Untangling the hybrid nature of modern pig genomes: a mosaic derived from biogeographically distinct and highly divergent Sus scrofa populations (original) (raw)
. Author manuscript; available in PMC: 2015 Feb 1.
Published in final edited form as: Mol Ecol. 2014 Jun 16;23(16):4089–4102. doi: 10.1111/mec.12807
Abstract
The merging of populations after an extended period of isolation and divergence is a common phenomenon, in natural settings as well as due to human interference. Individuals with such hybrid origins contain genomes that essentially form a mosaic of different histories and demographies. Pigs are an excellent model species to study hybridization because European and Asian wild boars diverged ~1.2 Mya and pigs were domesticated independently in Europe and Asia. During the Industrial Revolution in England, pigs were imported from China to improve the local pigs. This study utilizes the latest genomics tools to identify the origin of haplotypes in European domesticated pigs that are descendant from Asian and European populations. Our results reveal fine-scale haplotype structure representing different ancient demographic events, as well as a mosaic composition of those distinct histories due to recently introgressed haplotypes in the pig genome. As a consequence, nucleotide diversity in the genome of European domesticated pigs is higher when at least one haplotype of Asian origin is present, and haplotype length correlates negatively with recombination frequency and nucleotide diversity. Another consequence is that the inference of past effective population size is influenced by the background of the haplotypes in an individual, but we demonstrate that by careful sorting based on the origin of haplotypes both distinct demographic histories can be reconstructed. Future detailed mapping of the genomic distribution of variation will enable a targeted approach to increase genetic diversity of captive and wild populations, thus facilitating conservation efforts in the near future.
Keywords: Sus scrofa, Domestication, Introgression, Identity by descent, Hybridization, Conservation Genetics
Introduction
Separation and consecutive fusion of populations is common in both natural and managed populations. For instance, the waxing and waning continental ice sheets during the Pleistocene is known to have had a pronounced effect on shaping the population genetics of many species. While the glacial periods usually resulted in refugial populations and thereby promoted population differentiation, the interglacial periods that followed would result in renewed gene flow. Apart from natural causes, populations can also be reunited due to deliberate management. It is well known that the adaptive ability of a population or species to an ever changing environment is mainly determined by its standing variation, and susceptibility to a variety of diseases and environmental changes is assumed to increase if nucleotide diversity is low in the population (e.g. Jimenez et al. 1994, Lacy et al. 1996, Keller and Waller 2002). An increased probability of homozygosity for partially deleterious recessive mutations may lead to individuals with reduced fitness, i.e. inbreeding depression. Such inbreeding effects can be offset by directed population management aimed to facilitate outcrossing, which could result in higher haplotype diversity.
These patterns of reticulation can severely complicate the elucidation of population history. In the past decades, marker systems that have relative fast coalescence and do not or rarely undergo recombination (e.g. mtDNA, Y chromosome) have proven to be useful for phylo-geographic analysis. However, the ensuing pictures of population history that were thus constructed often turned out, or will turn out, to be literally only part of the demographic story. Because autosomes recombine, the genome of a single individual can contain haplotypes from distinct sources, each with another demographic history. Hybridization of populations therefore entails a great challenge to disentangle what has essentially become a mosaic of different demographies. In studies that focused on a relatively small number of nuclear DNA markers, results are usually concatenated to provide a “genome average”, for instance by doing Structure analysis. Although such analyses may provide insight in the degree of mixing of populations, they do not contain details of the distribution of the introgressed haplotypes over the genome. For instance, the number of generations since the last common ancestor influences the probability of haplotypes in the genome of two individuals to be Identical By Descent (IBD), since the size of the IBD segment declines over time due to recombination. Therefore, the length of IBD haplotypes as a function of local recombination frequency is a measure for the time since the last common ancestor (Palamara et al. 2012, Ralph et al. 2013, Henn et al. 2012) and signatures of introgression are revealed by coalescence times of haplotypes that are shorter than expected (Staubach et al. 2012). Current high-throughput genotyping and sequencing techniques enable such investigations on a whole-genome scale, providing information on how long ago the reticulation took place. Genomes have been studied in detail to elucidate population history for only a handful of species, e.g. Human (Harris and Nielsen 2013), polar bears (Miller, Schuster et al. 2012) and pigs (Groenen et al 2012). However, the effects of admixture in terms of nucleotide diversity on the genome but also on inferences of demographic parameters like past effective population size (Ne) are largely unknown.
Ever since Darwin, domesticated populations have served as important model organisms for evolutionary and population genetic questions (Megens and Groenen 2012). Sus scrofa – domesticated pigs and wild boars - is an excellent model species to examine the evolution of genome-wide patterns of haplotype sharing because of its complex but generally well documented demographic history, multiple domestication events and recent admixture between Asian and European breeds. The Eurasian wild boar has its origin in Southeast Asia where it diverged ~3-6 Mya from a clade that gave rise to several other species in the genus Sus that are mostly confined to Islands South East Asia (Frantz et al. 2013, Meijaard et al. 2011). Sus scrofa spread throughout the entire Eurasian mainland ~1.2 Mya and an Eastern and Western clade diverged soon after colonization of the West during the cold Calabrian period, in which especially the European population experienced severe bottlenecks (Fang & Andersson 2006, Fang et al. 2006, Alves et al. 2010, Groenen et al. 2012). Domestication of wild boars occurred independently in Europe and Asia, as early as 10,000 years ago and subsequent intensification of the pig breeding industry has led to a variety of breeds (Ottoni et al. 2012, Larson et al. 2005, Kijas et al. 2001, Megens et al. 2008, Groenen et al. 2012). Hybridization between wild and domesticated Sus scrofa occurs sporadically nowadays (Giuffra et al. 2000, Goedbloed et al. 2013), but is likely to have been common until pigs were kept in sties (e.g. Larson et al., 2007; Herrero et al., 2013). Around the late 18th, early 19th century, pigs were imported from Asia to improve local European pigs for key traits such as fertility, growth and fatness.
As a consequence of this hybridization, two very divergent populations, that were separated around 1.2 million years ago, have artificially become merged again. Each of these populations from the Eastern and Western regions of the Eurasian landmass had their own demographic history, with the European wild boar in particular being very much less variable compared to the East Asian wild boars (Groenen et al. 2012, Bosse et al 2012), due to founder effects during migration throughout Eurasia and the sequential marginalization in refugia during glaciations. It is historically well documented that pigs from the UK in particular were improved by hybridization with Asian pigs in the 18th, 19th century, and subsequently, due to superior production traits, became founders of a number of the modern commercial pig breeds such as the Large White breed (LW, formally established as a breed in 1868). Therefore, the LW breed serves as an excellent model for studying divergence and subsequent hybridization between populations, since it originated from two highly distinct source populations, that have even been called subspecies (a.o. Groves 2008, Genov 1999), and the hybridization events have been well documented.
