Comparative Genomic Analysis of slc39a12/ZIP12: Insight into a Zinc Transporter Required for Vertebrate Nervous System Development (original) (raw)
Abstract
The zinc transporter ZIP12, which is encoded by the gene slc39a12, has previously been shown to be important for neuronal differentiation in mouse Neuro-2a neuroblastoma cells and primary mouse neurons and necessary for neurulation during Xenopus tropicalis embryogenesis. However, relatively little is known about the biochemical properties, cellular regulation, or the physiological role of this gene. The hypothesis that ZIP12 is a zinc transporter important for nervous system function and development guided a comparative genetics approach to uncover the presence of ZIP12 in various genomes and identify conserved sequences and expression patterns associated with ZIP12. Ortholog detection of slc39a12 was conducted with reciprocal BLAST hits with the amino acid sequence of human ZIP12 in comparison to the human paralog ZIP4 and conserved local synteny between genomes. ZIP12 is present in the genomes of almost all vertebrates examined, from humans and other mammals to most teleost fish. However, ZIP12 appears to be absent from the zebrafish genome. The discrimination of ZIP12 compared to ZIP4 was unsuccessful or inconclusive in other invertebrate chordates and deuterostomes. Splice variation, due to the inclusion or exclusion of a conserved exon, is present in humans, rats, and cows and likely has biological significance. ZIP12 also possesses many putative di-leucine and tyrosine motifs often associated with intracellular trafficking, which may control cellular zinc uptake activity through the localization of ZIP12 within the cell. These findings highlight multiple aspects of ZIP12 at the biochemical, cellular, and physiological levels with likely biological significance. ZIP12 appears to have conserved function as a zinc uptake transporter in vertebrate nervous system development. Consequently, the role of ZIP12 may be an important link to reported congenital malformations in numerous animal models and humans that are caused by zinc deficiency.
Introduction
Zinc is required for enzyme catalysis, cell signaling, and DNA repair by all organisms and is vital for growth and development and multiple physiological processes including immune and brain function [1]–[3]. During pregnancy, maternal zinc deficiency can increase the frequency of congenital malformations across many animal species [4]. Symptoms of zinc deficiency in humans include weight loss, severe dermatitis, slow wound healing, male hypogonadism, and reduced immune function [5]. Zinc deficient experimental diets in laboratory animals [6] or plant protein-based feed with low zinc bioavailability in livestock [7], [8] lead to similar symptoms such as impaired growth, development, fertility, and epidermal health.
Members of the solute carrier 39 (SLC39) gene family encode zinc transport proteins that are critical mechanisms for maintaining zinc homeostasis across a wide range of species [9]. The SLC39 family, with similarity to iron transporter IRT1 [10], is present in Saccharomyces cerevisiae yeast [11], Arabidopsis plants [12], invertebrates [13], and vertebrates [14] including humans [15], [16]. The phenotypic similarities of zinc deficiency across different mammals due to impaired intestinal zinc transport caused by spontaneous or targeted mutations of slc39a4 (ZIP4) in humans [17], [18], mice [19], and cows [20] demonstrates how physiological functions for SLC39 members may be conserved across species.
The zinc transporter ZIP12 encoded by the slc39a12 gene was uncovered by analyses of gene expression across different human tissues [21]. ZIP12, which highly expressed in the brain, is required for multiple aspects of neuronal differentiation including cAMP response element binding protein (CREB) phosphorylation and activity, neurite outgrowth, and microtubule polymerization and stability [21]. ZIP12 is also necessary for embryogenesis because inactivation of ZIP12 by antisense morpholino knockdown halted neural tube closure in Xenopus tropicalis, resulting in arrested development and lethality during neurulation [21].
Comparative genomics can be useful for determining the functions of genes in varied and assorted contexts. The general assumption of comparative genomics is the conservation of genomic sequence due to evolutionary constraints that imply some kind of biological importance or function [22] encoded by the conserved sequence. Comparative genomics may predict the physiological role of genes due to the relationship between tissue expression profiles and biological function [23]. Models and approaches based upon comparative genomics have been used to determine possible functions for genes and their protein products at different levels. For example, comparative genomics can be used to identify possible interaction partners [24], protein folding and structure [25], and the evolution of protein phosphorylation sites [26]. Comparative genomics may be able to identify genomic differences between organisms that confer physiological differences and species identity [22]. For example, nonsynonymous changes in the human FOXP2 gene from non-human primates are hypothesized to be responsible for language development [27].
Because ZIP12 appears to be conserved and is highly expressed in the brain across humans, mice, and frogs [21], a comparative analysis of the slc39a12 gene across multiple species and the SLC39 gene family was conducted to uncover information about this transporter. These analyses indicate that there are conserved elements in the slc39a12 gene across various organisms that likely contribute towards important biochemical, physiological, and developmental functions for this zinc transporter. The presence of slc39a12 in nearly all vertebrate genomes examined and a possible lack of slc39a12 in other invertebrate organisms may indicate an association between ZIP12 and neurulation during vertebrate embryogenesis.
Materials and Methods
Sequence analyses, alignment, and phylogenetic tree
Nucleotide and amino acid sequences (Table 1) for slc39a12 were obtained from annotated entries, BLASTP or TBLASTN using human slc39a12 amino acid sequence, in Xenbase (for Xenopus laevis, 6.0 scaffolds) [28], Ensembl (for Ciona intestinalis and Petromyzon marinus) [29], Joint Genome Institute (for Branchiostoma floridae), or National Center for Biotechnology Information (NCBI) (all other organisms) [30]. UCSC Genome Browser, PhyloP, and PhastCons were used to display broad phylogenetic alignments and sequence conservation of slc39a12 in vertebrates [31]. BLASTP was used to compare ZIP12 amino acid sequences of various organisms to both human ZIP12 and ZIP4. Additional information about slc39a12 in Xenopus laevis was also obtained from daudin.icmb.utexas.edu. The amino acid sequence of ZIP12 from various organisms (see Table 1) was aligned by ClustalW [32] using Bioedit software version 7.0.9.0 [33]. The phylogenetic tree was drawn using TreeDyn [34] using Phylogeny.fr [35]. Because the existence of an exon, which is included in the long isoform, could not be confirmed in all organisms tested, the phylogenetic tree was drawn using only the putative short isoforms of ZIP12 from each organism. EST representations of slc39a12 in the nervous system (brain, eye, spinal cord) and other tissues of different organisms were accomplished using NCBI Unigene [30]. Bioedit was used to calculate the nucleotide composition of the 5′ and 3′ untranslated and upstream regions.
Table 1. Orthologs of ZIP12 (slc39a12) are more similar to human ZIP12 than paralog ZIP4.
