Coexpression of Neighboring Genes in the Genome of Arabidopsis thaliana (original) (raw)
Abstract
Large-scale analyses of expression data of eukaryotic organisms are now becoming increasingly routine. The data sets are revealing interesting and novel patterns of genomic organization, which provide insight both into molecular evolution and how structure and function of a genome interrelate. Our study investigates, for the first time, how genome organization affects expression of a gene in the Arabidopsis genome. The analyses show that neighboring genes are coexpressed. This pattern has been found for all eukaryotic genomes studied so far, but as yet, it remains unclear whether it is due to selective or nonselective influences. We have investigated reasons for coexpression of neighboring genes in Arabidopsis, and our evidence suggests that orientation of gene pairs plays a significant role, with potential sharing of regulatory elements in divergently transcribed genes. Using the data available in the KEGG database, we find evidence that genes in the same pathway are coexpressed, although this is not a major cause for the coexpression of neighboring genes.
Several large-scale analyses of expression data in higher eukaryotes have shown that neighboring genes tend to have similar expression patterns. Regional similarity in expression has been found in humans (Caron et al. 1995; Lercher et al. 2002), Drosophila (Cohen et al. 2000; Boutanaev et al. 2002; Spellman and Rubin 2002;), yeast (Cohen et al. 2000), and Caenorhabditis elegans (Lercher et al. 2003).
There are a number of potential causes for neighboring genes in a genome to have similar expression patterns. First, duplicated genes often remain neighbors for significant periods of evolutionary time, and given their common ancestry, are likely to have similar expression patterns. Second, neighboring genes in prokaryotic genomes, particularly those that are functionally related, are often found in operons. To date, operons have been found in Caenorhabditis elegans (Blumenthal et al. 2002), and there are also several examples of polycistronic genes in the human genome (Reiss et al. 1998; Gray et al. 1999). It is possible that genes involved in a particular metabolic pathway that requires coordinate regulation will be found to be clustered in other higher eukaryotes. For example, recent studies on Arabidopsis thaliana have identified clustered genes in relation to root development (Birnbaum et al. 2003) and mitochondrial function (Elo et al. 2003). Third, even in the absence of coordinate regulation, the close proximity of neighboring genes in eukaryotic genomes could lead to sharing of _cis_-regulatory elements such as enhancers or insulators, leading to a similarity in their expression patterns. Fourth, there may be a selective advantage for coexpressed genes to be in the same chromosomal domain.
The observations on coexpression of neighboring genes have been based on data gained from a variety of experimental techniques. These have included Serial Analysis of Gene Expression (SAGE; Lercher et al. 2002), DNA microarray data (Spellman and Rubin 2002), and data derived from gene annotation, such as Gene Ontology (GO terms; Spellman and Rubin 2002) and pathway assignation (Lee and Sonnhammer 2003).
Increasingly, data sets from DNA microarrays, which enable large numbers of genes to be analyzed simultaneously in a single experiment, are used for bioinformatics analysis. However, there are several different microarray technologies currently in use, including cDNA, oligo, and Affymetrix arrays. It is unclear as yet whether quantitative comparison of data sets from these different technologies is feasible. An example of this difficulty is illustrated in Kuo et. al. (2002), where a comparison of human microarray data sets using cDNA and Affymetrix technologies found no direct correlation.
This study describes the first analysis of the Arabidopsis genome to determine whether neighboring genes are coexpressed. Gene expression in Arabidopsis has been studied in-depth worldwide, and there are publicly available data sets for both cDNA and Affymetrix microarrays. This gives the added opportunity to directly compare the impact of these two technologies on the analysis. Our results from a pairwise comparison, show that coexpression of neighboring genes does exist in the Arabidopsis genome. There is significant disparity in the conclusions that can be drawn from data derived from the two different microarray technologies. The causes of coexpression have been explored, and evidence is provided to suggest that neither gene duplication nor common functionality are the main cause for coexpression of neighboring genes in the Arabidopsis genome.
