Principles of bacterial cell-size determination revealed by cell wall synthesis perturbations (original) (raw)
. Author manuscript; available in PMC: 2014 Dec 3.
Summary
Although bacterial cell morphology is tightly controlled, the principles of size regulation remain elusive. In Escherichia coli, perturbation of cell wall synthesis often results in similar morphologies, making it difficult to de-convolve the complex genotype-phenotype relationships underlying morphogenesis. Here, we modulated cell width through heterologous expression of sequences encoding the essential enzyme PBP2, and sub-lethal treatments with drugs that inhibit PBP2 and the MreB cytoskeleton. We quantified the biochemical and biophysical properties of the cell wall across a wide range of cell sizes. We find that while cell wall chemical composition is unaltered, MreB dynamics, cell twisting, and cellular mechanics exhibit systematic large-scale changes consistent with altered chirality and a more isotropic cell wall. This multi-scale analysis enabled identification of distinct roles for MreB and PBP2, despite having similar morphological effects when depleted. Taken together, our results highlight the robustness of cell wall synthesis and physical principles dictating cell-size control.
Introduction
The molecular, chemical, and physical mechanisms that control cell shape have been longstanding questions in all kingdoms of life. In bacteria, cell morphology affects many behaviors, such as cell division, motility, nutrient uptake, and biofilm formation (Justice et al., 2008; Young, 2006). Different species adopt a diverse set of morphologies (Young, 2006), although most species can robustly maintain a particular shape. Elucidating the perturbations that adjust morphology and the biophysical mechanisms that transduce these changes to the cellular scale is critically important for our understanding of bacterial physiology.
Bacterial cell shape is conferred by the peptidoglycan (PG) cell wall, a macromolecular polymer network surrounding the cytoplasmic membrane (Schleifer and Kandler, 1972) that is composed of repeating sugar (glycan) subunits cross-linked by short peptides. In Gram-negative bacteria such as E. coli, the cell wall is a predominantly single-layered, dynamic meshwork that maintains an approximately constant width as the cell elongates (Scheffers and Pinho, 2005). A major class of proteins involved in the insertion of new PG is the penicillin binding proteins (PBPs), many of whose biochemical activities (transpeptidation, transglycosylation, hydrolysis) have been characterized using liquid chromatography (Banzhaf et al., 2012; Popham and Young, 2003; Vollmer and Bertsche, 2008). Disrupting the function of the PBPs can cause morphologies such as filamentous, coccal, or branched cells (Popham and Young, 2003).
Spatiotemporal coordination of the PBPs has been linked to the cytoskeletal protein MreB, a homolog of eukaryotic actin that polymerizes into filaments that are co-localized with sites of growth (Ursell et al., 2014; White et al., 2010). Depletion of MreB (Carballido-López, 2006; Wachi et al., 1987) or inhibition of MreB polymerization by the small molecule A22 results in progressive cell rounding and eventual lysis (Bean et al., 2009). The recently discovered cell twisting during E. coli growth is MreB dependent, and is thought to result from chiral ordering of the PG in which the glycan strands have a right-handed orientation bias (Wang et al., 2012). In both E. coli and the Gram-positive rod-shaped bacterium Bacillus subtilis, MreB moves circumferentially in a directed manner dependent on cell wall synthesis (Domínguez-Escobar et al., 2011; Garner et al., 2011; van Teeffelen et al., 2011). MreB motion in E. coli is reduced by the addition of mecillinam (Lee et al., 2014; van Teeffelen et al., 2011), a beta-lactam antibiotic that specifically inhibits PBP2, an essential transpeptidase encoded by the gene mrdA that participates in glycan strand cross-linking. Depletion of wild-type PBP2 causes cell rounding and eventual lysis, similar to MreB depletion (Lee et al., 2014). Given the similar effects of PBP2 and MreB perturbation, these two proteins are often assumed to work in a conserved linear pathway despite the lack of direct evidence (Osborn and Rothfield, 2007).
Even with the identification of many genes and biochemical activities required for cell wall synthesis, it has been challenging to uncover the principles that unify related mechanisms of cell-shape maintenance and cell-size determination. Since morphogenesis inherently spans the molecular and cellular scales, a number of factors such as enzyme dynamics and activities, cell wall chemical composition, spatial organization, and mechanical anisotropy are all potentially important factors. Loss of function studies have been invaluable in identifying key necessary activities, but important distinctions between genotype-phenotype relationships are still unresolved. Perturbations that result in graded phenotypic changes to cell width are potentially more useful, as they allow for the discovery of systematic changes in emergent behaviors that suggest a common physical mechanism of cell-width determination.
Here, we aim to correlate changes in cell wall biophysical properties in response to changes in cell geometry that cover a wide, yet physiologically relevant range. We created a library of strains with varied cellular morphologies via heterologous expression of mrdA from a range of species. As a complementary means of exploring the morphological phase space, we use sublethal doses of A22 and mecillinam to systematically vary cell size within a single genotype. For these cells, we quantitatively characterize a diverse set of physical and chemical phenotypes, including cell width, elongation rate, response to osmotic shock, and cell wall composition. Our study demonstrates that heterologous expression and sub-lethal impairment of cell wall synthesis can result in subtle modulations in cell width, and that these changes are correlated with alterations in peptidoglycan insertion dynamics and cell wall mechanical properties.
