Break-Induced Replication is a Source of Mutation Clusters Underlying Kataegis (original) (raw)
. Author manuscript; available in PMC: 2015 Jun 12.
Summary
Clusters of simultaneous multiple mutations can be a source of rapid change during carcinogenesis and evolution. Such mutation clusters have been recently shown to originate from DNA damage within long single-strand (ss) DNA formed at resected double-strand breaks and dysfunctional replication forks. We identify here double-strand break (DSB)-induced replication (BIR) as another powerful source of mutation clusters that formed in nearly half of wild-type yeast cells undergoing BIR in the presence of alkylating damage. Clustered mutations were primarily formed along the track of DNA synthesis and were frequently associated with additional breakage and rearrangements. Moreover, the base specificity, strand coordination and strand bias of the mutation spectrum was consistent with mutations arising from damage in persistent ssDNA stretches within unconventional replication intermediates. Together, these features closely resemble kataegic events in cancers, suggesting that replication intermediates during BIR may be the most prominent source of mutation clusters across species.
Introduction
DNA mutations provide genetic variation that promotes evolution and carcinogenesis. Since most individual genetic changes result in neutral or deleterious effects, the establishment of novel protein characteristics often requires multiple mutations arising in concert within a single gene (Camps et al., 2007). Processes that over-produce scattered mutations throughout the genome (i.e., defects in mismatch repair and proofreading) are unlikely sources of spatially clustered multiple mutations since they increase the overall mutation load, which would negatively affect fitness. Therefore, processes that produce simultaneous mutations clustered within small sections of the genome are more likely to play a role in promoting rapid genetic change.
Recent genome studies of many cancer types have demonstrated that clustered mutations frequently occur during carcinogenesis (Roberts et al., 2012; Nik-Zainal et al., 2012; Alexandrov et al., 2013). In addition, analyses of mutations in cancer and experiments conducted in yeast have provided evidence that damage introduced into ssDNA is a primary source of clustered mutations. Among various environmental and intracellular DNA damaging agents that may contribute to cluster formation, a sub-family of AID/APOBEC cytidine deaminases known to target ssDNA, is a major source of DNA damage that leads to mutation clusters in cancer (Roberts et al., 2012; Nik-Zainal et al., 2012; Alexandrov et al., 2013; Roberts et al., 2013; Taylor et al., 2013; Burns et al., 2013; Chan et al., 2012; Lada et al., 2012; Lada et al., 2013). Importantly, when APOBECs or other damaging agents are present in a cell, it is the accumulation of ssDNA that becomes the limiting factor in cluster formation. Common mechanisms promoting the formation of ssDNA include dysfunctional replication forks and DSBs, both of which arise by various cellular processes and conditions including oncogene-induced replication stress (Halazonetis et al., 2008). ssDNA formed by 5′ to 3′ resection during DSB repair was suggested to be one source of kataegic events (Roberts et al., 2012; Nik-Zainal et al., 2012; Taylor et al., 2013; Yang et al., 2008). However, the switching pattern of strand coordinated mutations expected to result from bidirectional DSB resection was rarely observed suggesting that other sources of ssDNA may exist (Roberts et al., 2012). In addition, multi-kilobase resection tracts that have been demonstrated in yeast (Chung et al., 2010) have never been observed in mammalian systems, which further supports the idea that alternative sources of ssDNA likely exist.
