a (G37S) |
| KP06C18_a_ | 2011 | IT-3 | Blood | 258 | KPC-3 | 16 | g109a (G37S) | |
| KP06C07 | 2011 | IT-3 | LRS | 258 | KPC-2 | 32 | GEN, TGC | WT |
| KP207-2 | 2010 | IT-4 | CVC | 258 | KPC-3 | 16 | GEN, SXT, TGC | Insertional inactivation, IS_5_-like element at nt 75 (RW) |
| KP08C01 | 2011 | IT-5 | Urine | 512 | KPC-3 | 16 | SXT | WT |
| KP08C02 | 2011 | IT-5 | Urine | 512 | KPC-3 | 32 | SXT, TGC | WT |
| KP08C08 | 2011 | IT-5 | Urine | 512 | KPC-3 | 32 | SXT | WT |
| KP08C11 | 2011 | IT-5 | Urine | 512 | KPC-3 | 8 | SXT | WT |
| KP08C16 | 2011 | IT-5 | Urine | 512 | KPC-3 | 32 | SXT | a7t (nonsense, premature termination) |
| KP08C24 | 2011 | IT-5 | Urine | 512 | KPC-3 | 32 | SXT | Insertional inactivation, IS_5_-like element at nt 75 (FW) |
| KP08C32 | 2011 | IT-5 | LRS | 512 | KPC-3 | 8 | SXT | WT |
| KPKL1HU | 2010 | IT-6 | Bile | 258 | KPC-3 | 16 | SXT, TGC | Insertional inactivation, IS_5_-like element at nt 75 (FW) |
| KPKL52MG11 | 2011 | IT-7 | Blood | 512 | KPC-3 | 8 | SXT, TGC | Δ_mgrB locus_ f |
| KPL171 | 2011 | IT-8 | Urine | 512 | KPC-2 | 8 | GEN, SXT, TGC | Δg47 (frameshift and premature termination) |
| KP10C06 | 2011 | IT-8 | Urine | 258 | KPC-2 | 32 | TGC | Δg47 (frameshift and premature termination) |
| KP11C07 | 2011 | IT-9 | Urine | 512 | KPC-3 | 16 | SXT, TGC | Insertional inactivation, IS_5_-like element at nt 75 (FW) |
| KP07C07 | 2011 | IT-10 | LRS | 258 | KPC-2 | 16 | GEN, SXT | WT |
| KP12C48 | 2011 | IT-11 | Urine | 512 | KPC-3 | 32 | SXT | Δ109/119 (frameshift and premature termination) |
| KP12C69 | 2011 | IT-11 | WEX | 101 | KPC-2 | 8 | GEN | WT |
| KPAN-3 | 2011 | IT-12 | Blood | 512 | KPC-3 | 32 | SXT, TGC | WT |
| KP15C05 | 2011 | IT-12 | LRS | 512 | KPC-3 | 32 | SXT | WT |
| KP15C10 | 2011 | IT-12 | Urine | 258 | KPC-3 | 32 | SXT | WT |
| KP15C15 | 2011 | IT-12 | LRS | 512 | KPC-3 | 32 | SXT, TGC | WT |
| KP15C16 | 2011 | IT-12 | Urine | 512 | KPC-3 | 32 | SXT, TGC | WT |
| KP15C17 | 2011 | IT-12 | LRS | 512 | KPC-3 | 32 | SXT | WT |
| KP15C18 | 2011 | IT-12 | Urine | 258 | KPC-3 | 32 | SXT | WT |
| KP15C23 | 2011 | IT-12 | CVC | 258 | KPC-3 | 16 | SXT, TGC | WT |
| KP15C24 | 2011 | IT-12 | Urine | 512 | KPC-3 | 32 | SXT | WT |
| KP15C30 | 2011 | IT-12 | WEX | 258 | KPC-3 | 32 | SXT | WT |
| KP9 | 2011 | IT-13 | Urine | 258 | KPC-3 | 16 | GEN, SXT, TGC | Insertional inactivation, IS_5_-like element at nt 75 (RW) |
| KP16C05 | 2011 | IT-13 | Blood | 512 | KPC-3 | 32 | SXT | WT |
| KP16C08 | 2011 | IT-13 | Blood | 258 | KPC-3 | 16 | SXT, TGC | WT |
| KP16C12 | 2011 | IT-13 | WEX | 258 | KPC-3 | 8 | SXT | WT |
