Discovery of a Biomarker and Lead Small Molecules to Target r(GGGGCC)-Associated Defects in c9FTD/ALS (original) (raw)
. Author manuscript; available in PMC: 2015 Sep 3.
Summary
A repeat expansion in C9ORF72 causes frontotemporal dementia and amyotrophic lateral sclerosis (c9FTD/ALS). RNA of the expanded repeat (r(GGGGCC)exp) forms nuclear foci or undergoes repeat-associated non-ATG (RAN) translation producing “c9RAN proteins”. Since neutralizing r(GGGGCC)exp could inhibit these potentially toxic events, we sought to identify small molecule binders of r(GGGGCC)exp. Chemical and enzymatic probing of r(GGGGCC)8 indicate it adopts a hairpin structure in equilibrium with a quadruplex structure. Using this model, bioactive small molecules targeting r(GGGGCC)exp were designed and found to significantly inhibit RAN translation and foci formation in cultured cells expressing r(GGGGCC)66 and neurons trans-differentiated from fibroblasts of repeat expansion carriers. Finally, we show that poly(GP) c9RAN proteins are specifically detected in c9ALS patient cerebrospinal fluid. Our findings highlight r(GGGGCC)exp-binding small molecules as a possible c9FTD/ALS therapeutic, and suggest c9RAN proteins could potentially serve as a pharmacodynamic biomarker to assess efficacy of therapies that target r(GGGGCC)exp.
Keywords: FTD/ALS, GGGGCC repeat, RAN translation, foci, RNA-small molecule interaction
Introduction
Frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) are overlapping neurodegenerative diseases with no effective treatment. Success in developing a treatment will require a well-orchestrated effort that addresses multiple aspects of the drug discovery process, including target identification and validation, as well as the identification of biomarkers to assess efficacy of potential therapies in clinical trials. These endeavors have been hampered by an incomplete understanding of FTD and ALS pathogenesis. However, with the discovery that a GGGGCC repeat expansion in C9ORF72 is the most common genetic cause of FTD and ALS (DeJesus-Hernandez et al., 2011; Renton et al., 2011), a new therapeutic target has come to light.
Two putative pathomechanisms of “c9FTD/ALS” involve RNA transcribed from the expansion. First, these transcripts (termed r(GGGGCC)exp) may cause toxicity through the formation of nuclear RNA foci that sequester various RNA-binding proteins [for review, see (Gendron et al., 2014)]. Second, r(GGGGCC)exp undergoes repeat associated non-ATG (RAN) translation producing “c9RAN proteins” that form neuronal inclusions throughout the central nervous system (Ash et al., 2013; Mori et al., 2013b). Consequently, neutralizing or degrading r(GGGGCC)exp holds promise as a therapeutic approach for c9FTD/ALS. Indeed, antisense oligonucleotides to C9ORF72 transcripts suppress features associated with the repeat expansion in human induced pluripotent stem cell-derived neurons (Donnelly et al., 2013; Sareen et al., 2013). In light of pharmacological advantages, small molecules may offer an attractive option for targeting r(GGGGCC)exp. Capitalizing on our findings that r(GGGGCC)n adopts a hairpin structure in addition to a G-quadruplex one, we designed small molecules able to bind r(GGGGCC)exp and to significantly decrease RAN translation and foci formation in cultured cells expressing r(GGGGCC)66, and induced neurons (iNeurons) directly converted from fibroblasts of C9ORF72 repeat expansion carriers. These findings indicate that designer small molecules targeting r(GGGGCC)exp may prove promising as a c9FTD/ALS therapeutic. Furthermore, since we found that poly(GP) c9RAN proteins are detected in c9ALS cerebrospinal fluid (CSF), poly(GP) proteins may serve as a pharmacodynamic biomarker to assess efficacy of potential therapies that target r(GGGGCC)exp.