Results
Heterologous expression of PBP2 in E. coli gives rise to distinct cellular morphologies
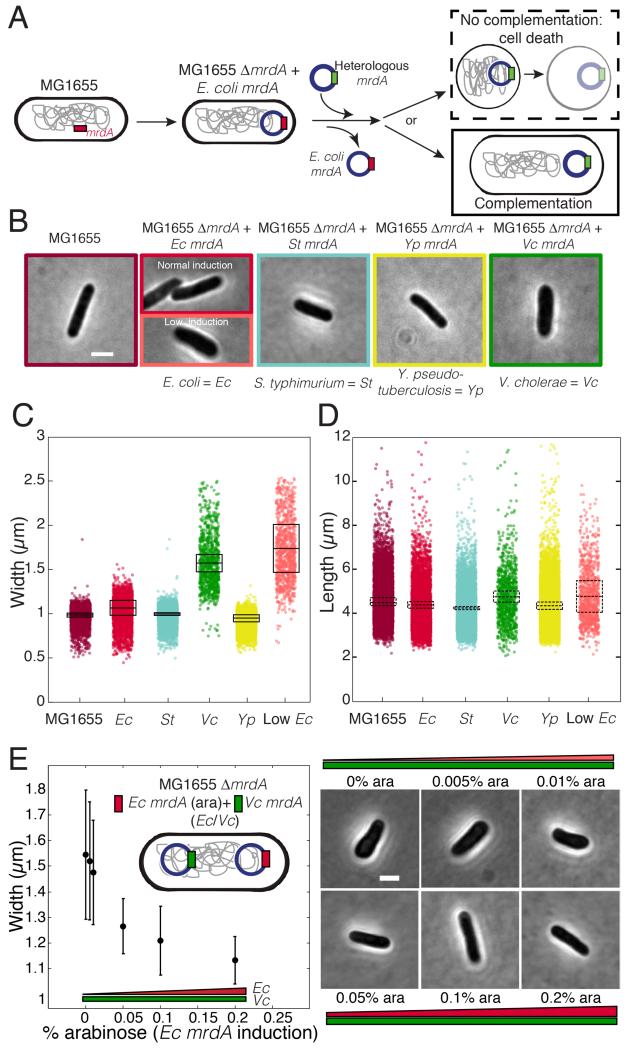
We created an E. coli (Ec) MG1655 strain deleted for mrdA, and complemented with Ec mrdA expressed from a low-copy plasmid and inducible promoter (Lee et al., 2014) (Table S1). In the absence of inducer, PPB2 levels become limiting after several cell divisions and lysis results (Lee et al., 2014). We then substituted the Ec mrdA plasmid with plasmids carrying mrdA from a number of species with varying sequence similarity to Ec PBP2 (Fig. 1A). mrdA homologs from Caulobacter crescentus (Cc) and Pseudomonas aeruginosa (Pa) (25% and 44% amino acid identity to Ec PBP2, respectively) did not complement viability as the sole copy of mrdA; depletion of Ec PBP2 in cells expressing Cc or Pa mrdA resulted in cell enlargement and lysis (Fig. S1C). However, mrdA homologs from Salmonella typhimurium (St), Yersinia pseudotuberculosis (Yp), and Vibrio cholerae (Vc) (96%, 81%, and 56% amino acid identity to Ec PBP2, respectively) as the sole source of mrdA complemented enough of the Ec PBP2 function to support viability (Fig. 1B-D).
Figure 1. Heterologous sequences can complement the essential PBP2 functions in an E. coli MG1655 mrdA knockout.
A) Schematic of experimental protocol. Heterologous mrdA genes from several species were introduced on an inducible plasmid in an E. coli MG1655 ΔmrdA strain. Since PBP2 is essential, only heterologous mrdA plasmids that successfully complement the native PBP2 function allow proliferation. B) Phase-contrast images of complementing heterologous mrdA strains (see also Fig. S1 for non-complementing strains). Complementing strains show changes in width (C) but no large differences in length distributions (D). In (C) and (D), boxes define the mean (central bar) and standard deviation (box edges) across 3 separate experiments. Dots represent single-cell measurements (_n_=906-10902 cells). E) Ec mrdA suppresses the effect of Vc mrdA when the two genes are co-expressed at high levels. Cells expressing Ec and Vc mrdA controlled by inducible and constitutive promoters, respectively, exhibit a range of stable widths dependent on the level of Ec PBP2. Error bars represent standard deviations from the mean. In (B) and (E), cell width and length closely match the population means. Scale bars: 2 μm.