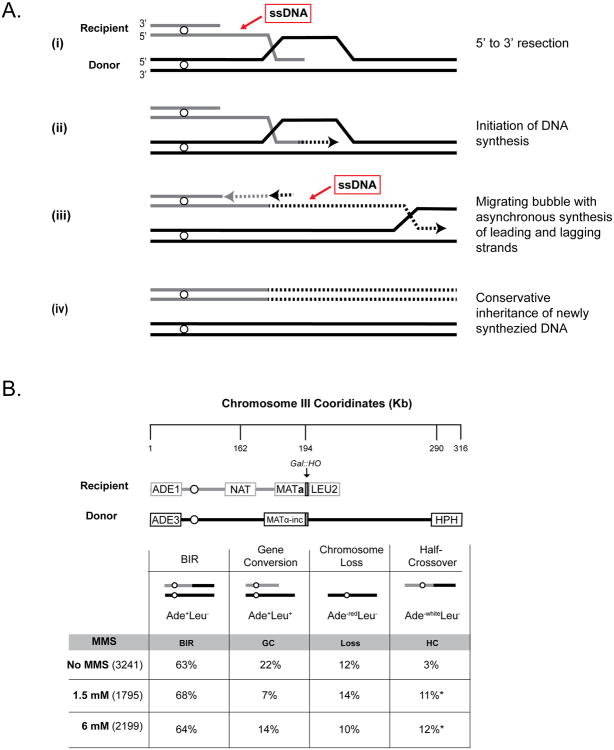
It has recently been demonstrated (Saini et al., 2013; Wilson et al., 2013) that ssDNA accumulates during one type of DSB repair - break-induced replication (BIR), which is conserved from viruses and bacteria to eukaryotes, including human cells (Costantino et al., 2014). BIR commonly repairs DSBs possessing only one repairable end that frequently occur at collapsed replication forks or at eroded telomeres (reviewed in (Malkova and Ira, 2013)). DNA synthesis during BIR is drastically different from S-phase replication. Instead of a replication fork, BIR is driven by a migrating bubble where lagging strand synthesis is substantially delayed as compared to leading strand synthesis, which results in the accumulation of ssDNA behind the replication bubble (Figure 1A) (Saini et al., 2013; Wilson et al., 2013).
Figure 1. DSB repair by BIR.
(A) Model of BIR. Dotted lines: newly synthesized DNA. (B) Experimental system to study BIR in yeast. The percent occurrence of the main DSB repair outcomes and their relevant phenotypes are indicated with total isolates scored in parenthesis. Asterisks: statistically significant increase of half-crossovers in MMS versus no-MMS. See Supplemental Experimental Procedures and Table S1 for details.
We hypothesized that the ssDNA accumulating during BIR could be a substrate for damage-induced clustered mutations. It remained unclear, however, whether the regions of ssDNA formed during BIR would be sufficiently long or stable enough to lead to clustered mutations recoverable as viable BIR repair outcomes. Here, we demonstrate in yeast that BIR is a novel source of damage-induced clustered mutations formed along the track of BIR, as well as mutation clusters associated with additional DNA breakage and chromosomal rearrangements, similar to those commonly found in human cancers.
Results
Half of BIR events completed in the presence of DNA damage result in mutation clusters
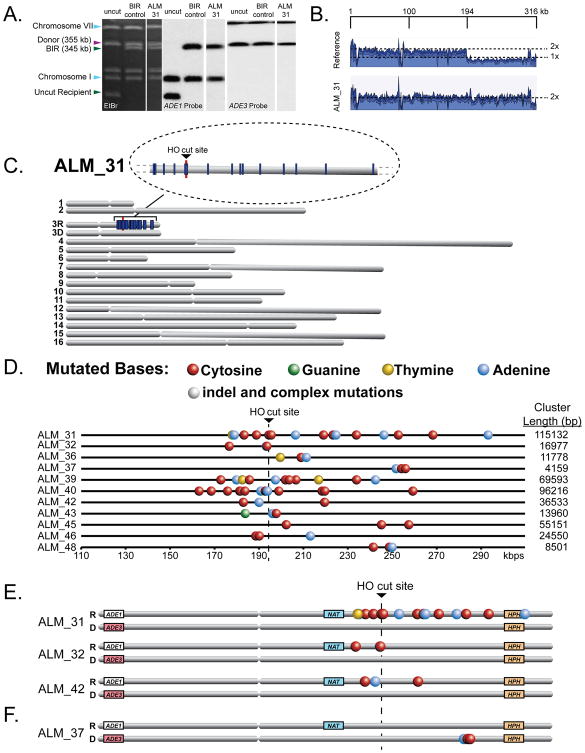
To test the hypothesis that BIR creates a substrate for the formation of damage-induced mutation clusters, we used a yeast strain where a galactose-inducible DSB is made at MAT in one copy of chromosome III (Deem et al., 2008). The DSB was predominately repaired by BIR (see Supplemental Results) using a second (uncut) copy of chromosome III as a template, while other outcomes including gene conversion (GC), chromosome loss (Loss) and half-crossovers (HC) occurred less frequently (Figure 1B, Table S1, see Supplemental Results for details). BIR was completed in liquid medium in the presence of 1.5mM MMS, a DNA alkylating agent with a ssDNA-specific mutation signature (Yang et al., 2010). There was practically no loss of viability after treatment with MMS and the majority of DSB repair outcomes displayed an Ade+Leu- NatR phenotype with a chromosomal structure expected from BIR (Figure 1B, 2A, Table S1). To determine whether BIR promoted the formation of damage-induced mutation clusters, we sequenced the genomes of 18 independent Ade+Leu- NatR BIR outcomes formed in the presence of 1.5 mM MMS. Coverage maps generated by whole-genome sequencing showed that in each isolate approximately 120kbs of the right-arm portion of chromosome III from MAT to the end of the chromosome (Figure 2B) was duplicated, as expected for BIR (Deem et al., 2008; Malkova et al., 2005). We found that over 50% of the analyzed BIR outcomes (11/18) contained mutation clusters varying in length from 4kbs to 115kbs in the area of BIR on the right arm of chromosome III (Figure 2C, 2D, Table S2A, S3). The association between BIR and mutation clusters could in principle be even higher, since some isolates that lacked clusters could have resulted from interrupted BIR that led to half-crossovers (HCs) which segregated with an intact copy of the donor chromosome during mitosis (Deem et al., 2008); an outcome that would be indistinguishable from BIR. Also, the interruption of BIR could generate varying lengths of ssDNA tracts providing one explanation for the variations in mutation cluster length.
