Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss (original) (raw)
Abstract
Despite the prominence of Caenorhabditis elegans as a major developmental and genetic model system, its phylogenetic relationship to its closest relatives has not been resolved. Resolution of these relationships is necessary for studying the steps that underlie life history, genomic, and morphological evolution of this important system. By using data from five different nuclear genes from 10 Caenorhabditis species currently in culture, we find a well resolved phylogeny that reveals three striking patterns in the evolution of this animal group: (i) Hermaphroditism has evolved independently in C. elegans and its close relative Caenorhabditis briggsae; (ii) there is a large degree of intron turnover within Caenorhabditis, and intron losses are much more frequent than intron gains; and (iii) despite the lack of marked morphological diversity, more genetic disparity is present within this one genus than has occurred within all vertebrates.
Caenorhabditis elegans is an important model system that allows great depth of study into how the genome is translated into a developing, functioning animal (1). To generalize from this model, a phylogenetic context and information about related species are essential. The genome of a close relative, Caenorhabditis briggsae, was recently sequenced, providing an important comparative genomics tool for annotating the C. elegans genome (2). However, genome comparisons for multiple species that are closely related can provide substantially more analytical power, as demonstrated recently by genome comparisons among several closely related yeast species (3). A well resolved phylogeny for closely related species provides the basis for selecting appropriate representatives for such comparisons, for distinguishing orthologous from paralogous genes, and for distinguishing ancestral versus derived states for characters (4).
Comparisons among phylogenetically closely related species, as opposed to comparisons among distantly related groups, are more likely to reveal finer detail about the steps that underlie life history, genomic, and morphological evolution. For example, in the absence of a phylogeny that includes additional closely related species, a feature that is actually convergent between two species may appear to be homologous, as we show below for the case of hermaphroditic reproduction. Also, the time and the frequency at which evolutionary events occurred, like the loss and gain of introns, may be obscured by comparing distantly related species or anciently duplicated genes. We therefore aimed to resolve the phylogenetic relationships of all Caenorhabditis species currently in culture by using gene sequence data.
Previous analyses with morphological (5) and small subunit (SSU) rRNA gene data (6) supported a monophyletic group called the Elegans group, consisting of C. elegans, C. briggsae, Caenorhabditis remanei, and an undescribed Caenorhabditis species (strain CB5161). However, the relationships of the Elegans group species to each other remained unresolved in these studies. Other molecular analyses included only three of the Elegans group species and no other Caenorhabditis species (7-10). These studies showed that C. briggsae and C. remanei are likely to be sister species, but the sister group of C. elegans remained unknown.
Here we present a completely resolved phylogeny of all 10 Caenorhabditis species currently in culture and four representative outgroup species from the closest species groups within family Rhabditidae (Nematoda) (11), inferred from the sequences of five nuclear genes: nearly complete sequences from SSU and large subunit (LSU) rRNA-encoding DNA (rDNA), part of the gene for the largest subunit of RNA polymerase II (RNAP2; also known as ama-1 in C. elegans), and portions of the par-6 and pkc-3 genes. We used this phylogeny to reevaluate the evolution of hermaphroditism within Caenorhabditis and the evolution of introns in RNAP2. Our data also allowed us to explore the range of genetic divergence that has occurred across the genus of Caenorhabditis and compare it to that in other organisms.
Materials and Methods
Strains. The following 12 nematode species were used in this study: Caenorhabditis drosophilae (strains DF5077 and SB225), Caenorhabditis japonica (SB339), Caenorhabditis plicata (SB355), Caenorhabditis remanei (SB146 and EM464), Caenorhabditis sp. (CB5161), Caenorhabditis sp. (DF5070), Caenorhabditis sp. (PS1010), Caenorhabditis sp. (SB341), Prodontorhabditis wirthi (DF5074), Protorhabditis sp. (DF5055), Oscheius myriophila (DF5020), Rhabditella axei (DF5006). All nematode strains were maintained by using standard methods (12), with the exception of P. wirthi, which was cultivated on plates with 1.5% agar in tap water supplied with a piece of brown algae.