Heterologous expression produced a range of cell sizes, with Vc mrdA in particular conferring a large increase in cell width and width variability (Fig. 1B,C); cell length was not substantially affected (Fig. 1D). The increased width phenotype of Vc mrdA was independent of its expression level (Fig. S1D). In a strain with inducible Ec mrdA and constitutive Vc mrdA (Ec/Vc), Ec mrdA suppressed the effects of Vc mrdA, and titrating the expression of Ec mrdA yielded graded, stable changes in cell width (Fig. 1E). Width also increased when Ec mrdA was under-expressed (Fig. 1B), similar to previous studies (de Pedro et al., 2001; Popham and Young, 2003). These data suggest that PBP2 interactions with other components of the cell wall synthesis machinery are sufficiently flexible to tolerate some degree of sequence divergence or fluctuations in expression, although cellular morphology may be altered. In addition, our results suggest that the morphological effects of heterologous expression are enacted through conserved pathways, since expression of Ec mrdA suppressed the effects of Vc mrdA.
Growth rate and cell wall composition are maintained as cell size changes
Despite the different cell widths resulting from heterologous mrdA expression or altered expression levels of Ec mrdA, cells grew at similar rates (Fig. S1D). These data indicate that the metabolic functions dictating growth rate were not greatly affected by shape changes resulting from perturbations to PBP2 function.
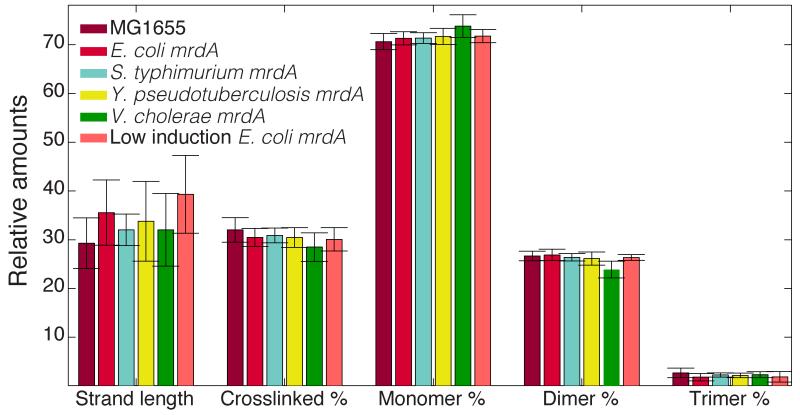
Given PBP2’s essential role in cell wall synthesis, we sought to determine whether altered PBP2 function affected the chemical composition or degree of cross-linking of the cell wall. We isolated sacculi from each viable heterologous expression strain and cells with low Ec mrdA expression, and analyzed muropeptide composition using ultra performance liquid chromatography (UPLC; Experimental Procedures) (Lee et al., 2014). Despite the large range of morphological changes (Fig. 1), PG composition remained virtually unchanged across all strains and conditions, with no significant differences in either cross-linking or average glycan strand length (Fig. 2). These data suggest that the biochemistry of cell wall synthesis is maintained in addition to cellular growth rate, although cell-wall ultrastructure could vary.
Figure 2. Quantification of PG content in heterologous and under-expressed E. coli mrdA strains.
UPLC of PG isolated from cells reveals no significant differences in overall crosslinking levels, average strand length, or relative concentrations of particular chemical species (Experimental Procedures, Tables S2). Data are represented as mean ± standard deviation over 3 separate experiments. The legend color scheme is consistent with that of Fig. 1.
In a previous study, we observed that PG crosslinking was unchanged by treatment with the PBP2 inhibitor mecillinam (Lee et al., 2014). We next investigated whether drug treatments that target the PG synthesis machinery could also result in changes in cell size, and serve as a complementary tuning knob to our genetic perturbations. Sub-lethal concentrations of A22 or mecillinam caused stable changes in average cell width across a population of wild-type E. coli MG1655 cells (Fig. S1E,F). Interestingly, mecillinam-treated cells lost their rod-shape at lower widths than A22-treated cells (~1.5 and 2 μm, respectively; Fig. S1E,F), indicating possible mechanistic differences.