Figure 2. Mutagenesis associated with BIR in the presence of 1.5mM MMS.
(A) The structure of one representative outcome (ALM_31). Left: ethidium bromide-stained PFGE gel. Middle and right: Southern blot analysis of PFGE gel using _ADE1_- and _ADE3_- specific probes, respectively. (B) Coverage of Illumina sequencing reads for BIR event (ALM_31) is increased 2x centromere distal to MAT (positions >194180bp) as compared to the parental strain. (C) MMS-induced mutations (blue lines) in ALM_31. Enlarged: mutation cluster on the track of BIR. (D) Clustered mutations in Ade+ Leu- BIR isolates. Positions of mutated bases (colored circles) are depicted along the chromosome III reference. (E and F) Select BIR-associated mutation clusters with mutations located in recipient (E) or donor (F) chromosomes.
The median density of mutations in clusters was 1 mutation per 6.3kb, which was ∼900 times higher than the density of scattered mutations in the rest of the genome (Table S2A, S3). In addition to 11 clusters found in a relatively small (<1% of the yeast genome) region surrounding the HO-break site on chromosome III, we identified only 4 clusters occurring on one of the other 15 chromosomes (Table S2A, S3) (P-value for random co-localization <1E-15). No mutation clusters were found in the 6 whole-genome sequenced clones that resulted from BIR repair in the absence of MMS (Table S2A). Taken together, our data indicate that the DNA region associated with BIR is a target for damage-induced clustered mutations.
Based on what is known about the mechanism of BIR, we hypothesized that clustered mutations resulted from lesions induced by MMS in ssDNA formed during BIR as a result of asynchronous leading and lagging strand synthesis (Saini et al., 2013) or during 5′to 3′ DSB resection preceding BIR (Chung et al., 2010) (Figure 1A). BIR synthesis begins on the 5′-side of the DSB at the point of strand invasion and can continue for more than 100kbs until it reaches the end of the chromosome. (5′- and 3′-sides relative to the DSB are defined along the top (Watson) DNA strand and correspond respectively to the left and right sides on Figure 2D). We observed that BIR-induced clusters occurred on both sides of the break (6/11) as well as only on one side i.e., either on the 3′-side (4/11) or on the 5′-side (1/11). Mutations clustered on the 5′-side of the break could have resulted from damage to ssDNA that was generated either by resection or by BIR synthesis. Importantly, the mutation clusters on the 3′-side of the break were located along the track of BIR, and therefore likely stemmed from BIR synthesis.
