TorsinA in the nuclear envelope (original) (raw)
Abstract
Early-onset torsion dystonia, a CNS-based movement disorder, is usually associated with a single amino acid deletion (ΔE302/303) in the protein torsinA. TorsinA is an AAA+ ATPase in the endoplasmic reticulum, but what it does is unknown. Here, we use torsinA mutants with defects in ATP hydrolysis (E171Q, ATP-bound) and ATP binding (K108A, ATP-free) to probe torsinA's normal cellular function. Surprisingly, ATP-bound torsinA is recruited to the nuclear envelope (NE) of transfected cells, where it alters connections between inner and outer nuclear membranes. In contrast, ATP-free torsinA is diffusely distributed throughout the endoplasmic reticulum and has no effect on the NE. Among AAA+ ATPases, affinity for substrates is high in the ATP-bound and low in the ATP-free state, leading us to propose that component(s) of the NE may be substrates for torsinA. We also find that the disease-promoting ΔE302/303 mutant is in the NE, and that this relocalization, as well as the mutant's previously described ability to induce membranous inclusions, is eliminated by the K108A ATP-binding mutation. These results suggest that changes in interactions involving torsinA in the NE could be important for the pathogenesis of dystonia and point to torsinA and related proteins as a class of ATPases that may operate in the NE.
Early-onset torsion dystonia, also known as Oppenheim's disease or DYT1 dystonia, is the most severe form of hereditary dystonia (1). The disease is inherited as an autosomal dominant trait with ≈30% penetrance and is characterized by CNS-driven muscle contractions in arms and/or legs that evolve during childhood and adolescence. No distinct neuropathology has been associated with this disease (2), although studies using positron emission tomography have established that there are changes in regional brain metabolic patterns (3).
Positional cloning of the DYT1 gene led to discovery of the protein torsinA (4). Most cases of DYT1 dystonia are associated with a 3-bp deletion (ΔGAG) in torsinA that removes one of a pair of glutamic acid residues (E302/E303) (4). TorsinA contains an ATP-binding domain characteristic of the AAA+ family of ATPases (ATPases associated with a variety of cellular activities) (4, 5). AAA+ ATPases induce conformational changes, sometimes as extreme as complete unfolding, in their substrates, often leading to dissociation of otherwise stable protein complexes or aggregates (6, 7). Examples include SNARE complex disassembly by NSF, microtubule-severing by katanin, and dissociation of protein aggregates by ClpB (reviewed in refs. 7 and 8). Most AAA+ ATPases operate as ring-shaped oligomers in which activity depends on cooperation between coassembled subunits. This provides one explanation for why mutations in genes encoding several of these proteins have dominant effects in vivo (6).
TorsinA is the first AAA+ ATPase found to reside in the lumen of the endoplasmic reticulum (ER) (9–11) and is widely expressed throughout the body, with highest levels in liver, muscle, pancreas, and regions of the brain (4). Related human proteins torsinB, torp2A, and torp3A also have N-terminal signal sequences to direct them to the lumen of the secretory pathway (12), as do three similar proteins in Caenorhabditis elegans (13, 14) and one in Drosophila melanogaster. No protein clearly related to torsinA has been found in yeast or in prokaryotes.
Given its localization in the ER, torsinA presumably functions as an ATP-dependent chaperone there. Among the many tasks of the ER are processing, sorting, and packaging membrane and secretory proteins; lipid biosynthesis; regulation of intracellular Ca2+; and assembly of specialized domains such as the nuclear envelope (NE) (15, 16). There are few clues to guide thinking as to which of these processes, or others, involve torsinA. TorsinA levels do not increase after inducing ER stress by treating cells with DTT or tunicamycin (10), as would be expected if it were important in general ER quality control. Expression of the mouse homologue of torp3a, ADIR, increases when cells are treated with IFN-α (17), pointing to a role for torsin proteins in antiviral responses. In C. elegans, one torsin homolog, OOC-5, is involved in nuclear rotation during early embryogenesis (13). Recent studies have identified roles for overexpressed torsinA in modulating cytoplasmic quality control systems (14, 18) and in protecting cells from oxidative stress (19, 20), but whether these are direct or indirect effects of expressing torsinA remains to be established.
What is wrong with ΔE302/303 torsinA, and why does it promote the development of dystonia? Until we know what torsinA normally does, we cannot definitively answer these questions. Results to date suggest that the deletion does not perturb overall protein structure or basal ATPase activity (11, 21, 22) but instead changes something about torsinA's behavior in the ER membrane (9, 11, 23). In cultured cells, overexpressed ΔE302/303 torsinA induces characteristic membranous inclusions apparently derived from the ER (9, 11). However, because there is no evidence of inclusion in cells of dystonia patients (2, 24), the relationship between them and early-onset torsion dystonia is uncertain.
In this study, we take advantage of conserved ATP-interacting motifs in the AAA+ domain of torsinA to design mutant proteins with predictable alterations in their binding of nucleotide and substrates. Using these mutants, we identify the NE as a likely site of torsinA's action. We conclude that changes in torsinA activity in the NE may be relevant to the development of dystonia, because we find that ΔE302/303 torsinA is also in the NE and recruits an NE marker into the inclusions that it induces. Taken together, these studies suggest an unexpected site of action for torsinA and raise the interesting possibility that DYT1 dystonia may be one of a growing number of diseases associated with defects in NE structure and function.