| Animal | Species | Taxonomic Category | NCBI Gene ID | NCBI Accession DNA | NCBI Accession Protein | Human ZIP12 | Human ZIP4 |
|---|---|---|---|---|---|---|---|
| Human | Homo sapiens | Mammalia-Primate | 221074 | NM_001145195, NM_152725 | NP_001138667, NP_689938 | 100% | 31% |
| Chimpanzee | Pan troglodytes | Mammalia-Primate | 739440 | XM_001153573, XM_001153510 | XP_001153573, XP_001153510 | 99% | 36% |
| Rhesus monkey | Macaca mulatta | Mammalia-Primate | 701128 | XM_001092136 | XP_001092136 | 97% | 37% |
| Bushbaby | Otolemur garnettii | Mammalia-Primate | 100948394 | XM_003786777, XM_003786778 | XP_003786825, XP_003786826 | 86% | 36% |
| Rat | Rattus norvegicus | Mammalia-Rodent | 291328 | NM_001106124 | NP_001099594 | 78% | 37% |
| Mouse | Mus musculus | Mammalia-Rodent | 277468 | NM_001012305 | NP_001012305 | 77% | 38% |
| Guinea pig | Cavia porcellua | Mammalia-Rodent | 100719835 | XM_003462762, XM_003462763 | XP_003462810, XP_003462811 | 84% | 32% |
| European rabbit | Oryctolagus cuniculus | Mammalia-Other | 100346557 | XM_002717411, XM_002717412 | XP_002717457, XP_002717458 | 83% | 32% |
| Domestic dog | Canis lupus familiaris | Mammalia-Other | 477989 | XM_843257, XM_535173 | XP_848350, XP_535173 | 84% | 31% |
| Feral pig | Sus scrofa | Mammalia-Other | 100523814 | XM_003130728, XM_003130727 | XP_003130776, XP_003130775 | 83% | 33% |
| Horse | Equus caballus | Mammalia-Other | 100056009 | XM_003364356, XM_001497406 | XP_003364404, XP_001497456 | 86% | 38% |
| Cow | Bos taurus | Mammalia-Other | 527210 | NM_001076878 | NP_001070346 | 84% | 36% |
| Elephant | Loxodonta africana | Mammalia-Other | 100671276 | XM_003410560, XM_003410561 | XP_003410608, XP_003410609 | 80% | 37% |
| Opossum | Monodelphis domestica | Mammalia-Other | 100026757 | XM_001377227 | XP_001377264 | 72% | 34% |
| Platypus | Ornithorhynchus anatinus | Mammalia-Other | 100078275 | XM_001509297 | XP_001509347 | 59% | 36% |
| Chicken | Gallus gallus | Aves (Birds) | 420514 | XM_004939251 | XP_004939308 | 62% | 34% |
| Anole lizard | Anolis carolinensis | Reptilia | 100552790 | XM_003222078 | XP_003222126 | 51% | 40% |
| Western clawed frog | Xenopus tropicalis | Amphibia | 100493177 | XM_002939000 | XP_002939046 | 51% | 31% |
| African clawed frog | Xenopus laevis | Amphibia | JGI Xenla 6.0 Scaffold 172106 | not available | not available | 51% | 37% |
| Nile tilapia | Oreochromis niloticus | Actinopterygii (Bony Fish) | 100695101 | XM_003439349 | XP_003439397 | 50% | 33% |
| European seabass | Dicentrarchus labrax | Actinopterygii (Bony Fish) | FQ310508 (chromosome) | (chromosome) | CBN82065 | 49% | 34% |
| Spotted green pufferfish | Tetraodon nigroviridis | Actinopterygii (Bony Fish) | CAAE01015017 (chromosome) | (chromosome) | CAG10206 | 48% | 37% |
| Japanese medaka | Oryzias latipes | Actinopterygii (Bony Fish) | NC_019878 | XM_004081302 | XP_004081350 | 50% | 33% |
| Zebrafish | Danio rerio | Actinopterygii (Bony Fish) | NC_007135, NC_007113, NC_007118 | not found | not found | N/A | N/A |
| Sea squirt | Ciona intestinalis | Ascidiacea (Tunicate) | 100178452; Chr 8: 148339 to 149331 | XM_002121310 | XP_002121346 | 39% | 40% |
| Sea lamprey | Petromyzon marinus | Agnatha | GL481648 | ENSPMAT00000007608 | N/A | 47% | 37% |
| Lancelet | Branchiostoma floridae | Cephalochordata (Amphioxus) | Scaffolds 92, 119, 158 | not found | not found | N/A | N/A |
| Purple sea urchin | Strongylocentrotus purpuratus | Echinodermata | NW_003578262, NW_003577087 | not found | not found | N/A | N/A |
| Fruit fly | Drosophila melanogaster | Insecta | NC_004354, NT_033779, NT_037436 | not found | not found | N/A | N/A |
| Roundworm | Caenorhabditis elegans | Chromadorea (Nematode) | NC_003279 | not found | not found | N/A | N/A |
Sequence motif scanning
Possible transcription factor binding sites were scanned using TRANSFAC (Match) Matrix Search for Transcription Factor Binding Sites [36]. Eukaryotic Linear Motif [37] was used to search for possible dileucine and tyrosine motifs associated with intracellular sorting and localization, as described previously by Huang and Kirschke for ZIP1 [38]. Positive matches for transcription factor binding sites and dileucine and tyrosine motifs were further examined for conservation across species in aligned amino acids sequences.
Synteny analyses
Local synteny for slc39a12 and neighboring genes (cubn, vim, stam, mrc1, cacnb2, nsun6, and arl5b) in most vertebrate organisms was discovered using NCBI Homologene [30]. Xenbase [28] was used for synteny analysis of slc39a12 in Xenopus tropicalis. Searches for slc39a12 and nearby genes in the genomes of zebrafish, Drosophila, C. elegans, and Ciona intestinalis were conducted using TBLASTN using translated protein versions of cubn, vim, stam, mrc1, cacnb2, nsun6, and arl5b in either tilapia or Xenopus tropicalis [30].
Detection of splice variants of ZIP12 in different species
Bioinformatic searches for exon 9, which is not present in annotated entries for slc39a12 in the chicken, cow, opossum, or platypus genomes, was detected by TBLASTN using a translated sequence (GLXLVNXHVGHXHHLXLNXELXDQXXXGKSXSTIQL) in exon 9 completely conserved across humans and mice.
Total brain RNA of human, cow, rat, and mouse origin was obtained from Zyagen (San Diego, CA). cDNA was reverse-transcribed from total RNA as described previously [21], using either a polyT primer or a ZIP12-specific primer (human and cow: acttatattttaatattttg; rat: atgtgaacatataaattcat; mouse: tgagtcatttcaggaagc). The different splice variants were detected using primers that flank exon 9, which is present in the long isoform and absent in the short isoform (Human- Fwd: acgctctgctccaccttatccctca, Rev: aaattatgcaggctgtccccaacca; Cow- Fwd: acagctgcgaggagaactacaggctca, Rev: ggttttgcattttctgttgggggtgtt; Rat- Fwd: cgaaagccaaagtcctatttggaagctg, Rev: gagcacagcaaagtctcccatttcatgt; Mouse- Fwd: taaccttgggctccatgcttgggacag, Rev: ggctggcacattgcctatgggtagcac) and the following PCR conditions: 94°C, 1 min; 37 cycles: 94°C, 30 s, 68°C, 1 min with Platinum Hifi Taq (Invitrogen, Carlsbad, CA). PCR products and DNA molecular weight markers (1 kb Trackit or 1kb+, Invitrogen) were separated by agarose gel electrophoresis and viewed as described previously [21]. From accompanying gels not exposed to ultraviolet light, bands corresponding to the different isoforms were excised (Qiagen, Hilden, Germany) and confirmed by DNA sequencing.
Detection of slc39a12 and igf1 genes by PCR
Comparisons of genomic sequences for slc39a12 of Japanese medaka, Nile tilapia, and European seabass by bl2seq were used to design PCR primers with some degeneracy. As a positive control, PCR primers were also designed for the igf1 (insulin growth factor-1) gene present in zebrafish (NC_007115) using similar comparisons between Japanese medaka, Nile tilapia, and European seabass. This region of igf1 was previously identified by Faircloth et al. [39] as containing an ultraconserved element among ray-finned fish (Node 267).
Genomic DNA was isolated from fish carcasses purchased at grocery stores (Nile tilapia and European seabass) using the QIAamp DNA Mini Kit (Qiagen). Zebrafish and Japanese medaka genomic DNAs were kindly provided as a gift by Bruce Draper (University of California-Davis) and Swee Teh (University of California-Davis). Using 100 ng of genomic DNA as a template, the slc39a12 and igf1 genes were amplified by PCR using the following primers (_slc39a12_- Fwd: ccantcanctggngganatt, Rev: attnccnagcaactgntga; _igf1_- Fwd: cccagctgtttcctgttgaa, Rev: ttccnactttgttccattgc; degeneracy underlined) and conditions: 94°C, 1 min; 40 cycles: 94°C, 20 s, 55°C, 20 s, 72°C, 40 s with GoTaq (Promega, Madison, WI), and PCR products were viewed as described above.
Results and Discussion
The human slc39a12 gene spans 13 exons across 91.4 kilobases (kb) on chromosome 10p12.33 (Figure 1, Figure 2). Orthologs to the human slc39a12 gene (Table 1) were identified by combinations of Homologene searches, reciprocal BLAST hits [40], and local synteny preservation [41]. Identified orthologs of ZIP12 shared amino acid identities with the human ZIP12 that correlated with the relatedness of the organisms to humans, ranging from 86 to 99 percent for non-human primates to 48 to 50 percent in fish (Table 1). In contrast, the amino acid identities of the ZIP12 orthologs to human ZIP4 ranged between 31 to 40 percent without any correlation to relatedness with humans (Table 1). Because there are many common elements between members of the SLC39 gene family, particularly between ZIP12 and ZIP4 [9], [42], it can be difficult to distinguish orthologs of ZIP12 from SLC39 paralogs solely based upon sequence similarity such as BLAST. Local synteny between ortholog candidates is extremely useful in confirming reciprocal BLAST hits [41], especially in cases of gene families with large numbers of paralogs, such as the case with the solute carrier (SLC) gene families [43], [44]. In general, synteny was observed across vertebrates (Figure 3, Figure S1), but possible disruptions of the syntenic block in more distant organisms corresponded with reduced relatedness to humans, possibly due to genomic rearrangements during evolution [45]. As with the case with most orthologous genes in vertebrates [46], the coding exon structure of slc39a12, including number of exons and exon size, is conserved across humans, mice, and Xenopus tropicalis (data not shown). An alignment of human ZIP12 with other related ZIP genes (Figure S2) and with other ZIP12 orthologs (Figure S3) shows that many C-terminus elements are conserved, especially those predicted to encode the transmembrane helices and zinc transport function, such as the HEXPHEGD motif that is present in the LIV-1 subfamily of ZIP transporters [47]–[49]. The phylogenetic tree (Figure 4) indicates that the relatedness of ZIP12 across different organisms is highly correlated with the relatedness of the whole genomes across organisms [50]–[52].