Based on the established signature of MMS mutagenesis in ssDNA (Yang et al., 2010; Roberts et al., 2012), we expected that clustered mutations would most frequently result from N3-methyl cytosine lesions in the ssDNA strand, and N1-methyl adenine lesions would be the second most frequent cause of clustered mutations. Note: the ssDNA specific MMS mutations of cytosines have no particular signature, such as TpC or CpG mutations reported in cancer genomes that are assigned to APOBEC and 5-methyl cytosine deamination in ssDNA, respectively (Nik-Zainal et al., 2012; Roberts et al., 2012; Burns et al., 2013; Nikolaev et al., 2012). During BIR, the same DNA strand (Watson or top strand) was expected to be single-stranded regardless of whether it was formed by either 5′→3′ resection or by asynchronous synthesis of leading and lagging strands (Figure 1A). Thus, cytosines in mutated C:G pairs (and possibly adenines in mutated A:T pairs) were expected to be found mostly in this strand of DNA. Indeed, mutations in BIR-induced clusters on either side of the DSB were biased towards C's located in the Watson strand (P=0.000017 and 8.02E-9, 5′ and 3′ of the break respectively, by two-sided goodness-of-fit test) (Table S2B). There was also an overall statistically significant bias towards A's in the Watson strand despite the smaller number of changes in A:T pairs (P=0.003249) (Table S2B). As expected for cytosine-specific MMS mutagenesis in ssDNA, changes in neighboring mutations of C:G pairs were strand-coordinated (i.e., mutations observed in cytosines in the same strand (P =2.60E-9 by goodness-of-fit test) (Table S4), and C to T and C to A mutations prevailed over C to G mutations (P=4.32E-13, by two-sided goodness-of-fit) (Table S2B), in good agreement with the known mutation signature of MMS in ssDNA (Delaney and Essigmann, 2004; Yang et al., 2010). We conclude that ssDNA formed during BIR synthesis is vulnerable to hyper-mutation by low levels of alkylating damage resulting in mutation clusters.
The conservative inheritance of newly synthesized DNA during BIR (Saini et al., 2013; Donnianni and Symington, 2013) should result in the majority of clustered mutations being heterozygous and located only in the recipient. Indeed, 61 out of 64 clustered mutations were heterozygous (Table S2A). In addition, the vast majority of BIR-associated clustered mutations occurred only on the recipient chromosome (55 out of 64 mutations; Figure 2E, 2F, Table S2A). Altogether our data show that ssDNA accumulated during uncoupled leading strand synthesis in the course of BIR is a source of damage-induced clustered mutations.
Complex mutation clusters associated with DNA breakage are formed by BIR in the presence of an increased level of DNA damage
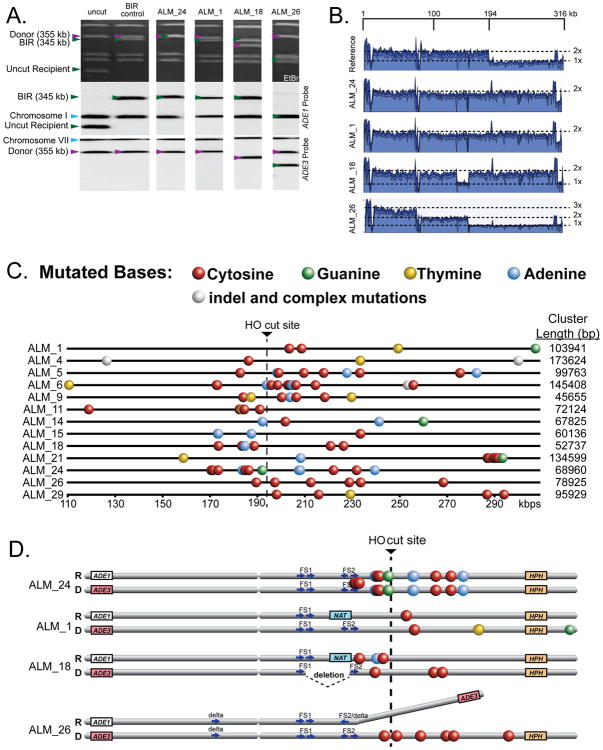
To further evaluate the role of DNA damage in BIR-induced mutation clusters, we sequenced 30 independent BIR (Ade+Leu-) isolates that were formed in the presence of higher concentrations of MMS (6 mM). In addition to Ade+Leu-, we used the information about other phenotypes including Nat status and the presence of additional rearrangements in chromosome III to choose isolates representing a complete spectrum of changes (see Supplemental Experimental Procedures for details). We found that approximately half (17/30) of the analyzed isolates contained mutation clusters, whose location strongly associated with the tract of BIR (P<0.0001 comparing 13 clusters on chromosome III (2.5% of the genome) to 8 clusters on all other chromosomes combined (97.5% of the genome) by binomial test; Figure 3, Table S2A). As in the case of 1.5mM MMS, the majority of clusters (10 out of 13) were located on both sides of the break (Table S2), contained mutations that were biased toward cytosines and adenines in the Watson strand (P = 6.26E-14 and 0.021, respectively by two-sided goodness-of-fit test) (Table S2B), and were strand coordinated (P = 2.38E-9 by goodness-of-fit test) (Figure 3C, Table S4). In addition, BIR-induced clusters were equally frequent among repair outcomes with and without chromosome III rearrangements.