Materials and Methods
Plasmids. Wild-type torsinA-GFP, ΔE302/303 torsinA-GFP, and torsinB-GFP were generated by amplifying human torsinA or -B, respectively, with primers containing _Xho_I and _Eco_RI restriction sites and ligating them into pEGFP-N1 or pECFP-N1 (Clontech). K108A and E171Q mutations were introduced into GFP-tagged, untagged (pcDNA3, ref. 9), and ΔE302/303 torsinA by QuikChange mutagenesis (Stratagene). The sequences of all constructs were verified by automated nucleotide sequencing. Sequences of all primers are available upon request. Lamin B-receptor yellow fluorescent protein (YFP) (25) was a kind gift from Jennifer Lippincott-Schwartz (National Institutes of Health, Bethesda).
Antibodies. The following antibodies were used: mouse anti-Nup153 (SA1, kind gift from Brian Burke, University of Florida, Gainesville), mouse antinucleoporin 414 (Covance, Berkeley, CA), mouse anti-LAP2 (BD Biosciences), mouse anti-PDI (StressGen Biotechnologies, Victoria, Canada), mouse and rabbit antitorsinA (9, 26), and rabbit anti-GFP (27). An additional rabbit polyclonal antibody against torsinA (rabbit B10) was raised and affinity purified by using His-6-tagged torsinA (63–332) expressed in and isolated from Escherichia coli. Secondary goat anti-mouse and goat anti-rabbit antibodies conjugated to Alexa 488 or Alexa 568 were purchased from Molecular Probes.
Cell Culture and Transfection. COS-7 and Chinese hamster ovary (CHO) cells from American Type Culture Collection were grown in DMEM supplemented with 10% FBS. PC12 cells from Tom Martin (University of Wisconsin, Madison) grew in DMEM containing 5% horse serum, 5% supplemented calf serum (HyClone), and 2% FBS. Transient transfections were performed by using Lipofectamine (Invitrogen) according to the manufacturer's instructions. Cells were analyzed 6–24 h after transfection. All transfections were repeated three or more times with equivalent results. CHO cell lines stably expressing torsinA-GFP proteins were selected by using 400 μg/ml G418 and expanded from single colonies. Expression levels relative to endogenous torsinA were determined by immunoblotting with torsinA-specific antibodies.
Immunoblotting and Glycosidase Treatment. Immunoblots were developed as described (27). Triton X-100 solubilized cell extracts were boiled in 1× SDS-sample buffer and treated with endoglycosidase H following the manufacturer's directions (New England Biolabs).
Immunofluorescence and Light Microscopy. Immunofluorescence was performed on cells fixed in 3.5% paraformaldehyde/4% sucrose and permeabilized with 0.2% Triton X-100. GFP fluorescence was visualized directly after fixation and mounting in Mowiol (Calbiochem). Epifluorescence images were captured with a Leica Diaplan microscope using a 63 × 1.4-numerical aperture (N.A.) objective and a Zeiss AxioCam MRm. Confocal microscopy was performed on a Bio-Rad Radiance 2000 microscope with a 63 × 1.4-N.A. objective by using 488- and 543-nm laser lines. All images were acquired with sequential scans by using lasersharp 2000. Where indicated, laser settings were kept constant to allow direct comparison of expression in different cells. Fluorescence intensity in regions of interest was measured by using metamorph 6.0 software (Universal Imaging, Downingtown, PA). Figures were prepared by using photoshop and illustrator (Adobe Systems, San Jose, CA).
Electron Microscopy. For thin sections, cells grown to ≈80% confluence were removed by trypsinization, fixed with 2.5% glutaraldehyde in Na-cacodylate, embedded, sectioned, and stained with uranyl acetate according to standard procedures. For freeze-fracture, cells pelleted from warm media were rapidly frozen and transferred to liquid nitrogen. Freeze-fracture, deepetching, and replica preparation were carried out as described (28). Thin sections and replicas were viewed in a JEOL transmission electron microscopy operating at 100 kV and photographed with an Advanced Microscopy Techniques (Danvers, MA) charge-coupled device camera system.