| KP18C37 | 2011 | IT-14 | Urine | 258 | KPC-3 | 4 | SXT | WT |
| KP20C14 | 2011 | IT-15 | Blood | 258 | KPC-3 | 32 | SXT | Insertional inactivation, IS_5_-like element at nt 75 (FW) |
| KP20C26 | 2011 | IT-15 | Blood | 258 | KPC-3 | 32 | SXT | Insertional inactivation, IS_5_-like element at nt 75 (FW) |
| KP19C07 | 2011 | IT-16 | CVC | 512 | KPC-3 | 32 | SXT | WT |
| KP22C22 | 2011 | IT-17 | Urine | 258 | KPC-3 | 32 | SXT | WT |
| KP23C07_a_ | 2011 | IT-18 | Urine | 512 | KPC-3 | 8 | Insertional inactivation, IS_1F_-like element at nt 105 (FW) | |
| KP23C12_a_ | 2011 | IT-18 | LRS | 512 | KPC-3 | 32 | SXT, TGC | Insertional inactivation, IS_1F_-like element at nt 105 (FW) |
| KPTRAV | 2010 | IT-18 | WEX | 258 | KPC-3 | 16 | SXT, TGC | Δ_mgrB_ (from −400 to +599) |
| KPIAC | 2010 | IT-18 | Blood | 258 | KPC-3 | 32 | GEN, SXT, TGC | t71a (L24H) |
| KPCAT | 2011 | IT-19 | Blood | 512 | KPC-3 | 4 | GEN, SXT, TGC | WT |
| KP4603_a_ | 2011 | GR-1 | Urine | 258 | KPC-2 | 64 | GEN, SXT | Δ_mgrB_ locus_f_ |
| KP4623_a_ | 2011 | GR-1 | Urine | 258 | KPC-2 | 64 | Δ_mgrB_ locus_f_ | |
| KP4566 | 2012 | GR-2 | LRS | 258 | KPC-2 | 32 | GEN, SXT | Insertional inactivation, IS_5_-like element at nt 75 (RW) |
| KP4495_a_ | 2012 | GR-2 | Blood | 258 | KPC-2 | 64 | SXT | Insertional inactivation, IS_5_-like element at nt 75 (RW) |
| KP4569_a_ | 2012 | GR-2 | Blood | 258 | KPC-2 | 8 | GEN, SXT | Insertional inactivation, IS_5_-like element at nt 75 (RW) |
| KP4579_a_ | 2012 | GR-2 | LRS | 258 | KPC-2 | 4 | GEN, SXT | Insertional inactivation, IS_5_-like element at nt 75 (RW) |
| KP4594_a_ | 2012 | GR-2 | Blood | 258 | KPC-2 | 4 | GEN, SXT | Insertional inactivation, IS_5_-like element at nt 75 (RW) |
| KP4619_a_ | 2012 | GR-2 | LRS | 258 | KPC-2 | 4 | GEN, SXT | Insertional inactivation, IS_5_-like element at nt 75 (RW) |
| KP4620_a_ | 2012 | GR-2 | LRS | 258 | KPC-2 | 4 | GEN, SXT | Insertional inactivation, IS_5_-like element at nt 75 (RW) |
| KP4657_a_ | 2012 | GR-2 | LRS | 258 | KPC-2 | 4 | GEN, SXT | Insertional inactivation, IS_5_-like element at nt 75 (RW) |
| KP4697 | 2012 | GR-3 | CVC | 258 | KPC-2 | 32 | SXT | Insertional inactivation, IS_5_-like element at nt 75 (RW) |
| KP4707 | 2012 | GR-3 | Blood | 258 | KPC-2 | 32 | SXT | g83a (C28Y) |
| KP4553_a_ | 2012 | GR-4 | CVC | 147 | KPC-2 | 16 | GEN, SXT | Insertional inactivation, IS_Kpn14_ at nt 124 (FW) |
| KP4554_a_ | 2012 | GR-4 | Blood | 147 | KPC-2 | 8 | GEN, SXT | Insertional inactivation, IS_Kpn14_ at nt 124 (FW) |
Antimicrobial susceptibility testing.