MreB dynamics shift systematically with increasing cell width
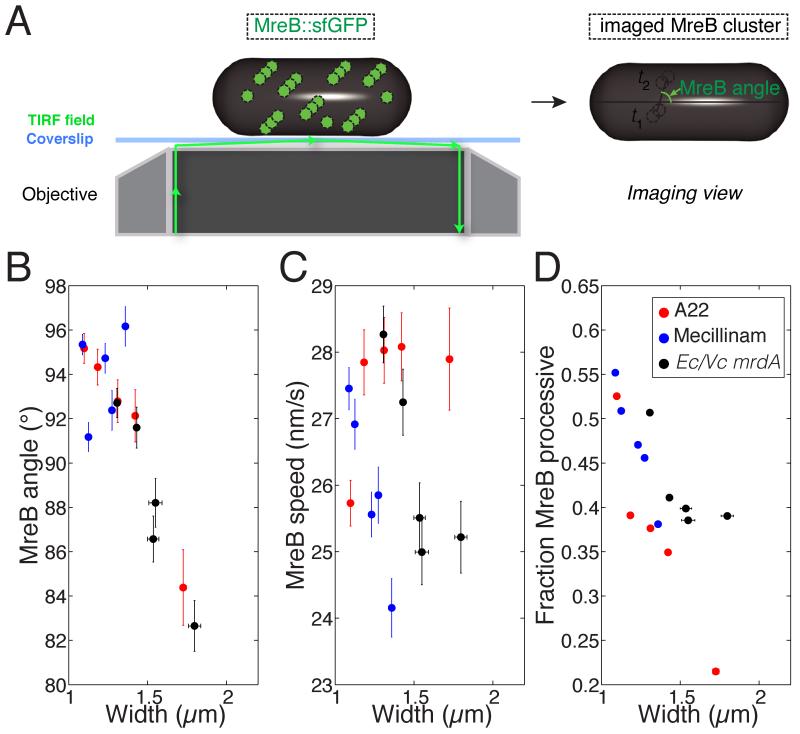
To gain insight into how the PG synthesis machinery patterns insertion into the walls of cells with varying widths, we measured the dynamics of MreB clusters by expressing a complementing fusion of E. coli MreB to super-folder GFP as the sole copy of MreB at the native chromosomal locus (Fig. 3A) (Ursell et al., 2014). We imaged single cells using total internal reflectance fluorescence (TIRF) microscopy and quantified MreB cluster movement based on angle with respect to the longitudinal axis of the cell, speed, and processivity (Fig. 3B-D, Fig. S2) (Lee et al., 2014). We defined a track as processive if the mean squared deviation fit well to a sum of linear and quadratic terms, as expected in the case of linear, directed motion (Experimental Procedures) (Lee et al., 2014); changing the threshold for goodness of fit did not affect our general conclusions (Fig. S2A-E). Under sub-lethal mecillinam treatment, MreB angle was not systematically affected (Figs. 3B, S2G). However, in agreement with our previous studies using single MreBsw-PAmCherry molecules (Lee et al., 2014), mecillinam treatment decreased MreB speed (Figs. 3C, S2G). A22 treatment does not affect MreB speed (van Teeffelen et al., 2011); however, the angle of MreB motion was correlated with cell width (Figs. 3B-C, S2F). Varying the levels of Ec PBP2 in Ec/Vc mrdA cells changed both MreB angle and speed (Figs. 3B-C, S2H). For both A22 treatment and induction of Ec PBP2 in Ec/Vc mrdA cells, average MreB angle shifted from >90° to <90° (indicating a change in the average handedness of trajectories), and the spread in the distribution of angles increased (Figs. 3B-C, S2F-H). Interestingly, in all cases, the fraction of processive MreB decreased with increasing width (Figs. 3D, S2F-H), indicating that the coordination of PG insertion may be less directed in wider cells; however, it is unknown if diffusive MreB clusters still participate in coordinating PG synthesis. Regardless, the correlations among width, MreB angle, and processive fraction (Fig. 3B,D) suggest that the spatiotemporal dynamics of cell wall synthesis shift systematically in wider A22-treated and Ec/Vc mrdA cells.
Figure 3. MreB dynamics are affected by cell width.
A) Cells with MreBsw-sfGFP as the sole copy of mreB were imaged via TIRF microscopy to quantify (B) MreB angle, (C) speed, and (D) fraction of processive MreB molecules (Experimental Procedures). Data in (B-D) are mean ± standard error (n = 84-185 cells). B) MreB angle is not affected by mecillinam treatment but does exhibit a decrease in average angle and increase in the width of the distribution of angles in both A22-treated and Ec/Vc cells. C) MreB speed decreases under mecillinam treatment and in Ec/Vc cells but is relatively constant under A22 treatment. D) In all cases, larger width is coupled to a decreased fraction of processive MreB. See Fig. S2 for detailed MreB dynamics.