Results
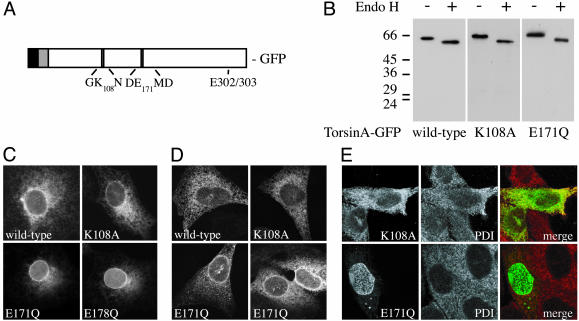
Cellular Behavior of TorsinA ATP Binding and Hydrolysis Mutants. AAA+ ATPases transmit conformational changes to substrate proteins by cycling through states with high and low affinity for substrates in parallel with ATP binding and hydrolysis (7). To explore the cellular function of torsinA, we took advantage of conserved sequence motifs within its ATPase domain to make mutations that should interfere with these cycles. An essential lysine in Walker A motifs interacts with β and γ phosphate groups on ATP to stabilize nucleotide binding, whereas a conserved acidic residue in Walker B motifs is important in activating ATP hydrolysis (5). Mutating these residues (K108A and E171Q) generated torsinA variants predicted to be primarily nucleotide-free (K108A, ATP-binding mutant) or ATP-bound (E171Q, ATP-hydrolysis mutant) (Fig. 1_A_). By analogy to other AAA+ ATPases (27, 29–31), we expected that the E171Q ATP-bound mutant would function as a substrate trap, binding tightly to torsinA's normal substrate(s) in living cells and possibly interfering with their function. In contrast, we expected that the K108A nucleotide-free mutant would bind substrates poorly or not at all. To facilitate analysis of these mutants, we fused GFP to torsinA. GFP-tagged torsinA and mutants were expressed, glycosylated, and, like untagged torsinA, remained sensitive to digestion with endonuclease H (Fig. 1_B_).
Fig. 1.
Expression and distribution of torsinA mutants. (A) Domain structure of torsinA, showing position of Walker A (K108A), Walker B (E171Q), and DYT1 (ΔE302/303) mutations. N-terminal signal sequence (black), the adjacent hydrophobic domain implicated in ER retention and membrane association (residues 21–40, gray), and the position of our GFP tag are also shown. GFP was added to the C terminus to avoid interfering with the N-terminal hydrophobic sequences. (B) TorsinA-GFP mutants expressed in CHO cells are ER glycoproteins sensitive to endoglycosidase H digestion (compare - and + EndoH). (C) Localization of GFP-tagged torsinA mutants. Epifluorescence images show the distribution in transiently transfected COS-7 cells of wild type, K108A, E171Q torsinA-GFP, and torsinB(E178Q)-GFP. (D) Localization of untagged torsinA mutants. Shown are confocal images of CHO cells transiently transfected with untagged torsinA, torsinA(K108A), or torsinA(E171Q). TorsinA was visualized by indirect immunofluorescence. The first three images were taken with the same settings on the confocal microscope, whereas the fourth had reduced gain to show the distribution of highly overexpressed mutant. (E) Confocal images comparing the ER in CHO cells stably expressing torsinA(K108A)-GFP or (E171Q)-GFP with that in untransfected cells. Shown are GFP (Left), the ER marker protein disulfide isomerase (Center), and a merger of the two (Right).
To study the behavior of torsinA containing these mutations, we examined torsinA's distribution in transiently transfected COS-7 cells. Using fluorescence microscopy, we found that both wild-type and K108A torsinA-GFP were present throughout the ER (Fig. 1_C_). In contrast, torsinA(E171Q)-GFP was concentrated around the nucleus, apparently in the NE (Fig. 1_C_). A comparable mutation in torsinA's closest homolog, torsinB, produced a protein again concentrated around the nucleus (Fig. 1_C_). We also made a double mutant containing both K108A and E171Q mutations and found that its distribution was closer to that of torsinA(K108A) than torsinA(E171Q) (data not shown).
To confirm that the effects of these mutations were intrinsic to torsinA, we looked at the distribution of untagged mutants in transfected CHO cells (chosen because they have relatively low levels of endogenous torsinA). We again found enrichment of torsinA(E171Q) in the NE (Fig. 1_D_). As the level of expression increased, the amount of torsinA(E171Q) in the peripheral ER increased (compare Fig. 1_D_ Left with Right), suggesting possible saturation of factor(s) responsible for recruiting the mutant to the NE. At high expression levels, one difference between untagged and GFP-tagged torsinA was that the GFP-tagged proteins formed spheroid inclusions (see dots in Fig. 1_C_ wild-type torsinA-GFP, also in Figs. 1_E_ and 2). These inclusions, like those associated with expression of untagged ΔE302/303 torsinA (9, 11), stained weakly with markers of the ER and not at all with antibodies recognizing ubiquitin or vimentin as cytoplasmic aggresomes would (data not shown) and thus seem to represent outgrowths of the ER or NE. Only cells with few or no inclusions were used for analysis.
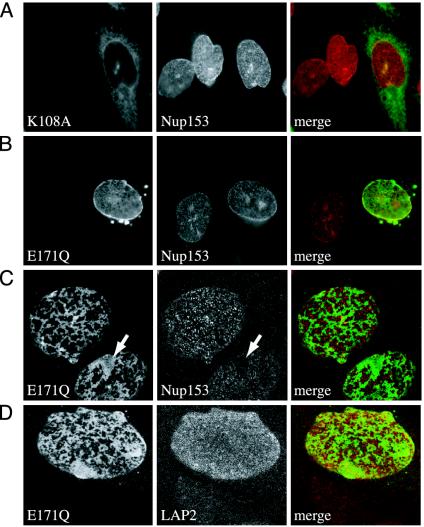
Fig. 2.