MICs of colistin and other antimicrobial agents (including carbapenems, cephalosporins, trimethoprim-sulfamethoxazole, ciprofloxacin, levofloxacin, amikacin, gentamicin, and tigecycline) were determined by reference broth microdilution (12) and interpreted according to the EUCAST guidelines (EUCAST breakpoint tables for interpretation of MICs and zone diameters, version 4.0, 2014) (see http://www.eucast.org). Escherichia coli ATCC 25922 and K. pneumoniae KKBO-4 (colistin resistant) (10) were used as control strains for antimicrobial susceptibility testing.
Bacterial genotyping.
Genotyping of the K. pneumoniae isolates by pulsed-field gel electrophoresis (PFGE) was carried out as described previously (9), and results were interpreted as recommended by van Belkum et al. (13). Multilocus sequence typing (MLST) was performed as previously described (14), and sequence type (ST) was assigned according to the K. pneumoniae MLST database (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html).
Analysis of the mgrB locus.
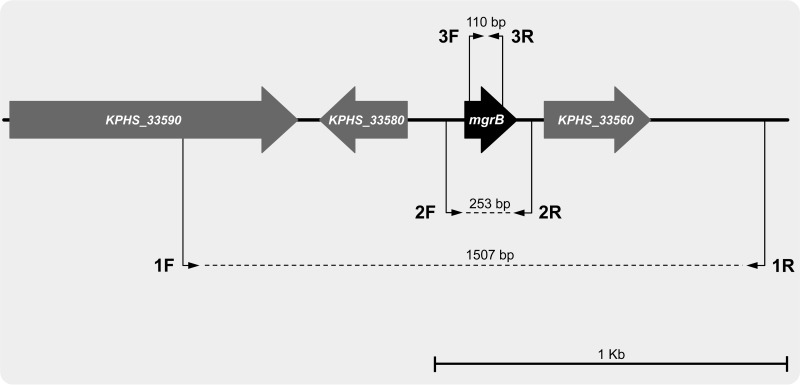
PCR amplification of the mgrB locus was carried out using primers mgrB_Ext_F and mgrB_Ext_R, which target amplification of the mgrB coding sequence and some flanking regions (Fig. 2 and Table 2). A set of primers external to mgrB, covering a larger portion of the mgrB locus, and a set of primers targeting amplification of an internal region of the mgrB coding sequence were also used with some strains (Fig. 2 and Table 2). K. pneumoniae KKBO-1 (10) was used as a control strain carrying wild-type mgrB in PCR mapping experiments. DNA sequences were determined on both strands of the amplification products at an external sequencing facility (Macrogen, Seoul, South Korea).
FIG 2.
Map of the K. pneumoniae chromosomal region carrying the mgrB gene. The map is drawn based on sequence information from the K. pneumoniae HS11286 chromosome (GenBank accession no. CP000649) (19). Open reading frames are indicated by arrows (arrow direction indicates orientation). The locations of primers used for PCR mapping of the mgrB locus are indicated by thin black arrows (1F, EE_mgrB_F; 1R, EE_mgrB_R; 2F, mgrB_Ext_F; 2R, mgrB_Ext_R; 3F, Int_mgrB_F; 3R, Int_mgrB_R). The expected sizes of the PCR products are also shown.
TABLE 2.