The aim of our study is to investigate the consequences of hybridization on genome-wide variation and on disentangling demographic parameters. On a genome-wide segment-by-segment basis we elucidate the origin of the haplotypes in LW pigs, investigating whether they have a Western Eurasian origin or an Eastern Eurasian origin. By this we aim to investigate patterns of introgression and to unravel genomic consequences of isolation and outbreeding. Because the time of divergence between Eastern and Western Sus scrofa has been estimated to be around ~1.2Mya (Frantz et al. 2013, Groenen et al. 2012), Asian wild haplotypes in European commercial pigs are expected to be shorter and less abundant than European wild haplotypes. Since the European population suffered a severe bottleneck, genomic regions for which pigs have one haplotype of Western origin and one of Eastern origin, are likely to show a higher degree of nucleotide diversity than regions for which pigs have two haplotypes that both are of European origin. For comparison purposes, we also investigated the haplotype patterns in an Asian breed, Meishan (MS), as a representative of East Asian pigs. Not only do these pigs represent a domestication event independent from the Western Eurasian pigs, they also represent the demographic history of the East Asian wild boars (up until domestication). Because introgression of Asian haplotypes into European pigs has occurred fairly recently (White et al. 2011, Merks et al. 2012), it is expected that haplotypes shared by European and Asian pigs are longer compared to haplotypes shared by common ancestry in the Western pigs and Asian wild boar. Finally we investigate the effect of the composite nature of the LW genome on demographic inferences like Ne. This analysis of haplotype patterns in pigs provides a detailed insight into the genomic distribution of variation after recent hybridization.
Materials and Methods
The genomes of 70 domesticated pigs and wild boars were re-sequenced for this study (Table 1S). These individuals originate from Asia and Europe and form four different functional and geographical groups; Asian wild boars (ASWB), Asian domesticated pigs (ASDom), European wild boars (EUWB) and European domesticated pigs (EUDom). We sequenced two wild boars from Sumatra as outgroup (Groenen et al., 2012). The other Asian wild individuals come from North China (3), South China (4) and Japan (1). The 18 European wild boars originate from the Netherlands, France, Switzerland, Greece and Italy. We sequenced 13 Asian domesticated pigs from the MS, Jianquhai and Xiang breeds and 29 European domesticated pigs from the Duroc, Hampshire, Pietrain, Landrace and LW breeds.
Sampling and preparation
DNA was extracted from whole blood samples from all 70 individuals using the QIAamp DNA blood spin kit (Qiagen Sciences). Quality and quantity of DNA extraction was checked on the Qubit 2.0 fluorometer (Invitrogen). 1-3 ug of genomic DNA was used for the construction of the sequencing library (insert size range 300-500 bp), according to the Illumina library preparation protocol (Illumina Inc.). All samples were 100 bp paired-end sequenced on 1-3 ug of genomic DNA on Illumina HiSeq sequencing systems to a targeted ~10× depth of coverage. Details on all used samples can be found in Table S1.
Alignment and variant calling
Reads were quality trimmed to a phred quality >20 for both mates over 3 consecutive bases, and read length were >44 bp after trimming for each mate. Trimmed reads were aligned with the unique alignment option of Mosaik aligner (V. 1.1.0017) to the porcine reference genome build 10.2. SNPs were called for each sample individually with Samtools mpileup 0.1.12a (Li et al. 2009), with the alternative base covered at least 2 times. We filtered the SNPs with VCFtools for a read-depth between 7× and twice the average depth, and discarded SNP sites with a genotype quality<20. We constructed a genotype matrix for all 70 individuals, for those sites that were heterozygous or non-reference in at least 1 individual. We included only sites that were covered >=4× in all the individuals to reduce biases, resulting in a total of 2,377,607 autosomal markers.
IBD detection
We phased all 70 individuals for each chromosome separately, based on the 2,377,607 markers, with Beagle fastPhase (V. 3.3.2). IBD detection between individuals was executed with Beagle fastIBD for each chromosome, as described in Browning and Browning (2011). We ran 10 independent cycles of phasing and pairwise IBD detection, and merged the identified IBD tracts based on the Beagle probability scores, as suggested (Browning and Browning, 2011). Since fastIBD was originally designed for human data, we tested different thresholds for IBD detection to examine which threshold fits our pig data best. We empirically determined that the relative IBD size and number of recorded IBD tracts remained stable with different thresholds, although absolute numbers varied. Our aim was to identify haplotypes that are IBS or IBD, and reflect demographic history over a relatively large time frame. Asian haplotypes are expected to be more diverse and fragmented, and therefore comparatively small in size. Because a higher threshold will enable us to identify Asian wild haplotypes within the genome of the LW pigs, we decided on a threshold of 5.0−6. This is higher than that used in the original paper (Browning and Browning 2011), but this threshold fits our data best.
Haplotype classification
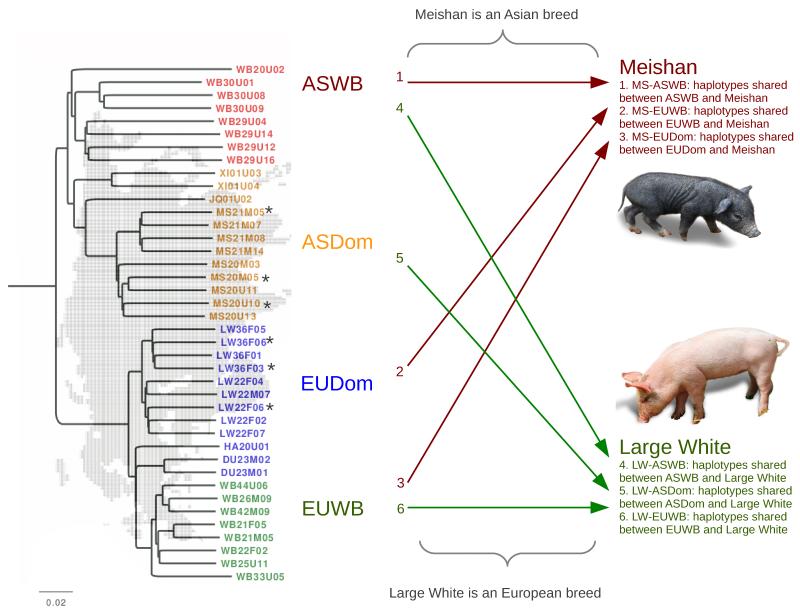
The purpose of the haplotype classification is to be able to infer the geographic origin of the haplotypes that are present in the LW and MS pigs. Shared haplotype tracts were recorded for all pairwise comparisons between the individuals in our matrix. Then, only those haplotypes were extracted from this dataset that were shared between any individual and an individual belonging to either the MS or the LW breed. The haplotypes shared with a MS were grouped into one of three classes, i.e. haplotypes shared between MS and either a) European wild boars, b) European domestics and c) Asian wild boars. The haplotypes shared with a LW pig were also grouped into one of three classes, i.e. haplotypes shared between LW and either a) European wild boars, b) Asian domestics and c) Asian wild boars (figure 1). In total there are four reference groups of pigs, but the MS and LW pigs were only compared to three groups because they were not compared to the same group as they belong to themselves. With this setup, we have a total of 6 group comparisons and 351 unique pairwise comparisons between individuals. The rationale of the pairwise comparisons is further described in figure S1. The group of Asian breeds included 3 individuals of the MS group, and the group of European breeds included 3 individuals of the LW group. Only six individuals were used from the EUDom group, even though a larger number has been used for the phasing step, to keep number of animals in all four reference groups similar. Because the analysis is based on pairwise comparisons, any individual may share a haplotype with multiple individuals from different pig groups. The average length and number of shared haplotype tracts between the LW or MS pigs and the members of the four pig groups were computed and significant levels were calculated with a two-sample Kolmogorov-Smirnov test in R version 2.13.1. Recombination frequency was obtained from Tortereau et al. (2012) and correlation with IBD length was calculated with a Pearson’s product-moment correlation test in R.