TIRF imaging after photobleaching reveals increased cell twisting in wider cells
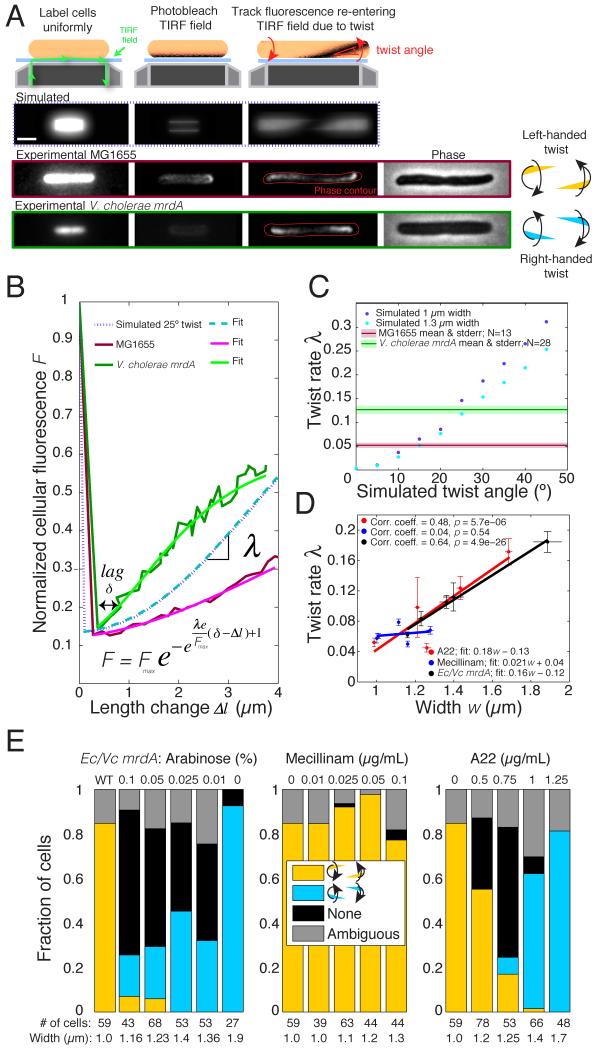
We recently showed that a left-handed pattern of PG insertion (which is guided by MreB) gives rise to a right-handed bias of the glycan strands, which produces the left-handed twisting we observed during the growth of E. coli cells (Wang et al., 2012). These twist measurements were obtained by labeling the cell poles with beads and tracking their relative motion as the cell elongates. This method is technically challenging and laborious due to the requirement for bead placement and attachment via optical trapping. We therefore developed a novel technique (Twist ‘n TIRF) to measure growth twist based on bleaching of fluorescent cell wall label using TIRF microscopy. We previously showed that fluorescent wheat germ agglutinin (flWGA) is a high-affinity, locally stationary cell surface marker that enables probing of cell wall growth patterns (Ursell et al., 2014). After uniform flWGA labeling, we used TIRF illumination to selectively photobleach the thin section of the cell wall lying within the TIRF field (extending ~150 nm from the coverslip) (Experimental Procedures, Fig. 4A). The cell was then imaged in TIRF as it grew in the absence of label. Twisting of the cell wall during growth results in gradual rotation of the unbleached stain into the field of view, causing an increase in fluorescence. In wild-type cells, we observed progressive fluorescence recovery starting from opposite sides on the two longitudinal halves of the cell, corresponding to left-handed twist (_n_=33/33 cells in which twisting was apparent; Fig. 4A, Movie S1). We then examined Vc mrdA cells using our assay, and surprisingly, we always observed right-handed twist (_n_=60/60 twisting cells; Fig. 4A, Movie S1).
Figure 4. Cell twisting properties change with cell width.
A) Cell surfaces were uniformly labeled with flWGA, washed, and imaged in TIRF in the absence of the label and in the presence of cephalexin to prevent cell division (Experimental Procedures). At the beginning of the time-lapse experiment, cells were photobleached. Due to twisting, the unbleached cell wall re-entered the TIRF field during elongation; the rate of the fluorescence increase was measured (Movie S1). Scale bar is 2 μm. B) Cell outlines were determined from phase-contrast images (A) and used to integrate TIRF fluorescence intensity relative to the pre-bleached state at each time point. Single-cell fluorescence recovery curves were fitted to a Gompertz equation (Experimental Procedures), where Fmax is the plateau F level and δ is the lag; the rate of recovery λ was determined for each cell. C) Simulated cell twisting and generation of simulated fluorescence images (Experimental Procedures) were used to determine how the cell-twisting angle affects twist rate (w), which is not very sensitive to width at low twist angles. Experimentally, Vc mrdA cells (dashed mean and shaded standard error) had a higher twist angle than wild-type MG1655 cells. D) Single-cell twist measurements show a systematic increase in cell twist rate with width (w) in heterologous Ec/Vc mrdA and A22-treated cells, but not in mecillinam-treated cells. Data are shown as mean ± standard error in both width and twist rate, with linear fits. Pearson’s correlation coefficients are shown. E) Twisting chirality changes in heterologous Ec/Vc mrdA and A22-treated strains but not in mecillinam-treated strains, in the manner predicted by the distribution of MreB angles (Fig. 3B). Wild-type (WT or 0 μg/ml A22/mecillinam) cells twist with left-handed chirality, while wider Ec/Vc or A22-treated cells tend to exhibit right-handed twist. See also Fig. S3 and Movie S1 for detailed information.