RESULTS
Neighboring Genes Are Coexpressed
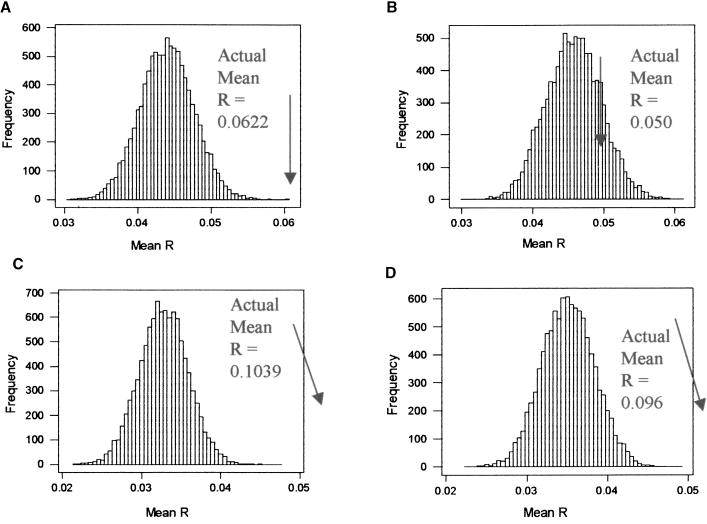
The data sets used for this analysis were derived from cDNA and Affymetrix microarrays. For each data set, as shown in Figure 1, the mean Pearsons correlation coefficient (R) of all pairs of neighboring genes was calculated to give a measure of the similarity in their expression pattern. The significance of this value was confirmed using a Monte-Carlo simulation, which compares the value obtained to a distribution of random mean R-values derived from the same set of data. Surprisingly, the mean R from the random distribution was positive rather than being zero, as would be expected by chance. A possible explanation for this effect may be the influence of housekeeping genes showing common patterns of expression in many different tissues and experimental conditions, thereby shifting the mean value into the positive. There was clear evidence for significant coexpression of neighboring genes across the genome. This was obtained for data sets from both cDNA and Affymetrix microarrays (cDNA arry: P < 0.0001, +4.99 standard deviations from the random mean, Affymetrix array, +23.1 standard deviations; Fig. 1A,C). Tandem duplicates, defined as gene pairs with a BLAST e-value <0.2 and within 10 genes of one another on the chromosome, were found to have a higher degree of coexpression than that of neighboring genes that were not tandem duplicates. This was obtained using a Mann-Whitney U-test (both data sets: _P_ < 0.0001, Table 1). The result suggested that tandem duplicates could be a significant cause of coexpression of neighboring genes. Therefore, to determine the extent of this effect, one member of each pair of tandem duplicates was removed, and the mean coexpression was recalculated and again compared with randomized data sets. The results of these analyses are shown in Figure 1, B and D, and clearly demonstrate that the impact of tandem duplicates on the coexpression of neighboring genes is different between data obtained from the two technologies. The cDNA array data set free of tandem duplicates showed no evidence of coexpression of neighboring genes (_n_ = 2109; _P_ > 0.10, +1.08 standard deviations; Fig. 1B), whereas the Affymetrix data set continued to show a significant pattern (n = 1367; P < 0.0001, +18.6 standard deviations; Fig. 1D).
Figure 1.
Histogram of mean R generated from 10,000 randomized pairs of genes. For each randomized genome, the mean R of pairwise comparisons was calculated. The Mean R calculated from the original set of neighboring gene pairs is marked with an arrow. For both data sets, including tandem duplicates, there was a significant degree of coexpression of neighboring pairs. After the removal of tandem duplicates only, the Affymetrix data set showed evidence that neighboring genes were more likely to be coexpressed. (A) cDNA array including tandem duplicates. Mean R = 0.04397, σ = 0.00365. (B) cDNA array not including tandem duplicates. Mean R = 0.04574, σ = 0.00394. (C) Affymetrix array including tandem duplicates. Mean R = 0.03275, σ = 0.00307. (D) Affymetrix array not including tandem duplicates. Mean R = 0.03510, σ = 0.00328.
Table 1.
Descriptive Statistics for the Pairwise Comparisons of Neighboring Gene Pairs
| Number of gene pairs (missing values) | Mean R ± se | Median R | ||
|---|---|---|---|---|
| Stanford cDNA array | All genes +td | 2497 (95) | 0.0622 ± 0.0042 | 0.0226 |
| All genes -td | 2109 (71) | 0.050 ± 0.0042 | 0.01769 | |
| Tandem duplicates | 140 (6) | 0.240 ± 0.026 | 0.1441 | |
| NASC Affymetrix array | All genes +td | 18908 (8656) | 0.1039 ± 0.0033 | 0.08070 |
| All genes -td | 14959 (6304) | 0.096 ± 0.0035 | 0.07387 | |
| Tandem duplicates | 1307 (787) | 0.271 ± 0.016 | 0.2638 |
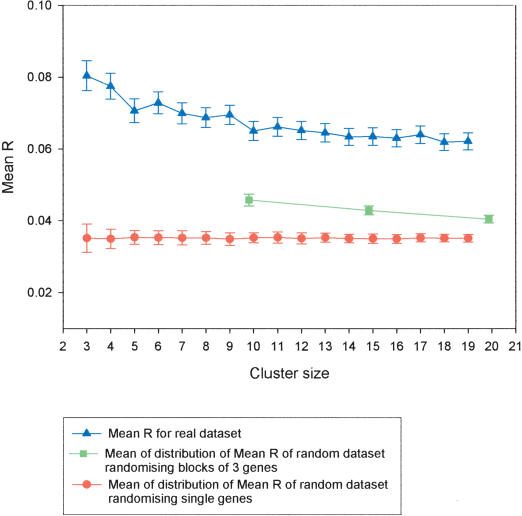
To investigate whether the correlation continues beyond neighboring gene pairs into clusters of increasing size, nonoverlapping blocks of three to 20 genes were compared, and the results are shown in Figure 2. Previous analyses in Drosophila suggested that blocks of genes up to 20 in size showed significant clustering of coexpressed genes. Data from only the Affymetrix arrays minus tandem duplicates are shown. The difference in degree of coexpression between real and randomized data sets remained significant for all block sizes. For nonoverlapping blocks of three to 10 genes, there is a clear, gradual decrease in coexpression. Beyond this, there is no further decrease in coexpression, and this continued for block sizes of up 20 genes. This implies that in the Arabidopsis genome, there may be clusters of up to 20 genes that are coexpressed, with an overall median cluster size of 100 kb. It was possible that the statistical significance of these results was inflated by genes that are only one, two, or three genes apart. To investigate this possibility, the randomizations were repeated, but rather than randomizing single genes in each block, groups of three genes were used. When these additional analyses were carried out, the mean R for the randomized data sets increased, but as shown in Figure 3, no random data set produced a higher mean R value than the real data set. This confirmed the significance of the finding that blocks of genes are coexpressed in the genome.