PCR Amplifications and Sequencing. Genomic DNA was amplified from worm lysates with primers for SSU rDNA, LSU rDNA, part of RNAP2, the par-6 gene, and the pkc-3 gene. For amplification of RNAP2 cDNA, total RNA was isolated (RNeasy mini kit, Qiagen, Valencia, CA) and RT-PCR was performed with the Qiagen OneStep RT-PCR kit. The primer sequences are available as Supporting Materials and Methods, which is published as supporting information on the PNAS web site. PCR products were sequenced directly by using the ABI BigDye Terminator Version 3.0 or Version 3.1 cycle sequencing kit and an ABI 377 DNA Sequencer (Applied Biosystems). A list of primers used to generate overlapping sequences of both strands can be obtained from the authors. Sequences were assembled with sequencher, Version 4.1.2 (Gene Codes, Ann Arbor, MI). The sequences for C. elegans and C. briggsae genes were obtained from WormBase (www.wormbase.org). A list of GenBank accession numbers for the sequences obtained in this study and all other sequences used are available in Table 1, which is published as supporting information on the PNAS web site. Data files are also available from TreeBase (www.treebase.org, accession no. SN1907), the NemAToL database (http://nematol.unh.edu), and the authors (www.nyu.edu/projects/fitch).
Alignment and Identification of Introns. SSU rDNA sequences were aligned manually by using secondary structure predictions as described in ref. 6. LSU rDNA sequences were aligned with clustalx (13) and improved manually with macclade 4.05 (14). Previous analyses with rhabditid rDNA showed that alternative alignments do not affect the phylogenetic conclusions, because alignment ambiguous sites do not contribute greatly to the phylogenetic signal (6). RNAP2, pkc-3, and par-6 sequences were aligned manually. For all three protein-coding genes, the alignment was unambiguous and introns were easily recognized and removed. Intron positions in RNAP2 genes of several species were also confirmed by sequencing cDNA. For the purposes of analysis, two introns were hypothesized to be homologous when both occurred in the identical position in the coding sequence alignment. Sequences of these introns varied greatly in size and were unalignable, even between close relatives. All sequence alignments with intron positions indicated for the protein coding genes can be obtained from the authors.
Phylogenetic Analyses. Except as described, phylogenetic analyses were performed by using weighted maximum parsimony as implemented in paup* 4.0b10 (15), where a transversion was weighted twice a transition. This weighting scheme takes into account the observation that transitions occur twice as frequently as transversions in most eukaryote nuclear genomes (16) This observation applies approximately to the nematode sequences analyzed here: Considering only the most closely related five species (to reduce homoplasy effects) and all five genes, excluding third codon positions from the protein-coding genes, we obtained a likelihood-estimated transition/transversion (Ti/Tv) ratio of 1.87 (under the HKY85+I+Γ model). Lower Ti/Tv estimates are obtained when third-codon positions were included (1.73) or when all taxa were included (1.49), probably because of higher accumulation of superimposed substitutions in the more divergent lineages or more quickly evolving sites. That is, as sites become saturated with transversions and transitions, the observed Ti/Tv ratio should decrease toward 0.5. Weighted parsimony with such a weighted transformation scheme is more computationally efficient than likelihood and has been shown to perform considerably better than an equal-weighting scheme or neighbor-joining (17). For rDNA, stem sequences and loop sequences were both included and weighted equally. In previous analyses of SSU rDNA for some of these and related nematodes, differential weighting or exclusion of loops had no effect on tree topology, and the information content of loops alone was low (6).