To relate the cellular fluorescence F to the increase in cell length Δ_l_ (Fig. 4B), we fit the recovery phase to an experimental growth model function (Experimental Procedures) to extract the rate of fluorescence increase λ (Experimental Procedures, Fig. 4B). To predict how fluorescence recovery should be affected by changes in cell width and/or twist angle, we developed a computational model to simulate the spatial dynamics of cell wall labels due to twisting growth with a fixed angle after photobleaching (Fig. 4A, Experimental Procedures). To compare these labeling distributions with our experimental data, we generated simulated fluorescence microscopy images from the 3D positions of wall labels (Experimental Procedures) and then analyzed the simulated images with the same algorithms as our experimental data. We extracted predicted values for λ across a range of widths and twist angles. As expected, simulated cells with higher twist angles had higher values of λ (Fig. 4C), since twist dictates the amount of fluorescence entering the field of view as cell length increases. Based on our simulated data, we estimated that Vc mrdA cells have a higher twist angle (26±4°) than wild-type E. coli (13±3°) (Fig. 4C), in addition to the previously mentioned reversal in handedness. We also quantified the twist rate in A22- or mecillinam-treated and Ec/Vc mrdA cells across a broad range of cell widths (Figs. 4D and S3). In A22-treated and Ec/Vc mrdA cells, twist angle increased with width in a quantitatively similar manner, suggesting that cell wall organization is generally altered in these wider cells. Interestingly, mecillinam-treated cells did not show a strong correlation between λ and cell width (Figs. 4D and S3B). Consistent with this, mecillinam treatment also affected MreB angle far less than the other treatments. The fact that a variety of perturbations to MreB angle result in corresponding changes to twist rate and cell width suggests that MreB angle is mechanistically coupled to these variables.
Given the differential changes in MreB angle and twist rate as cell width increased, we measured the chirality of twist under various conditions (Fig. 4E). Consistent with our observed changes in MreB angle (Fig. 3B) relative to the circumferential direction (90°), we detected a width-dependent chirality of twist in A22-treated cells and Ec/Vc mrdA cells but not in mecillinam-treated cells (Fig. 4E). The fraction of right-handed cells progressively increased as Ec mrdA expression was reduced in Ec/Vc cells (Fig. 4E, left). We also observed a surprising switch from left- to right-handed chirality at ~0.75 μg/mL A22 (Fig. 4E, right). In contrast, mecillinam-treated cells showed the same left-handed chirality as wild-type E. coli (Fig. 4E, middle) across the entire range of accessible widths. These results suggest that changes in MreB angle are strongly coupled with cell wall twisting and chirality, independent of whether the perturbation is to MreB (A22) or PBP2 (Vc/Ec). This also illustrates the important point that although perturbing cell-shape pathways may result in similar morphological effects (e.g., cell rounding), the underlying mechanisms are potentially quite different.
Mechanical response of wider cells suggests differences in cell wall organization
Since PG composition remained fixed in our heterologous expression mutants (Fig. 2), we hypothesized that changes in cell width might be coupled to alterations in the spatial architecture of the cell wall. We therefore investigated how cellular mechanical properties vary with width by measuring the longitudinal and transverse mechanical strain (fractional stretching) of the cell wall due to turgor pressure. We labeled the cell surface with flWGA (Ursell et al., 2014) and exposed cells to a large (0.5 M) hyperosmotic shock in a microfluidic flow cell (Experimental Procedures, Fig. S4). This magnitude of shock caused measureable shrinkage in both length and width (Fig. S4A) and resulted in visible plasmolysis (Fig. S4B), indicating that the turgor pressure was reduced to zero (Pilizota and Shaevitz, 2012; 2013). Mechanical strain was computed from the fluorescence outline of the cell wall before and after hyperosmotic shock (Fig. S4). For wild-type cells, the ratio of circumferential to longitudinal strain was ≈0.5 rather than the value of 2 expected for an isotropic cylindrical shell (Supp. Info.), indicating that more stretching occurs along the longitudinal direction than expected (Fig. S4D-H). Interestingly, we observed that the ratio increased and approached 1 for wider cells, as would be expected for an isotropic sphere (Supp. Info., Fig. S4C). Taken together, our results suggest that cell widening is coupled to a loss of cell wall anisotropy.
Discussion
In this study, we systematically varied E. coli cell width through heterologous expression of PBP2 sequences and by treating wild-type cells with sub-lethal levels of drugs that target PBP2 (mecillinam) or MreB (A22). We then examined how biochemical, organizational, and physical attributes of the cell wall scale across a broad range of morphologies and perturbations. This analysis identified general features underlying changes in cell width, such as wider cells exhibiting a more isotropic cell wall organization and a lower fraction of processive MreB. In addition, we also identified differences in cell-wall chirality that distinguish between MreB and PBP2 perturbation, a key development given that at the morphological level their genotype-phenotype relationships are not resolvable.
The ability to replace an essential enzyme such as PBP2 with homologous proteins implies a remarkable flexibility in the interactions among the components of the cell wall synthesis machinery. Given that heterologous expression induces changes in morphology (Fig. 1), we infer that some aspect of PBP2 functionality must be affected, such as biochemical activity, expression, and/or interactions with other components. How, then, is growth rate maintained, particularly in strains large increases in cell width? Although it has been assumed that the cell wall synthesis machinery assembles into a stable complex (Cabeen and Jacobs-Wagner, 2005), we recently demonstrated that PBP2 molecules rapidly diffuse across the cell surface, unlike the directed motion of MreB, indicating that the kinetics of PBP2 activity are not rate-limiting for growth (Lee et al., 2014). Thus, growth rate is robust to large changes in both PBP2 abundance and kinetics, consistent with our current observations (Fig. S1D).