Figure 3. Mutagenesis associated with BIR in the presence of 6mM MMS.
(A) Chromosome structures of representative BIR outcomes. Upper panel: ethidium bromide stained PFGE gel. Middle and lower panels: Southern blot analysis with _ADE1_- or _ADE3_- specific probes, respectively. (B) Coverage of Illumina sequencing reads derived from representative Ade+ Leu- isolates (2x and 3x: fold increases as compared with the parental strain). See Supplemental Experimental Procedures for details. (C) Clustered mutations on chromosome III in Ade+ Leu- isolates indicated as in Figure 2D. (D) Distribution of mutations between the recipient and donor chromosomes for the representative isolates indicated in (C). Complete information about cluster structure and formation is given in Figures S1-S4.
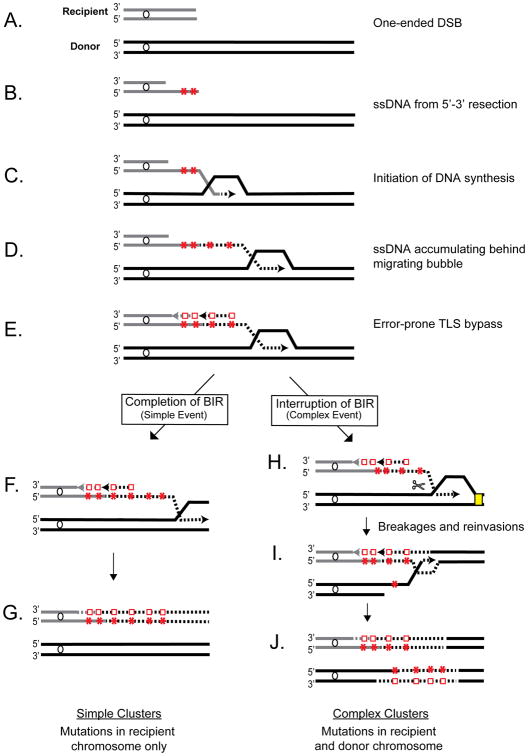
Surprisingly though, we observed that a high concentration of MMS changed the distribution of mutation clusters between the donor and recipient molecules. In particular, unlike clusters from 1.5mM MMS exposure, only 2 out of 13 clusters (in ALM_4 and ALM_11) had heterozygous mutations limited to the recipient chromosome, while the majority (10 out of 13) of BIR clusters had heterozygous and/or homozygous mutations in both the recipient and donor chromosomes (Figure 3D, Figure S1, Table S2A). We called such clusters complex and proposed that they formed as a result of a disruption during BIR by MMS-induced damage in dsDNA located in front of the BIR bubble. This event could lead to resolution of the HJ migrating behind the BIR bubble causing the donor chromosome to break, which could then initiate a second round of BIR where clustered mutations in the donor could form (Figure 4H-J).
Figure 4. Model of BIR-induced cluster formation.
DNA lesions (red stars) from MMS are shown in ssDNA formed by resection and BIR synthesis. Red squares: mutations from TLS bypass. Scissors: Resolution of HJ-like structure. Details of resolution are unknown. Yellow rectangle: damage in dsDNA. One possible scenario of lagging strand synthesis is shown. See text and Supplemental Figures S2, S3 and S4 for details.