Colocalization of torsinA mutants with NE markers. (A) Epifluorescence image of torsinA(K108A)-GFP and untransfected CHO cells stained for nucleoporin Nup153 (red). (B) Epifluorescence image of torsinA(E171Q)-GFP and untransfected CHO cells stained for Nup153 (red). (C) Confocal image of lower nuclear surface in (E171Q)-GFP expressing CHO cell stained for Nup153. Arrow highlights region lacking Nup153. (D) Confocal image of similar nuclear surface stained for LAP2.
To enable study of torsinA mutants in a homogeneous cell population, we generated clonal CHO cell lines stably expressing K108A or E171Q torsinA-GFP at levels ranging from a few to as many as 50-fold above that of endogenous torsinA. We selected cell lines with comparable expression for further analysis. Both K108A and E171Q torsinA-GFP displayed the same localization they had after transient transfection, and neither noticeably perturbed the structure of the ER marked by protein disulfide isomerase (Fig. 1_E_).
TorsinA(E171Q) Affects NE Structure. To examine in more detail the specific relationship between torsinA mutants and the NE, which consists of inner and outer nuclear membranes, nuclear pores, and the nucleoskeletal lamina (32), we carried out immunofluorescence using markers for NE components. We monitored colocalization of torsinA-GFP fusion proteins with these markers and also looked for any unusual effects of the mutants on organization of the NE. This was particularly important because we anticipated that the Walker B ATP-bound mutant, by prolonged binding to torsinA's substrates, might perturb their normal function. To facilitate comparison between cells, we examined nuclei in mixtures of stably transfected and untransfected CHO cells. TorsinA(K108A)-GFP, although present around the nucleus, was not enriched there and did not colocalize well with markers of the NE, including the nucleoporin Nup153 (Fig. 2_A_). In contrast, torsinA(E171Q)-GFP colocalized well with NE markers except where it was present in occasional inclusions (Fig. 2_B_).
To more closely define the relationship between torsinA(E171Q)-GFP and components of the NE, we switched to confocal microscopy. Tangential views of the nuclear surface showed that torsinA(E171Q)-GFP forms honeycomb-like arrays around the nucleus (Fig. 2 C and D). As expected, Nup153 was in punctate structures on the nuclear surface. Interestingly, at this higher level of resolution, Nup153 no longer overlapped well with torsinA(E171Q) but instead was present everywhere that torsinA was not. In areas of confluent torsinA(E171Q)-GFP, nuclear pores were typically absent (arrows in Fig. 2_C_). This alternating distribution contrasts with that of the inner nuclear membrane and lamina-associated polypeptide LAP2, which was more evenly spread over the nuclear surface and unaffected by torsinA(E171Q)-GFP (Fig. 2_D_).
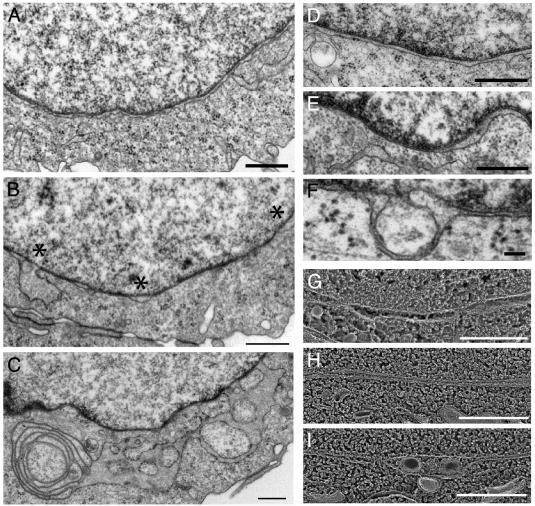
To look for more subtle changes in the structure of the NE, we turned to electron microscopy. The NE in CHO cells displays typical uniform spacing between outer and inner nuclear membranes, evenly distributed nuclear pores bridging these membranes, and occasional tubular connections to the peripheral ER (Fig. 3 A and D). In CHO cells that overexpress torsinA(E171Q)-GFP, several aspects of NE structure were abnormal (Fig. 3 B, E, and F). Inner and outer nuclear membranes were often tightly apposed to each other, forming stretches of NE with abnormally narrow perinuclear space and no nuclear pores. Such closely spaced membranes were interspersed with regions in which the membranes were unduly separated from each other, ballooning into large bubbles in the NE (* in Fig. 3_B_). Also, at many sites the inner nuclear membrane separated from the nuclear lamina, producing membrane-delimited herniations into the perinuclear space (Fig. 3_F_). Depending on the plane of sectioning, these often looked like isolated vesicles or vacuoles. Similar changes in the NE were seen in three independent cell lines. The cell line expressing the highest level of torsinA(E171Q)-GFP (≈50-fold above endogenous torsinA) also had abnormal structures in the peripheral ER (interconnected ribosome-decorated membranes, Fig. 3_C_) that probably correspond to the spheroid inclusions seen by light microscopy.
Fig. 3.
NE ultrastructure in torsinA(E171Q)-GFP expressing CHO cells. (A) Control CHO NE (nucleus on top, cytoplasm below). (B) NE in torsinA(E171Q)-GFP cell line. Note regions of tight membrane apposition alternating with separated areas indicated by *. (C) Membranous inclusion in high-expressing torsinA(E171Q)-GFP cell line. (D) Control CHO NE. (E) Abnormal spacing in torsinA(E171Q)-GFP cell line. (F) Herniation of inner nuclear membrane into perinuclear space in torsinA(E171Q)-GFP cell line. (G) Quick-freeze/deep-etch image of CHO NE (nucleus on top, cytoplasm below). (H and I) Same in torsinA(E171Q)-GFP cell line. [Bars = 500 nm (A–E) and 100 nm (F).]