Figure 2.
Using the Affymetrix data set lacking tandem duplicates, the mean R for nonoverlapping windows of neighboring genes (three to 20 genes in size) was plotted against cluster size (blue line). The Mean R from 100 random sets of gene clusters (three to 20 genes in size) was also plotted (red line).
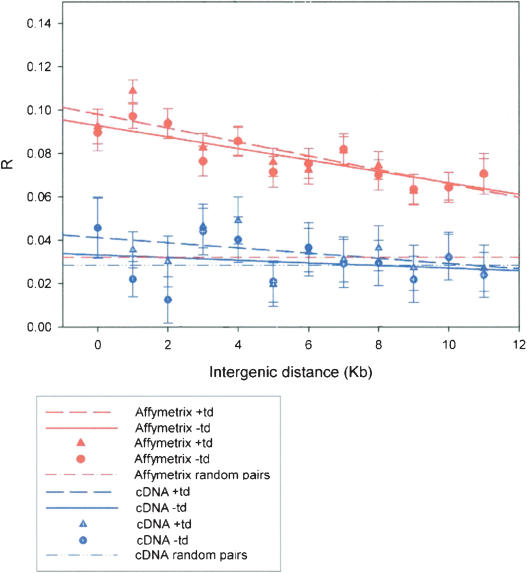
Figure 3.
Gene pairs up to 12 kb apart were binned according to their intergenic distance for both the data set containing and lacking tandem duplicates. The Mean R for all pairs within each bin was calculated using the cDNA microarray data and the Affymetrix data. (Red) Results obtained using Affymetrix data set; (blue) results obtained using cDNA microarray data. Data points using triangles are those obtained including tandem duplicates; circles indicate results obtained after removal of tandem duplicates. The regression lines are plotted, full lines those with all genes included, dashed lines those without tandem duplicates. Also plotted is the mean R value for all gene pairs (dashed lines with dots).
It was interesting to determine whether there was a direct correlation between distance and degree of coexpression. Thus, each pair of genes was placed in bins according to their intergenic distance (0–1 kb, 1–2 kb, 2–3 kb, etc.). If there is a relationship between proximity and degree of coexpression, then it could be expected that genes that are closer together would have a greater degree of coexpression than genes that are further away. For the Affymetrix data set, as shown in Figure 3, a significant correlation was observed between coexpression and intergenic distance of gene pairs up to 12 kb apart (with tandem duplicates: R2 = 0.73; P < 0.005, without tandem duplicates: R2 = 0.69; _P_ < 0.005). Interestingly, when gene pairs in intergenic blocks >12 kb were considered, the correlation between coexpression and gene distance was no longer found to be significant. No correlation was observed for the cDNA array data sets, with or without tandem duplicates. Given this lack of correlation, it is unclear whether the quantitative results from cDNA microarrays are useful for bioinformatic analysis, and therefore, further work focused only on the Affymetrix data sets.
Genes Thought to Be Involved in the Same Biological Process Are Coexpressed
The KEGG database defines genes that are thought to function in the same biological process, such as in a metabolic or regulatory pathway. Recently, a study of several genomes, including that of Arabidopsis, has used the KEGG database to demonstrate that genes functioning in the same pathway are often clustered in the genome (Lee and Sonnhammer 2003). It was important, therefore, to determine whether genes in the same pathway are coexpressed and whether this could be the causal reason for the coexpression of neighboring genes. Currently, 1891 genes in the Arabidopsis genome are assigned to pathways listed in the KEGG database. Of these, 912 gene pairs can be defined as near neighbors (within 10 genes of each other). The mean R is three times higher for gene pairs assigned to the same pathway, compared with those that were not (Mann-Whitney; P < 0.001; Table 2). On removing those gene pairs that were in the same pathway from the remainder, there continued to be significant coexpression of neighboring genes (Monte Carlo simulation: P < 0.0001). Thus, using the limited data currently available, coexpression of neighboring gene pairs is not only caused by clustering of genes in the same pathway.
Table 2.