The dynamic association of PBP2 with MreB-directed glycan insertion (Lee et al., 2014) predicts that muropeptide composition of the cell wall is more robust against perturbations to PBP2 activity. UPLC measurements of cell wall composition from this study (Fig. 2) lend support to this prediction. MreB dynamics in wider cells support the hypothesis that the orientation of newly inserted material is more variable than in wild-type cells, indicating that growth may become less coordinated as width increases (Fig. 3B). Furthermore, the increase in diffusive MreB at larger widths (Fig. 3D) is consistent with a possible increase in non-directed insertion of peptidoglycan material. Our results also indicate that the effects of PBP2 and MreB inhibition by mecillinam and A22, respectively, lead to larger cells by different mechanisms, despite the similar resulting phenotypes. In the case of A22 treatment, the speed of processive MreB clusters was not affected, while the orientation of motion and hence presumably PG insertion changes systematically (Fig. 3B), leading to opposite twisting chirality at larger widths (Fig. 4E); we note that this observation is consistent with our previous finding that cell twisting is abolished at high A22 concentrations, which almost completely depolymerize MreB (Wang et al., 2012). Conversely, mecillinam-treated cells displayed slowed MreB dynamics (Fig. 3C) but no change in MreB angle (Fig. 3B) or cell wall twisting chirality (Fig 4E) as width increases. Last, co-expression of heterologous Vc PBP2 with varying levels of Ec mrdA caused consistent changes to MreB angle (Fig. 3B), MreB speed (Fig. 3C), and twisting chirality (Fig. 4E), indicating that perturbations to PBP2 alone can affect MreB dynamics in multiple ways.
Our novel TIRF-based assay for measuring twist has higher throughput than our previous optically trapped-bead measurements (Wang et al., 2012), making it feasible to scan collections of mutants. Our measurements of the twist angle for wild-type cells are somewhat higher than our previous bead-based measurements with E. coli MC4100 cells at room temperature (~6.5°), which may be due to differences in growth media, temperature, and/or background strain (Wang et al., 2012). Although the physiological function of twisting is currently unknown, we note that it could help to separate daughter cells after cytokinesis or penetrate surfaces. A previous study demonstrated that mixed populations of fluorescent protein-expressing E. coli cells separate into sectors as they expand on agar plates, and the sector boundaries exhibit left-handed chirality (Hallatschek et al., 2007). Twisting at the cellular level was hypothesized to be magnified into the handedness of sector boundaries (Hallatschek et al., 2007); our Vc mrdA mutant and A22-treated cells could serve as a test of this hypothesized link between cellular handedness and the chirality of colony growth.
Our study also disentangled effects on the chemical construction of the cell wall from downstream effects on cell shape, indicating that changes in PBP2 function perturb cell shape by changing the dynamics of MreB and PG insertion (Fig. 4, S4), rather than PG composition (Fig. 2). We cannot exclude the possibility that other enzymes with transpeptidase activity offset any reduction in PBP2 activity in our heterologous PBP2 and mecillinam-treated cells (Popham and Young, 2003); regardless, cross-linking levels were quantitatively maintained even in cells that experienced large changes in morphology (Fig. 2, Table S2). Our comparison of the longitudinal and transverse stretching in cells of different sizes suggested that this change might be due to changes in cell wall molecular organization and anisotropy (Fig. S4).
Deconstructing the processes required for establishment of cellular dimensions requires investigations that span many length scales, especially given our results demonstrating that similar morphological phenotypes can belie distinct molecular causation. Future discoveries will rely on the rapid, precise quantification of the physical and dynamic features of morphogenesis, thereby providing a rich, multi-dimensional phenotype. Ultimately, there are likely several mechanisms to establish a given shape, and the ability to tune cell size through a multitude of chemical and genetic perturbations will be critical for distinguishing between general principles and specific phenomena.
Experimental Procedures
Strains and plasmids used in this study are described in Table S1. For routine culturing, cells were grown in lysogeny broth consisting of tryptone (1% w/v), yeast extract (0.5% w/v), and NaCl (0.5% w/v), and supplemented with the appropriate inducer and antibiotic.
For fluorescence imaging, cells were grown in EZ-RDM+0.2% glucose. Strains were grown at 37 °C unless stated otherwise. Custom MATLAB (MathWorks, Natick, MA, USA) image-processing code was used to segment cells and to identify cell outlines from phase-contrast microscopy images (Ursell et al., 2014).
Ultra performance liquid chromatography of peptidoglycan composition was performed as previously described (Brown et al., 2012; Desmarais et al., 2014).
Descriptions of other Experimental Procedures can be found in the Supplemental Information.