To explain the formation of complex clusters in individual outcomes, we utilized the results from a combination of analyses (phenotype, chromosome size, whole genome sequencing and Sanger sequencing). Our results suggested that the formation of complex mutation clusters in all repair outcomes proceeded in general, through the following steps (Figure 4, Figures S2-S4): (i) DSB resection and invasion of the recipient into the donor chromosome initiating BIR synthesis (primary BIR) producing clustered mutations in the recipient from damage in ssDNA, (ii) interruption of primary BIR leading to a half-crossover event (similar to (Deem et al., 2008; Smith et al., 2009)) and a fragmented donor chromosome, (iii) resection and invasion of the fragmented donor into homologous DNA initiating a secondary BIR event and generating clustered mutations in the donor chromosome from damage in ssDNA. The exact details of how these steps were completed varied among different repair outcomes, which can explain the variety of phenotypes observed in the outcomes (see Supplemental Results for details). For example, the fact that ALM_24 was NatS and had homozygous clustered mutations while ALM_1 was NatR and had heterozygous clustered mutations suggested that differences existed between these outcomes with respect to the extent of DSB resection and the location of strand invasion (Figure S2). We propose that primary BIR in ALM_24 was preceded by extensive resection and strand invasion into the donor chromosome at a position centromere proximal to NAT leading to a NatS phenotype, while strand invasion in ALM_1 occurred telomere proximal to NAT, which resulted in a NatR phenotype. Furthermore, in ALM_24 the broken donor invaded the HC product at a position centromere proximal to its mutation cluster allowing the mutation cluster to be copied during secondary BIR, thus forming homozygous mutations. For outcome ALM_1, which had heterozygous mutations instead of homozygous mutations, we propose that the broken donor invaded the HC at a position centromere distal to the clustered mutations in the recipient, thus precluding the mutation cluster from being copied. Heterozygous mutations could result from MMS damage introduced in ssDNA formed during DNA resection of the donor and/or during secondary BIR. Alternatively, heterozygous mutations in ALM_1 could have also formed by the invasion of the broken donor into another homologous DNA template (i.e., sister chromatid) instead of the HC product when initiating secondary BIR. The use of a sister chromatid as a template for secondary BIR was likely to occur in other isolates and is strongly supported by the analysis of ALM_18 (Figure 3D, S1C, S3), where a 20kb deletion between two pairs of Ty elements, FS1 and FS2, likely resulted from recombination between FS1 of the broken donor chromosome and FS2 of a sister chromatid (see Supplemental Results for details).
Thus far, our data indicate that complex clusters form by at least one HC followed by a secondary BIR event. Additional rounds of HCs and BIR events could however lead to even more complex outcomes. This was supported by the analysis of ALM_26, which likely resulted from the following events: formation of 6 clustered mutations, two HC events, and an ectopic recombination event that led to a chromosomal rearrangement (Figure 3D, S1D, S4; also see Supplemental Results for details).
Overall our data suggest that increased levels of DNA damage can disrupt BIR and result in multiple rounds of repair leading to the formation of complex clusters that can be associated with chromosomal rearrangements.
Discussion
Recent studies have revealed that cancer genomes frequently contain clusters of strand-coordinated mutations (also called kataegis) caused by ssDNA specific enzymes – APOBEC cytidine deaminases (Roberts et al., 2012; Nik-Zainal et al., 2012; Alexandrov et al., 2013; Roberts et al., 2013; Burns et al., 2013). A fraction of kataegic clusters co-localized with breakpoints of chromosomal rearrangements (Nik-Zainal et al., 2012; Roberts et al., 2012). However, the source of ssDNA and the mechanistic connection between clusters and chromosomal rearrangements remained unexplained. Here, we demonstrate that an unusual type of DNA synthesis, break-induced replication (BIR) completed in the presence of MMS, leads to repair outcomes containing clustered mutations 50% of the time. Importantly these clusters were frequently associated with DNA breakage and chromosomal rearrangements, suggesting that DNA damage to BIR intermediates may account for both clustering of mutations and associated rearrangements that characterize kataegis in human cancer.
We propose that the formation of mutation clusters and chromosomal rearrangements associated with BIR result from the interplay between DNA damage and two specific structural features of BIR replication (Figure 4B-E). First, ssDNA accumulated during BIR from asynchronous leading and lagging strand synthesis and from long resection prior to BIR initiation provides the substrate for clustered mutagenesis. Second, the HJ migrating behind the BIR bubble is a substrate for resolution, which is promoted by stalling of BIR synthesis and leads to breaking and re-invading events that can manifest as half-crossovers, HC-initiated cascades, and other chromosomal rearrangements (Deem et al., 2008; Smith et al., 2009; Vasan et al., 2014). We propose that the formation of mutation clusters in this study were promoted by MMS-induced damage in ssDNA (N3-me C and to a lesser extent N1-me A) (Yang et al., 2010). We further propose that stalling of the replication bubble and the consequent breakage of DNA through HJ resolution was likely induced by either N3-me A, a common MMS-induced lesion in dsDNA, or abasic sites created by unfinished repair of MMS damage (Yang et al., 2010).