Figure 1. Experimental setup for the IBD detection.
Arrows indicate all the pairwise comparisons between groups that are used for the IBD detection. Individuals from two breeds (LW: Large white and MS: Meishan) are used for all comparisons with four geographical and functional groups: European wild boars (EUWB), European domesticated pigs (EUDom), Asian wild boars (ASWB) and Asian domesticated pigs (ASDom). All individuals in the phylogenetic tree are used for the pairwise IBD detections. Three individuals from the LW and MS group (indicated with*) are also used for the representation of the EUDom and ASDom group, respectively, which means that they are used for IBD detection in both groups. Total numbers of IBD detection between 2 individuals for the 6 pairwise comparisons are: 1-72; 2-72; 3-54; 4-72; 5-54; 6-72 (see also figure S1).
Nucleotide variation
To estimate nucleotide diversity within the individuals on a genome-wide scale, the genome was divided into bins of 10,000 bp and within each bin SNPs were called according to the criteria mentioned above. Nucleotide diversity was calculated as SNPs per called base in the bin (read-depth of 7× to 2 times the average coverage). To compute the nucleotide diversity in the LW or MS pigs within regions that are IBD with the 4 different pig groups, the IBD tracts that were recorded during the IBD detection were likewise divided into bins of 10,000 bp. Nucleotide diversity within the LW and MS individuals was extracted for these bins as described above. Significance levels were calculated with a two-sample Kolmogorov-Smirnov test in R. We also computed the average nucleotide diversity for entire IBD tracts (without dividing the tracts into bins). The correlation of the nucleotide diversity and length of IBD tracts was calculated with Pearson’s product-moment correlation test in R.
Fst analysis
We calculated pairwise Fst as defined by Weir and Cockerham (1984) in bins of 10,000 bp over the full genome with Genepop 4.2 (Rousset 2008), based on the 2,377,607 SNPs. The pairwise Fst between the LW and two wild boar groups (EUWB and ASWB) as well as pairwise Fst between the MS and the two wild boar groups (ASWB and EUWB) was computed.
Phylogenetic analysis
A phylogenetic tree was constructed for all the 42 re-sequenced individuals that were used in the pairwise comparisons (figure 1, figure S1) with the Sumatran Sus scrofa INDO22 as an outgroup. A distance matrix was constructed in PLINK (Purcell et al 2007) for all 2,377,607 genotypes spanning the full genome and a neighbor-joining tree was created in Phylip (Felsenstein 2005). The tree was depicted in FIGTREE (http://tree.bio.ed.ac.uk/software/figtree/).
Admixture analysis
For the admixture analysis, the outgroup individuals from Sumatra were removed, and all bi-allelic sites in the matrix were LD-pruned with the PLINK option -indep with a window size of 50, steps of 5 SNPs and a variance inflation factor of 1.5 and the remaining SNPs were filtered for MAF<0.05. Then an Admixture (Alexandre et al 2009) analysis, which uses the same statistical model as STRUCTURE (Pritchard, Stephens and Donnelly, 2000), was computed for the remaining 68 individuals with K between 2 and 5.
PSMC analysis
The consensus sequence for one LW (LW22F07), one MS (MS20U10) and one European wild boar (WB25U11) was constructed using samtools mpileup and vcftools (Li et al. 2009). To estimate past effective population sizes, we performed a Pairwise Sequential Marcovian Coalescent (PSMC) analysis (Li and Durbin 2011) on these consensus sequences. Generation time was set at 5 years and mutation rate at μ=2.5*10−8 as used in previous analyses (Groenen et al., 2012, Bosse et al., 2012 and Frantz et al., 2013). The PSMC analysis was also performed for all three individuals on only those regions of the genome in which the LW contains at least one haplotype shared with ASDom. The same analysis was done for those regions where the LW did not contain an Asian haplotype, but did have a shared haplotype with an European wild boar. These genomic fragments were filtered for regions that contained only a EUWB signal for at least 100 kbp in length, because we expect these calls to be more reliable. Smaller IBD fragments are more difficult to detect and therefore these are more prone to false positives and negatives of Asain heritage, which in turn may influence the effective population size estimates.
Results
The Asian and European Sus scrofa in our dataset formed two distinct clades (figure 1). Our two focal populations, the European Large Whites (LW) and the Asian Meishans (MS), both represent the domesticated form on their continent. We show however that the LW contained a proportion of Asian haplotypes in their genome, indicative of the recent admixture probably stemming from the late 18th, early 19th century (figure 2). Although the Admixture analysis with K=4 had the highest likelihood (figure S2), the analysis with K=2 assigned the Asian or European heritage of the alleles (figure 2). The genetic differentiation between the LW and MS populations, measured as Wrights fixation index (Fst, Weir and Cockerham 1984), is 0.383(+/− 0.217). Fst between LW and ASWB is higher than the Fst between LW and the EUWB (p<0.001, figure 2). By contrast, the Fst between the MS and ASWB is lower than between MS and EUWB. The overall Fst between LW and the ASWB is lower than Fst between the MS and EUWB, which corroborates the Asian introgression. This phenomenon is the initial concept behind our further analyses.
Figure 2. Genomic structure and Fst of the four main pig groups used for analysis.
2A. The percentage of Asian (red) and European (blue) genetic material is displayed on the y-axis, and each individual is displayed on the x-axis. We examined the genetic structure of all individuals when we force two populations (K=2) for four groups: Asian wild boars (ASWB), an Asian domesticated breeds (including MS), European wild boars (EUWB) and the European domesticated breeds (including LW). Asian wild boars are the most diverse group in terms of both within-individual and between-individual variation. 2B. Distribution of Fst between MS and EUWB (blue) and MS and ASWB (red). Fst was calculated for each bin of 10,000 bp over the full genome. 2C. Distribution of Fst between LW and EUWB (blue) and LW and ASWB (red).