Figure 1. Human slc39a12 gene structure.
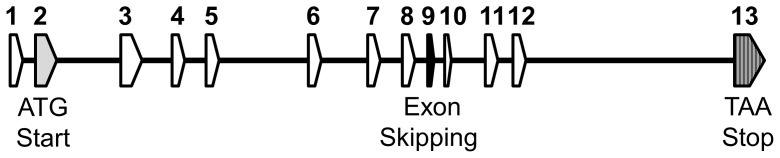
Exon-intron structure drawn to approximate scale. Exon 2 (shaded gray) contains the translation start codon. Exon 9 (shaded black) contains a variable exon depending on splice variation that leads to exon inclusion or exclusion. Exon 13 (striped) contains the stop codon (ochre). The exon structure (number of exons, relative exon size) of slc39a12 is conserved in mice and Xenopus tropicalis.
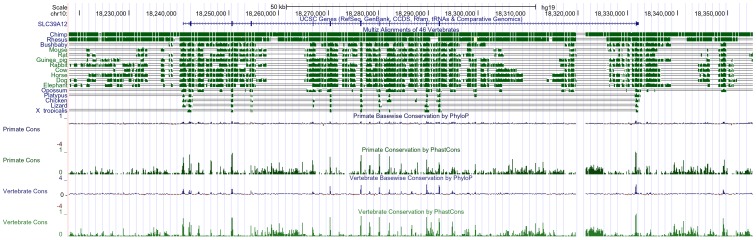
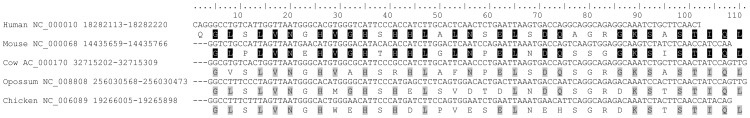
Figure 2. Phylogenetic alignment and nucleotide sequences of slc39a12 show conservation across vertebrates.
Human slc39a12 gene structure is indicated. Multiz phylogenetic alignment of slc39a12 orthologs in 16 vertebrate genomes show conservation in exons and some extra-exonic regions. Primate and vertebrate exonic and intronic regions of conservation are indicated by peaks following analysis by PhyloP and PhastCons. Scale bar at top indicates 50 kb.
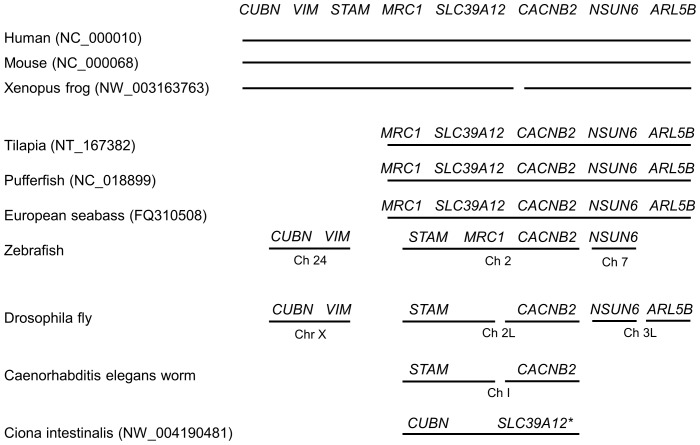
Figure 3. Synteny of slc39a12 is preserved across nearly all vertebrates examined.
Xenopus refers to tropicalis species of frog. NCBI accession numbers are indicated in parentheses where slc39a12 is present. Gaps between solid lines within the same chromosome indicate that genes may be distant from each other. Chromosome number (Ch) is noted to indicate chromosome location of genes. The asterisk indicates that the putative slc39a12 gene in Caenorhabditis elegans could not be confirmed using reciprocal BLAST hits.
Figure 4. Phylogenetic tree based upon ZIP12 amino acid sequences in different species.
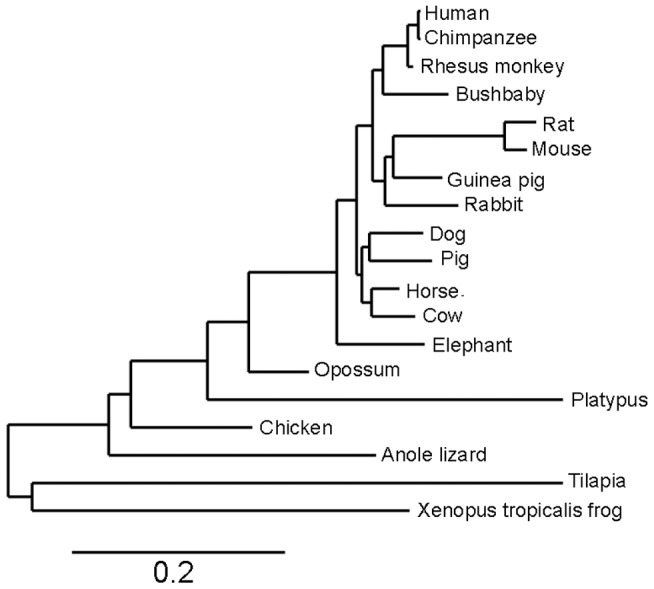
Common names of organisms are listed. Scale bar indicates 0.2 amino acid substitutions per site.
Xenopus laevis embryology has a long history as a developmental model that precedes the use of Xenopus tropicalis in biological research, but the availability of a genome sequence for Xenopus tropicalis supports its use in genetic applications [53]. The larger size of Xenopus laevis embryo relative to Xenopus tropicalis can facilitate micromanipulations, such as microdissections, explants, tissue transplants, and targeted microinjections at advanced cleavage stages [54], [55]. Although Xenopus laevis has a pseudotetraploid genome [56] with some gene duplication, only a single match for slc39a12 in Xenopus laevis [Joint Genome Institute (JGI) scaffold 000029451: 376741-407094] was detected, and the expected protein shares 85% amino acid identity with ZIP12 in Xenopus tropicalis. However, because the Xenopus laevis genome is still being assembled [57], the possibility of another duplicate slc39a12 gene in the Xenopus laevis genome cannot be ruled out. The combination of a single ZIP12 ortholog in Xenopus laevis and larger embryo size may ease the study of ZIP12 in this organism and could complement further studies of ZIP12 in Xenopus tropicalis.
In agreement with Feeney et al. [58], attempts using different strategies to detect the slc39a12 gene in zebrafish were unsuccessful. The slc39a12 gene is present in other teleost fish, including tilapia, European sea bass, and green spotted pufferfish (Figure 3). Local synteny near slc39a12 is preserved across humans, mice, European sea bass, and green spotted pufferfish (Figure 3). In contrast, for slc39a12 and genes such as cacnb2 and nsun6 that surround slc39a12 in other species (Figure S1), the genomic arrangement is disrupted in zebrafish. Furthermore, the slc39a12 gene could be detected in Japanese medaka, European sea bass, and tilapia by PCR using degenerate primers, but slc39a12 was not detectable in zebrafish (Figure S4). Because ZIP12 is required for embryonic viability in Xenopus [21], it is unclear how neurulation in zebrafish [59] proceeds without the presence of ZIP12. One possibility is that slc39a12 resides in a region of the zebrafish genome that is still being resolved [60], but the disruption of microsynteny in surrounding genes in zebrafish relative to other teleosts and inability to detect slc39a12 by PCR in zebrafish do not support this explanation. Although there are morphological differences in neurulation between amphibians and teleosts [61], slc39a12 is present in other fish species and the cellular mechanisms in neurulation are conserved between frogs and fish. A likely explanation is that the function of ZIP12 has been substituted with another SLC39 family member, possibly due to genome duplication [62]. Studies have shown that the zebrafish has retained many duplicated genes, whereas the pufferfish has lost many of the duplicated genes [63]. More research is required to resolve this issue and to provide an explanation for the apparent absence of the slc39a12 gene in zebrafish.