Results
r(GGGGCC)8 preserves a hairpin structure with periodically repeating 1×1 nucleotide GG internal loops in equilibrium with a G-quadruplex
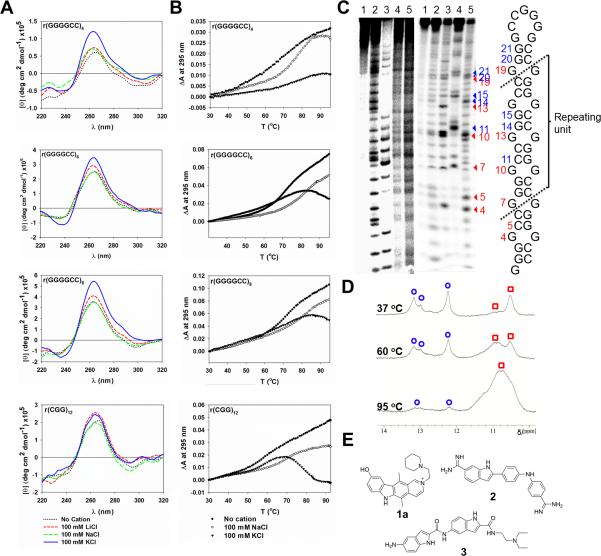
With the goal of designing small molecule modulators of r(GGGGCC), we investigated its structure. Using evidence from gel mobility shift assays and spectroscopic methods, previous reports suggest r(GGGGCC) forms intra- and intermolecular G-quadruplex structures (Fratta et al., 2012; Reddy et al., 2013), with another suggesting r(GGGGCC) repeats adopt both G-quadruplex and hairpin structures (Haeusler et al., 2014). To further probe the structure of r(GGGGCC), we completed spectroscopic (circular dichroism (CD) and optical melting), chemical (modification with dimethyl sulfate (DMS)), and enzymatic analyses (for a full description of our biophysical studies see Supplementary Information). CD studies of r(GGGGCC)4, r(GGGGCC)6 and r(GGGGCC)8 revealed these RNAs likely fold into a G-quadruplex structure in the presence of K+ but not Na+, which promotes a hairpin structure (Fig. 1A). We next studied the structures of r(GGGGCC) by optical melting, as G-quadruplexes have signature melting curves (large hypochromic transition of UV absorbance at 295 nm) (Mergny et al., 1998). In agreement with CD studies, optical melts completed in the presence of Na+ indicated that r(GGGGCC)4, r(GGGGCC)6 and r(GGGGCC)8 form intramolecular hairpins. In contrast, optical melts completed in the presence of K+ indicate the presence of both hairpin and G-quadruplex structures (Fig. 1B, Table S1).
Fig. 1. Probing r(GGGGCC)n secondary structures.
A) CD spectra of r(GGGGCC)4,6,8 and r(CGG)12 in the presence of various monovalent cations. B) Optical melting curves for r(GGGGCC)4,6,8 and r(CGG)12 monitored at 295 nm. RNA samples (4 μM) were folded prior to recording CD spectra (20°C) and optical melting curves. C) DMS mapping data (left), enzymatic mapping data (middle) and predicted hairpin structure of r(GGGGCC)8 using mapping restraints (right). DMS mapping lanes: 1, untreated RNA; 2, alkaline hydrolysis; 3, T1 digestion under denaturing conditions; 4, DMS modification in the presence of 185 mM Li+; 5, DMS modification in the presence of 185 mM K+. Enzymatic mapping lanes: 1, untreated RNA; 2, T1 digestion under denaturing conditions; 3, T1 digestion under native conditions; 4, V1 digestion in which base paired nucleotides are cleaved; 5, S1 digestion in which all single-stranded nucleotides are cleaved. D) 1D 1H NMR spectra of r(GGGGCC)8 in the presence of 100 mM K+ annealed at 37°C, 60°C, or 95°C. Blue circles are imino proton resonances from G residues present as base pairs and red squares are imino proton resonances from non-canonically paired G residues. E) Structures of lead compounds 1a, 2, and 3 that bind to r(GGGGCC)8. See also Figure S1.
The folding of r(GGGGCC)8 was next examined using enzymatic and chemical mapping in the presence of Li+ or K+, the latter known to stabilize G-quadruplexes (Hardin et al., 1992). Enzymatic mapping revealed an alternating pattern of cleavage by enzymes that specifically cleave paired or non-canonically paired nucleotides (Fig. 1C), suggesting that some populations form a hairpin structure. These findings were confirmed using the chemical modification reagent DMS.
We additionally explored the structure of r(GGGGCC)8 by analyzing its 1D 1H NMR spectra. At low annealing temperatures, the NMR spectra indicate r(GGGGCC)8 folds into a hairpin with non-canonically paired Gs in the stem (spectrum collected at 37°C; Fig. 1D). As the annealing temperature increases, however, NMR peaks become broad, indicating increased population of a G-quadruplex. The existence of both conformations is not surprising as other studies have suggested RNAs that form quadruplexes can form alternative structures, including hairpins (Bugaut et al., 2012).