IBD haplotype occurrence
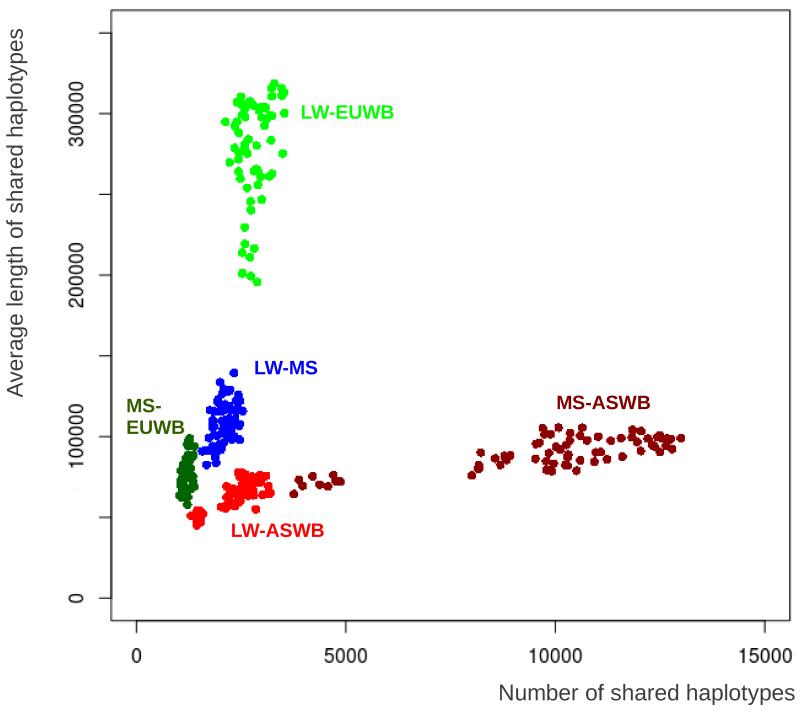
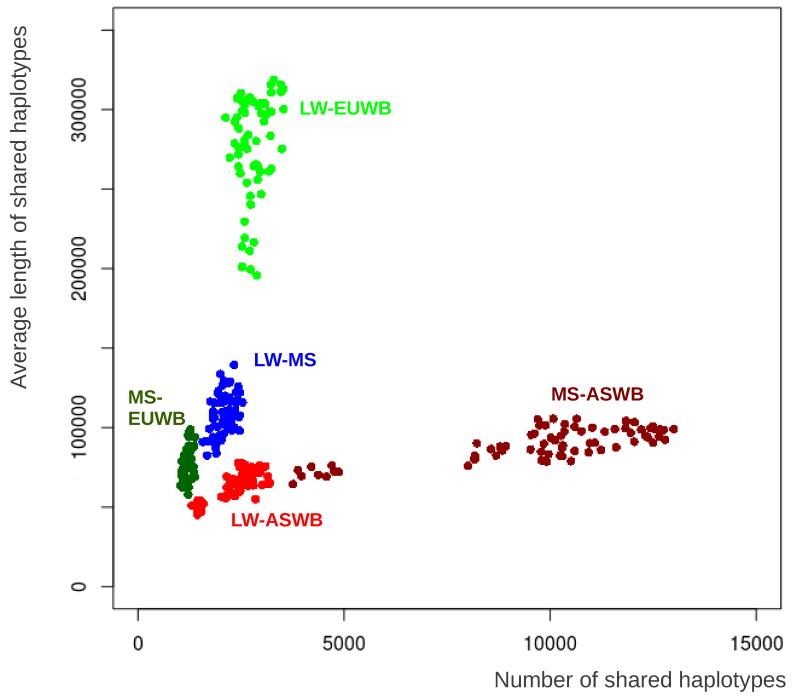
We extracted shared haplotype tracts between LW or MS pigs and pigs originating from the four wild and domesticated pig groups from Asia and Europe. An example of the distribution of IBD haplotypes in the genome of one LW pig is shown in figure S3. This example clearly shows a large proportion of Asian-derived haplotypes in the genome of the LW, sometimes in homozygous state, sometimes occurring together with a European haplotype. The length and number of shared haplotypes shows a distinct pattern for each of the pairwise comparisons as described in figure 1. Size and number of all shared haplotype groups differ significantly (p<0.001 for all; figure 3); the LW share more and longer haplotypes with the European wild boars than with both Asian Sus scrofa groups (p<0.001 for both). Likewise, the MS share more and longer haplotypes with the Asian pigs than with the European domestics and wild boars (p<0.001 for both), in agreement with their independent domestication history. The average size of LW haplotypes that were found to be IBD with the Asian domesticated pigs is significantly larger than the haplotypes shared with the Asian wild boars (p<0.01). In addition, the MS-LW haplotypes are, on average, longer than the MS-EUWB haplotypes. Haplotypes that are shared between LW and EUWB are longer than haplotypes shared between MS and ASWB, but the number of the relatively smaller MS-ASWB haplotypes is higher in the MS genome than the number of LW-EUWB haplotypes in the genome of the LW. The occurrence of all IBD haplotypes in the genome of LW pigs is not randomly distributed. For all three groups of IBD haplotypes in the LW, we found a negative correlation between length of the IBD haplotype and recombination frequency (r=−0.3 +/− 0.12, p<0.001, Pearson’s product-moment correlation, example in figure 4).
Figure 3. Size and number of Pairwise IBD tracts.
The average size of an IBD haplotype is plotted against the number of pairwise IBD haplotypes for each pairwise comparison between individuals. Coloration is based on five different groups of pairwise comparisons: The LW individuals compared with ASWB (red) and EUWB (light green); the MS individuals compared with ASWB (brown), and EUWB (dark green); and both domesticated groups compared with each other (LW-MS, in blue).
Figure 4. Recombination frequency and average length of IBD haplotypes over chromosome 1.
Example of the distribution of recombination frequency and the average length of IBD haplotypes over the full length of chromosome 1 in bins of 1Mb. 3A. Recombination frequency per bin of 1Mb (Tortereau et al. 2012), the x-axis displays location on chromosome 1. 3B. Length of IBD fragments in LW shared with EUWB(green), ASDom (blue) and ASWB (red) over chromosome1. 3C. Correlation between log-transformed recombination frequency (x-axis) and log-transformed haplotype length (y-axis) over all autosomes for haplotypes in LW shared with EUWB(green), ASDom (blue) and ASWB (red).
We compared the distribution of shared haplotypes with ASDom over the full genome for all 9 LW pigs. On a population wide scale most parts of the genome contain at least one Asian haplotype (Figure S4), but some parts of the genome contain no Asian haplotype and others are relatively high in Asian haplotype frequency. The regions in the genome without Asian haplotypes are longer in the middle of the chromosomes, which is in line with the observed correlation of haplotype length and recombination frequency.