The identification and comparison of genes in invertebrates and vertebrates can provide significant insight into the origins of genes and their role in development. Neurulation is limited to chordates, including Ciona [64] and Petromyzon [65], but Drosophila [66], C. elegans [67], and Strongylocentrotus [68] have nervous systems but do not undergo neural tube closure. Neurulation in Branchiostoma proceeds with some distinct differences compared to vertebrates [69]. Regulatory subfunctionalization, which occurs through changes in cis-regulatory domains of duplicated genes, is recognized as a possible mechanism for the development of paralogs following a presumed gene duplication event [70]. For zinc transporter genes of the SLC30 family, Gustin et al. [71] proposed that the retention of duplicated genes occurs through changes in the expression patterns leading to eventual neofunctionalization or subfunctionalization. To test for a role of ZIP12 in neurulation, searches for ZIP12 were conducted in invertebrates with no or limited aspects of neural tube closure during embryogenesis. If ZIP12 functions in a critical role during neural tube closure, then positive selection may apply to slc39a12 in vertebrate genomes, whereas organisms that do not undergo neurulation may lack the evolutionary pressures that led to the emergence of slc39a12 in vertebrates. The criteria used to successfully identify ZIP12 in vertebrates, including reciprocal BLAST, conserved synteny, and conserved HNFAD and HEIPHE motifs in predicted transmembrane helices (Figure S2), was used to detect ZIP12 in other organisms.
There are possible matches for ZIP12 in Ciona intestinalis and Petromyzon marinus with similarity to human ZIP12, but a slc39a12 gene could not be conclusively defined in these genomes. The putative sea squirt and lamprey genes have the motifs HNFTD-HEIPHE and HNFAD-HEVPHE that are near-matches for conserved transmembrane helices of ZIP12 in vertebrates. Although BLAST searches using human and tilapia ZIP12 amino acid sequences identify putative ZIP12 genes in tunicates and sea lampreys, reciprocal BLAST searches were unable to distinguish the putative tunicate and sea lamprey slc39a12 genes as orthologs distinct from slc39a4. The putative Ciona slc39a12 gene is located within 20 kb of an ortholog for cubulin, a gene that neighbors slc39a12 in many vertebrate species (Figure 3). The limited size of the genome scaffold and putative transcripts for ZIP12 available in the sea lamprey restricted the analysis of slc39a12 and did not enable synteny analysis. Two intriguing questions are whether tunicates and lampreys contain slc39a12 genes with a similar function to ZIP12 in vertebrate neurulation. Future experiments may shed light on the developmental regulation of zinc transport through functional characterization of putative zinc transporters of the SLC39 gene family, and both gain-of-function and loss-of-function approaches are possible in tunicate and lamprey embryos [71], [72].
Attempts to find ZIP12 in fruit flies, nematodes, sea urchins, and lancelets using BLAST were unsuccessful. Local synteny in Drosophila, C. elegans, Strongylocentrotus, and Branchiostoma was not conserved for the genes that neighbor slc39a12 in vertebrates (Figure 3). ZIP12 is required for both neurulation during embryogenesis and neurite outgrowth in Neuro-2a cells and primary mouse neurons [21]. However, the inability to conclusively identify ZIP12 in these invertebrates, despite the presence of a nervous system, suggests a possible link to neural tube closure as opposed to later stages of nervous system development. It is possible that the origins of slc39a12 come from the requirement for zinc transport during neurulation in vertebrates (and possibly chordates), and that a role of ZIP12 in vertebrate nervous system function evolved following neurulation.
There may be some plausible alternate explanations for the observed lack of slc39a12 in invertebrates. For example, the inability to detect ZIP12 may be due to inadequate or overly stringent search criteria or incomplete genome coverage. It is possible that a similar ZIP transporter such as ZIP4 or other metal-permeable transporters may substitute for ZIP12 in other organisms. For example, a TRP channel mediates zinc transport and is critical for development in Drosophila [73], an organism in which ZIP12 was not detected. However, the lack of slc39a12 in zebrafish, a teleost that undergoes neurulation [59], represents an important observation that conflicts with the putative association between ZIP12 and vertebrate neurulation. Functional characterization across various organisms such as mice, frogs, and other developmental model organisms may shed light about the role of ZIP12, other ZIP transporters, and zinc regulation during nervous system development.
Many of the predicted sequences for slc39a12 (Table 1) were performed through computational analyses [30], and some sequences lack biological evidence such as cDNA support. Bioinformatic and computational analyses are extremely useful, especially for genome-wide annotation of genes, but there can be some discrepancies or unresolved regions [74]. Accordingly, some differences were detected between annotated versions of slc39a12 solely derived from computation and other versions of slc39a12 with additional bases including expressed sequence tags (ESTs) and other biological experimentation. Comparisons with other ZIP12 sequences showed that the rat ZIP12 protein sequence [GenBank: NP_001099594] was likely missing the N-terminus (Figure S3). As a result, the N-terminus of ZIP12 in this report was reconstituted from the translated sequences of a rat EST [GenBank: FM065041] and 12 nucleotides of the rat genome [GenBank: NW_047496 3955687-3955698] (Figure 4, Figure S3). The 5′ untranslated region (UTR) of the annotated cow ZIP12 [GenBank: NM_001076878] (Figure 5) is missing at least 60 bp, based upon EST data and the observation that the conserved exon-intron structure of human, mouse, and chicken slc39a12 has the start codon in exon 2 (Figure 1, Figure 2). A comparison of the 5′ end of 2 EST clones [GenBank: EV626550 and EE901356] aligns to a separate exon in the cow genome [GenBank: AC_000170, 32666526-32666609]. The refinement of draft genome sequences and the correct annotation of gene sequences is important because genome sequences often provide the initial foundation for biological experimentation, particularly in reverse genetics.
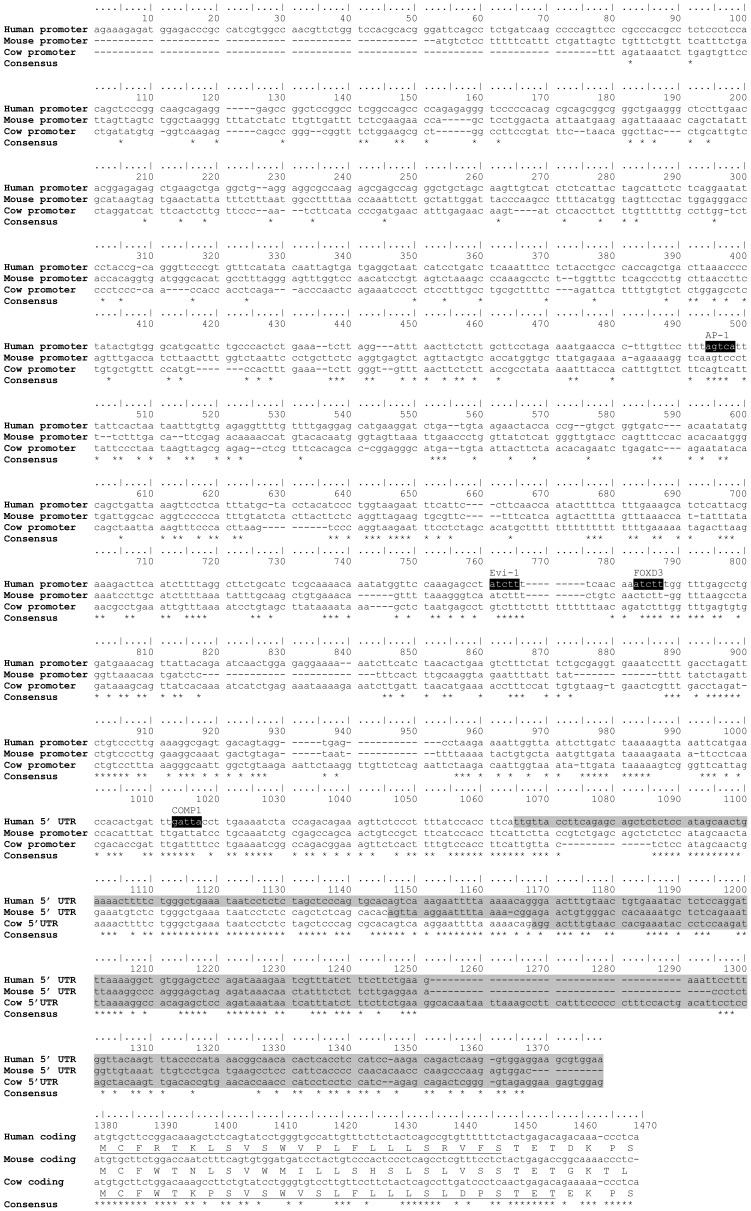
Figure 5. Alignment for 5′ UTR, first 90 bp of coding region, and the proximal promoter (1000 bp upstream of transcription start site) of human, mouse, and cow slc39a12.