Identification of small molecules that bind r(GGGGCC)exp
Exploiting the findings above, we sought to identify small molecules that bind r(GGGGCC)exp and determine whether they improve c9FTD/ALS-associated defects. It was reported that TMPyP4, a known G-quadruplex binder, binds r(GGGGCC)8 in vitro (Zamiri et al., 2014). Although the bioactivity of TMPyP4 was not explored, these studies indicate it is possible to identify small molecules that bind r(GGGGCC) repeats. We previously developed a strategy to design small molecules that bind an RNA target using information about RNA-small molecule interactions (Velagapudi et al., 2014). Small molecule leads can be further optimized by chemical similarity searching, which identifies compounds that are chemically similar to the leads. We reported that small molecule 1a binds 1×1 GG internal loops present in r(CGG)exp and improves fragile X-associated tremor/ataxia syndrome (FXTAS)-associated defects (Disney et al., 2012). Given the structural similarity between r(CGG)exp and r(GGGGCC)exp, we hypothesized that 1a and compounds chemically similar to it might bind r(GGGGCC)exp. We collected 132 such small molecules and screened them for binding to r(GGGGCC)8. Three lead compounds (1a, 2 and 3) were identified (Fig. 1E, S1A; Table S2) and further characterized. Kinetic binding studies showed that 1a, 2, and 3 bind to r(GGGGCC)8 with Kd's of 9.7, 10, and 16 μM, respectively, similar to those observed for r(CGG)12. In contrast, 1a, 2, and 3 bind more weakly to a hairpin with a fully paired stem, suggesting the compounds are at least modestly selective (Fig. S1B). We perturbed the equilibrium between hairpin and G-quadruplex structures by folding r(GGGGCC)8 in the presence of an additional 100 mM NaCl (favors hairpin) or KCl (favors quadruplex). The observed Kd's for 1a and 3 were 3- to 10-fold weaker in the presence of Na+ and K+, indicating that ionic strength affects binding. Of interest, the affinity of 2 for r(GGGGCC)8 was not significantly affected by addition of Na+, but became >6-fold weaker in K+. These results indicate that compound 2 recognizes the hairpin structure over the G-quadruplex (Fig. S1B).
Given that small molecule binders of r(GGGGCC)exp may influence the thermodynamic stability of the RNA, which could in turn influence foci formation and RAN translation, optical melting was used to study whether compounds increase r(GGGGCC)8 stability. While compound 3 did not significantly affect r(GGGGCC)8's stability or melting temperature, 1a and 2 stabilized the RNA by 0.95 and 0.63 kcal/mol, respectively, and increased the Tm by 3.1 and 1.9 °C, respectively (Fig. S1C & Table S3).
Small molecule binders of r(GGGGCC)exp inhibit RAN translation and foci formation in (GGGGCC)66-expressing cells
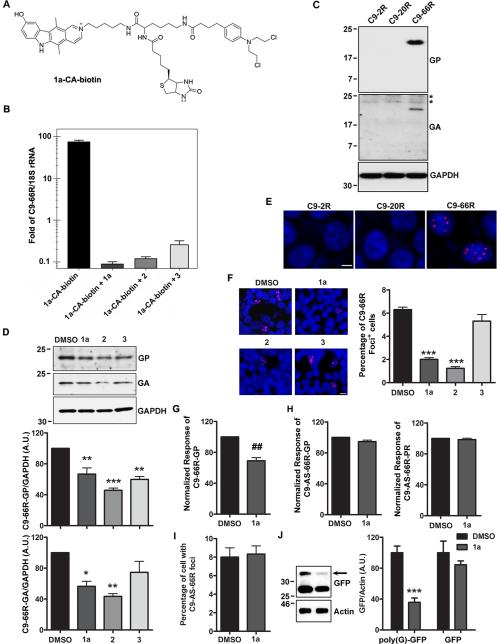
To determine whether compounds 1a, 2 and 3 bind r(GGGGCC)exp in cells, we employed COS7 cells transfected to express 66 GGGGCC repeats with no upstream ATG, and our previously reported strategy to identify small molecule cellular targets. In this strategy, small molecules are conjugated to: (i) a reactive module that forms a covalent cross-link with the target (chlorambucil; CA); and (ii) biotin for facile isolation of small molecule-biomolecule adducts (Guan and Disney, 2013). First, a biotin-chlorambucil conjugate of 1a was synthesized (1a-CA-biotin; Fig. 2A, Fig. S2A,B), added to (GGGGCC)66-expressing cells, and allowed to react with its cellular targets. Biomolecule-small molecule adducts were then isolated with streptavidin-functionalized resin. qRT-PCR analysis of the isolated fractions showed an 80-fold enrichment of r(GGGGCC)66 compared to 18S rRNA (normalized to untreated lysate; Fig. 2B). To determine whether 1a, 2 and 3 bind r(GGGGCC)exp directly, we completed a competitive profiling experiment by co-treating (GGGGCC)66-expressing cells with 1a-CA-biotin and the compound of interest. That is, the targets of 1a, 2 and 3 can be inferred by their depletion in pull-down fractions. Indeed, the amount of r(GGGGCC)66 that forms an adduct with 1a-CA-biotin was significantly depleted in the presence of each compound (Fig. 2B).
Fig. 2. Small molecules targeting r(GGGGCC) inhibit RAN translation and foci formation in (GGGGCC)66-expressing cells.