Nucleotide diversity
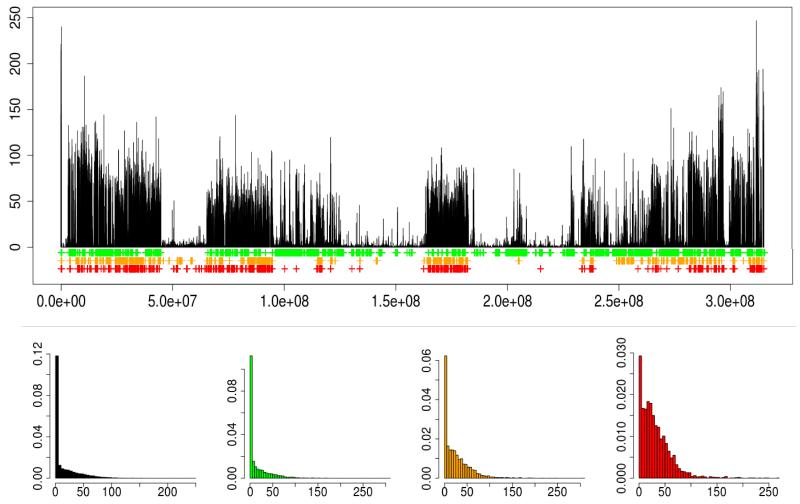
We define nucleotide diversity in this paper as the proportion of SNPs between the two haplotypes of an individual in a particular region of the genome, relative to all the sites called in that region. The nucleotide diversity within the genome of an individual was computed for all LW and MS pigs. Average nucleotide diversity was higher within the genomes of the MS pigs than within the LW pigs (p<0.001). The geographic origin of the haplotypes influenced the local nucleotide diversity in the genome. Figure 5A shows an overview of the nucleotide diversity within one LW pig, over the full length of chromosome 1. Relatively recent consanguineous matings are reflected as Regions of Homozygosity (ROH) on this chromosome. The diversity between two haplotypes on chromosome 1 for this pig was significantly higher when at least one haplotype was shared with an Asian pig or Asian wild boar (p<0.001, figure 5A-E). The same pattern was observed when we extrapolated this to a genome-wide scale for all LW pigs (p<0.001, figure S5A). Those genomic regions in the LW that share at least one haplotype with an European wild boar are relatively less diverse than the regions that share at least one haplotype with the European domesticated pigs (p<0.001), but note that these regions are not mutually exclusive. All distributions of genome-wide nucleotide diversity contain multiple peaks at low nucleotide diversity, showing the presence of homozygosity in the genome, regardless the origin of the present haplotypes (figure S5A-B). A negative correlation can be observed between length of the IBD fragment and nucleotide diversity in the fragment (r=−0.26, p<0.0001, figure S5B). The strongest correlation was found for LW-EUWB haplotypes (r=−0.35).
Figure 5. Example of nucleotide diversity and shared haplotypes for a LW pig over chromosome 1.
5A. The nucleotide diversity between two haplotypes per bin of 10Kbp is plotted against the physical position of the bin on chromosome 1. Regions in the genome in bins of 10Kbp where one of the two haplotypes is IBD with EUWB (green), ASDom (orange) or ASWB (red) are indicated in bars beneath the original plot. 5B. Histogram of nucleotide diversity between the two haplotypes in this individual in bins of 10Kbp. 5C-E. Histogram of nucleotide diversity between the two haplotypes in this individual in bins of 10Kbp, where at least one of the two haplotypes is IBD with EUWB (green, 5C), ASDom (orange, 5D) or ASWB (red, 5E).
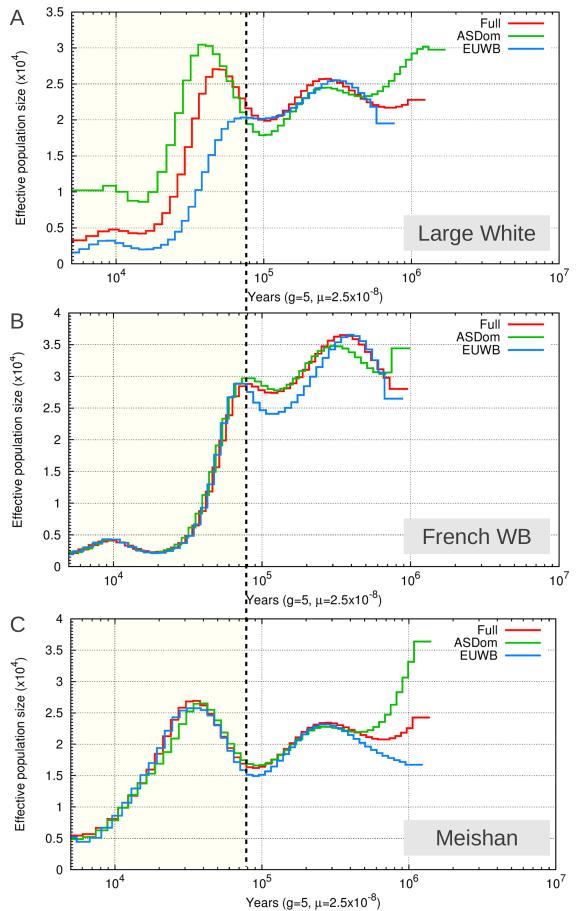
The past effective population size was estimated for the full genome of one LW pig, one French wild boar and one MS (figure 6). Although the LW breed is known to be domesticated from the European wild population, its past effective population size is estimated to be larger than that of the French wild boar (figure 6A,B). When the same analysis is done for regions where the genome of this LW pig has a European haplotype (and no Asian), the population size for the LW is lower than for regions where the LW has an Asian haplotype (figure 6A). However, the French wild boar and the MS have the same estimated population size when estimated for these regions as compared to their full genome (figure 6B,C), which suggests there is no effect on the estimate of Ne due to the regions in the genome that these haplotypes were extracted from.
Figure 6. Estimates of past effective population size based on parts of the genome with different demographic history for three pigs.
Red lines indicate the effective population size when a pairwise sequential marcovian coalescent analysis is run on the consensus sequence for the full genome. Green lines are estimates for Ne based on only those segments in the genome where the LW pig contains an Asian haplotype, and blue lines indicate the Ne when the PSMC is run on only those segments where the LW contains an European wild haplotype and no Asian haplotype. These regions are filtered for a length of at least 100 kb and therefore we consider only the left side of the dotted black line to be accurate Ne estimates. 6A. Ne estimated for the LW individual LW22F07. 6B. Ne estimated for the French wild boar WB25U11. 6C. Ne estimated for the MS pig MS20U10.
Discussion
IBD haplotype occurrence
Multiple studies have shown that domestication of pigs took place at least twice, and independently, in Western and Eastern Eurasia (Larson et al. 2005, Kijas et al. 2001). In both cases, it was the local wild boar that was domesticated, and subsequent hybridizations with local wild boar populations as agricultural practices spread have been documented from ancient DNA studies (e.g. Larsen et al., 2007; Larsen et al., 2010). Pigs, therefore, represent a subset of the natural variation present in the east and west of the natural range of the wild boar that encompasses the Eurasian supercontinent. The LW breed used in this study serves as a model in which these diverged populations have been reunited. Incidentally, an ideal model for ancestral European pigs is not available, as it is presumed that even most of the traditional heritage breeds in Europe may have been influenced by Asian pigs over the past two centuries albeit probably indirectly through improving pigs using popular commercial stock. Intriguingly, the European wild boar, despite representing the non-domesticated form, may therefore be the best model for the “original”, pre-18th century European pigs alive today.