To confirm that the abnormalities in nuclear membrane structure were not merely the result of chemical fixation, we quick-froze living cells and carried out freeze-fracture/deepetch electron microscopy. Once again, stretches of abnormally close nuclear membranes were readily apparent in cells expressing torsinA(E171Q)-GFP (compare Fig. 3 G and H). In addition, vesicles (presumably corresponding to herniated membranes) were present in areas where the nuclear membranes were separated (Fig. 3_I_).
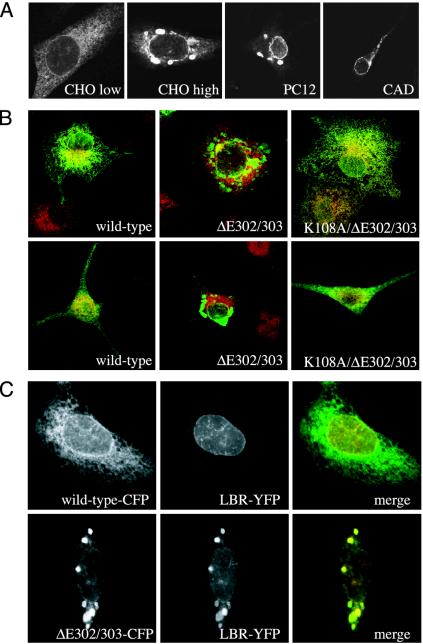
ΔE302/303 TorsinA Is Enriched in the NE When It Can Bind ATP. While studying the behavior of disease-linked ΔE302/303 torsinA transfected into various cell lines (including CHO, PC12, and CAD), we noticed that ΔE302/303 torsinA accumulated not only in inclusions but also in the NE (Fig. 4_A_). This was particularly true at lower expression levels, where the only abnormality in torsinA distribution was often the enhanced NE localization (Fig. 4_A_ Left). At higher levels of expression, cells showed both inclusions and sometimes discontinuous NE staining (Fig. 4_A_ Right). Staining in the peripheral ER was usually decreased. If torsinA(E171Q) concentrates in the NE because it is trapped in the ATP-bound state, we reasoned that the ΔE302/303 mutant might be similarly trapped. If so, localization to the NE (and perhaps also the ability to induce inclusions) should depend on the mutant's ability to bind ATP. To determine whether this is the case, we combined ΔE302/303 and K108A ATP-binding mutations and examined the distribution of the double mutant in transfected cells. Strikingly, the K108A/ΔE302/303 double mutant remained diffusely distributed throughout the ER in both COS-7 and PC12 cells (Fig. 4_B_).
Fig. 4.
ΔE302/303-torsinA in the NE. (A) Indirect immunofluorescence showing localization of ΔE302/303-torsinA (untagged) in transfected CHO, PC12, and CAD cells. CHO low and CHO high show representative low and high expressing cells, taken with 4-fold differences in gain on the microscope. (B) Effects of K108A mutation on localization of ΔE302/303-torsinA. (Upper) COS-7 cells. (Lower) PC12 cells expressing wild-type torsinA (Left), ΔE302/303 torsinA (Center), and K108A/ΔE302/303 torsinA (Right). Cells were stained for torsinA (green) and PDI (red). (C) Inclusions formed by ΔE302/303-torsinA are enriched in lamin-B receptor-YFP. Shown are wild-type (Upper) and ΔE302/303 (Lower) torsinA-CFP coexpressed with lamin-B receptor-YFP. For merge, torsinA-CFP is green, and lamin-B receptor-YFP is red.
Having seen that ΔE302/303 torsinA is present in the NE as well as in inclusions, we wondered what the relationship between the inclusions and components of the NE might be. In particular, if the inclusions develop from the NE, they might contain proteins characteristic of NE membranes. We therefore examined the distribution of a well studied inner nuclear membrane marker, lamin B receptor-YFP (25), coexpressed with either wild-type or ΔE302/303 torsinA-cyan fluorescent protein (CFP). As expected, when cotransfected with torsinA-CFP, lamin B receptor-YFP was seen primarily around the nucleus (Fig. 4_C_). In contrast, lamin B receptor-YFP was present in membranous inclusions induced by ΔE302/303 torsinA-CFP. Its concentration in the inclusions was paralleled only by that of ΔE302/303-torsinA-CFP and was much more pronounced than what we had previously seen with other ER markers (e.g., protein disulfide isomerase in Fig. 4_B_). The presence of an inner nuclear membrane marker in ΔE302/303-induced inclusions raises the possibility that the inner nuclear membrane might contribute to torsinA-induced inclusions, although further study with endogenous markers of the NE will be necessary to confirm this idea.