Lowercase and uppercase in sequences indicate nucleotide and amino acid sequences, respectively. Black shaded text indicates possible transcription factor binding sites in largely conserved regions. Gray shaded text indicates 5′ UTR. For coding regions, possible signal peptide is underlined. Asterisks indicate nucleotides conserved in all sequences following alignment.
ZIP12 is highly expressed in human, mouse, and Xenopus tropicalis brain tissue [21]. In support of this observation, the majority of expressed sequence tags (ESTs) matching ZIP12 is derived from brain or other nervous system tissues. Over-representation of ZIP12 in the brain occurred in almost all mammalian and avian species examined, including human, mouse, cow, pig, rhesus monkey, chicken, rat, and crab-eating macaque (Table 2). Only in dogs (Table 2) were ESTs not derived from brain tissue, and the pool of ESTs that match ZIP12 in dogs has few samples. This observation largely supports the notion that ZIP12 function in the nervous system is widely conserved across vertebrates. The conserved expression of ZIP12 in the brain across species is significant because the rate of evolution in tissue-specific genes appears to be faster and genetic disorders with Mendelian inheritance are more likely to be caused by mutations in tissue-specific genes [75]. Consistent with this observation, inhibition of ZIP12 function in mouse neurons and Xenopus tropicalis embryos leads to impaired neuronal differentiation and neural tube defects, respectively [21].
Table 2. Tissue origin of ESTs matching ZIP12 is predominantly brain and nervous system.
| Organism | Unigene ID | ESTs from brain | 3′ UTR A/U |
|---|---|---|---|
| Human (Homo sapiens) | 193909 | 129/135 (95.6%) | A:32.9% U:40.3% |
| Mouse (Mus musculus) | 271009 | 31/47 (66.0%) | A:31.8% U:41.8% |
| Cow (Bos taurus) | 1209583 | 30/42 (71.4%) | A:33.3% U:41.2% |
| Pig (Sus scrofa) | 3109541 | 13/14 (92.9%) | A:32.0% U:39.0% |
| Chicken (Gallus gallus) | 1236550 | 5/8 (62.5%) | A:30.0% U:34.4% |
| Macaque (Macaca fascicularis) | 2484482 | 5/5 (100.0%) | A:30.9% U:40.4% |
| Rat (Rattus norvegicus) | 3092273, 1534920 | 4/5 (80.0%) | A:31.6% U:41.5% |
| Rhesus monkey (Macaca mulatta) | 5927016 | 2/2 100.0% | A:31.2% U:40.3% |
| Dog (Canis lupus familiaris) | 1193028 | 0/2 0% | A:31.5% U:38.8% |
There are possible mechanisms outside the coding sequence which may control the tissue-specific expression of slc39a12 in vertebrates and are likely conserved. The 3′ untranslated region of ZIP12 in many species is adenine/uracil (A/U) rich (Table 2). The 3′ UTRs of many brain-specific genes are A/U rich [76]. There are proteins that can affect mRNA stability by binding to these A/U rich regions [77], which may account for the high expression of ZIP12 in the nervous system. An alignment of the 5′ UTR, the first 90 bp of the coding sequence, and the proximal promoter (1000 bp upstream of the transcription start site) (Figure 5) shows that there are areas of sequence conservation. A scan of the human proximal promoter combined with an alignment of the sequences with cow and mouse shows that there are putative transcription binding sites for AP-1, Evi-1, FoxD3, and COMP1 (Figure 6). It is possible that the N-terminus encodes a signal peptide, and the prediction software SignalP [78] indicates that there may be a cleavage site between amino acids 23–24 or 26–27 (Figure 5). In contrast to many genes important for neuronal development [79], the 5′ UTR and proximal promoter are not guanine/cytosine (GC) rich (GC content: human 44.8%; mouse 39.3%; cow 39.8%), and there was no difference in GC content between the 5′ UTR (41.8%) and proximal promoter (45.6%) of the human slc39a12 gene. More analyses, possibly combined with biological experimentation, will be needed to determine if the 5′ UTR, 3′ UTR, or nearby upstream portions contribute towards the high expression of ZIP12 in the nervous system. As an example of how regulatory elements can control zinc uptake transporter expression, active metal-response elements have been identified in the 5′ UTR of zinc transporter genes in mice and zebrafish [80], [81]. Because there are possible conserved elements in the distal promoter and some intronic regions, as indicated by PhyloP and PhastCons (Figure 2), these areas may also contribute towards the distinct pattern of ZIP12 expression in the vertebrate nervous system and the possible regulatory subfunctionalization of ZIP12 and close paralogs. The tissue specificity of slc39a12 in these species and previous findings showing enriched expression of slc39a12 in vertebrate brains [21] support a role for ZIP12 in the central nervous system.
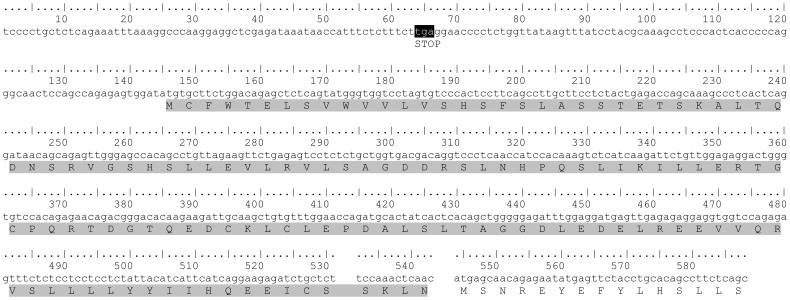
Figure 6. Rat slc39a12 mRNA and translated protein sequences derived from EST and genome analyses.
Full sequence was formed from rat EST [GenBank: FM065041], genome sequence [GenBank: NW_047496, nucleotides 3955687-3955698], and current annotated entry for rat slc39a12 [GenBank: NM_001106124]. The additional N-terminus amino acid sequence is shaded gray. Stop codon (opal) upstream of putative start codon is shaded black.
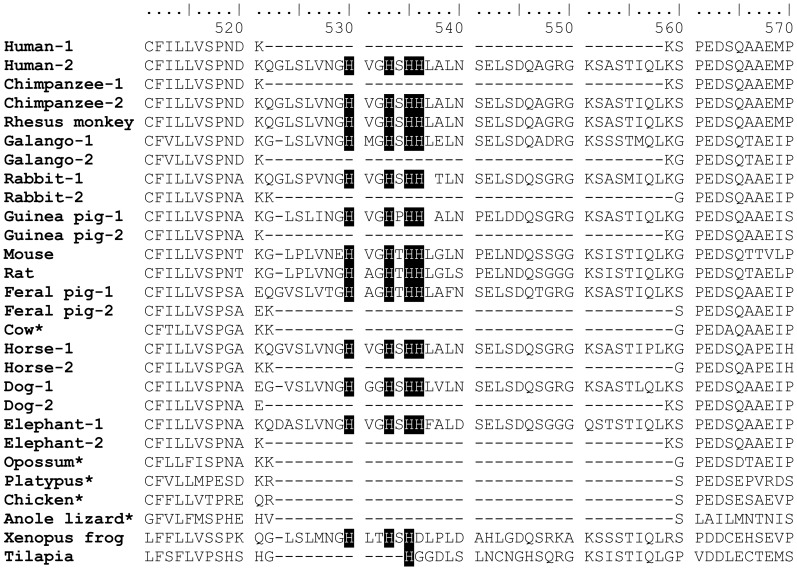
The alignment (Figure S3) uncovered 2 different isoforms of ZIP12 frequently detected across many species which correspond to the inclusion or exclusion of exon 9. Furthermore, this exon was present in at least two non-mammalian organisms, Xenopus tropicalis and tilapia (Figure S1), which supports an ancestral history for this exon in slc39a12 that precedes the split with birds and mammals [82]. Although this exon is likely present in other mammalian organisms, the longer isoform has not been described previously in chicken, cow, opossum, or platypus. TBLASTN searches for exon 9 showed that the sequence is present in cow, chicken, and opossum genomes (Figure 7). This sequence was not detected in the platypus, but this may be due to gaps in the genomic sequence [GenBank: NW_001594582, 1525899-1525899, 1527878-1528642]. Reverse transcriptase-polymerase chain reaction (RT-PCR) was used to determine that both isoforms are expressed in humans, cows, and rats (Figure 8A-8C). However, the shorter isoform in mice (Figure 8D) could not be detected by RT-PCR despite repeated attempts with different primer sequences and PCR cycling conditions (data not shown). The reading frame in this exon appears to be conserved across species (111 bp in humans and 108 bp in other species, Figure 9). It is possible that the reading frame of this exon is conserved, so that inclusion or exclusion of this exon does not affect the downstream reading frame. The translated product of this exon is expected to increase the length of a cytoplasmic loop between transmembrane domains 3 and 4 of ZIP12 [21]. The transcript variation of ZIP12 is likely due to exon skipping, which is the most common form of splice variation [83]. The intron-exon structure flanking exon 9 is conserved across multiple vertebrate and mammalian species (Figure 2), which supports the notion that this variation has biological significance. Because this region of ZIP12 encodes a histidine-rich segment that is expected to lengthen a cytoplasmic loop [21], [48], this region could be important for post-translational regulation of ZIP12. In support of this possible function, ZIP4 contains a similarly located histidine-rich, cytoplasmic-facing motif that is sensitive to zinc and required for ubiquitin-mediated protein degradation in response to excess zinc [47]. Wide-scale global approaches have used comparative genomics to discover novel human exons that were previously unidentified because of weak or lacking cDNA support due to low transcript levels or restricted tissue specificity [84].