The Asian breed for which we were able to obtain the largest number of sequenced individuals was the MS. MS pigs, as far as is known, have never been crossed with European pigs, and therefore serve as a good model for the imported Asian pigs. The Fst between MS and European Sus scrofa is higher than between MS and ASWB, confirming two independent domestication centers. Incidental exchange of genetic material between European and Chinese pigs has been suggested to happen as early as during Roman times (Porter 1993). However, during the intensification of Northern European agriculture in the eighteenth century, pig breeding expanded from forested areas to more urban environments, resulting in a changing selection pressure on multiple traits. Since in particular the European breeds that were crossed with Asian breeds seemed to perform best in this relatively new environment, there was an extensive period of genetic exchange between multiple European breeds in the early nineteenth century (White 2011). It was during this time of experimental crossing and breeding that the first modern pig breeds with mixed English and Asian origin emerged. Therefore it is not surprising that the proportion of Asian material that we identified in these breeds is roughly similar. The genetic signature of introgression from Asian into European pigs was first discovered using mtDNA sequence data (Giuffra et al., 2000), and many pig populations, particularly certain commercial breeds derived from British breeds such as LW, were found to contain large proportions (>>50%) of Asian-derived mitochondrial haplotypes. Interestingly, Asian Y-chromosome haplotypes appear to be very rare in European pigs, which suggests that the introgression was predominantly female-driven (Ramirez et al, 2009). our Fst analysis shows a greater divergence between LW and Asian individuals than between LW and EUWB, confirming that indeed there is an asymmetry in the hybridization event. The Asian component is less than half, which is consistent with earlier findings based on full genome data that suggested that the Asian fraction in European modern pigs can be up to 35% (Groenen et al., 2012), and mitochondrial studies that estimate the average proportion of Asian mitochondrial (mt) haplotypes in European breeds at ~29% (Fang and Andersson 2006). However, the proportion of Asian mt haplotypes can vary considerably between breeds with Duroc and Hampshire containing less Asian mt material than Large White. In this study we show that the average Asian component in the autosomes is very similar in individuals belonging to different European breeds, suggesting that autosomal and mtDNA tell different stories regarding the introgression history as has been proposed previously (Ramirez et al 2009). Our Admixture analysis confirms the global introgression of Asian material into European breeds, although the estimated fraction is somewhat lower (~20%). The introgression of Asian domesticated breeds into European breeds may have reduced the genetic differentiation (Fst) between the LW and ASWB compared to the MS and EUWB.
If no hybridization had taken place since the original split of European and Asian Sus scrofa, one would expect that the shared haplotypes between all European and Asian Sus scrofa are similar in size and abundance, regardless of the domestication status of the individuals. The fact that haplotypes shared between LW and ASDom are, on average, larger than haplotypes shared between LW and ASWB, is an indication that indeed Asian domesticated haplotypes have been introgressed at a later stage. These differences in length in particular indicate a more recent common ancestor between these haplotypes, since haplotypes are broken up in time due to recombination. There is, however, a proportion of LW-ASDom haplotypes that have lengths overlapping the length distribution of the LW-ASWB haplotypes. These haplotypes may represent the original split between European and Asian wild boars, i.e. incomplete lineage sorting between Eastern and Western Sus scrofa. More IBD tracts are found between MS and ASWB than between LW and EUWB and haplotypes shared between EUWB and LW are, on average, longer. The difference in length between MS-ASWB haplotypes and LW-EUWB haplotypes can be a signal of smaller effective population size in the EUWB and the LW (i.e all European Sus scrofa), resulting in a smaller haplotype diversity in European wild boars, compared to Asian Sus scrofa.
Nucleotide diversity
Because the effective population size in Asia was larger and the latest glacial bottlenecks was not as severe in Asia compared to Europe, on average, two Asian haplotypes drawn from a population are thought to be more divergent than two European haplotypes (Groenen et al. 2012). Secondly, the relative old divergence (~1.2 Mya) between Asian and European Sus scrofa, is expected to result in more variation between haplotypes of Asian and European origin than any two European haplotypes. Therefore, those parts in the genome of LW pigs where an Asian haplotype has been detected were expected to be more diverged, as was corroborated in this study. It has been shown previously that European domesticated pigs contain more variation than European wild boars (Groenen et al. 2012). The higher nucleotide diversity in the regions that contain an Asian haplotype in the LW pigs compared to regions that contain two European haplotypes, suggests that the higher diversity is due to hybridization with Asian individuals, rather than a post-domestication bottleneck in the European wild boar population. Both breeds show some regions of low variation, probably due to recent inbreeding resulting in ROH formation. ROHs tend to be longer and more abundant at the center of the chromosomes, chiefly following the distribution of recombination frequency (Bosse et al. 2012). In our IBD analysis we show that length of shared haplotypes follows the same pattern and correlates negatively with recombination frequency. The negative correlation between length of the IBD fragment and nucleotide diversity in the fragment might also be influenced by the recombination frequency, since higher nucleotide diversity tends to be found in regions of high recombination (Bosse et al 2012). Less recombination results in longer haplotypes, and in many species there is a positive correlation between recombination rate and nucleotide diversity [ i.a. Begun and Aquadro 1992, Fang et al 2008, Lercher and Hurst 2002], which may explain the negative correlation in pigs as well. Another explanation for this observation can be that long IBD fragments are an indication of (recent) selection for a particular haplotype, resulting more often in homozygosity, regardless the background of the haplotype. Figure S4 shows that some regions in the genome of the LW pigs are enriched in Asian haplotypes, while other parts do not contain any Asian material at all. Such variation of Asian haplotypes may also hint towards selection, which can be expected in a hybrid population that carries very divergent haplotypes. After introgression, Asian haplotypes may either have had a neutral effect, could have been beneficial or have had a negative effect. This study focuses on the consequences for nucleotide diversity in the genome and inference of demographic history under a neutral introgression scenario, which is observed in the majority of the genome resulting in the general patterns that are described. However, introgression mapping could also be used to screen for regions with an excess of heterozygosity within individuals with introgressed and non-introgressed haplotypes, in order to detect regions under balancing selection, as has been shown for the major histocompatibility complex [Charbonnel and Pemberton 2005, Castric et al 2008, Abi-Rached et al 2011]. Also regions with a lack of introgressed haplotypes or with more introgressed haplotypes than expected could answer interesting questions about selection in the focal population after introgression, as has been shown for i.a. mouse [Song et al 2011], and human [Jeong et al 2014]. Our results clearly show that the genomes of LW and MS pigs are a mosaic of haplotypes, representing a variety of demographic and selection events.