Descriptive Statistics for Gene Pairs in the Same Metabolic Pathway and Those Not in the Same Metabolic Pathway
| R (Pearsons correlation coefficient) | Intergenic distance (bp) | ||||
|---|---|---|---|---|---|
| Gene pairs | N | Mean R ± se | Median R | Mean bp ± se | Median bp |
| In same metabolic pathway | 72 | 0.2268 ± 0.0448 | 0.2566 | 19115 ± 1441 | 19180 |
| Not known to be in same metabolic pathway | 840 | 0.0756 ± 0.0111 | 0.0422 | 19160 ± 464 | 17614 |
The mean R value, that is, degree of coexpression, was calculated for genes in each pathway listed in the KEGG database. The results are shown in Table 3, and illustrate several interesting features. First, the degree of coexpression shows considerable variation between different pathways. Second, the degree of coexpression is extremely high for some pathways, particularly those in which there is a known molecular interaction between gene products, such as components of the proteosome, ribosome, and replicon. Third, genes encoding enzymes of metabolic pathways are not so highly coexpressed, with some exceptions, such as those involved in the TCA cycle and fatty acid biosynthesis.
Table 3.
Degree of Coexpression of Genes Within the Same Pathway as Defined by the KEGG Database
| Pathway no. | Pathway id | Pathway description | Total comparisons | R | No. genes |
|---|---|---|---|---|---|
| 1 | ath03050 | Proteasome | 946 | 0.436 | 47 |
| 2 | ath03010 | Ribosome | 24504 | 0.385 | 249 |
| 3 | ath00580 | Phospholipid degradation | 25 | 0.378 | 9 |
| 4 | ath03030 | DNA polymerase | 89 | 0.360 | 16 |
| 5 | ath00960 | Alkaloid biosynthesis II | 10 | 0.349 | 5 |
| 6 | ath03032 | Replication complex | 36 | 0.300 | 10 |
| 7 | ath00020 | Citrate cycle (TCA cycle) | 670 | 0.264 | 39 |
| 8 | ath00860 | Porphyrin and chlorophyll metabolism | 190 | 0.254 | 22 |
| 9 | ath00061 | Fatty acid biosynthesis (path 1) | 78 | 0.240 | 13 |
| 10 | ath03020 | RNA polymerase | 491 | 0.214 | 37 |
| 11 | ath00720 | Reductive carboxylate cycle (CO2 fixation) | 170 | 0.213 | 20 |
| 12 | ath00195 | Photosynthesis | 1646 | 0.198 | 63 |
| 13 | ath00510 | N-Glycans biosynthesis | 262 | 0.189 | 25 |
| 14 | ath00521 | Streptomycin biosynthesis | 21 | 0.189 | 7 |
| 15 | ath00193 | ATP synthesis | 525 | 0.184 | 37 |
| 16 | ath03022 | Basal transcription factors | 326 | 0.175 | 34 |
| 17 | ath00970 | Aminoacyl-tRNA biosynthesis | 629 | 0.174 | 38 |
| 18 | ath03014 | Other translation factors | 15 | 0.171 | 7 |
| 19 | ath00150 | Androgen and estrogen metabolism | 10 | 0.167 | 6 |
| 20 | ath03034 | Other replication, recombination and repair factors | 66 | 0.158 | 14 |
| 21 | ath00300 | Lysine biosynthesis | 66 | 0.143 | 13 |
| 22 | ath00360 | Phenylalanine metabolism | 2502 | 0.138 | 78 |
| 23 | ath00760 | Nicotinate and nicotinamide metabolism | 2894 | 0.136 | 86 |
| 24 | ath00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 629 | 0.135 | 40 |
| 25 | ath00632 | Benzoate degradation via CoA ligation | 3208 | 0.132 | 89 |
| 75 | ath00750 | Vitamin B6 metabolism | 10 | 0.050 | 5 |
| 76 | ath00561 | Glycerolipid metabolism | 1032 | 0.049 | 48 |
| 77 | ath00511 | N-Glycan degradation | 35 | 0.048 | 9 |
| 78 | ath00252 | Alanine and aspartate metabolism | 494 | 0.045 | 36 |
| 79 | ath00330 | Arginine and proline metabolism | 560 | 0.041 | 36 |
| 80 | ath00910 | Nitrogen metabolism | 378 | 0.037 | 29 |
| 81 | ath00220 | Urea cycle and metabolism of amino groups | 136 | 0.036 | 20 |
| 82 | ath00410 | β-Alanine metabolism | 120 | 0.034 | 17 |
| 83 | ath00670 | One carbon pool by folate | 78 | 0.034 | 16 |
| 84 | ath00362 | Benzoate degradation via hydroxylation | 36 | 0.033 | 10 |
| 85 | ath00340 | Histidine metabolism | 91 | 0.029 | 15 |
| 86 | ath00472 | D-Arginine and D-ornithine metabolism | 21 | 0.027 | 8 |
| 87 | ath00251 | Glutamate metabolism | 818 | 0.025 | 43 |
| 88 | ath00351 | 1,1,1-Trichloro-2.2-bis(4-chlorophenyl)ethane (DDT) degradation | 21 | 0.022 | 8 |
| 89 | ath00361 | γ-Hexachlorocyclohexane degradation | 587 | 0.021 | 40 |
| 90 | ath00053 | Ascorbate and aldarate metabolism | 836 | 0.020 | 47 |
| 91 | ath00100 | Sterol biosynthesis | 449 | 0.018 | 34 |
| 92 | ath03060 | Protein export | 377 | 0.018 | 31 |
| 93 | ath00530 | Aminosugars metabolism | 153 | 0.018 | 20 |
| 94 | ath00628 | Fluorene degradation | 587 | 0.017 | 40 |