A) Structure of 1a-CA-biotin; the chlorambucil moiety (CA) forms a covalent bond with the cellular target once the small molecule is bound. Biotin is used to isolate biomolecule-small molecule adducts from cells. B) Normalized enrichment of r(GGGGCC)66 from biomolecules isolated with streptavidin beads as compared to 18S rRNA determined by qRT-PCR. Cells were treated with 1a-CA-biotin alone or with 1a, 2 or 3 (competitive profiling). In competitive profiling experiments, the unreactive compound inhibits reaction of the target with 1a-CA-biotin and thus the target is depleted in pull-down fractions. Data presented as mean±SD (n=2). C) Cells overexpressing 66 (GGGGCC) repeats (C9-66R), but not 2 (C9-2R) or 20 (C9-20R), express poly(GP) and poly(GA) proteins. Asterisks demark non-specific bands. D) Western blot and densitometry of poly(GP) and poly(GA) proteins in (GGGGCC)66-expressing cells treated with DMSO or compounds 1a, 2 or 3 (100 μM, 24 h). Data represents mean+SEM (n=3). E) RNA foci (red) are detected in the nucleus (Hoechst; blue) of C9-66R cells. F) Evaluation of the percentage of foci-positive cells by RNA FISH post-treatment. Data represents mean+SEM in 10 fields. G-H) Compound 1a decreases RAN translation of poly(GP) proteins from sense (GGGGCC)66 repeats in C9-66R cells (G), but does not influence levels of poly(GP) or poly(PR) proteins RAN translated from antisense (CCCCGG)66 repeats in C9-AS-66R cells (H), as assessed by GP and PR immunoassays. Data represents mean+SEM (n=3 or 4). I) The percentage of r(CCCCGG)-containing foci in C9-AS-66R cells is not affected by 1a. J) 1a decreases RAN translation of poly(G) in cells expressing (CGG)88 upstream of GFP. Data presented as mean+SEM (n=3). *P<0.05, **P<0.01, ***P<0.001, as assessed by One-way ANOVA followed by Dunnett's Multiple Comparison Test. ##P<0.01, as assessed by t-test. See also Figure S2.
Having established that all three compounds bind r(GGGGCC)66, we evaluated their effect on RAN translation. While no evidence of RAN translation was seen in HEK293 cells expressing only 2 or 20 (GGGGCC) repeats, expression of (GGGGCC)66 resulted in the synthesis of poly(GP) and poly(GA) proteins (Fig. 2C), but not poly(GR) proteins (not shown). Compound 3 (100 μM, 24 h) modestly inhibited synthesis of poly(GP) proteins, but did not influence poly(GA) protein production (Fig. 2D). In contrast, compounds 1a and 2 significantly decreased both poly(GP) and poly(GA) protein levels (Fig. 2D). Given that 1a and 2 have similar effects on RAN translation, and that 1a also inhibits this event in iNeurons (shown below), we tested additional concentrations of 1a and found it affords a dose-dependent effect on RAN translation; statistically significant decreases in poly(GP) of 10, 18 and 47% were detected by immunoassay of lysates from (GGGGCC)66-expressing cells treated with 25, 50 or 100 μM, respectively (Fig. S2C).
In addition to the accumulation of c9RAN proteins, nuclear foci are detected in (GGGGCC)66-expressing cells (Fig. 2E). Consistent with their effect on RAN translation, compounds 1a and 2, but not 3, significantly decreased the percentage of foci-positive cells (Fig. 2F). This was likely caused by inhibition of foci formation, and not a result of impaired binding of the probe to r(GGGGCC)66 in the presence of compound, given that conducting RNA FISH on fixed, non-treated (GGGGCC)66-expressing cells with a probe co-incubated with 1a did not prevent detection of foci (Fig. S2D).
Since the C9ORF72 repeat expansion is bidirectionally transcribed in c9FTD/ALS, and since antisense transcripts containing (CCCCGG) repeats are also RAN translated and form foci (Gendron et al., 2013; Mori et al., 2013a; Zu et al., 2013), we evaluated the effect of 1a in r(CCCCGG)66-expressing cells previously shown to express poly(PR) and poly(GP) proteins (Gendron et al., 2013). Whereas 1a (100 μM, 24 h) significantly decreased poly(GP) proteins RAN translated from sense transcripts (Fig. 2G), it had no effect on poly(GP) or poly(PR) proteins RAN translated from antisense r(CCCCGG)66, as assessed by immunoassay (Fig. 2H; see Fig. S2E and Fig. S4A for assay validation). Likewise, no change in the percentage of cells bearing r(CCCCGG) foci was detected following 1a treatment (Fig. 2I). In contrast, we reported that 1a does reduce foci in r(CGG)60-expressing cells (Disney et al., 2012), and we show here that 1a also inhibits RAN translation in cells expressing (CGG)88 placed in the 5'UTR of GFP, but does not affect downstream canonical translation (Fig. 2J). These results confirm the structural similarity between r(CGG)exp and r(GGGGCC)exp in cells, and selectivity of 1a towards this structure.