Figure 7. Histidine-rich exon 9 present in human and mouse ZIP12 is also present in cow, opossum, and chicken genomes.
Organism common names are accompanied by accession numbers, corresponding nucleotides, and translated amino acid sequence. Amino acids conserved between humans and mice are shaded in black. Corresponding amino acids that are conserved in cow, opossum, and chicken are shaded in grey.
Figure 8. Splice variation of ZIP12 confirmed in the brain of multiple species.
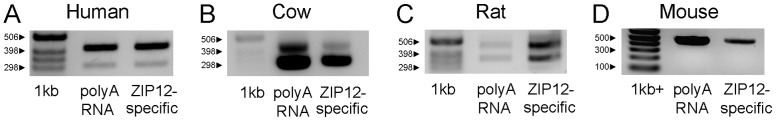
Different splice variants of ZIP12 are present in (A) humans, (B) cow, and (C) rat, but the short variant is not detectable in (D) mice. PCR was conducted on cDNA reverse-transcribed from polyadenylated or ZIP12-specific RNA using primers spanning the exon that is present and absent in the long and short isoform, respectively. The corresponding molecular weights of the DNA markers are indicated.
Figure 9. Amino acid alignment of ZIP12 demonstrates splice variation due to variable inclusion of a exon which contains a histidine-rich motif.
Where indicated, -1 and -2 indicate annotated entries for splice variants of ZIP12 from inclusion or exclusion of exon 9. Shown alignment performed by ClustalW corresponds to amino acids 464–523 of the longer human ZIP12 variant (Human-2) [GenBank: NP_001138667]. Conserved histidine residues are shaded in black. Asterisks indicate organisms with a single annotated amino acid sequence for ZIP12 that lacks exon 9. Full sequence alignment is provided in Figure S3.
ZIP12 is present at both the plasma membrane and in intracellular compartments in a similar fashion to other SLC39 transporters [21], [38], [85], but it is unclear what mechanisms control the localization of ZIP12. Human ZIP12 has possible di-leucine motifs [86] at L94-L95 (EPDALLI), L116-L118 (QRVSLLL), and L255-L256 (ELDQLLL) and a possible tyrosine motif at Y120 (YYII) that are conserved in many mammalian species (Figure S3). Adaptor protein complexes recognize both di-leucine and tyrosine motifs and bind to transmembrane proteins to dynamically alter their cellular localization [86]. Human ZIP1 has a di-leucine motif that is important for endocytosis from the plasma membrane [38], [86], which likely controls zinc transport activity by withdrawing the zinc transporter from the plasma membrane [85] so that zinc can no longer be transported from the extracellular media by ZIP1. ZIP4 contains an ectodomain that is proteolytically cleaved during zinc deficiency, which could be due to a metalloprotease cleavage site [87]. The ectodomain of ZIP6 (LIV-1) can also be separated from its transmembrane-spanning C-terminus [88], [89]. Kambe and Andrews previously reported that the putative metalloproteinase site in ZIP4 is conserved at amino acids 338–341 (PGII) in human ZIP12 [87]. It is possible that these motifs that are conserved in ZIP12 and present in other SLC39 transporters are important for regulating the biochemical and cellular properties of this zinc transporter.
Conclusion
The conservation of ZIP12 expression in the nervous system of vertebrates, combined with the previous report of the critical role of ZIP12 in Xenopus embryogenesis and mouse neuronal development supports the notion that zinc transport by ZIP12 is crucial for many vertebrate organisms. Zinc deficiency in numerous animal models is associated with congenital malformations and impaired development including neural tube defects [4], [6], [19]. Conserved gene co-expression analysis [23] and genotype-phenotype relationships (phenologs) across species [90] have been hypothesized by other researchers to predict candidate genes in human disease. These observations further strengthen the notion that slc39a12 may be a candidate gene for neural tube defects or neurodevelopmental disorders [21].
The role of zinc in the evolution of embryogenesis is not currently well-studied, but the wide presence of ZIP12 in vertebrates and possibly chordates suggests a physiological importance of zinc transporters in these pathways. Cellular zinc homeostasis and free zinc availability is tightly regulated [91], even in bacteria, but there is emerging evidence shows that zinc may affect cell signaling [21], [49] and post-transcriptional mechanisms [13]. It is possible that the origins of zinc transporter functions in cell signaling originated from the role of related paralogs and their roles in zinc homeostasis and detoxification. Further investigation is required in order to determine the origins of ZIP12 in relation to the evolution of developmental processes and the likely significant role of ZIP12 in nervous system function in vertebrate organisms.
Supporting Information
Figure S1
Synteny of slc39a12 is preserved across nearly all vertebrates examined. NCBI accession numbers are indicated in parentheses. Solid lines indicate that neighboring genes on the same chromosome.
(TIF)
Figure S2
Sequence alignments of different members of the SLC39 (ZIP) family. The amino acid sequences (and NCBI accession numbers) are as follows: human ZIP12 [GenBank: NP_001138667] and ZIP1 [GenBank: NP_055252], Saccharomyces cerevisiae (yeast) ZRT1 [GenBank: NP_011259], Arabidopsis ZIP2 [GenBank: NP_200760], and mouse ZIP4 [GenBank: NP_082340], ZIP6 [GenBank: NP_631882], and ZIP14 [GenBank: NP_001128623]. The consensus sequence is represented below the alignment, ranging from no (blank), low (.), medium (:), to high conservation (*). Conserved amino acids from the putative transmembrane domains 4 and 5 are indicated by gray shading.
(PDF)
Figure S3
Sequence alignments of ZIP12 orthologs from different vertebrates. NCBI accession numbers for amino acid sequences are provided in Table 1. The consensus sequence is represented below the alignment, ranging from no (blank), low (.), medium (:), to high conservation (*).
(PDF)
Figure S4
The slc39a12 gene is detectable in Japanese medaka, Nile tilapia, and European seabass but not zebrafish. The slc39a12 and igf1 genes were detected by PCR using genomic DNA from zebrafish, Japanese medaka, Nile tilapia, and European seabass and primers with degeneracy. In zebrafish, only the igf1 gene was detected, whereas both slc39a12 and igf1 genes were detected in the other fish. The expected PCR product sizes for slc39a12 and igf1 are 106 bp and 130 bp, respectively.
(TIF)
Acknowledgments
The author thanks Bruce Draper and Swee Teh for generously providing zebrafish and Japanese medaka genomic DNA. The author thanks Robert Rucker for support of this research and thanks Yalin Liao for critical reading of the manuscript.