Supplementary Material
1
2
3
4
Acknowledgments
The authors thank Daniel Fisher, Julie Theriot, Steve Quake, members of the Huang lab, Ned Wingreen and Joshua Shaevitz for helpful discussions. We also thank Mats Hidestrand, Krystal St. Julien, and Danielle Swem for help with strain construction and the Dueber lab for providing plasmid backbones. This work was supported by a Stanford Interdisciplinary Graduate Fellowship and a Stanford Graduate Fellowship (to C.T.), a Siebel Scholars Graduate Fellowship (to T.K.L.), support from a National Institutes of Health (NIH) Biotechnology Training Grant (to T.K.L.), a Bio-X Senior Postdoctoral Fellowship (to R.D.M.), NIH Ruth L. Kirschstein National Research Service Award 1F32GM100677-01A1 (to J.H.), a Stanford School of Medicine Dean’s Postdoctoral Fellowship (to J.H.), and NIH Director’s New Innovator Award DP2OD006466 (to K.C.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
C.T., T.K.L., J.H., S.M.D., R.D.M., and K.C.H. designed the experiments. C.T. and T.K.L. executed the experiments. C.T., T.K.L., J.H., S.M.D., and T.U. performed the analyses. All authors contributed to writing the paper.
The authors declare no conflict of interests.
References
- Banzhaf M, van den Berg van Saparoea B, Terrak M, Fraipont C, Egan A, Philippe J, Zapun A, Breukink E, Nguyen-Distèche M, Blaauwen, Den T, et al. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol Microbiol. 2012 doi: 10.1111/j.1365-2958.2012.08103.x. [DOI] [PubMed] [Google Scholar]
- Bean GJ, Flickinger ST, Westler WM, McCully ME, Sept D, Weibel DB, Amann KJ. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry. 2009;48:4852–4857. doi: 10.1021/bi900014d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJBP, de Pedro MAM, Kysela DTD, Van der Henst CC, Kim JJ, De Bolle XX, Fuqua CC, Brun YVY. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci USA. 2012;109:1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. Bacterial cell shape. Nat Rev Microbiol. 2005;3:601–610. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- Carballido-López R. The bacterial actin-like cytoskeleton. Microbiology and Molecular Biology. 2006 doi: 10.1128/MMBR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro MAM, Donachie WDW, Höltje JVJ, Schwarz HH. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J Bacteriol. 2001;183:4115–4126. doi: 10.1128/JB.183.14.4115-4126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmarais SM, Cava F, de Pedro MA, Huang KC. Isolation and Preparation of Bacterial Cell Walls for Compositional Analysis by Ultra Performance Liquid Chromatography. JoVE. 2014:e51183–e51183. doi: 10.3791/51183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Söldner R, Carballido-López R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallatschek OO, Hersen PP, Ramanathan SS, Nelson DRD. Genetic drift at expanding frontiers promotes gene segregation. Proc Natl Acad Sci USA. 2007;104:19926–19930. doi: 10.1073/pnas.0710150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice SSS, Hunstad DAD, Cegelski LL, Hultgren SJS. Morphological plasticity as a bacterial survival strategy. Nat Rev Microbiol. 2008;6:162–168. doi: 10.1038/nrmicro1820. [DOI] [PubMed] [Google Scholar]
- Lee TK, Tropini C, Hsin J, Desmarais SM, Ursell TS, Gong E, Gitai Z, Monds RD, Huang KC. A dynamically assembled cell wall synthesis machinery buffers cell growth. 2014:4554–4559. doi: 10.1073/pnas.1313826111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MJM, Rothfield LL. Cell shape determination in Escherichia coli. Curr Opin Microbiol. 2007;10:606–610. doi: 10.1016/j.mib.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Pilizota T, Shaevitz JW. Fast, Multiphase Volume Adaptation to Hyperosmotic Shock by Escherichia coli. PLoS ONE. 2012;7:e35205. doi: 10.1371/journal.pone.0035205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilizota T, Shaevitz JW. Plasmolysis and Cell Shape Depend on Solute Outer-Membrane Permeability during Hyperosmotic Shock in E. coli. Biophys J. 2013;104:2733–2742. doi: 10.1016/j.bpj.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham DL, Young KD. Role of penicillin-binding proteins in bacterial cell morphogenesis. Curr Opin Microbiol. 2003;6:594–599. doi: 10.1016/j.mib.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Scheffers D-J, Pinho MG. Bacterial Cell Wall Synthesis: New Insights from Localization Studies. Microbiol Mol Biol Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriological Reviews. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell TS, Nguyen J, Monds RD, Colavin A, Billings G, Ouzounov N, Gitai Z, Shaevitz JW, Huang KC. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc Natl Acad Sci USA. 2014;111:E1025–E1034. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proceedings of the National Academy of Sciences. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Bba. 2008;1778:21–21. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Wachi M, Doi M, Tamaki S, Park W, Nakajima-Iijima S, Matsuhashi M. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. Journal of …. 1987 doi: 10.1128/jb.169.11.4935-4940.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Furchtgott L, Huang KC, Shaevitz JW. Helical insertion of peptidoglycan produces chiral ordering of the bacterial cell wall. Proceedings of the National Academy of Sciences. 2012;109:E595–E604. doi: 10.1073/pnas.1117132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Kitich A, Gober JW. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07108.x. [DOI] [PubMed] [Google Scholar]
- Young KD. The selective value of bacterial shape. Microbiol Mol Biol Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4