| 95 | ath04710 | Circadian rhythm | 496 | 0.014 | 32 |
| 96 | ath00120 | Bile acid biosynthesis | 91 | 0.006 | 14 |
| 97 | ath00900 | Terpenoid biosynthesis | 95 | 0.002 | 17 |
| 98 | ath00460 | Cyanoamino acid metabolism | 65 | -0.007 | 15 |
| 99 | ath02052 | Other ion-coupled transporters | 45 | -0.074 | 10 |
| 100 | ath00550 | Peptidoglycan biosynthesis | 10 | -0.172 | 5 |
The Effect of Gene Orientation on Coexpression of Neighboring Genes
Genes in a genome can be transcribed in one of two directions and therefore pairs of genes can be orientated in three alternative combinations as follows: divergent transcription (← →), convergent transcription (→ ←), or parallel transcription (→ →/← ←). Using the Affymetrix data set minus tandem duplicates, those pairs of genes with divergent (← →) or parallel (→ →/← ←) orientation were found to have a higher degree of coexpression than those genes with convergent (→ ←) orientation oftranscription (Table 4; Kruskal-Wallis, P < 0.0001). Interestingly, the pairs of genes with convergent orientation were found to have shorter intergenic distance than those with divergent or parallel orientation (Table 4; Kruskal-Wallis, P < 0.0001).
Table 4.
Descriptive Statistics for Pairwise Comparison of Neighboring Genes According to Orientation of Transcription
| R (Pearson correlation coefficient) | Intergenic distance (bp) | |||||
|---|---|---|---|---|---|---|
| Orientation | N | Mean R ± se | Median R | Mean bp ± se | Median bp | |
| Complete dataset without tandem duplicates (Affymetrix data) | ←→ | 2212 | 0.106 ± 0.007 | 0.08866 | 2770 ± 65.3 | 1872 |
| →→/←← | 4201 | 0.104 ± 0.0051 | 0.07831 | 2093.7 ± 33.7 | 1351 | |
| →← | 2241 | 0.071 ± 0.0068 | 0.05515 | 1147.6 ± 37.4 | 597 | |
| Tandem duplicates only (Affymetrix data) | ←→ | 38 | 0.391 ± 0.055 | 0.4734 | 7758 ± 600 | 7621 |
| →→/←← | 445 | 0.27 ± 0.018 | 0.2563 | 5427 ± 142 | 4625 | |
| →← | 36 | 0.158 ± 0.064 | 0.2519 | 5377 ± 491 | 4879 |
The above analysis excluded tandem duplicates. The same analysis was performed on a data set of neighboring genes that consisted only of tandem duplicates. As a basis for this analysis, the transcriptional orientation of tandem duplicates was first investigated, and as predicted, most were found to be in the parallel (→→/←←) orientation (χ2 test, P < 0.0001). However, it was the tandem duplicates existing in the divergent (← →) orientation of transcription that showed the greatest degree of coexpression (Table 4; Kruskal-Wallis, P < 0.05).
DISCUSSION
Many technologies are now available to determine the different patterns of gene expression exhibited in cells and tissues of an organism. Often, the entire genomes of these organisms have also been sequenced. This provides the opportunity to analyze gene expression in the context of genome organization. For A. thaliana, the genome sequencing program was completed in 2000 (The Arabidopsis Genome Initiative 2000), and it is fast becoming routine to apply a variety of microarray technologies to the model plant to define global patterns of gene expression. Despite the availability of these data, very few detailed global gene expression analyses have been published on this organism. This study has explored, for the first time, the possibility of coexpression of neighboring genes in Arabidopsis and the reasons that this might occur.
Our results show that neighboring genes in the Arabidopsis genome are indeed coexpressed. We have observed this coexpression from two different sources of data for the statistical analysis, Affymetrix and cDNA microarray technologies. Tandem duplicates were found to have a higher degree of coexpression than other neighboring genes in our analysis, but interestingly, the impact of their removal was found to be different when the data from the two technologies were compared. Only the Affymetrix data set continued to show a significant pattern of coexpression. The loss of significance from the cDNA microarray data sets can readily be understood given the known problem of cross-hybridization arising from highly homologous genes such as tandem duplicates. This leads to a higher overall level of noise and unreliability when using cDNA arrays. In contrast, the Affymetrix technology bypasses this problem by using multiple oligonucleotides unique for each gene.