Funding Statement
Funding for this research was provided by a grant from the University of California, Davis Center for Health and Nutrition Research (to Robert B. Rucker). No additional funding was received for this study. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ho E, Ames BN (2002) Low intracellular zinc induces oxidative DNA damage, disrupts p53, NFkappa B, and AP1 DNA binding, and affects DNA repair in a rat glioma cell line. Proc Natl Acad Sci U S A 99: 16770–16775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frederickson CJ, Koh JY, Bush AI (2005) The neurobiology of zinc in health and disease. Nat Rev Neurosci 6: 449–462. [DOI] [PubMed] [Google Scholar]
- 3.Ibs KH, Rink L (2003) Zinc-altered immune function. Journal of Nutrition 133: 1452S–1456S. [DOI] [PubMed] [Google Scholar]
- 4.Swenerton H, Shrader R, Hurley LS (1969) Zinc-deficient embryos: reduced thymidine incorporation. Science 166: 1014–1015. [DOI] [PubMed] [Google Scholar]
- 5.Prasad AS (1988) Zinc in growth and development and spectrum of human zinc deficiency. Journal of the American College of Nutrition 7: 377–384. [DOI] [PubMed] [Google Scholar]
- 6.Forbes RM (1984) Use of laboratory animals to define physiological functions and bioavailability of zinc. Federation Proceedings 43: 2835–2839. [PubMed] [Google Scholar]
- 7.Luecke RW (1984) Domestic animals in the elucidation of zinc's role in nutrition. Federation Proceedings 43: 2823–2828. [PubMed] [Google Scholar]
- 8.Hidiroglou M (1979) Trace element deficiencies and fertility in ruminants: a review. Journal of Dairy Science 62: 1195–1206. [DOI] [PubMed] [Google Scholar]
- 9.Eide DJ (2004) The SLC39 family of metal ion transporters. Pflugers Archiv European Journal of Physiology 447: 796–800. [DOI] [PubMed] [Google Scholar]
- 10.Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proceedings of the National Academy of Sciences of the United States of America 93: 5624–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao H, Eide D (1996) The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proceedings of the National Academy of Sciences of the United States of America 93: 2454–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grotz N, Fox T, Connolly E, Park W, Guerinot ML, et al. (1998) Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proceedings of the National Academy of Sciences of the United States of America 95: 7220–7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathews WR, Ong D, Milutinovich AB, Van Doren M (2006) Zinc transport activity of Fear of Intimacy is essential for proper gonad morphogenesis and DE-cadherin expression. Development 133: 1143–1153. [DOI] [PubMed] [Google Scholar]
- 14.Dufner-Beattie J, Langmade SJ, Wang F, Eide D, Andrews GK (2003) Structure, function, and regulation of a subfamily of mouse zinc transporter genes. J Biol Chem 278: 50142–50150. [DOI] [PubMed] [Google Scholar]
- 15.Gaither LA, Eide DJ (2000) Functional expression of the human hZIP2 zinc transporter. J Biol Chem 275: 5560–5564. [DOI] [PubMed] [Google Scholar]
- 16.Costello LC, Liu Y, Zou J, Franklin RB (1999) Evidence for a zinc uptake transporter in human prostate cancer cells which is regulated by prolactin and testosterone. Journal of Biological Chemistry 274: 17499–17504. [DOI] [PubMed] [Google Scholar]
- 17.Wang K, Zhou B, Kuo YM, Zemansky J, Gitschier J (2002) A novel member of a zinc transporter family is defective in acrodermatitis enteropathica. Am J Hum Genet 71: 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kury S, Dreno B, Bezieau S, Giraudet S, Kharfi M, et al. (2002) Identification of SLC39A4, a gene involved in acrodermatitis enteropathica. Nat Genet 31: 239–240. [DOI] [PubMed] [Google Scholar]
- 19.Dufner-Beattie J, Weaver BP, Geiser J, Bilgen M, Larson M, et al. (2007) The mouse acrodermatitis enteropathica gene Slc39a4 (Zip4) is essential for early development and heterozygosity causes hypersensitivity to zinc deficiency. Human Molecular Genetics 16: 1391–1399. [DOI] [PubMed] [Google Scholar]
- 20.Yuzbasiyan-Gurkan V, Bartlett E (2006) Identification of a unique splice site variant in SLC39A4 in bovine hereditary zinc deficiency, lethal trait A46: An animal model of acrodermatitis enteropathica. Genomics 88: 521–526. [DOI] [PubMed] [Google Scholar]
- 21.Chowanadisai W, Graham DM, Keen CL, Rucker RB, Messerli MA (2013) Neurulation and neurite extension require the zinc transporter ZIP12 (slc39a12). Proceedings of the National Academy of Sciences of the United States of America 110: 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alfoldi J, Lindblad-Toh K (2013) Comparative genomics as a tool to understand evolution and disease. Genome Research 23: 1063–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ala U, Piro RM, Grassi E, Damasco C, Silengo L, et al. (2008) Prediction of human disease genes by human-mouse conserved coexpression analysis. PLoS Comput Biol 4: e1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabaldon T (2008) Comparative genomics-based prediction of protein function. Methods in Molecular Biology 439: 387–401. [DOI] [PubMed] [Google Scholar]
- 25.Te Velthuis AJ, Bagowski CP (2008) Linking fold, function and phylogeny: a comparative genomics view on protein (domain) evolution. Curr Genomics 9: 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearlman SM, Serber Z, Ferrell JE Jr (2011) A mechanism for the evolution of phosphorylation sites. Cell 147: 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, et al. (2002) Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418: 869–872. [DOI] [PubMed] [Google Scholar]
- 28.Bowes JB, Snyder KA, Segerdell E, Gibb R, Jarabek C, et al. (2008) Xenbase: a Xenopus biology and genomics resource. Nucleic Acids Res 36: D761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, et al. (2013) Ensembl 2013. Nucleic Acids Res 41: D48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, et al. (2012) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 40: D13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, et al. (2013) The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 41: D64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95–98. [Google Scholar]
- 34.Chevenet F, Brun C, Banuls AL, Jacq B, Christen R (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7: 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, et al. (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34: D108–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, et al. (2012) ELM–the database of eukaryotic linear motifs. Nucleic Acids Res 40: D242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang L, Kirschke CP (2007) A di-leucine sorting signal in ZIP1 (SLC39A1) mediates endocytosis of the protein. Febs J 274: 3986–3997. [DOI] [PubMed] [Google Scholar]
- 39.Faircloth BC, Sorenson L, Santini F, Alfaro ME (2013) A Phylogenomic Perspective on the Radiation of Ray-Finned Fishes Based upon Targeted Sequencing of Ultraconserved Elements (UCEs). PLoS ONE 8: e65923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno-Hagelsieb G, Latimer K (2008) Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 24: 319–324. [DOI] [PubMed] [Google Scholar]
- 41.Jun J, Mandoiu II, Nelson CE (2009) Identification of mammalian orthologs using local synteny. BMC Genomics 10: 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bin BH, Fukada T, Hosaka T, Yamasaki S, Ohashi W, et al. (2011) Biochemical characterization of human ZIP13 protein: a homo-dimerized zinc transporter involved in the spondylocheiro dysplastic Ehlers-Danlos syndrome. Journal of Biological Chemistry 286: 40255–40265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seve M, Chimienti F, Devergnas S, Favier A (2004) In silico identification and expression of SLC30 family genes: an expressed sequence tag data mining strategy for the characterization of zinc transporters' tissue expression. BMC Genomics 5: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, et al. (2004) The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins Introduction. Pflugers Archiv European Journal of Physiology 447: 465–468. [DOI] [PubMed] [Google Scholar]
- 45.Zhao H, Bourque G (2009) Recovering genome rearrangements in the mammalian phylogeny. Genome Research 19: 934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas JW, Touchman JW, Blakesley RW, Bouffard GG, Beckstrom-Sternberg SM, et al. (2003) Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424: 788–793. [DOI] [PubMed] [Google Scholar]
- 47.Mao X, Kim BE, Wang F, Eide DJ, Petris MJ (2007) A histidine-rich cluster mediates the ubiquitination and degradation of the human zinc transporter, hZIP4, and protects against zinc cytotoxicity. Journal of Biological Chemistry 282: 6992–7000. [DOI] [PubMed] [Google Scholar]
- 48.Rogers EE, Eide DJ, Guerinot ML (2000) Altered selectivity in an Arabidopsis metal transporter. Proc Natl Acad Sci U S A 97: 12356–12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor KM, Nicholson RI (2003) The LZT proteins; the LIV-1 subfamily of zinc transporters. Biochimica et Biophysica Acta 1611: 16–30. [DOI] [PubMed] [Google Scholar]
- 50.Miller W, Rosenbloom K, Hardison RC, Hou M, Taylor J, et al. (2007) 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Research 17: 1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W (2007) Using genomic data to unravel the root of the placental mammal phylogeny. Genome Research 17: 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janecka JE, Miller W, Pringle TH, Wiens F, Zitzmann A, et al. (2007) Molecular and genomic data identify the closest living relative of primates. Science 318: 792–794. [DOI] [PubMed] [Google Scholar]