Primers and PCR conditions used in this work
| PCR and primer | Sequence (5′ to 3′) | Cycling conditions (°C/s)a | Reference or source |
|---|---|---|---|
| Conventional PCR | |||
| mgrB_ext_F | AAGGCGTTCATTCTACCACC | D (95/30), A (54/30), E (72/105) | 10 |
| mgrB_ext_R | TTAAGAAGGCCGTGCTATCC | ||
| EE_mgrB_F | GGCTATGGCGAGGATAATGAG | D (95/30), A (54/30), E (72/120) | This work |
| EE_mgrB_R | GCTGTGATGTAAGCGTCTGGTG | ||
| Int_mgrB_F | CGGTGGGTTTTACTGATAGTCA | D (95/30), A (54/30), E (72/30) | This work |
| Int_mgrB_R | ATAGTGCAAATGCCGCTGA | ||
| RT-qPCR | |||
| pmrK_F2 | CGCTGAATATGCTCGACCCAGAAG | D (95/10), A (52/5), E (72/5) | This work |
| pmrK_R2 | GCTGGCGGTAATCGTCTGTACG | ||
| phoQ_F | ATATGCTGGCGAGATGGGAAAACGG | D (95/10), A (52/5), E (72/5) | 10 |
| phoQ_R | CCAGCCAGGGAACATCACGCT | ||
| rpsL13_F | GCCGTACTTGGAGCGAGCCTG | D (95/10), A (52/5), E (72/5) | 10 |
| rpsL14_F | CCGTGGCGGTCGTGTTAAAGA |
Complementation experiments.
For complementation experiments, a pACYC184 derivative carrying the wild-type mgrB gene with its putative promoter and terminator (named pACYC-mgrB) was used (10). The pACYC-mgrB plasmid was introduced into the K. pneumoniae strains by electroporation, and transformants were selected on Mueller-Hinton agar plates supplemented with 40 μg/ml of tetracycline.
Transcriptional analysis by real-time quantitative PCR.
RNA extraction and retrotranscription were carried out as previously described (10). Real-time quantitative PCR (RT-qPCR) was used to measure the expression of the phoQ and pmrK genes using the primers and conditions reported in Table 2. Expression of the rpsL gene was used as an internal standard (primers and conditions reported in Table 2). Normalization was performed against rpsL gene using the ΔΔ_CT_ method (relative) (15), and the obtained values were then normalized against those obtained with the colistin-susceptible strain KKBO-1 (10).
Nucleotide sequence accession numbers.
The nucleotide sequences of the altered mgrB genes described in this work have been deposited at DDBJ/EMBL/GenBank under the accession numbers KJ129594 (isolate KP04C62), KJ129595 (isolate KP04C83), KJ129596 (isolate KP06C01), KJ129597 (isolate KP08C16), KJ129598 (isolate KP08C24), KJ129599 (isolate KPL171), KJ129600 (isolate KP23C12), KJ129602 (isolate KP4707), KJ129603 (isolate KPIAC), KJ129604 (isolate KP4554), and KJ586580 (isolate KP12C48).
RESULTS
Characteristics of the investigated clinical isolates.
All isolates exhibited complex multidrug-resistant phenotypes, including carbapenems and other β-lactams, fluoroquinolones, and amikacin. Some of them retained susceptibility to gentamicin (71%), tigecycline (64%), and/or trimethoprim-sulfamethoxazole (12%). Colistin MICs varied from 4 to 64 μg/ml (MIC90, 32 μg/ml; median MIC, 32 μg/ml) (Table 1).
Most of the isolates belonged in ST258 and ST512, but isolates of other STs (ST101 and ST147) were also represented (Table 1).
Occurrence of mgrB alterations in colistin-resistant clinical isolates of KPC-KP.
A PCR mapping and sequencing strategy for the mgrB locus was carried out, using primers designed on regions flanking the mgrB gene for each strain (Fig. 2).
Using primers mgrB_Ext_F and mgrB_Ext_R, an amplification product was obtained with 62 of the 66 KPC-KP strains. Of these, 35 strains exhibited nonsilent alterations in the mgrB coding sequence, while 27 strains carried a wild-type mgrB gene (Table 1). Of the four strains that yielded no amplification product with the former primers (KPKL52MG11, KPTRAV, KP4603, and KP4623), one exhibited a large deletion of the mgrB locus that was mapped with the EE_mgrB_F and EE_mgrB_R primers, while the remaining three strains did not yield any amplification product even with the latter primers, suggesting a larger deletion of the locus (Table 1). In these four strains, the absence of mgrB sequences from genomic DNA was confirmed by PCR with the internal Int_mgrB_F and Int_mgrB_R primers.