Small molecule binders of r(GGGGCC) inhibit RAN translation and foci formation in (GGGGCC)exp-expressing iNeurons
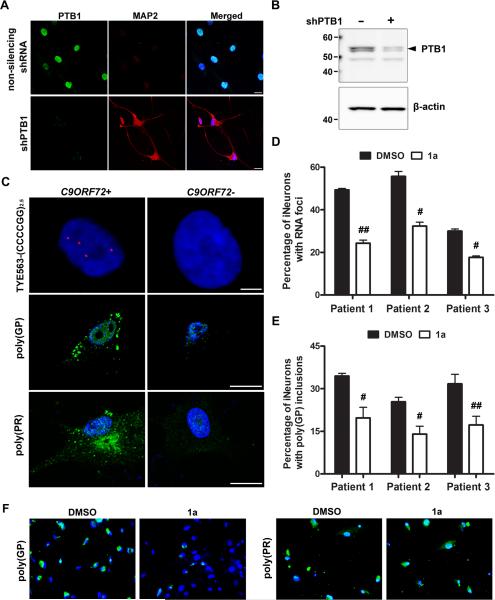
To establish a more physiological disease cell model, fibroblasts with or without the C9ORF72 repeat expansion were directly converted to a neuronal lineage by repressing polypyrimidine-tract-binding protein (PTB1), as recently described (Xue et al., 2013). PTB1 depletion caused fibroblasts to adopt a neuronal morphology with reduced soma size and neurite formation (Fig. 3A,B and S3A). These iNeurons expressed neuronal and synaptic markers, including MAP2, TUJ1, PSD95, Synapsin I and Drebrin (Fig. 3A, S3B). Nuclear foci, degraded by RNase A but resistant to DNase I (Fig. S3C), were present in both C9ORF72+ fibroblasts (Fig. S3D) and iNeurons (Fig. 3C). Cytoplasmic poly(GP) inclusions, as well as poly(PR) inclusions, were also present in C9ORF72+ iNeurons (Fig.3C), but were not found in parental fibroblasts (Fig. S3E). No foci or poly(GP) inclusions were detected in iNeurons lacking the expanded repeat (Fig. 3C). Of importance, in three C9ORF72+ iNeuron lines, compound 1a significantly decreased the percentage of cells with RNA foci (Fig. 3D) and poly(GP) inclusions (Fig. 3E,F), while having no effect on C9ORF72 mRNA levels (Fig. S3F). Consistent with findings in (GGGGCC)66-expressing cells, a dose-dependent decrease in RAN translation of poly(GP) was observed in 1a-treated C9ORF72+ iNeurons, but no change in poly(PR) expression, which is synthesized from the antisense transcript, was detected (Fig. S3G). Due to toxicity associated with compound 2 in iNeurons, it was not tested in this model. Taken together, our data indicate that our strategy to design small molecule modulators of r(GGGGCC) led to the successful identification of a compound that mitigates abnormal events initiated by r(GGGGCC)exp.
Fig. 3. Small molecules targeting r(GGGGCC) inhibit RAN translation and foci formation in iNeurons with the C9ORF72 expansion.
A) Immunofluorescence staining of fibroblasts transduced with non-silencing shRNA or shPTB1 for PTB1 and the neuronal marker, MAP2. Scale bar, 20 μm. B) PTB1 Western blot analysis of fibroblasts transduced or not with shPTB1. C) RNA foci accumulate in iNeurons with the C9ORF72 repeat expansion (C9ORF72+) but not in iNeurons lacking the expansion (C9ORF72-), as assessed by RNA FISH. Immunofluorescence staining with anti-GP or anti-PR illustrates the accumulation of c9RAN proteins specifically in C9ORF72+ iNeurons. Scale bars, 5 μm. D) Treatment of C9ORF72+ iNeurons with compound 1a (2 μM, 4 d) significantly decreased the percentage of cells with foci. For each iNeuron line, data reflects the average percentage of foci-positive cells+SEM in 3 separate fields. E-F) Compound 1a significantly decreased the percentage of cells with anti-GP, but not anti-PR, immunoreactive inclusions (magnification: 20x). For each line, data represent the average percentage of inclusion-positive cells+SEM counted in 9 wells of a 96-well plate. #P<0.05, ##P<0.01, as assessed by t test. See also Figure S3.
Poly(GP) proteins are detected in c9ALS CSF
Our findings above indicate that r(GGGGCC)exp-targeting small molecules can inhibit foci formation and RAN translation. Consequently, if c9RAN proteins are detected in CSF, they have the potential to serve as a measurable indicator of therapeutic efficacy. To test this notion, we developed a poly(GP) protein immunoassay (Fig. S4A) and validated it as a sensitive means to measure endogenous poly(GP) using soluble fractions of frontal cortex tissues. As expected, poly(GP) was specifically detected in c9FTD/ALS samples (Fig. S4B).