Discussion
TorsinA is normally found throughout the ER but, as described here, accumulates in the NE when mutated to impair ATP hydrolysis (E171Q Walker B mutation) and when it lacks the E302/303 residue deleted in DYT1 dystonia. Why are these mutants recruited to the NE? We propose that the simplest explanation is that torsinA has a substrate or substrates in the NE, and that its normally transient interaction with them is prolonged by these mutations. The NE, a subdomain of the ER that typically represents ≈10% of its total area (15, 33), has recently been shown to contain as many as ≈80 specific proteins (34). The localization and effects of overexpressed torsinA mutants in the NE shown here, together with the finding that endogenous torsinA is present among the proteins identified in rat liver NEs (34), should help guide future functional and biochemical studies aimed at defining torsinA's molecular targets and the cellular etiology of DYT1 dystonia. These findings do not exclude the presence of additional substrates for torsinA elsewhere in the ER, as suggested by its presence throughout the ER (35).
Our hypothesis that torsinA has substrate(s) in the NE is supported by several observations. (i) Most important is the distinct cellular behavior of torsinA mutants with changes in conserved features of their ATPase domains, the Walker A and B motifs (Figs. 1 and 2). Comparable Walker B mutations have been shown to stabilize interactions between other AAA+ ATPases and their substrates in vitro, whereas Walker A mutations do not (29, 30, 36). In transfected cells, Walker B mutants of NSF, p97, and Vps4/SKD1 bind to membranes containing their respective target molecules and act as substrate traps (27, 29). For several of these enzymes, biochemical studies have confirmed that Walker A and B mutations differentially affect ATP binding and hydrolysis (29). In the case of torsinA, it is not yet clear that conditions for reconstituting efficient ATPase activity outside of the ER have been defined, which is a prerequisite for comparable analyses (ref. 22 and our unpublished observations). (ii) The effects of torsinA(E171Q)-GFP on NE spacing and nuclear pore distribution (Figs. 2 and 3) indicate that accumulation of this mutant in the NE disturbs the arrangement of one or more of its components. Distortions in the NE are a common result of perturbations in lamins, nucleoporins, and other membrane proteins (32, 37), leaving many candidates for the factor(s) affected by torsinA. Based on the structural effects seen here, we propose that proteins involved in maintaining connections between inner and outer membranes are among the likely targets. (iii) Independent evidence pointing to localization and possible function of torsinA and related proteins in the NE comes from studies of the C. elegans torsinA homolog OOC-5, which is present in the NE and plays a role in enabling nuclear rotation in the embryo (13, 38), immunolocalization studies that show that endogenous torsinB is concentrated around the nucleus (26), and another study of transfected torsinA mutants showing NE localization that was published while our paper was under review (39).
The finding that the disease-associated ΔE302/303 torsinA mutant, like the Walker B mutant, is present in the NE, especially at low expression levels that reflect what might be seen in the physiological setting of DYT1 mutant cells (Fig. 4_A_) (40), suggests that a common change might move these mutants to the NE. Prolonged interaction with a substrate protein is again an attractive hypothesis, particularly because the ΔE302/303 mutant accumulates in the NE only when it can bind ATP (Fig. 4_B_). E302 and 303 lie within the C-terminal α-helical subdomain of torsinA's AAA+ fold, where a deletion could impair ATP hydrolysis by directly altering interactions with ATP or changing the interface between adjacent subunits (5). A recent study comparing ATPase activity of wild-type and ΔE302/303 torsinA from a baculovirus expression system found no significant difference between the two (22) but used monomeric enzyme with low specific activity and would likely not have seen changes in the activity of putative torsinA oligomers.
Whereas the simplest explanation for the NE accumulation of Walker B and DYT1 torsinA mutants is that they bind specifically to a substrate protein or proteins in the NE, until we have identified this substrate we cannot rule out other less likely explanations for the observed relocalization. Among these might be some preference of the mutant proteins for the parallel membranes of the NE over the parallel membranes of the rest of the ER. In this case, relocalization of ΔE302/303 torsinA to the NE could adversely affect cellular torsinA function by depleting mutant torsinA (and possibly associated wild-type enzyme in a heterooligomer) from sites of action elsewhere in the ER. Direct identification of torsinA's substrates will be necessary to further understand what recruits torsinA to the NE.
The torsinA mutants studied here varied in their tendency to induce formation of spheroid membranous inclusions when highly overexpressed, with untagged ΔE302/303 and GFP-tagged wild type, E171Q, and ΔE302/303 all noted to form inclusions at some level of expression. Many ER and NE proteins, when overexpressed, generate spheroid inclusions that appear similar to those induced by torsinA (25, 34, 41). There are a variety of different mechanisms by which inclusions form, and it is not yet clear precisely how overexpressed torsinA, present in the ER and NE lumen, induces inclusions. It is also not clear that the inclusions formed in different cells and in response to different torsinA constructs are all the same. An interesting recent study from Lippincott-Schwartz and coworkers (41) showed that GFP attached to the cytoplasmic end of various ER membrane proteins induced inclusions by a mechanism involving low affinity GFP-GFP interactions between ER membranes. Although we cannot exclude that similar low-affinity interactions between the GFP moieties attached to torsinA in the lumen affect the behavior of our fusion proteins, the specificity of the changes we see (NE enrichment only with Walker B and ΔE302/303 mutants) and the similar behavior of GFP-tagged and untagged torsinA proteins suggest that any effects of GFP are secondary. Differences in torsinA's tendency to oligomerize (something that might be affected by GFP) (41) could play a part in determining whether and at what expression level a particular torsinA variant induces inclusions. Further study of the oligomeric state of different torsinA proteins in ER and NE membranes will be needed to test this hypothesis.