We have shown that the genomes of LW pigs have a composite origin in which European and Asian haplotypes are combined. This phenomenon has important implications for demographic analyses on these genomes, since a single individual essentially represents multiple, distinct, demographies. By running a Pairwise Sequential Marcovian Coalescent analysis (PSMC, Li and Durbin 2011) on different fragments of the genome of one LW pig, we showed that it is possible to disentangle these separate demographies if the origin of the haplotypes can be properly assigned. If no introgressed haplotypes are included in this inference, the effective population size indeed resembles that of the source of domestication (the European wild boar). The effective population size is however greatly overestimated if one of the two haplotypes originated from an Asian pig. The effective population size of the LW resembles that of the MS in those genomic regions where European and Asian haplotypes are combined in the LW and used for the PSMC analysis. Since the European wild boars are descendants from Asian wild boar, the majority of genetic variation that is present in the European wild boar population has its origin in Asia. Therefore, one European and one Asian haplotype could indeed approximate the Ne estimates for two Asian haplotypes, as is found in the MS. Newly arrisen mutations in the European and Asian clades after the original split will probably have resulted in a slightly higher Ne estimate when an Asian and European haplotype are combined, as we see in the LW, than when the Ne is based on two Asian haplotypes. These findings highlight the importance of knowledge on the background of samples when these types of analyses are used to infer the demographic history of a population.
A combination of recombination, genetic drift, selection and introgression has resulted in a complex distribution of haplotypes in the two breeds. Knowing the genomic footprints of admixture can be used in commercial breeding and conservation management to increase the variation within populations. Introducing new haplotypes from one inbred population to another, highly divergent but also inbred population, may result in a strong increase in variation within the genomes of hybrid individuals. Detailed mapping of the genomic distribution of variation enables a targeted approach to increase genetic diversity of captive and wild populations, by selecting individuals that contain particular desired haplotypes in breeding programs. However, the identification of introgressed haplotypes may also be used in breeding efforts that intend to “purify” a particular breed or population. The integrity of a population can be very important for branding of particular regional products for example (e.g. Herrero et al., 2012, 2013), but also for species conservation (e.g. Frantz, 2013). When the contribution of introgressed haplotypes to future generations can be actively managed, this approach may facilitate conservation and breeding efforts in the near future.
Supplementary Material
Supplementary Material Captions
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Table 1
Acknowledgements
DNA samples were provided by Dr. Ning Li; China Agricultural University, China; Dr. Alain Ducos, UMR INRA-ENVT, France; Sem Genini, Parco technologico Padano, Italy; Dr. Gono Semiadi, Puslit Biologi, Indonesia; Dr. Naohiko Okumura, Staff Institute 446-1 Ippaizuka, Japan; Dr. Alan Archibald, Roslin Institute and the Royal (Dick) School of Veterinary Studies, University of Edinburgh, Scotland; Institute of pig genetics TOPIGS BV, The Netherlands; Dr. Oliver Ryder, San Diego Zoo, USA; Cheryl L. Morri, Ph.D., Omaha’s Henry Doorly Zoo, USA. This project is financially supported by the European Research Council under the European Community’s 256 Seventh Framework Programme (FP7/2007-2013) / ERC Grant agreement n° 249894. We thank Barbara Harlizius and Naomi Duijvesteijn from TOPIGS for valuable discussion.
Footnotes
Data Accessibility: DNA sequences: All BAMfiles are available from the ENA repository under accession number ERP001813. The phylogenetic treefile is available on Dryad under doi:10.5061/dryad.33982.
References
- Abi-Rached L, Jobin MJ, Kulkarni S, et al. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 2011;334:89–94. doi: 10.1126/science.1209202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar A, Roemer G, Debenham S, et al. High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proceedings of the National Academy of Sciences of the United States of America. 2004 Mar 9;101(10):3490–4. doi: 10.1073/pnas.0306582101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Research. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves PC, Pinheiro I, Godinho R, et al. Genetic diversity of wild boar populations and domestic pig breeds (Sus scrofa) in South-western Europe. Biological Journal of the Linnean Society. 2010;101:797–822. [Google Scholar]
- Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–20. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- Bosse M, Megens H-J, Madsen O, et al. Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape. PLoS Genetics. 2012;8(11):e1003100. doi: 10.1371/journal.pgen.1003100. doi:10.1371/journal.pgen.1003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A fast, powerful method for detecting identity by descent. American Journal of Human Genetics. 2011;88(2):173–182. doi: 10.1016/j.ajhg.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric V, Bechsgaard J, Schierup MH, Vekemans X. Repeated Adaptive Introgression at a Gene under Multiallelic Balancing Selection. PLoS Genetics. 2008;4(8):e1000168. doi: 10.1371/journal.pgen.1000168. doi:10.1371/journal.pgen.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnel N, Pemberton J. A long-term genetic survey of an ungulate population reveals balancing selection acting on MHC through spatial and temporal fluctuations in selection. Heredity. 2005;95:377–388. doi: 10.1038/sj.hdy.6800735. [DOI] [PubMed] [Google Scholar]
- Fang L, Ye J, Li N, et al. Positive correlation between recombination rate and nucleotide diversity is shown under domestication selection in the chicken genome. Chinese Science Bulletin. 2008;53(5):746–750. [Google Scholar]
- Fang MY, Andersson L. Mitochondrial diversity in European and Chinese pigs is consistent with population expansions that occurred prior to domestication. Proc Biol Sci. 2006;273:1803–1810. doi: 10.1098/rspb.2006.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Berg F, Ducos A, Andersson L. Mitochondrial haplotypes of European wild boars with 2n=36 are closely related to those of European domestic pigs with 2n=38. Animal Genetics. 2006;37:459–464. doi: 10.1111/j.1365-2052.2006.01498.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington; Seattle: 2005. [Google Scholar]
- Frantz L, Schraiber J, Madsen O, et al. Genomic sequencing provides fine scale inference of evolutionary history. Genome Biology. 2013;14:R107. doi: 10.1186/gb-2013-14-9-r107. doi:10.1186/gb-2013-14-9-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genov PV. A review of the cranial characteristics of the wild boar (Sus scrofa Linnaeus, 1758), with systematic conclusions. Mammal Review. 1999;29:205–238. [Google Scholar]
- Giuffra E, Kijas JM, Amarger V, et al. The origin of the domestic pig: independent domestication and subsequent introgression. Genetics. 2000;154:1785–1791. doi: 10.1093/genetics/154.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedbloed D, Megens H, van Hooft P, et al. Genome-wide SNP analysis reveals recent genetic introgression from domestic pigs into Northwest European wild boar populations. Molecular Ecology. 2013;22:856–866. doi: 10.1111/j.1365-294X.2012.05670.x. [DOI] [PubMed] [Google Scholar]