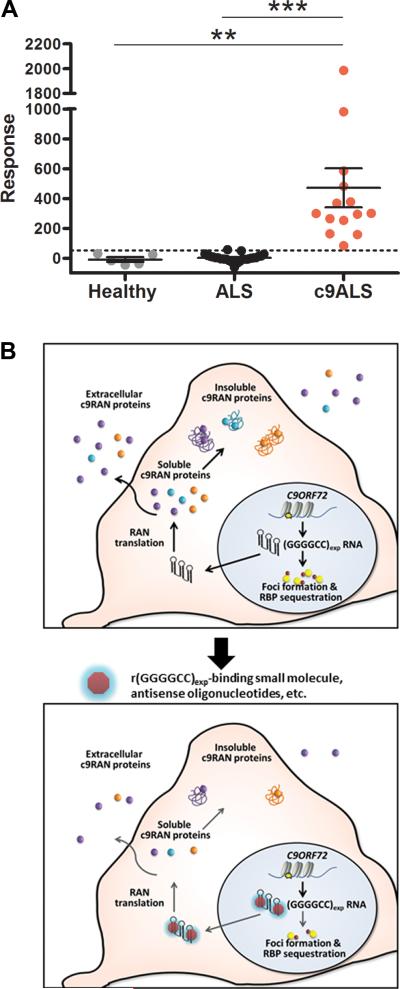
To test poly(GP) proteins as clinically relevant biomarkers, we evaluated whether they are detectable in CSF. We analyzed CSF from 14 c9ALS patients in comparison to CSF from 25 ALS patients without the C9ORF72 mutation and 5 healthy subjects (see Table S4 for patient details). As shown in Fig. 4A, poly(GP) proteins were detected only in c9ALS CSF. Using a (GP)8 peptide standard curve, we estimate the median concentration of poly(GP) in c9ALS CSF to be 0.67 ng/ml (Fig. S4C, Table S4). These exciting findings provide an important first step in identifying pharmacodynamic biomarkers for c9FTD/ALS (Fig. 4B).
Fig. 4. Poly(GP) proteins are detected in c9ALS cerebrospinal fluid.
A) Poly(GP) expression in CSF from 5 healthy individuals, 25 ALS patients negative for the C9ORF72 repeat expansion, and 14 c9ALS patients were assessed by GP immunoassay. Response values correspond to the intensity of emitted light from which the background response was subtracted. Dashed line represents the limit of detection [LoD=LoB+1.645(SDblank); where LoB (limit of blank)=meanblank+1.645(SDblank)]. **P<0.001, ***P<0.001 as assessed by one-way ANOVA followed by Tukey's Multiple Comparison Test. B) Schematic representation of the effect of r(GGGGCC)exp-targeting small molecules on foci formation and RAN translation, and the potential use of extracellular c9RAN proteins as a pharmacodynamic marker of treatment efficacy. See also Figure S4.
Discussion
In the present study, we provide evidence that: 1) r(GGGGCC)exp adopts both hairpin and G-quadruplex structures – information important for the design of r(GGGGCC)-binding small molecules; 2) human fibroblasts can be directly converted to neurons that recapitulate salient features of disease; 3) small molecules that bind r(GGGGCC)exp inhibit RAN translation and foci formation; and 4) c9RAN proteins are detectable in c9ALS CSF.
Recent studies show that r(GGGGCC) repeats forms G-quadruplexes (Fratta et al., 2012; Reddy et al., 2013), with another reporting they adopt both G-quadruplex and hairpin structures (Haeusler et al., 2014). In agreement with the latter, our chemical and enzymatic probing studies reveal that r(GGGGCC)8 do form a hairpin structure. NMR studies suggest that the hairpin structure predominates at low refolding temperatures while the quadruplex predominates at higher ones. To understand the finer details of this equilibrium in cells, chemical mapping could be completed as described in yeast (Wells et al., 2000), although readout using reverse transcriptase could be challenging. Alternatively, antibodies that recognize RNA quadruplexes could be employed (Lam et al., 2013).
We identified three compounds that bind r(GGGGCC) (1a, 2 and 3), two of which significantly inhibited RAN translation and foci formation in a novel r(GGGGCC)66 cell model developed for the rapid screening of drugs. In addition, we found that 1a inhibits RAN translation and foci formation in C9ORF72+ iNeurons. This report shows that adult human fibroblasts can be directly converted to neurons that mirror disease-specific defects and that these defects can be blocked by pharmacological manipulation. Of interest, RNA foci were observed in both _C9ORF72_+ fibroblasts and iNeurons, while poly(GP) and poly(PR) inclusions were observed in iNeurons but not fibroblasts. These findings are consistent with our observation that poly(GP) inclusions are restricted to neurons in c9FTD/ALS (Ash et al., 2013).
There are at least two mechanisms by which our small molecules may affect RAN translation. In the first model, their binding to r(GGGGCC)exp increases the thermodynamic stability of the RNA and is thus an impediment for ribosomal read-through of the transcript. Indeed, 1a and 2 stabilize r(GGGGCC)8 in vitro. In a second model, the binding of small molecules to the repeats impedes initiation of translation at these sites. In a similar fashion, the decrease in foci observed upon treatment could result from the inability of 1a- or 2-bound r(GGGGCC)exp to bind RBPs that promote foci formation.