The idea that two destabilizing outcomes of BIR- mutation clusters and DNA breakage promoting chromosomal rearrangements- are stimulated by different DNA lesions, predicts that these two outcomes of BIR could be independently modulated and therefore together can give rise to a variety of outcomes. In agreement with this idea, different doses of MMS resulted in clusters with varying length and complexity.
We propose that BIR is not only a prominent source of damage-induced mutation clusters in yeast, but also in cancer genomes including clusters associated with DNA breakage and rearrangements. The variety of mutation clusters found in cancer such as mutation clusters with and without various chromosomal rearrangements (i.e., LOH, CNV and translocations), could each result from combining DNA damage induced by APOBEC family enzymes (modeled in our experiments by MMS) with DNA breakage resulting from other sources of DNA damage or from special features of cancer cells such as checkpoint defects. Supporting this, our recent data obtained in yeast demonstrated that checkpoint defects can promote cascades of chromosomal breakage and rearrangements in cells undergoing BIR even in the absence of additional DNA lesions (Vasan et al., 2014). In addition, Costantino and colleagues (Costantino et al., 2014) have recently demonstrated that a massive collapse of replication forks resulting from the overexpression of an oncogene in human cancer cells led to the initiation of BIR, which in turn promoted a burst of chromosomal rearrangements known to lead to cancer. Our results predict that the exposure of such cells to DNA damage will lead to kataegic mutation clusters associated with chromosomal rearrangements. Moreover, short stretches of ssDNA resulting from interrupted BIR could contribute to some of the genome-wide mutagenesis in individual cancers where APOBEC mutagenesis in clusters often represents only a small fraction (e.g. less than 10%) of all mutations in APOBEC motifs (Roberts et al., 2012; Alexandrov et al., 2013). Indeed the rates of single mutations, including mutations with APOBEC motifs were increased in the immediate vicinity of rearrangement breakpoints in many cancer samples (Drier et al., 2013), suggesting DSB repair processes including BIR may be targets for scattered mutations as well as for mutation clusters. Additional APOBEC-induced single mutations that are not located close to the positions of rearrangement breakpoints could also result from BIR since not all BIR events are associated with detectable rearrangements.
All together, this study suggests that BIR could be one of the most powerful sources of destabilizing genomes of somatic cells, capable of producing a wide range of multiple simultaneous genomic changes. We expect that future studies will reveal the scale to which BIR contributes to genome destabilization in both normal and cancer cells as well as uncover how such instability is regulated.
Experimental Procedures
Yeast strains and growth conditions
All experiments were performed using yeast strain AM1003 (Deem et al., 2008). Rich medium (yeast extract-peptone-dextrose [YEPD]) and synthetic complete medium were made as described in (Guthrie and Fink, 1991). YEP-lactate (YEP-Lac) and YEP-galactose (YEP-Gal) used for DSB induction were similar to (Deem et al., 2008). MMS was added to YEP-Gal to the final concentrations of 1.5mM and 6mM. See Supplemental Experimental Procedures for further details.
Characterization of DSB repair isolates
DSB repair isolates were characterized by phenotype, similar to (Deem et al., 2008). The chromosomal structure of BIR (Ade+Leu-) outcomes to determine the presence of chromosomal rearrangements was analyzed using PFGE, similar to (Deem et al., 2008) (see Supplemental Experimental Procedures for details). The preparation of yeast genomic DNA for sequencing, library construction, mapping of reads to a reference genome, and identification of mutations were performed similar to (Roberts et al., 2012). See Supplemental Experimental Procedures for details of whole genome sequencing, reference sequence construction, and mutation calling. The identification of mutation clusters and analyses of strand bias, strand-coordination, and co-localization of mutation clusters with breakpoints were performed similar to (Roberts et al., 2012). Coverage maps for chromosome III were created using CLC Genomics Workbench 6.0.
The assignment of mutations to the recipient or donor copy of chromosome III was performed similar to (Saini et al., 2013). See Supplemental Experimental Procedures for further details.
Supplementary Material
01
02
03
04
05
Highlights.
- Damage in ssDNA formed during BIR can cause simultaneous clustered mutations.
- Mutation clusters occur in ssDNA formed during uncoupled conservative DNA synthesis.