- 53.Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, et al. (2010) The genome of the Western clawed frog Xenopus tropicalis. Science 328: 633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sive HL, Grainger RM, Harland RM (2007) Microdissection: Explant and Transplant Assays in Xenopus laevis. CSH Protoc 2007: pdb top10. [DOI] [PubMed]
- 55.Mimoto MS, Christian JL (2011) Manipulation of gene function in Xenopus laevis. Methods in Molecular Biology 770: 55–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bisbee CA, Baker MA, Wilson AC, Haji-Azimi I, Fischberg M (1977) Albumin phylogeny for clawed frogs (Xenopus). Science 195: 785–787. [DOI] [PubMed] [Google Scholar]
- 57.Gilchrist MJ (2012) From expression cloning to gene modeling: the development of Xenopus gene sequence resources. Genesis 50: 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feeney GP, Zheng D, Kille P, Hogstrand C (2005) The phylogeny of teleost ZIP and ZnT zinc transporters and their tissue specific expression and response to zinc in zebrafish. Biochimica et Biophysica Acta 1732: 88–95. [DOI] [PubMed] [Google Scholar]
- 59.Lowery LA, Sive H (2004) Strategies of vertebrate neurulation and a re-evaluation of teleost neural tube formation. Mechanisms of Development 121: 1189–1197. [DOI] [PubMed] [Google Scholar]
- 60.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, et al. (2013) The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong E, Brewster R (2006) N-cadherin is required for the polarized cell behaviors that drive neurulation in the zebrafish. Development 133: 3895–3905. [DOI] [PubMed] [Google Scholar]
- 62.Amores A, Force A, Yan YL, Joly L, Amemiya C, et al. (1998) Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711–1714. [DOI] [PubMed] [Google Scholar]
- 63.Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y (2003) Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Research 13: 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Colas JF, Schoenwolf GC (2001) Towards a cellular and molecular understanding of neurulation. Developmental Dynamics 221: 117–145. [DOI] [PubMed] [Google Scholar]
- 65.Richardson MK, Wright GM (2003) Developmental transformations in a normal series of embryos of the sea lamprey Petromyzon marinus (Linnaeus). Journal of Morphology 257: 348–363. [DOI] [PubMed] [Google Scholar]
- 66.Bellen HJ, Tong C, Tsuda H (2010) 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat Rev Neurosci 11: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chalfie M, Jorgensen EM (1998) C. elegans neuroscience: genetics to genome. Trends in Genetics 14: 506–512. [DOI] [PubMed] [Google Scholar]
- 68.Sawyer JM, Harrell JR, Shemer G, Sullivan-Brown J, Roh-Johnson M, et al. (2010) Apical constriction: a cell shape change that can drive morphogenesis. Developmental Biology 341: 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benito-Gutierrez E (2006) A gene catalogue of the amphioxus nervous system. Int J Biol Sci 2: 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, et al. (2013) Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nature Genetics 45: : 415–421, 421e411–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nikitina N, Bronner-Fraser M, Sauka-Spengler T (2009) Microinjection of RNA and morpholino oligos into lamprey embryos. Cold Spring Harb Protoc 2009: pdb prot5123. [DOI] [PubMed]
- 72.Christiaen L, Wagner E, Shi W, Levine M (2009) Microinjection of morpholino oligos and RNAs in sea squirt (Ciona) embryos. Cold Spring Harb Protoc 2009: pdb prot5347. [DOI] [PubMed]
- 73.Hofmann T, Chubanov V, Chen X, Dietz AS, Gudermann T, et al. (2010) Drosophila TRPM channel is essential for the control of extracellular magnesium levels. PLoS One 5: e10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nagy A, Hegyi H, Farkas K, Tordai H, Kozma E, et al. (2008) Identification and correction of abnormal, incomplete and mispredicted proteins in public databases. BMC Bioinformatics 9: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Winter EE, Goodstadt L, Ponting CP (2004) Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Research 14: 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bolognani F, Perrone-Bizzozero NI (2008) RNA-protein interactions and control of mRNA stability in neurons. Journal of Neuroscience Research 86: 481–489. [DOI] [PubMed] [Google Scholar]
- 77.Kao HT, Ghafoori S, Porton B, Wong DL, Ciaranello RD (1996) Brain specific proteins binding to the 3' UTR of the 5-HT2C receptor mRNA. Brain Research Molecular Brain Research 43: 174–184. [DOI] [PubMed] [Google Scholar]
- 78.Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- 79.Long YS, Qin JM, Su T, Zhao QH, Yi YH, et al. (2011) Human transcription factor genes involved in neuronal development tend to have high GC content and CpG elements in the proximal promoter region. J Genet Genomics 38: 157–163. [DOI] [PubMed] [Google Scholar]
- 80.Lichten LA, Ryu MS, Guo L, Embury J, Cousins RJ (2011) MTF-1-Mediated Repression of the Zinc Transporter Zip10 Is Alleviated by Zinc Restriction. PLoS One 6: e21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zheng D, Feeney GP, Kille P, Hogstrand C (2008) Regulation of ZIP and ZnT zinc transporters in zebrafish gill: zinc repression of ZIP10 transcription by an intronic MRE cluster. Physiol Genomics 34: 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kumar S, Hedges SB (1998) A molecular timescale for vertebrate evolution. Nature 392: 917–920. [DOI] [PubMed] [Google Scholar]
- 83.Sammeth M, Foissac S, Guigo R (2008) A general definition and nomenclature for alternative splicing events. PLoS Comput Biol 4: e1000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Siepel A, Diekhans M, Brejova B, Langton L, Stevens M, et al. (2007) Targeted discovery of novel human exons by comparative genomics. Genome Research 17: 1763–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang F, Dufner-Beattie J, Kim BE, Petris MJ, Andrews G, et al. (2004) Zinc-stimulated endocytosis controls activity of the mouse ZIP1 and ZIP3 zinc uptake transporters. Journal of Biological Chemistry 279: 24631–24639. [DOI] [PubMed] [Google Scholar]
- 86.Bonifacino JS, Traub LM (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72: 395–447. [DOI] [PubMed] [Google Scholar]
- 87.Kambe T, Andrews GK (2009) Novel proteolytic processing of the ectodomain of the zinc transporter ZIP4 (SLC39A4) during zinc deficiency is inhibited by acrodermatitis enteropathica mutations. Molecular and Cellular Biology 29: 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ehsani S, Salehzadeh A, Huo H, Reginold W, Pocanschi CL, et al. (2012) LIV-1 ZIP ectodomain shedding in prion-infected mice resembles cellular response to transition metal starvation. Journal of Molecular Biology 422: 556–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hogstrand C, Kille P, Ackland ML, Hiscox S, Taylor KM (2013) A mechanism for epithelial-mesenchymal transition and anoikis resistance in breast cancer triggered by zinc channel ZIP6 and STAT3 (signal transducer and activator of transcription 3). Biochemical Journal 455: 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McGary KL, Park TJ, Woods JO, Cha HJ, Wallingford JB, et al. (2010) Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proceedings of the National Academy of Sciences of the United States of America 107: 6544–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Outten CE, O'Halloran TV (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292: 2488–2492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Synteny of slc39a12 is preserved across nearly all vertebrates examined. NCBI accession numbers are indicated in parentheses. Solid lines indicate that neighboring genes on the same chromosome.
(TIF)
Figure S2
Sequence alignments of different members of the SLC39 (ZIP) family. The amino acid sequences (and NCBI accession numbers) are as follows: human ZIP12 [GenBank: NP_001138667] and ZIP1 [GenBank: NP_055252], Saccharomyces cerevisiae (yeast) ZRT1 [GenBank: NP_011259], Arabidopsis ZIP2 [GenBank: NP_200760], and mouse ZIP4 [GenBank: NP_082340], ZIP6 [GenBank: NP_631882], and ZIP14 [GenBank: NP_001128623]. The consensus sequence is represented below the alignment, ranging from no (blank), low (.), medium (:), to high conservation (*). Conserved amino acids from the putative transmembrane domains 4 and 5 are indicated by gray shading.
(PDF)
Figure S3
Sequence alignments of ZIP12 orthologs from different vertebrates. NCBI accession numbers for amino acid sequences are provided in Table 1. The consensus sequence is represented below the alignment, ranging from no (blank), low (.), medium (:), to high conservation (*).
(PDF)
Figure S4
The slc39a12 gene is detectable in Japanese medaka, Nile tilapia, and European seabass but not zebrafish. The slc39a12 and igf1 genes were detected by PCR using genomic DNA from zebrafish, Japanese medaka, Nile tilapia, and European seabass and primers with degeneracy. In zebrafish, only the igf1 gene was detected, whereas both slc39a12 and igf1 genes were detected in the other fish. The expected PCR product sizes for slc39a12 and igf1 are 106 bp and 130 bp, respectively.
(TIF)