The results of this study suggest that DYT1 dystonia may be a disease associated with changes in the NE. A number of diseases, often with tissue-specific phenotypes analogous to those of early-onset torsion dystonia, are known to arise from mutations in proteins of the NE. These include Emery–Dreifuss muscular dystrophy, recessive Charcot–Marie–Tooth neuropathy, and at least 11 others (32, 42). To determine whether an abnormality in the NE or another problem elsewhere in the ER is responsible for DYT1 dystonia will require identification and functional analysis of torsinA's substrates.
Acknowledgments
We thank Robyn Roth and Marilyn Levy for excellent assistance with electron microscopy sample preparation and imaging, members of the Hanson lab for useful discussions and technical assistance, and Didier Hodzic and Cris Bragg for helpful discussions. This work was supported by grants from the Dystonia Medical Research Foundation (to P.I.H.), the W. M. Keck Foundation Distinguished Young Scholars Program (to P.I.H.), the McKnight Foundation (to P.I.H.), and the Jack Fasciana Fund for Support of Dystonia Research (to X.O.B.), and by National Institutes of Health Grants NS28384 (to X.O.B.) and GM29647 (to J.E.H.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER, endoplasmic reticulum; NE, nuclear envelope; CHO, Chinese hamster ovary; YFP, yellow fluorescent protein; CFP, cyan fluorescent protein.
References
- 1.Fahn, S., Bressman, S. B. & Marsden, C. D. (1998) Adv. Neurol. 78**,** 1-10. [PubMed] [Google Scholar]
- 2.Rostasy, K., Augood, S. J., Hewett, J. W., Leung, J. C., Sasaki, H., Ozelius, L. J., Ramesh, V., Standaert, D. G., Breakefield, X. O. & Hedreen, J. C. (2003) Neurobiol. Dis. 12**,** 11-24. [DOI] [PubMed] [Google Scholar]
- 3.Ghilardi, M. F., Carbon, M., Silvestri, G., Dhawan, V., Tagliati, M., Bressman, S., Ghez, C. & Eidelberg, D. (2003) Ann. Neurol. 54**,** 102-109. [DOI] [PubMed] [Google Scholar]
- 4.Ozelius, L. J., Hewett, J. W., Page, C. E., Bressman, S. B., Kramer, P. L., Shalish, C., de Leon, D., Brin, M. F., Raymond, D., Corey, D. P., et al. (1997) Nat. Genet. 17**,** 40-48. [DOI] [PubMed] [Google Scholar]
- 5.Neuwald, A. F., Aravind, L., Spouge, J. L. & Koonin, E. V. (1999) Genome Res. 9**,** 27-43. [PubMed] [Google Scholar]
- 6.Ogura, T. & Wilkinson, A. J. (2001) Genes Cells 6**,** 575-597. [DOI] [PubMed] [Google Scholar]
- 7.Vale, R. D. (2000) J. Cell Biol. 150**,** F13-F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weibezahn, J., Bukau, B. & Mogk, A. (2004) Microb. Cell Fact. 3**,** 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hewett, J., Gonzalez-Agosti, C., Slater, D., Ziefer, P., Li, S., Bergeron, D., Jacoby, D. J., Ozelius, L. J., Ramesh, V. & Breakefield, X. O. (2000) Hum. Mol. Genet. 9**,** 1403-1413. [DOI] [PubMed] [Google Scholar]
- 10.Hewett, J., Ziefer, P., Bergeron, D., Naismith, T., Boston, H., Slater, D., Wilbur, J., Schuback, D., Kamm, C., Smith, N., et al. (2003) J. Neurosci. Res. 72**,** 158-168. [DOI] [PubMed] [Google Scholar]
- 11.Kustedjo, K., Bracey, M. H. & Cravatt, B. F. (2000) J. Biol. Chem. 275**,** 27933-27939. [DOI] [PubMed] [Google Scholar]
- 12.Ozelius, L. J., Page, C. E., Klein, C., Hewett, J. W., Mineta, M., Leung, J., Shalish, C., Bressman, S. B., de Leon, D., Brin, M. F., et al. (1999) Genomics 62**,** 377-384. [DOI] [PubMed] [Google Scholar]
- 13.Basham, S. E. & Rose, L. S. (2001) Development (Cambridge, U.K.) 128**,** 4645-4656. [DOI] [PubMed] [Google Scholar]
- 14.Caldwell, G. A., Cao, S., Sexton, E. G., Gelwix, C. C., Bevel, J. P. & Caldwell, K. A. (2003) Hum. Mol. Genet. 12**,** 307-319. [DOI] [PubMed] [Google Scholar]
- 15.Voeltz, G. K., Rolls, M. M. & Rapoport, T. A. (2002) EMBO Rep. 3**,** 944-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattaj, I. W. (2004) Nat. Rev. Mol. Cell Biol. 5**,** 1-5. [DOI] [PubMed] [Google Scholar]