Several different nonsilent alterations of the mgrB coding sequence were detected, including inactivation by an insertion sequence (IS), missense or nonsense point mutations, and small deletions (Table 1). The presence of an IS-inactivated mgrB was detected in 22 of the 35 strains with alterations in the mgrB coding sequence. The ISs were of three different types, namely, an IS_5_-like element identical to that previously found to inactivate mgrB in strain KKBO-4 (10), an IS_1F_-like element 98% identical to IS_1F_ (GenBank accession no. X52538) (16), and an IS_Kpn14_ element (accession no. CP000649, from 28285 to 29052) (17). The different ISs were inserted at different sites, and the _IS5_-like element was found to be inserted at two different positions and in different orientations (Table 1). The presence of nonsilent point mutations or small deletions was detected in 13 of the 35 strains with alterations in the mgrB coding sequence. In some cases they resulted in truncated proteins, while in other isolates the alterations yielded a mutated MgrB protein with amino acid substitutions at different positions (Table 1).
Analysis of the distribution of colistin MICs among strains with different resistance mechanisms did not reveal significant differences. Geometric mean MICs were 18.1 and 19.6 μg/ml, MIC50s were 16 and 32 μg/ml, MIC90s were 32 and 32 μg/ml, and MIC ranges were 4 to 64 and 4 to 32 μg/ml in strains with or without mgrB alterations, respectively (Table 1).
Significance of the mgrB alterations detected in colistin-resistant strains.
The significance of the nonsilent alterations of the mgrB gene detected in colistin-resistant strains was investigated by complementation experiments with plasmid pACYC-mgrB, which carries a cloned copy of wild-type mgrB and its putative promoter. These experiments were carried out with 15 strains, representative for the various types of mgrB alteration, including the four isolates with large deletions (Table 3)
TABLE 3.
Different mgrB lesions detected in the colistin-resistant strains and results of complementation experiments with strains representative of each type of lesion_a_
| Strain | mgrB lesion_b_ | Colistin MIC (μg/ml) | Colistin MIC after complementation (μg/ml)c | pmrK expression (mean ± SD) | pmrK expression after complementation (mean ± SD) | phoQ expression (mean ± SD) | phoQ expression after complementation (mean ± SD) |
|---|---|---|---|---|---|---|---|
| KKBO-1 | WT | 0.125 | 0.064 | 1 | 1.24 ± 0.51 | 1 | 0.93 ± 0.18 |
| KP207-2 | Insertional inactivation, IS_5_-like element at nt 75 (RW) | 16 | 0.5 | 6.19 ± 0.19 | 1.34 ± 0.13 | 3.56 ± 0.15 | 0.92 ± 0.17 |
| KP04C62 | Insertional inactivation, IS_5_-like element at nt 126 (FW) | 32 | 0.03 | 7.97 ± 0.12 | 1.81 ± 0.11 | 4.42 ± 0.25 | 1.46 ± 0.35 |
| KP23C12 | Insertional inactivation, IS_1F_-like element at nt 105 (FW) | 32 | 0.25 | 8.54 ± 0.09 | 2.04 ± 0.23 | 8.81 ± 0.44 | 1.11 ± 0.14 |
| KPIAC | t71a (L24H) | 32 | 0.03 | 3.25 ± 0.23 | 1.87 ± 0.37 | 2.98 ± 0.77 | 1.24 ± 0.26 |
| KP04C83 | Δ18/27 (frameshift and premature termination) | 32 | 0.03 | 5.15 ± 0.04 | 1.59 ± 0.34 | 7.23 ± 0.30 | 2.18 ± 0.45 |
| KP06C01 | g109a (G37S) | 16 | 0.03 | 6.61 ± 0.07 | 1.97 ± 0.14 | 6.65 ± 0.21 | 0.48 ± 0.34 |
| KP08C16 | a7t (nonsense, premature termination) | 32 | 0.03 | 5.44 ± 0.11 | 1.82 ± 0.22 | 4.22 ± 0.29 | 2.18 ± 0.33 |
| KP10C06 | Δg47 (frameshift and premature termination) | 32 | 0.03 | 10.39 ± 1.12 | 1.10 ± 0.04 | 11.45 ± 1.23 | 1.08 ± 0.45 |
| KP4707 | g83a (C28Y) | 32 | 0.03 | 9.51 ± 0.32 | 1.48 ± 0.20 | 14.02 ± 1.17 | 3.71 ± 0.75 |
| KP12C48 | Δ109/119 (frameshift and premature termination) | 32 | 0.03 | 5.37 ± 0.15 | 1.69 ± 0.17 | 9.61 ± 0.55 | 1.59 ± 0.15 |