- BIR-generated mutation clusters co-localize with additional breaks and rearrangements.
- BIR-generated mutation clusters closely resemble kataegic clusters in cancer.
Acknowledgments
This work was funded by NIH grants R01GM084242 to AM, and 5R01GM052319-17 to P.A.M, and by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, (project ES065073 to M.A.R). S.A.R. was supported by NIH Pathway to Independence Award K99ES022633-01. C.J.S. was supported in part by the Department of Biology at IUPUI.
Footnotes
Author Contributions: C.J.S., S.A.R., D.A.G., and A.M. designed the experiments. C.J.S. performed BIR experiments. E.M. and P.A.M performed whole-genomic sequencing. S.A.R., C.J.S., M.A.R., D.A.G., and A.M. analyzed the data. C.J.S., S.A.R., D.A.G., and A.M. wrote the manuscript. A.M. and D.A.G. supervised the project. C.J.S. and S.A.R. contributed equally to the work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps M, Herman A, Loh E, Loeb LA. Genetic constraints on protein evolution. Crit Rev Biochem Mol Biol. 2007;42:313–326. doi: 10.1080/10409230701597642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Sterling JF, Roberts SA, Bhagwat AS, Resnick MA, Gordenin DA. Base damage within single-strand DNA underlies in vivo hypermutability induced by a ubiquitous environmental agent. PLoS Genetics. 2012;8:e1003149. doi: 10.1371/journal.pgen.1003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WH, Zhu Z, Papusha A, Malkova A, Ira G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genetics. 2010;6:e1000948. doi: 10.1371/journal.pgen.1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi O, Halazonetis TD. Break-Induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:99–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A, Barker K, Vanhulle K, Downing B, Vayl A, Malkova A. Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics. 2008;179:1845–1860. doi: 10.1534/genetics.108.087940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnianni RA, Symington LS. Break-induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci USA. 2013;33:13475–13480. doi: 10.1073/pnas.1309800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drier Y, Lawrence MS, Carter SL, Stewart C, Gabriel SB, Lander ES, Meyerson M, Beroukhim R, Getz G. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome research. 2013;23:228–235. doi: 10.1101/gr.141382.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink G. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Lada AG, Dhar A, Boissy RJ, Hirano M, Rubel AA, Rogozin IB, Pavlov YI. AID/APOBEC cytosine deaminase induces genome-wide kataegis. Biology direct. 2012;7:47. doi: 10.1186/1745-6150-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lada AG, Stepchenkova EI, Waisertreiger IS, Noskov VN, Dhar A, Eudy JD, Boissy RJ, Hirano M, Rogozin IB, Pavlov YI. Genome-wide mutation avalanches induced in diploid yeast cells by a base analog or an APOBEC deaminase. PLoS Genetics. 2013;9:e1003736. doi: 10.1371/journal.pgen.1003736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Molecular and cellular biology. 2005;25:933–944. doi: 10.1128/MCB.25.3.933-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Ira G. Break-induced replication functions and molecular mechanism. Current Opinion in Genetics and Development. 2013;23:271–279. doi: 10.1016/j.gde.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev SI, Sotiriou SK, Pateras IS, Santoni F, Sougioultzis S, Edgren H, Almusa H, Robyr D, Guipponi M, Saarela J, Gorgoulis VG, Antonarakis SE, Halazonetis TD. A single-nucleotide substitution mutator phenotype revealed by exome sequencing of human colon adenomas. Cancer Res. 2012;72:6279–6289. doi: 10.1158/0008-5472.CAN-12-3869. [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. Mutational Processes Molding the Genomes of 21 Breast Cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Molecular Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–392. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Lam AF, Symington LS. Aberrant double-strand break repair resulting in half-crossovers in mutants defective for Rad51 or the DNA polymerase delta complex. Mol Cell Biol. 2009;29:1432–1441. doi: 10.1128/MCB.01469-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan S, Deem A, Ramakrishnan S, Argueso J, Malkova A. Cascades of genetic instability resulting from compromised break-induced replication. PLoS genetics. 2014;10:e1004119. doi: 10.1371/journal.pgen.1004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, et al. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–396. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gordenin DA, Resnick MA. A single-strand specific lesion drives MMS-induced hyper-mutability at a double-strand break in yeast. DNA repair. 2010;9:914–921. doi: 10.1016/j.dnarep.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS genetics. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05