- 17.Dron, M., Meritet, J. F., Dandoy-Dron, F., Meyniel, J. P., Maury, C. & Tovey, M. G. (2002) Genomics 79**,** 315-325. [DOI] [PubMed] [Google Scholar]
- 18.McLean, P. J., Kawamata, H., Shariff, S., Hewett, J., Sharma, N., Ueda, K., Breakefield, X. O. & Hyman, B. T. (2002) J. Neurochem. 83**,** 846-854. [DOI] [PubMed] [Google Scholar]
- 19.Kuner, R., Teismann, P., Trutzel, A., Naim, J., Richter, A., Schmidt, N., von Ahsen, O., Bach, A., Ferger, B. & Schneider, A. (2003) Neurosci. Lett. 350**,** 153-156. [DOI] [PubMed] [Google Scholar]
- 20.Shashidharan, P., Paris, N., Sandu, D., Karthikeyan, L., McNaught, K. S., Walker, R. H. & Olanow, C. W. (2004) J. Neurochem. 88**,** 1019-1025. [DOI] [PubMed] [Google Scholar]
- 21.Liu, Z., Zolkiewska, A. & Zolkiewski, M. (2003) Biochem. J. 374**,** 117-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kustedjo, K., Deechongkit, S., Kelly, J. W. & Cravatt, B. F. (2003) Biochemistry 42**,** 15333-15341. [DOI] [PubMed] [Google Scholar]
- 23.Bragg, D. C., Slater, D. J. & Breakefield, X. O. (2004) Adv. Neurol. 94**,** 87-93. [PubMed] [Google Scholar]
- 24.Walker, R. H., Brin, M. F., Sandu, D., Good, P. F. & Shashidharan, P. (2002) Neurology 58**,** 120-124. [DOI] [PubMed] [Google Scholar]
- 25.Ellenberg, J., Siggia, E. D., Moreira, J. E., Smith, C. L., Presley, J. F., Worman, H. J. & Lippincott-Schwartz, J. (1997) J. Cell Biol. 138**,** 1193-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hewett, J. W., Kamm, C., Boston, H., Beauchamp, R., Naismith, T., Ozelius, L., Hanson, P. I., Breakefield, X. O. & Ramesh, V. (2004) J. Neurochem., in press. [DOI] [PubMed]
- 27.Dalal, S., Rosser, M. F., Cyr, D. M. & Hanson, P. I. (2004) Mol. Biol. Cell 15**,** 637-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heuser, J. (1981) Methods Cell Biol. 22**,** 97-122. [DOI] [PubMed] [Google Scholar]
- 29.Babst, M., Wendland, B., Estepa, E. J. & Emr, S. D. (1998) EMBO J. 17**,** 2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weibezahn, J., Schlieker, C., Bukau, B. & Mogk, A. (2003) J. Biol. Chem. 278**,** 32608-32617. [DOI] [PubMed] [Google Scholar]
- 31.Whiteheart, S. W., Rossnagel, K., Buhrow, S. A., Brunner, M., Jaenicke, R. & Rothman, J. E. (1994) J. Cell Biol. 126**,** 945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burke, B. & Stewart, C. L. (2002) Nat. Rev. Mol. Cell Biol. 3**,** 575-585. [DOI] [PubMed] [Google Scholar]
- 33.Bannykh, S. I., Rowe, T. & Balch, W. E. (1996) J. Cell Biol. 135**,** 19-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schirmer, E. C., Florens, L., Guan, T., Yates, J. R., III, & Gerace, L. (2003) Science 301**,** 1380-1382. [DOI] [PubMed] [Google Scholar]
- 35.Breakefield, X. O., Kamm, C. & Hanson, P. I. (2001) Neuron 31**,** 9-12. [DOI] [PubMed] [Google Scholar]
- 36.Nagiec, E. E., Bernstein, A. & Whiteheart, S. W. (1995) J. Biol. Chem. 270**,** 29182-29188. [DOI] [PubMed] [Google Scholar]
- 37.Suntharalingam, M. & Wente, S. R. (2003) Dev. Cell 4**,** 775-789. [DOI] [PubMed] [Google Scholar]
- 38.Basham, S. E. & Rose, L. S. (1999) Dev. Biol. 215**,** 253-263. [DOI] [PubMed] [Google Scholar]
- 39.Goodchild, R. E. & Dauer, W. T. (2004) Proc. Natl. Acad. Sci. USA 101**,** 847-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bragg, D. C., Camp, S. M., Kaufman, C. A., Wilbur, J. D., Boston, H., Schuback, D. E., Hanson, P. I., Sena-Esteves, M. & Breakefield, X. O. (2004) Neuroscience 125**,** 651-661. [DOI] [PubMed] [Google Scholar]
- 41.Snapp, E. L., Hegde, R. S., Francolini, M., Lombardo, F., Colombo, S., Pedrazzini, E., Borgese, N. & Lippincott-Schwartz, J. (2003) J. Cell Biol. 163**,** 257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ostlund, C. & Worman, H. J. (2003) Muscle Nerve 27**,** 393-406. [DOI] [PubMed] [Google Scholar]