| KP4553 | Insertional inactivation, IS_Kpn14_ at nt 124 (FW) | 16 | 0.12 | 5.97 ± 0.18 | 1.99 ± 0.18 | 4.72 ± 0.35 | 1.20 ± 0.39 |
| KPTRAV | Δ_mgrB_ (from −400 to +599) | 16 | 0.06 | 7.86 ± 0.87 | 2.08 ± 0.34 | 4.08 ± 0.23 | 0.97 ± 0.11 |
| KPKL52MG11 | Δ_mgrB locus_ | 8 | 0.12 | 3.26 ± 0.07 | 0.78 ± 0.24 | 7.54 ± 0.54 | 1.75 ± 0.65 |
| KP4603 | Δ_mgrB locus_ | 64 | 0.12 | 4.21 ± 0.13 | 1.77 ± 0.13 | 5.05 ± 0.05 | 1.02 ± 0.34 |
| KP4623 | Δ_mgrB locus_ | 64 | 0.12 | 5.75 ± 0.52 | 1.08 ± 0.36 | 4.51 ± 0.39 | 0.77 ± 0.43 |
In all cases complementation with wild-type mgrB restored full colistin susceptibility and reduced the level of pmrK expression to a value similar to those of colistin-susceptible strains (Table 3), supporting a key role of the corresponding mgrB alterations in determining colistin resistance.
Distribution of the mgrB alterations associated with colistin resistance.
The mgrB alterations associated with colistin resistance were detected in strains from 13 of the 19 Italian centers and from 4 of 4 Greek centers (Table 1, Fig. 1). Insertional inactivation by IS_5_-like elements was detected in strains from several different centers in Italy and Greece, while the other alterations exhibited a more restricted geographical distribution (Table 1).
Identical alterations were sometimes detected in strains of the same ST and PFGE profile from the same center (Table 1), suggesting the occurrence of clonal expansion and cross-transmission of the resistant strain within a center. However, identical alterations were also detected in strains of different STs isolated from different centers (Table 1), suggesting an independent origin of these alterations.
DISCUSSION
We have previously shown that inactivation of the mgrB gene, encoding a negative-feedback regulator of the PhoQ-PhoP signaling system, is responsible for acquired colistin resistance in KPC-KP by activation of the Pmr system responsible for modification of the lipopolysaccharide polymyxin target (10). In this work, we investigated a large collection of nonreplicate KPC-KP strains of clinical origin obtained from several different Greek and Italian centers, and we found that the majority of them (39 of 66, 59%) carried alterations of the mgrB gene that were apparently responsible for the colistin-resistant phenotype. These findings underscore the role of this genetic mechanism in acquired colistin resistance by KPC-KP in the studied clinical setting. It will be of interest to investigate whether a similar situation is also found in other epidemiological settings.
A number of different genetic alterations were actually observed, including insertional inactivation by various insertion sequences, point mutations, and small or even large deletions of the mgrB locus. These findings reflect the occurrence of several independent mutational events and suggest that genetic alteration of mgrB may occur at relatively high frequency and without major consequences for fitness and virulence of the KPC-KP strain. A similar hypothesis is further underscored by the phenomena of clonal expansions of colistin-resistant strains carrying mgrB alterations observed in some centers and will be the subject of future investigation.
Altogether, inactivation of mgrB by insertion sequences, and especially by IS_5_-like elements, appeared to be the most common mechanism of mgrB alteration in KPC-KP. This observation reinforces the hypothesis regarding the existence of a specific hot spot for IS_5_ insertion in the mgrB gene (10), although IS_5_-like elements were also detected at different positions and in different orientations. It will also be interesting to assess whether this mechanism of mgrB inactivation is facilitated by the presence of similar ISs on KPC-encoding plasmids.