A further difference shown by the analyses of the data sets from the two technologies relates to the effect of intergenic distance, as one could predict that genes closer together would have a greater degree of coexpression than those that are more distant in the genome. A significant correlation between distance and coexpression was only found for the Affymetrix data set, either with or without the inclusion of tandem duplicates. This finding also questions the general utility of cDNA microarrays for this type of quantitative analysis. Some discrepancies have been found previously between cDNA and Affymetrix data sets, such as, for example, in the study of gene expression patterns in 56 cell lines from the National Cancer Institute (Kuo et al 2002), as well as in a study using human neuroblastoma cells (Li et al. 2002). Given the potential problems associated with data sets from cDNA microarrays and the inherent problems of gene duplication in Arabidopsis, all further analyses in this study used only Affymetrix data sets omitting tandem duplicates.
We have addressed several possible explanations for the observed coexpression of neighboring genes. For example, MARS are thought to influence gene expression through changing chromatin conformation patterns (Mishra and Karch 1999; Gerasimova and Corces 2001). To explore this possibility, we used bioinformatic tools to identify MARS in the Arabidopsis genome (Glazko et al. 2000). However, this approach has considerable limitations, as there is no experimental certainty that the MARS identified are functional or operational under the conditions of plant growth and development used to gain expression data. Within these limitations, we found no positive or negative evidence that the presence of MARS correlates with coexpression of neighboring gene pairs (E.J.B. Williams and D.J. Bowles, unpubl.).
Gene orientation has been examined in a number of studies for its relationship to degree of coexpression. Studies on yeast have shown that divergently transcribed genes have a higher degree of coexpression than genes in convergent orientation (Kruglyak and Tang 2000). It has been suggested that the underlying cause for these observations may be due to sharing of common regulatory elements. Although several bidirectional promoters have been found in mammalian genomes (Adachi and Lieber 2002), few examples have been found in plants. Significantly, recent experimental data from Capsicum annuum have discovered two coexpressed homologous genes that are neighbors and are divergently transcribed (Shin et al. 2003). The authors demonstrated that a single promoter, situated between the genes, is responsible for driving their expression. In our analysis of Arabidopsis, we found clear evidence that gene pairs transcribed in divergent or parallel orientations showed a higher degree of coexpression than those gene pairs in the convergent orientation. Interestingly, tandem duplicates in the divergent orientation have a higher coexpression than those in the parallel orientation, despite most tandem duplicates being in the parallel orientation. These findings may indicate that bidirectional promoters may be more common than expected in plant genomes and may be particularly important in the coexpression of duplicate gene pairs. Our data provide the basis for undertaking experimental studies to investigate how the expression of defined gene pairs is regulated.
Coexpression of neighboring genes could arise through the genes sharing a common function. For example, one could readily predict that genes encoding enzymes in a common metabolic pathway may be coordinately regulated and therefore coexpressed, particularly if the entire pathway is responsive to environmental or developmental cues. To gain an insight into the role of shared function in coexpression, we used the KEGG database to analyze gene expression in the context of gene function (Kanehisa 2002). The database encompasses genes of annotated function, which currently only represents a small subset of genes in the Arabidopsis genome. An additional problem with the KEGG database is subjectivity of the annotation. Within these constraints, high degrees of coexpression were observed between pairs of genes in Arabidopsis thought to share a role in common biological processes (PATHWAYS, as defined by the KEGG database), implying that commonality of function does explain some degree of the coexpression observed. When these pairs of genes were removed and the analysis repeated, neighboring genes in the genome continued to be coexpressed. Thus, on the basis of the limited information currently available, the data suggest that the phenomenon of coexpression of neighboring genes in the Arabidopsis genome does not rely only on genes functioning in a common biological process. However, as the KEGG database is not comprehensive, not all pairs of genes involved in the same pathway can be definitively removed. It would be interesting to repeat these analyses at a later date when more information is available and a far greater proportion of genes in the Arabidopsis genome have been assigned a definite function.
Interestingly, when coexpression of genes across the entire genome was analyzed in the context of the KEGG database, particularly high degrees of correlation were observed for genes encoding proteins that are known to function in multicomponent complexes, such as the proteosome, ribosome, and replicon. Often, these complexes contain a high level of protein–protein interactions and our conclusions from the Arabidopsis data are supported by studies in yeast, in which genes encoding interacting proteins tend to be coexpressed (Ge et al. 2001; Grigoriev 2001; Jansen et al. 2002). In contrast, the degree of coexpression of genes encoding enzymes in metabolic pathways in Arabidopsis was low, with the exception of several key primary metabolic pathways, such as the TCA cycle and fatty acid metabolism. Given these findings, it is an interesting possibility that the number of interactions between proteins may be an important predictor of the degree of coexpression between their corresponding genes. Additionally, as more data emerges from studies of gene function in Arabidopsis, it will be important to determine whether protein–protein interactions play a role in the coexpression of neighboring genes.