Two different analyses were performed with different taxon and character sets. In the first, we aimed to resolve the relationships of all taxa, but sites likely to be saturated with substitutions were excluded to minimize homoplasy. This analysis included the sequences of SSU rDNA, LSU rDNA, and RNAP2 for all 14 species, but with the third-codon positions from the latter gene excluded. Third-codon positions were saturated with substitutions in pairwise comparisons involving several species less closely related to C. elegans than Caenorhabditis sp. PS1010. That is, some of these comparisons resulted in undefined distance values with the Tamura-Nei method implemented in paup*. In this analysis, O. myriophila and R. axei together were treated as outgroup representatives. The second analysis used all five genes and all positions but only from the following most closely related species: C. briggsae, C. elegans, C. japonica, C. remanei, and Caenorhabditis sp. CB5161, with C. drosophilae and Caenorhabditis sp. PS1010 as outgroup representatives. One indication that the third-codon positions were not saturated for these comparisons was that the parsimony Ti/Tv ratio was nearly 1.45 (unambiguous changes only, calculated using macclade). However, substitutions appear to be fully saturated at 4-fold synonymous sites, even for the most closely related five taxa (Ti/Tv = 0.53). All bootstrap and jackknife analyses used at least 2,000 replications, with heuristic searches in the first analysis and complete branch-and-bound searches in the second analysis. The second dataset also was used in a jackknife maximum likelihood analysis implemented in paup*. Parameters for a general time-reversible model, accounting for invariant sites and rate variation across sites (GTR+I+Γ), were estimated from the data by using the neighbor-joining cladogram (which was identical to the most parsimonious cladogram) and used to evaluate trees in a jackknife analysis employing 2,000 replications with branch-and-bound searches.
Divergence Analysis. By using RNAP2, SSU rDNA, or LSU rDNA sequence data, divergence analyses were performed separately for the 14 nematode species, a selection of deuterostome species, and a selection of protostome species, or for all species together. There was hardly any difference between treating the groups separately and treating them together, and only the separate analysis is reported. (Taxa and GenBank accession numbers are found in Table 1.) Likelihood, implemented by paup* 4.0b10 (15), was used to reconstruct the amount of nucleotide change along the lineages of an assumed phylogeny (18, 19) for each of the three groups of taxa represented by each molecule (see Figs. 3-5, which are published as supporting information on the PNAS web site). For these estimations of evolutionary distance, likelihood was used instead of parsimony to allow correction for superimposed substitutions. Separately for each gene, parameters for a general time-reversible model of evolution were estimated from the data, with corrections for nucleotide composition, proportion of invariant sites, and differential rates at different codon positions (GTR+I+SS). We used a phylogeny-based estimation of divergences because it allowed us to determine which divergence values were unusually large because of long-branch effects (increased rate of accumulation of change). Such an effect was not problematic for RNAP2, but it was problematic for Diptera in rDNA comparisons, for example. Calculating pairwise divergences alone does not allow such a distinction. The divergence between a pair of species was calculated as the sum of branch lengths separating them; the SD of a pairwise divergence was calculated as the square root of the sum of the squares of the SDs of these branches.
Results and Discussion
Taxon Representation. There are a maximum of 21 known (described and undescribed) species in the Caenorhabditis clade; possible synonymizations could reduce this number to 17 (5). The group of 10 representatives we include in the present phylogenetic analysis comprises all Caenorhabditis species currently in culture and encompasses the overall phylogenetic and morphological diversity of this genus (cf. 5). As outgroup taxa, we include two taxa, P. wirthi and Protorhabditis sp. (DF5055), from a clade likely to be a sister taxon to Caenorhabditis based on SSU rDNA data (11) and two taxa, O. myriophila and R. axei, from less closely related groups within the “Eurhabditis” clade of family Rhabditidae (6).