As a pathological hallmark of c9FTD/ALS, and one that is influenced by r(GGGGCC)exp- targeting small molecules, c9RAN proteins have potential to serve as clinically relevant biomarkers. Our discovery that poly(GP) is detectable specifically in c9ALS CSF could facilitate identification of C9ORF72 repeat expansion carriers in the course of diagnostic work-ups, and pave the way in determining whether changes in c9RAN protein levels in CSF correlate with disease severity or progression. Of importance, CSF c9RAN proteins could serve as an enrollment stratification tool in clinical trials, and a pharmacodynamic biomarker to assess efficacy of therapies that target r(GGGGCC)exp (Fig. 4B). While these critical questions are investigated, it should be kept in mind that the C9ORF72 expansion is bidirectionally transcribed (Gendron et al., 2013; Mori et al., 2013a; Zu et al., 2013); as such, therapeutics may have to target both r(GGGGCC)exp and r(CCCCGG)exp. Because poly(GP) proteins are produced by RAN translation of sense and antisense transcripts, poly(GP) immunoassays, such as the one described herein, could be of great use in testing therapeutics toward both r(GGGGCC)exp and r(CCCCGG)exp.
Materials and Methods
For 1H NMR spectroscopy, r(GGGGCC)8 (600 μM) was prepared in 10 mM Tris HCl, pH 7.4 and 100 mM KCl and annealed at the appropriate temperature for 5 min and allowed to cool to room temperature. After equilibration for 2 h, NMR spectra were recorded at 10°C.
To identify RNA targets of 1a, 2 and 3, COS7 cells expressing r(GGGGCC)66 were treated with 1a-CA-Biotin and compound of interest. Total RNA was extracted 24 h later and incubated with streptavidin beads. cDNA was generated from bound RNA-1a-CA-Biotin adducts released from beads for quantitation of r(GGGGCC)66 by qPCR. To detect products of RAN translation, lysates were prepared from (GGGGCC)n-expressing HEK293 cells treated or not with compound for Western blotting using anti-GP, anti-GA or anti-GR antibodies. The effect of 1a was also tested in cells expressing r(CGG)88 embedded in the 5’ UTR of an open reading frame encoding GFP to allow detection of RAN translated products by Western blot using anti-GFP. Poly(GP) or poly(PR) expression in (GGGGCC)n-expressing cells, human brain lysates and CSF were evaluated by sandwich immunoassays. Foci in (GGGGCC)n-expressing cells were visualized by RNA FISH using a Cy3-(GGCCCC)4 probe.
Fibroblasts derived from patient skin samples were converted to iNeurons by transduction with pLKO.1 coding for shPTB1 (Xue et al., 2013). Poly(GP) and poly(PR) expression in iNeurons treated or not with 1a were evaluated by immunofluorescence staining, and foci visualized by FISH using a locked nucleic acid probe (5′TYE563-CCCCGGCCCCGGCCCC-3’). RNA extracted from iNeurons was utilized for qRT-PCR analysis of C9ORF72 mRNA expression.
Data is presented as the mean±SEM from a minimum of three experiments unless otherwise indicated. Statistical comparisons were made by Student's t-test or ANOVA. Statistical significance was inferred at P<0.05.
For more details on all methods, please see Supplemental Information.
Supplementary Material
01
02
Highlights.
- (GGGGCC) RNA forms a hairpin structure in equilibrium with a G-quadruplex structure.
- Neurons directly converted from C9ORF72+ fibroblasts express c9RAN proteins and foci.
- Small molecule binders of (GGGGCC)exp RNA ameliorate c9FTD/ALS-associated defects.
- c9RAN proteins are detected in c9ALS patient cerebrospinal fluid.