METHODS
Data Sources
Microarray Data
Data was collected from two sources. The Stanford data set is a collection of microarray experiments using cDNA microarrays. The data was downloaded from the Stanford Web site (ftp://genome-ftp.stanford.edu/pub/smd/organisms/AT). A total of 233 experiments were used and the total number of genes across all experiments was 7627 genes. Not all genes were present in each array. As an indicator of the expression level, the normalized ratio was used (channel 1/channel 2 ratio normalized). The Affymetrix data was obtained using the Nottingham Arabidopsis Stock Centre (NASC) Affywatch service (http://arabidopsis.info/prototype/; Craigon et al 2004). The data set contained 175 experiments, 28 of which used 8300 chips; the remainder were full-genome chips. Expression level was defined as the normalized signal values where the detection call was 1, indicating that the signal value was statistically significant. If any one gene was represented more than once on a chip, then the mean expression level across the chip for that gene was used. Both sets of data contained experiments using various tissue types and sample sources.
Detecting Local Similarity in Expression
The level of coexpression between two genes was defined as the Pearson's correlation coefficient (R) of the expression level for these genes across all experiments.
To test for pairwise local similarity in expression in the Arabidopsis genome, the mean R (Pearson's correlation coefficient) of the expression profiles for neighboring pairs of genes was calculated for both the affymetrix and cDNA data sets. Neighbors were defined as genes that were immediately adjacent in the Arabidopsis genome according to each gene's AGI name, that is, gene pairs with an AGI name (of the form At[chr]g[xxxxx]), differing by 10 or less (e.g., At1g10020 and At1g10030 are defined as neighbors). The mean R calculated from the real data set was then compared with the mean R calculated from 10,000 data sets, in which the order of genes in the Arabidopsis genome was randomized. To ensure that the R-value calculated was statistically valid for each pairwise comparison, there had to be at least 10 experiments in which both genes had valid values. For the Affymetrix data in particular, this resulted in many comparisons being rejected, due to an insufficient number of experiments in which the transcript was identified. The number of gene pair comparisons was conserved between the randomized and the real data sets (Stanford n = 2498; NASC n = 7388).
When analyzing blocks of genes, the mean of all possible comparisons within the block was used as the level of coexpression for that block. Therefore, for a block of five genes, 10 different correlations were carried out, and the mean R was used as a measure of the level of coexpression for that particular block. The mean R was then compared with means calculated from randomized data sets. One hundred randomizations were carried out for each simulation. Where sub-blocks were used, the number of genes in a randomized block were varied. For example, when there were three genes in a sub-block, the Arabidopsis genome was split into blocks of three ordered neighboring genes. These blocks were then randomized. For each random distribution, the genes were split into blocks of 15 genes, from which the mean Pearson correlation coefficient was calculated using the Affymetrix array data. Tandem duplicates were excluded.
Distance between genes was defined as the distance in base-pairs between the last coding position, on either strand, of the first gene to the first coding position of the second gene.
Removal of Tandem Duplicates
All Arabidopsis protein sequences from the May 2003 build were downloaded from MIPS (http://mips.gsf.de/proj/thal/db/index.html). The protein sequences were compared using an allagainst-all BLAST algorithm. Any pair of genes within 100 genes of each other that showed sequence similarity (e-value cut off = 0.2) was counted as a tandem duplicate. This cut off value removes ∼90% of related genes from a data set, and has a false positive rate of about 10% (Lercher et al. 2002). One member of each pair of tandem duplicates was removed from the analysis. This gave 8890 pairs of tandem duplicates in the entire Arabidopsis genome. This compares favorably with the 17% of the Arabidopsis genome claimed to be in tandem arrays quoted in the Arabidopsis genome paper (The Arabidopsis Genome Initiative 2000).
Identification of Genes in the Same Metabolic Pathway
The KEGG database (http://www.genome.ad.jp/kegg/), downloaded August 2003, was used to assign 1891 genes to 117 PATHWAYS, resulting in 4048 gene-PATHWAY assignations. The KEGG database annotates only a small proportion of the Arabidopsis genome, and the ontology is biased toward mammalian metabolic pathways. For each pair of nonduplicate genes in which there was known pathway information and Affymetrix data associated with both genes (n = 912), where the pair was within 10 genes of each other, a Pearson's correlation coefficient was calculated. Of these, 101 pairs were classified as neighboring genes; eight of these were pairs classified as being in the same metabolic pathway. To increase the number of gene pairs for comparison against neighboring pairs of genes that were not in the same metabolic pathway, all gene pairs for which data had been calculated were used in the analysis (Nmetabolic = 72, Nnot metabolic = 840).
PERL scripts that carry out the methods described in this work are available from the authors on request.
Acknowledgments
We thank Yi Li, Kathryn Madagan, Fabian Vaistij, Eng-Kiat Lim, and Chris Winefield for their helpful comments and discussion. E.J.B.W. is funded by the BBSRC Exploiting Genomics Initiative (grant no. EGA16205).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2131104.
Footnotes
[Supplemental material is available online at www.genome.org. All of the raw microarray data and metabolic pathway data will be made available as additional information. Also, all programs used to analyze data will be made available on request as well as any other data used in the analyses.]