The Caenorhabditis Phylogeny. Weighted-parsimony analysis of concatenated sequences of SSU rDNA, LSU rDNA, and the single-copy nuclear gene RNAP2 resulted in a well resolved phylogeny for all 10 Caenorhabditis species (Fig. 1). For this analysis, the third-codon position of RNAP2 was not used because substitutions at these sites between the more distantly related taxa were close to saturation (see Materials and Methods). Jacknife analysis with 2,000 replications resulted in high support for most branches. The branching pattern of this tree agrees with a previous phylogenetic tree based on morphological characters (5), except for the position of C. plicata, which was uncertain in that study.
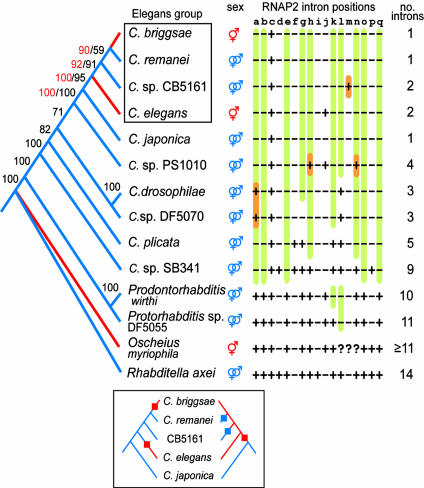
Fig. 1.
Phylogenetic tree of all Caenorhabditis species currently in culture and four outgroup representatives. The black numbers on the branches denote percentage jackknife support from 2,000 replicates for an analysis incorporating all taxa by using the two rRNA genes and RNAP2 with the third-codon positions deleted. The red numbers denote jackknife support in 5,000 replicates for an analysis incorporating the seven most closely related species and all nucleotide positions of five genes (including par-6 and pkc-3). The colors of the branches represent the most parsimonious distribution of reproductive modes in ancestral lineages (sex column: red for hermaphroditism and blue for gonochorism). Our analysis supports the hypothesis that hermaphroditism evolved independently in C. briggsae and C. elegans from gonochorism. This scenario (two changes represented by red blocks in the left-hand tree of Inset) is more parsimonious than if, for example, hermaphroditism evolved once (red block in the right-hand tree of Inset) and was reversed to gonochorism in C. remanei and Caenorhabditis sp. CB5161 (blue blocks; three changes total). The presence (+) or absence (-) of introns at particular orthologous sites in the RNAP2 gene are depicted as a matrix (question marks indicate missing data). Under the conservative assumption that gain and loss of an intron are equally likely, 12 intron losses (green highlights) and 4 gains (orange highlights) were unequivocally inferred at 11 sites. At the other six sites, there are two or more ways that changes could be distributed across the tree such that the total number of gains and losses cannot be determined unambiguously at these sites.
When the different data partitions are analyzed separately, they give incongruent results with regard to the relationship of Caenorhabditis sp. PS1010 and the pair of sister species, C. drosophilae and Caenorhabditis sp. DF5070. Whereas rDNA supports the branching pattern shown in Fig. 1, RNAP2 supports a sister group relationship between Caenorhabditis sp. PS1010 and the pair C. drosophilae plus Caenorhabditis sp. DF5070, with a jackknife value of 97% (2,000 replications, data not shown). This conflict is significant (P = 0.0001 in a partition homogeneity test (20) implemented in paup* with 10,000 replicates), but only because of these three taxa (P = 0.7827 in the homogeneity test when these taxa are removed). Such conflicts are known to occur (21), are likely due to homoplasy or lineage sorting, and can only be resolved by adding data from other independently evolving genes. Thus, only the relationship between Caenorhabditis sp. PS1010 and C. drosophilae plus Caenorhabditis sp. DF5070 remains ambiguous, but is not relevant to resolving the relationships of the Elegans group. For the Elegans group itself, the data partitions show no significant conflict in the partition homogeneity test (P = 1.000), although RNAP2 alone is unable to resolve the relationships of this clade. Even when the genes are concatenated and analyzed, the important branch leading to C. remanei and C. briggsae has only low jackknife support (black numbers in Fig. 1).