Regarding the missense mutations in the mgrB gene, observed in 7 isolates, the complementation of the mutant restored the susceptibility to colistin in all cases, suggesting that the mutated protein was not or was only partially functional. However, given the experimental approach performed in this work (complementation was carried out with a plasmid-borne wild-type mgrB gene expressed under its own promoter), we cannot completely rule out that, in these cases, the mutated MgrB protein was functional and the resistance was due to an alteration of the mgrB promoter leading to downregulated expression.
Although many colistin-resistant strains exhibited an alteration of mgrB, more than one-third of them carried a wild-type mgrB gene, revealing that mgrB alterations are not the sole mechanism of colistin resistance in the clinical setting. Additional mechanisms of colistin resistance might be related to promoter mutations leading to a reduction of mgrB expression or to mutations in other loci leading to upregulation of the Pmr system responsible for modification of the lipopolysaccharide polymyxin target. A recent report on in vitro selection of colistin-resistant K. pneumoniae isolates has described the occurrence of pmrB and phoQ mutations associated with colistin-resistant phenotypes (18), although the role of the observed mutations was not experimentally confirmed. Other recent reports have demonstrated the role of pmrB mutations in colistin resistance among clinical isolates of KPC-KP (20, 21). Further studies are ongoing to characterize the colistin resistance mechanisms in strains carrying a wild-type mgrB gene.
Alterations of mgrB were associated with a relatively broad range of colistin MICs (4 to 64 μg/ml), and no significant differences in the distribution of colistin MICs were observed between strains carrying mgrB alterations or other resistance mechanisms. Further investigations will be necessary to understand how the expression of colistin resistance is modulated in mutants carrying similar or different resistance mechanisms.
ACKNOWLEDGMENTS
This work was supported by a grant from FP7 project EvoTAR (no. HEALTH-F3-2011-2011-282004) (to G.M.R).
The COLGRIT Study Group participants were Erminia Casari (Humanitas Research Hospital, Milan, Italy), Antonella Navarra (Salvatore Maugeri Foundation, Pavia, Italy), Lucina Fossati (Città della Salute e della Scienza Hospital, Turin, Italy), Francesco Luzzaro (A. Manzoni Hospital, Lecco, Italy), Luisa Santoriello (Santa Corona Hospital, Pietra Ligure, Italy), Mario Sarti (S. Agostino Estense Hospital, Modena, Italy), Simone Ambretti (Sant'Orsola-Malpighi University Hospital, Bologna, Italy), Esther Manso (Ospedali Riuniti di Ancona University Hospital, Ancona, Italy) Teresa Spanu (A. Gemelli Hospital, Catholic University of Rome, Rome, Italy), Pier Giulio Conaldi (Mediterranean Institute of Transplantation [ISMETT], Palermo, Italy), Stefania Stefani (Department of Bio-Medical Sciences, University of Catania, Catania, Italy), Varvara Galanopoulou-Giouzepa (General Hospital of Thessaloniki Agios Pavlos, Salonika, Greece), Helen Souki (General Hospital of Agrinio, Agrinio, Greece), Evangelia Platsouka (General Hospital of N. Ionia Konstantopoulion-Patission, Athens, Greece), and Helen Vagiakou (General Hospital of Athens Gennimatas, Athens, Greece).