- Groenen MAM, Archibald AL, Uenishi H, et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491:393–398. doi: 10.1038/nature11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves C, Albarella U. Current views on the taxonomy and zoogeography of the genus Sus. In: Dobney K, Ervynck A, Rowley-Conwy P, editors. Pigs and Humans: 10,000 Years of Interaction. Oxford University Press; Oxford, UK: 2008. pp. 15–29. [Google Scholar]
- Harris K, Nielsen R. Inferring Demographic History from a Spectrum of Shared Haplotype Lengths. PLoS Genetics. 2013;9(6):e1003521. doi: 10.1371/journal.pgen.1003521. doi:10.1371/journal.pgen.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Molecular Ecology. 2013;22:4606–4618. doi: 10.1111/mec.12415. [DOI] [PubMed] [Google Scholar]
- Henn BM, Botigue LR, Gravel S, et al. Genomic Ancestry of North Africans Supports Back-to-Africa Migrations. PLoS Genetics. 2012;8(1):e1002397. doi: 10.1371/journal.pgen.1002397. doi:10.1371/journal.pgen.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero-Medrano JM, Megens HJ, Crooijmans RP, et al. Farm-by-farm analysis of microsatellite, mtDNA and SNP genotype data reveals inbreeding and crossbreeding as threats to the survival of a native Spanish pig breed. Animal Genetics. 2013;44:259–266. doi: 10.1111/age.12001. [DOI] [PubMed] [Google Scholar]
- Herrero-Medrano JM, Megens HJ, Groenen MAM, et al. Conservation genomic analysis of domestic and wild pig populations from the Iberian Peninsula. BMC Genetics. 2013;14:106. doi: 10.1186/1471-2156-14-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C, Alkorta-Aranburu G, Basnyat B, et al. Admixture facilitates genetic adaptations to high altitude in Tibet. Nature Communications. 2014;5:3281. doi: 10.1038/ncomms4281. doi:10.1038/ncomms4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez JA, Hughes KA, Alaks G, et al. An experimental study of inbreeding depression in a natural habitat. Science. 1994;266:271–273. doi: 10.1126/science.7939661. [DOI] [PubMed] [Google Scholar]
- Keller LF, Waller DM. Inbreeding effects in wild populations. Trends in Ecology and Evolution. 2002;17(5):230–241. [Google Scholar]
- Kijas JMH, Andersson L. A Phylogenetic Study of the Origin of the Domestic Pig Estimated from the Near-Complete mtDNA Genome. Journal of Molecular Evolution. 2001;52:302–308. doi: 10.1007/s002390010158. [DOI] [PubMed] [Google Scholar]
- Lacy RC, Alaks G, Walsh A. Hierarchical analysis of inbreeding depression in Peromyscus polionotus. Evolution. 1996;50:2187–2200. doi: 10.1111/j.1558-5646.1996.tb03609.x. [DOI] [PubMed] [Google Scholar]
- Larson G, Dobney K, Albarella U, et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science. 2005;307:1618–1621. doi: 10.1126/science.1106927. [DOI] [PubMed] [Google Scholar]
- Larson G, Albarella U, Dobney K, et al. Ancient DNA, pig domestication, and the spread of the Neolithic into Europe. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15276–15281. doi: 10.1073/pnas.0703411104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Hurst LD. Human SNP variability and mutation rate are higher in regions of high recombination. Trends in Genetics. 2002;18:337–340. doi: 10.1016/s0168-9525(02)02669-0. doi:10.1016/S0168-9525(02)02669-0. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megens HJ, Crooijmans R, San Cristobal M, et al. Biodiversity of pig breeds from China and Europe estimated from pooled DNA samples: differences in microsatellite variation between two areas of domestication. Genetics, Selection, Evolution. 2008;40:103–128. doi: 10.1186/1297-9686-40-1-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megens HJ, Groenen MAM. Domesticated species form a treasure-trove for molecular characterization of Mendelian traits by exploiting the specific genetic structure of these species in across-breed genome wide association studies. Heredity. 2012;109:1–3. doi: 10.1038/hdy.2011.128. doi:10.1038/hdy.2011.128; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijaard E, d’Huart JP, Oliver WLR. In: Handbook of the Mammals of the World. Wilson DE, Mittermeier RA, editors. Vol. 2. Lynx Edicions; Barcelona, Spain: 2011. pp. 248–291. [Google Scholar]
- Merks JWM, Mathur PK, Knol EF. New phenotypes for new breeding goals in pigs. Animal. 2012;6:535–543. doi: 10.1017/S1751731111002266. [DOI] [PubMed] [Google Scholar]
- Miller W, Schuster S, Welch A, et al. Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2382–E2390. doi: 10.1073/pnas.1210506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottoni C, Girland Flink L, Evin A, et al. Pig Domestication and Human-Mediated Dispersal in Western Eurasia Revealed through Ancient DNA and Geometric Morphometrics. Molecular Biology and Evolution. 2012;30:824–832. doi: 10.1093/molbev/mss261. doi:10.1093/molbev/mss261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palamara PF, Lencz T, Darvasi A, Pe’er I. Length Distributions of Identity by Descent Reveal Fine-Scale Demographic History. American Journal of Human Genetics. 2012;91(5):809–822. doi: 10.1016/j.ajhg.2012.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter V. Pigs – a Handbook to the Breeds of the World. Helm Information, Mountfield; East Sussex, UK: 1993. p. 256. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel Y, Madsen O, Megens HJ, et al. Evolutionary dynamics of copy number variation in pig genomes in the context of adaptation and domestication. BMC Genomics. 2013;14:449. doi: 10.1186/1471-2164-14-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JE, Visscher PM, Goddard ME. Reconciling the analysis of IBD and IBS in complex trait studies. Nature Reviews. Genetics. 2010;11:800–805. doi: 10.1038/nrg2865. [DOI] [PubMed] [Google Scholar]
- Ralph P, Coop G. The Geography of Recent Genetic Ancestry across Europe. PLoS Biology. 2013;11(5):e1001555. doi: 10.1371/journal.pbio.1001555. doi:10.1371/journal.pbio.1001555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez O, Ojeda A, Tomas A, et al. Integrating Y-chromosome, mitochondrial, and autosomal data to analyze the origin of pig breeds. Molecular Biology Evolution. 2009 Sep;26(9):2061–72. doi: 10.1093/molbev/msp118. [DOI] [PubMed] [Google Scholar]
- Ramos AM, Crooijmans RPMA, Affara NA, et al. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One. 2009;4:e6524. doi: 10.1371/journal.pone.0006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources. 2008;8:103–6. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Song Y, Endepols S, Klemann N, et al. Adaptive introgression of anticoagulant rodent poison resistance by hybridization between Old World mice. Current Biology. 2011 doi: 10.1016/j.cub.2011.06.043. doi:10.1016/j.cub.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staubach F, Lorenc A, Messer PW, et al. Genome Patterns of Selection and Introgression of Haplotypes in Natural Populations of the House Mouse (Mus musculus) PLoS Genetics. 2012;8(8):e1002891. doi: 10.1371/journal.pgen.1002891. doi:10.1371/journal.pgen.1002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortereau F, Servin B, Frantz LAF, et al. A high density recombination map of the pig reveals a correlation between sex-specific recombination and GC content. BMC Genomics. 2012;13:586. doi: 10.1186/1471-2164-13-586. doi:10.1186/1471-2164-13-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-Statistics for the Analysis of Population Structure. Evolution. 1984;38(6):1358. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- White S. From globalized pig breeds to capitalist pigs: a study in animals cultures and evolutionary history. Environmental History. 2011;16:94–120. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material Captions
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Table 1