We therefore performed a second analysis, this time only including the species of the Elegans group and C. japonica, with C. drosophilae and Caenorhabditis sp. PS1010 as outgroup representatives. For this reduced taxon set, we increased the number of characters by adding partial sequences from the single-copy nuclear genes _par-_6 and pkc-3, in addition to the other three genes, and including all codon positions. A weighted-parsimony jackknife analysis (5,000 replications with a branch-and-bound search) resulted in the same branching pattern with robust statistical support for each branch (red numbers in Fig. 1). That third-codon positions provided additional phylogenetic information and resolving power for this reduced set of taxa was indicated by lower jackknife support values when third-codon positions were excluded (e.g., 76% instead of 90% for the top node in Fig. 1 and 82% and 92% instead of 92% and 100%, respectively, for the next nodes down the tree). For this second dataset as well, there was no significant incongruence between protein-coding and rDNA data partitions (P = 0.99 for 1,000 replications). The inferred peptide sequences alone provide no or very low resolution for relationships in the Elegans group (bootstrap/jackknife values range 40-73% in a parsimony analysis with a blosum62 stepmatrix; other standard stepmatrices provided similar results).
Other methods confirm the robustness of these data. A maximum likelihood jackknife analysis (GTR+I+Γ model, 2,000 replications; see Materials and Methods) also provided high support for the same nodes (from the top down in Fig. 1: 84%, 82%, 87%, and 100%), as did a neighbor-joining bootstrap analysis [Tamura-Nei+I(I = 0.6)+Γ(α = 0.6) model, 10,000 replicates] for the same nodes (89%, 91%, 100%, 100%, respectively).
The phylogeny supports the monophyly of the Elegans group and the position of C. japonica as its sister species, which was proposed based on morphological characters (22). More importantly, the phylogeny shows that there is no single species most closely related to C. elegans. Instead, C. elegans is the outsider to the three other Elegans-group species; the most closely related species are C. briggsae and C. remanei. These relationships are consistent with studies of other genes that only included the three species C. elegans, C. remanei, and C. briggsae (7-10). The importance of including all Caenorhabditis species in this analysis, and Caenorhabditis sp. CB5161 in particular, becomes evident when the phylogeny is used to investigate character evolution, such as the evolution of reproductive modes.
Evolution of Hermaphroditism. Arguably the most important feature of C. elegans regarding its usefulness as a genetic model system is its peculiar form of protandrous hermaphroditism, which is quite different from the kind of hermaphroditism in other animals like snails or earthworms. In C. elegans, the gonads of individuals that are morphologically females first produce a limited number of sperm and then switch to oogenesis (23). A switch back to spermatogenesis never occurs. Hermaphrodites use their sperm only to fertilize their own eggs. Because they lack male organs, hermaphrodites cannot transfer the sperm to cross-fertilize another hermaphrodite's eggs. However, cross progeny can be obtained when hermaphrodites mate with the rarely occurring males. This sexual system, comprising self-fertile hermaphrodites and males, is rare in animals (24). Because both C. elegans and C. briggsae are hermaphroditic species, one might expect this feature to be shared through common ancestry. However, hermaphroditism probably evolved convergently from gonochorism (a male-female mating system) in the separate lineages to C. elegans and C. briggsae (Fig. 1). According to the phylogeny, this scenario is more parsimonious than if, for example, hermaphroditism evolved once and was reversed to gonochorism in C. remanei and Caenorhabditis sp. CB5161 (see Fig. 1 Inset). Note that it would not be possible to distinguish whether hermaproditism evolved once or twice independently without including Caenorhabditis sp. CB5161, thus suggesting it will be important to include in future comparative research on reproductive modes.