Footnotes
Published ahead of print 14 July 2014
REFERENCES
- 1.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17:1791–1798. 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25:682–707. 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canton R, Akova M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Miriagou V, Naas T, Rossolini GM, Samuelsen O, Seifert H, Woodford N, Nordmann P, the European Network on Carbapenemases 2012. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 18:413–431. 10.1111/j.1469-0691.2012.03821.x [DOI] [PubMed] [Google Scholar]
- 4.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13:785–796. 10.1016/S1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrosillo N, Giannella M, Lewis R, Viale P. 2013. Treatment of carbapenem-resistant Klebsiella pneumoniae: the state of the art. Expert Rev. Anti Infect. Ther. 11:159–177. 10.1586/eri.12.162 [DOI] [PubMed] [Google Scholar]
- 6.Bogdanovich T, Adams-Haduch JM, Tian GB, Nguyen MH, Kwak EJ, Muto CA, Doi Y. 2011. Colistin-resistant, Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae belonging to the international epidemic clone ST258. Clin. Infect. Dis. 53:373-376. 10.1093/cid/cir401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zagorianou A, Sianou E, Iosifidis E, Dimou V, Protonotariou E, Miyakis S, Roilides E, Sofianou D. 2012. Microbiological and molecular characteristics of carbapenemase-producing Klebsiella pneumoniae endemic in a tertiary Greek hospital during 2004–2010. Euro Surveill. 17(7):pii=20088http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=2008 [PubMed] [Google Scholar]
- 8.Mammina C, Bonura C, Di Bernardo F, Aleo A, Fasciana T, Sodano C, Saporito MA, Verde MS, Tetamo R, Palma DM. 2012. Ongoing spread of colistin-resistant Klebsiella pneumoniae in different wards of an acute general hospital, Italy, June to December 2011. Euro Surveill. 17(33):pii=20248http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20248 [PubMed] [Google Scholar]
- 9.Giani T, Pini B, Arena F, Conte V, Bracco S, Migliavacca R, AMCLI-CRE Survey Participants. Pantosti A, Pagani L, Luzzaro F, Rossolini GM. 2013. Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 May to 30 June 2011. Euro Surveill. 18(22):pii=20489http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20489 [PubMed] [Google Scholar]
- 10.Cannatelli A, D'Andrea MM, Giani T, Di Pilato V, Arena F, Ambretti S, Gaibani P, Rossolini GM. 2013. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57:5521–5526. 10.1128/AAC.01480-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.López-Camacho E, Gómez-Gil R, Tobes R, Manrique M, Lorenzo M, Galván B, Salvarelli E, Moatassim Y, Salanueva IJ, Pareja E, Codoñer FM, Alvarez-Tejado M, Garcillán-Barcia MP, De la Cruz F, Mingorance J. 2014. Genomic analysis of the emergence and evolution of multidrug resistance during a Klebsiella pneumoniae outbreak including carbapenem and colistin resistance. J. Antimicrob. Chemother. 69:632–636. 10.1093/jac/dkt419 [DOI] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 9th ed. CLSI document M07-A9 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 13.van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl 3):1–46 [DOI] [PubMed] [Google Scholar]
- 14.Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182. 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 16.Umeda M, Ohtsubo E. 1991. Four types of IS_1_ with differences in nucleotide sequence reside in the Escherichia coli K-12 chromosome. Gene 98:1–5. 10.1016/0378-1119(91)90096-T [DOI] [PubMed] [Google Scholar]
- 17.Carattoli A, Villa L, Poirel L, Bonnin RA, Nordmann P. 2012. Evolution of IncA/C _bla_CMY-2-carrying plasmids by acquisition of the _bla_NDM-1 carbapenemase gene. Antimicrob. Agents Chemother. 56:783–786. 10.1128/AAC.05116-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi M-J, Ko KS. 2014. Mutant prevention concentrations of colistin for Acinetobacter baumannii, Pseudomonas aeruginosa and Klebsiella pneumoniae clinical isolates. J. Antimicrob. Chemother. 69:275–277. 10.1093/jac/dkt315 [DOI] [PubMed] [Google Scholar]
- 19.Liu P, Li P, Jiang X, Bi D, Xie Y, Tai C, Deng Z, Rajakumar K, Ou HY. 2012. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J. Bacteriol. 194:1841–1842. 10.1128/JB.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cannatelli A, Di Pilato V, Giani T, Arena F, Ambretti S, Gaibani P, D'Andrea MM, Rossolini GM. 2014. In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC producing Klebsiella pneumoniae is associated with low-dosage colistin treatment. Antimicrob. Agents Chemother. 58:4399–4403. 10.1128/AAC.02555-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayol A, Poirel L, Brink A, Villegas MV, Yilmaz M, Nordmann P. 2014. Resistance to colistin associated to a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob. Agents Chemother. 58:4762–4766. 10.1128/AAC.00084-14 [DOI] [PMC free article] [PubMed] [Google Scholar]