References
- Adachi, N. and Lieber, M.R. 2002. Bidirectional gene organization: A common architectural feature of the human genome. Cell 109**:** 807–809. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408**:** 796–815. [DOI] [PubMed] [Google Scholar]
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. 2003. A gene expression map of the Arabidopsis root. Science 302**:** 1956–1960. [DOI] [PubMed] [Google Scholar]
- Blumenthal, T., Evans, D., Link, C.D., Guffanti, A., Lawson, D., Thierry-Mieg, J., Thierry-Mieg, D., Chiu, W.L., Duke, K., Kiraly, M., et al. 2002. A global analysis of Caenorhabditis elegans operons. Nature 417**:** 851–854. [DOI] [PubMed] [Google Scholar]
- Boutanaev, A.M., Kalmykova, A.I., Shevelyov, Y.Y., and Nurminsky, D.I. 2002. Large clusters of co-expressed genes in the Drosophila genome. Nature 420**:** 666–669. [DOI] [PubMed] [Google Scholar]
- Caron, H., Peter, M., Vansluis, P., Speleman, F., Dekraker, J., Laureys, G., Michon, J., Brugieres, L., Voute, P.A., Westerveld, A., et al. 1995. Evidence for 2 tumor-suppressor loci on chromosomal bands-1p35–36 involved in neuroblastoma—one probably imprinted, another associated with n-myc amplification. Hum. Mol. Genet. 4**:** 535–539. [DOI] [PubMed] [Google Scholar]
- Cohen, B.A., Mitra, R.D., Hughes, J.D., and Church, G.M. 2000. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat. Genet. 26**:** 183–186. [DOI] [PubMed] [Google Scholar]
- Craigon, D.J., James, N., Okyere, J., Higgins, J., Jotham, J., and May, S. 2004. NASCArrays: A repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res. 32**:** D575–D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elo, A., Lyznik, A., Gonzalez, D.O., Kachman, S.D., and Mackenzie, S.A. 2003. Nuclear genes that encode mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome. Plant Cell 15**:** 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, H., Liu, Z., Church, G.M., and Vidal, M. 2001. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat. Genet. 29**:** 482–486. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T.I. and Corces, V.G. 2001. Chromatin insulators and boundaries: Effects on transcription and nuclear organization. Annu. Rev. Genet. 35**:** 193–208. [DOI] [PubMed] [Google Scholar]
- Glazko, G.V., Rogozin, I.B., and Glazkov, M.V. 2000. Computer prediction of DNA sites of attachment to different nuclear matrix elements. Mol. Biol. 34**:** 1–5. [PubMed] [Google Scholar]
- Gray, T.A., Saitoh, S., and Nicholls, R.D. 1999. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc. Natl. Acad. Sci. 96**:** 5616–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev, A. 2001. A relationship between gene expression and protein interactions on the proteome scale: Analysis of the bacteriophage T7 and the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 29**:** 3513–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R., Greenbaum, D., and Gerstein, M. 2002. Relating whole-genome expression data with protein–protein interactions. Genome Res. 12**:** 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. 2002. The KEGG database. Novartis. Found. Symp. 247**:** 91–101. [PubMed] [Google Scholar]
- Kruglyak, S. and Tang, S. 2000. Regulation of adjacent yeast genes. Trends Genet. 16**:** 109–111. [DOI] [PubMed] [Google Scholar]
- Kuo, W.P., Jenssen, T.K., Butte, A.J., Ohno-Machado, L., and Kohane, I.S. 2002. Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics. 18**:** 405–412. [DOI] [PubMed] [Google Scholar]
- Lee, J.M. and Sonnhammer, E.L. 2003. Genomic gene clustering analysis of pathways in eukaryotes. Genome Res. 13**:** 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher, M.J., Urrutia, A.O., and Hurst, L.D. 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 31**:** 180–183. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J., Blumenthal, T., and Hurst, L.D. 2003. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res. 13**:** 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Pankratz, M., and Johnson, J.A. 2002. Differential gene expression patterns revealed by oligonucleotide versus long cDNA arrays. Toxicol. Sci. 69**:** 383–390. [DOI] [PubMed] [Google Scholar]
- Mishra, R.K. and Karch, F. 1999. Boundaries that demarcate structural and functional domains of chromatin. J. Biosci. 24**:** 377–399. [Google Scholar]
- Reiss, J., Cohen, N., Dorche, C., Mandel, H., Mendel, R.R., Stallmeyer, B., Zabot, M.T., and Dierks, T. 1998. Mutations in a polycistronic nuclear gene associated with molybdenum cofactor deficiency. Nat. Genet. 20**:** 51–53. [DOI] [PubMed] [Google Scholar]
- Shin, R., Kim, M.J., and Paek, K.H. 2003. The CaTin1 (Capsicum annuum TMV-induced Clone 1) and CaTin1-2 genes are linked head-to-head and share a bidirectional promoter. Plant Cell Physiol. 44**:** 549–554. [DOI] [PubMed] [Google Scholar]
- Spellman, P.T. and Rubin, G.M. 2002. Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 1**:** 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://genome-ftp.stanford.edu/pub/smd/organisms/AT; Stanford database.
- http://arabidopsis.info/prototype; NASC Affymetrix database.
- http://mips.gsf.de/proj/thal/db/index.html; MIPS Web site.
- http://www.genome.ad.jp/kegg/; Kegg database.