Consistent with our conclusion that hermaphroditism is convergent in C. elegans and C. briggsae, the same type of hermaphroditism has evolved independently from gonochorism at least 10 times in rhabditid nematodes (e.g., in Oscheius; Fig. 1); conversely, there is only one instance in which gonochorism could have evolved from hermaphroditism (D.H.A.F. and K.K., unpublished data). Recent molecular data suggest that the mechanisms producing hermaphroditism are different in C. elegans and C. briggsae. First, fog-2, a gene required for making sperm in C. elegans hermaphrodites, appears to be missing in C. briggsae (S. Nayak and T. Schedl, personal communication). Second, specification of sperm requires fem-3 in C. elegans hermaphrodites, but is independent of the orthologous Cbr-fem-3 in C. briggsae (8).
Because changes in development can underlie homologous features, differences in mechanisms by themselves cannot be used to prove that hermaphroditism originated independently in C. elegans and C. briggsae. A classic example for a homologous feature with different underlying developmental mechanisms is the amphibian lens. Despite the obvious homology of the lens in all amphibians, its development is induction-dependent in some species (e.g., Ambystoma maculatum and Rana fusca) and induction-independent in other species, even close relatives (e.g., Ambystoma mexicanum and Rana esculenta) (25). In this case, as in the case of hermaphroditism in Caenorhabditis, the phylogeny provides the strongest evidence for homology or convergence.
Intron Evolution. Having sequences of the RNAP2 genes from closely related species also allowed us to map the evolution of introns on our phylogeny, revealing a striking picture of unusually frequent intron loss in Caenorhabditis. The 1,860-bp coding region of the RNAP2 gene we sequenced from the Caenorhabditis species and several outgroup representatives has 17 different sites that are occupied by introns (Fig. 1, columns a-q), but different sites are occupied in different species. The number of introns present in a species ranges between 1 and 14. To evaluate intron evolution, we first applied the most conservative a priori assumption that intron losses and gains are equiprobable. Under this simplistic assumption, we inferred the intron losses and gains by using parsimony reconstruction in macclade 4.05 (14). Introns were lost 12 times and gained 4 times at the 12 sites in which all reconstructions are unequivocal (a-c, e, g, h, k-o, and q). At the other five sites, there are two or more ways that changes could be distributed across the tree such that the number of gains and losses cannot be determined unambiguously. At these sites, gains could have occurred between 4 and 11 times, and losses could have occurred between 7 and 0 times, again assuming that both are equally likely to occur. Overall, the maximum number of intron gains could have been 15, and the maximum number of losses could have been 19. Of all of the possible gains, only one is in a unique position (site m in Caenorhabditis sp. CB5161) and is thusly unambiguously a gain. All of the other intron gains must have occurred at least twice in the exact same position. Whether or not multiple intron gains can occur in the same position is a matter of much debate (26, 27); a discussion of this subject is beyond the scope of this study. Adopting a different view, we can apply the model that introns in one position can be gained only once. Under this assumption, we infer at least 27 intron losses and at most 3 gains over all 17 sites. Thus, no matter what a priori assumption is used about relative probabilities of intron loss and gain, we conclude that introns are lost in this region more frequently than gained. More importantly, our analysis reveals a striking degree of intron turnover within these phylogenetically closely related species.
This phylogenetic analysis with orthologous genes from close relatives allows us to assign intron evolution events to particular species lineages in Caenorhabditis, which cannot be accomplished in two-taxon comparisons, in comparisons involving paralogous genes, or in comparisons by using very distantly related model systems. For example, frequent intron loss and only a few gains were also observed within the large families of chemoreceptor genes in C. elegans (28, 29). However, because the gene duplications leading to these gene families were much more ancient than the _C. elegans_-C. briggsae divergence, intron losses could be “dated” relative to the species divergences only in the few cases in which C. briggsae orthologs were available and both differed in their introns. Comparisons of whole genome data for distantly related model organisms suggested that many introns were lost and gained in the nematode lineage (30). However, this kind of data does not allow one to differentiate whether the intron turnover happened early in the lineage separating C. elegans from the other animals tested (Drosophila melanogaster and Homo sapiens) or within Caenorhabditis. With our RNAP2 dataset for many closely related species for which the phylogenetic relationships are known, we can assign many intron losses and gains to specific lineages. Our data suggest that introns in this gene were lost gradually during the evolution of Caenorhabditis, leading to strikingly few introns in the species of the Elegans group.