Acknowledgements
We are grateful to patients who donated CSF, skin samples, and postmortem tissue. We acknowledge Lillian Daughrity, Luc Pregent, and Mary Davis for technical support, Dr. Xiangming Kong for help with 1D 1H NMR experiments. This work was supported by the National Institutes of Health (NIH)/National Institute on Aging (R01GM097455 [M.D.D.]; R01AG026251 [L.P.]); NIH/National Institute of Neurological Disorders and Stroke (R21NS074121 [K.B.B., T.F.G.], R21NS079807 [Y.Z.], R21NS084528 [L.P.], R01NS088689 [R.H.B., L.P.], R01NS063964 [L.P.]; R01NS077402 [L.P.]; R01NS050557 [R.H.B.]; ARRA Award RC2-NS070-342 [R.H.B.], P01NS084974 [L.P., D.D., R.R., K.B.B.]); National Institute of Environmental Health Services (R01ES20395 [L.P.]); Department of Defense (ALSRP AL130125 [L.P., M.D.D.]); the Intramural Program of the NIH, the National Institute on Aging (Z01-AG000949-02 [B.J.T.]), and the National Institute of Neurological Disorders and Stroke (Z01 NS002976-16 to M.F.K. and B.J.T.); Mayo Clinic Foundation (L.P.); Mayo Clinic Center for Regenerative Medicine (P.O.B.), Mayo Clinic Center for Individualized Medicine (T.F.G., L.P., K.B.B.), ALS Association (K.B.B., P.O.B., L.P., T.F.G., Y.Z., and M.E.D.), Alzheimer’s Association (Y.Z.), Robert Packard Center for ALS Research at Johns Hopkins (L.P., J.D.R.), Target ALS (L.P., M.D.D.), Canadian Institutes of Health Research (V.V.B.), Siragusa Foundation (V.V.B.), Robert and Clarice Smith & Abigail Van Buren Alzheimer’s Disease Research Foundation (V.V.B.), Digiovanni Sundry Fund (M.E.D.), ATA (M.E.D.), Project ALS (R.H.B.), Angel Fund (R.H.B.), AriSLA cofinanced with support of ‘5x1000’-Healthcare research of the Italian Ministry of Health (grants EXOMEFALS 2009 [V.S.] and NOVALS 2012 [V.S.]), the Italian Ministry of Health (RF-2009-1473856 [C.M., A.R., V.S.]), and the European Commission (JNPD SOPHIA and STRENGTH [C.M., A.R., V.S.]). Z.S. and V.B. are Milton Safenowitz Postdoctoral Fellows funded by the ALS Association. W-Y.Y is a FRAXA Postdoctoral Fellow funded by FRAXA Research Foundation. Antibodies to c9RAN proteins have been licensed to a commercial entity. R.R. holds a patent on methods to screen for the hexanucleotide repeat expansion in the C9ORF72 gene.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Co-first author contributions
All co-first authors participated in the design of the study. Dr. Su performed experiments relating to the determination of r(GGGGCC) secondary structure, the identification of small molecule binders of r(GGGGCC), and the evaluation of cellular targets of lead compounds. Dr. Zhang led studies developing and characterizing the (GGGGCC)66-expressing cell model, and testing the effect of compounds on foci formation and RAN translation in this model. Dr. Gendron developed immunoassays for the detection of poly(GP) and poly(PR) proteins, collaborated with Dr. Zhang in evaluating RAN translation in (GGGGCC)66-expressing cells, assisted in the coordination of CSF sample collection, and assessed whether poly(GP) proteins are detectable in patient CSF. Dr. Bauer developed and characterized iNeurons directly converted from patient fibroblasts, and tested the effect of compounds on foci formation and RAN translation in this model.
References
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaut A, Murat P, Balasubramanian S. An RNA hairpin to G-quadruplex conformational transition. J Am Chem Soc. 2012;134:19953–19956. doi: 10.1021/ja308665g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney MD, Liu B, Yang WY, Sellier C, Tran T, Charlet-Berguerand N, Childs-Disney JL. A small molecule that targets r(CGG)(exp) and improves defects in fragile X-associated tremor ataxia syndrome. ACS Chem Biol. 2012;7:1711–1718. doi: 10.1021/cb300135h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, et al. RNA Toxicity from the ALS/FTD C9ORF72 Expansion Is Mitigated by Antisense Intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G, Isaacs AM. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Belzil VV, Zhang YJ, Petrucelli L. Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol. 2014;127:359–376. doi: 10.1007/s00401-013-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L, Disney MD. Covalent Small-Molecule-RNA Complex Formation Enables Cellular Profiling of Small-Molecule-RNA Interactions. Angewandte Chemie. 2013;52:10010–10013. doi: 10.1002/anie.201301639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014 doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin CC, Watson T, Corregan M, Bailey C. Cation-dependent transition between the quadruplex and Watson-Crick hairpin forms of d(CGCG3GCG). Biochemistry. 1992;31:833–841. doi: 10.1021/bi00118a028. [DOI] [PubMed] [Google Scholar]
- Lam EY, Beraldi D, Tannahill D, Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat Commun. 2013;4:1796. doi: 10.1038/ncomms2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- Mori K, Arzberger T, Grasser FA, Gijselinck I, May S, Rentzsch K, Weng SM, Schludi MH, van der Zee J, Cruts M, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013a;126:881–893. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013b;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Reddy K, Zamiri B, Stanley SY, Macgregor RB, Jr., Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem. 2013;288:9860–9866. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, O'Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, et al. Targeting RNA Foci in iPSC-Derived Motor Neurons from ALS Patients with a C9ORF72 Repeat Expansion. Sci Transl Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi SP, Gallo SM, Disney MD. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat Chem Biol. 2014;10:291–297. doi: 10.1038/nchembio.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Hughes JM, Igel AH, Ares M., Jr. Use of dimethyl sulfate to probe RNA structure in vivo. Methods in enzymology. 2000;318:479–493. doi: 10.1016/s0076-6879(00)18071-1. [DOI] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamiri B, Reddy K, Macgregor RB, Jr., Pearson CE. TMPyP4 Porphyrin Distorts RNA G-quadruplex Structures of the Disease-associated r(GGGGCC)n Repeat of the C9orf72 Gene and Blocks Interaction of RNA-binding Proteins. J Biol Chem. 2014;289:4653–4659. doi: 10.1074/jbc.C113.502336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110:E4968–4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02