A comparison of the C. elegans and C. briggsae genome sequences revealed that in ≈12,000 putative orthologous gene pairs, there are twice as many _C. elegans_-specific introns as there are _C. briggsae_-specific introns (2). Previous data (28, 29, 31) led to the hypothesis that the “species-specific” introns in the C. elegans and C. briggsae genomes were more likely to have resulted from intron loss in one or the other ancestral lineage than from species-specific intron gain. Our results for several closely related species support and further extend this interpretation to additional species in the Caenorhabditis group.
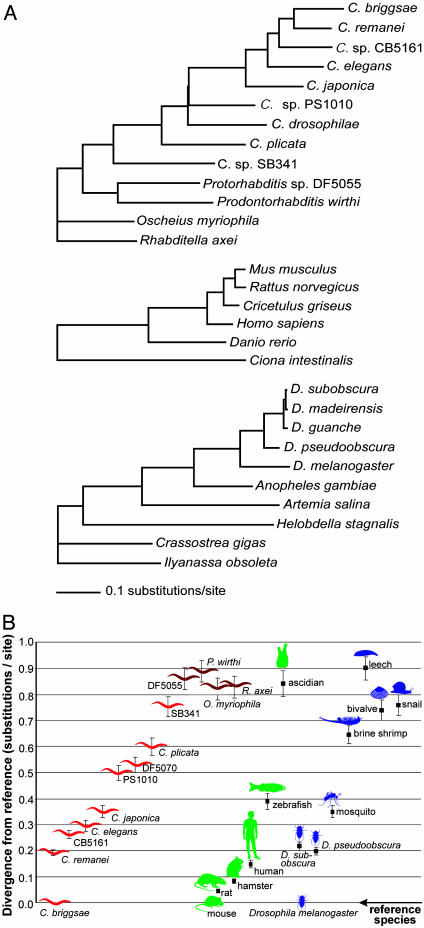
Genetic and Morphological Divergence. When analyzing our sequence data, we were surprised to observe that genetic divergences among these closely related Caenorhabditis species were so large. Not only do the rRNA genes show a large degree of divergence, as observed earlier (6), but RNAP2 also accumulated a large number of substitutions. Thus, we were interested to compare the range of genetic divergence that has occurred in Caenorhabditis with that in vertebrates, other deuterostomes, and in protostomes. As a measure of genetic divergence, we determined the branch lengths on assumed phylogenies by using maximum likelihood (Figs. 2_A_, 3, and 5). We then plotted the divergences between each of the nematode species and the reference species C. briggsae, the divergences between each of the deuterostome representatives and mouse, and the divergences between each of the representative protostomes and D. melanogaster (Fig. 2_B_ and Fig. 6, which is published as supporting information on the PNAS web site). We find that C. briggsae and C. elegans are more divergent than are human and mouse at the RNAP2 locus (likelihood distance estimates of 0.29 ± 0.02 substitutions per site between these two Caenorhabditis species and 0.14 ± 0.01 substitutions per site between human and mouse; Fig. 2_B_). Similar relative divergence comparisons have been made with genome-wide data (2, 32). Here, we extend such comparisons across the Caenorhabditis genus. For instance, the divergence between C. briggsae and C. japonica (0.35 ± 0.02 substitutions per site) is comparable with that between mouse and zebrafish (0.39 ± 0.03 substitutions per site).
Fig. 2.
RNAP2 divergences between pairs of taxa. (A) Likelihood estimates of branch lengths in three assumed topologies for RNAP2 genes from the nematodes considered in this paper (Top), representative deuterostomes (Middle), and representative protostomes (Bottom). A general time-reversible model was assumed with different rates for each of the three codon positions, as detailed in Materials and Methods. Numerical values for branch lengths with SDs are shown in Fig. 4. (B) Comparisons of evolutionary divergences in RNAP2 genes between species pairs within three different groups: nematodes (red), vertebrates and other deuterostomes (green), and arthropods and other protostomes (blue). Pairwise divergences were calculated by summing the lengths of the branches between species in A; error bars show SDs calculated as the square root of the sum of the squares of the SDs for each of the component branches. Shown are divergences between C. briggsae (red icon at bottom) and the other Caenorhabditis species (light-red icons) or related rhabditid nematodes (dark-red icons), divergences between mouse (Mus musculus, green icon at bottom) and other deuterostomes [green icons: Rattus norvegicus (rat), Cricetulus griseus (Chinese hamster), H. sapiens, Danio rerio (zebrafish), and Ciona intestinalis (ascidian)], and between D. melanogaster (blue icon at bottom) and other protostomes [blue icons: Drosophila subobscura, Drosophila pseudoobscura, Anopheles gambiae (mosquito), A. salina (brine shrimp), Helobdella stagnalis (leech), Crassostrea gigas (oyster), and Ilyanassa obsoleta (snail)]. Note that the distribution of icons along the horizontal axis is arbitrary.
Perhaps even more unexpectedly, the genetic distances across arthropods was less than the distances spanned by Caenorhabditis alone. For example, the greatest genetic distance between the arthropods compared (D. melanogaster and the brine shrimp Artemia salina) was 0.65 ± 0.03 substitutions per site, less than the 0.75 ± 0.04 divergence between C. briggsae and Caenorhabditis sp. SB341 (Fig. 2). Divergences between the major taxonomic groups (e.g., 1.27 ± 0.05 substitutions per site for _C. briggsae_-mouse; 1.38 ± 0.05 substitutions per site for _C. briggsae_-D. melanogaster) are large enough to suggest that the smaller divergences within the three major taxonomic groups are not affected significantly by saturation effects. Very similar relative divergences are obtained by using the SSU or LSU rRNA genes, although some divergences are unusually high because of the high rates of accumulation of substitutions in particular branches (see Figs. 3, 5, and 6).
Surprisingly, the large genetic divergence in these nematodes is not accompanied by large morphological divergence. Beyond the difference in reproductive biology mentioned above, there are only subtle morphological differences between most Caenorhabditis species (5, 11, 33). In contrast, at similar genetic divergences, deuterostomes and protostomes have evolved a large range of different morphologies (indicated by icons in Figs. 2_B_ and 6). It had been noted previously (34) that the genetic distance between humans and chimpanzees seemed too small to account for their substantial organismal differences. However, the situation is strikingly reversed in Caenorhabditis.
One possible explanation for the remarkable differences in the ratio of morphological to molecular change in deuterostomes and protostomes versus Caenorhabditis is that stabilizing selection has prevented major changes to body plan in these nematodes. For example, the efficiency of their dispersal by using insects or other small invertebrates may require these nematodes to be small. Alternatively, developmental constraints may limit the “evolvability” of Caenorhabditis. For example, the hydro-static “skeleton,” a pressurized body cavity wrapped with a flexible but inelastic cuticle, might prevent the evolution of compartmentalization or the outgrowth of appendages. Certainly appendages and segmental modularity have been preadaptive to an astounding diversification of body plans in arthropods and other animal groups.
Molecular Clocks and Estimations of Divergence Times. One possible explanation for large genetic divergence between Caenorhabditis species is that in these nematodes the molecular clock “ticks” at a faster rate than in vertebrates or arthropods. Indeed, it has been estimated that two-thirds of the genes in C. elegans evolved more rapidly than their putative orthologs in D. melanogaster (35). This rate difference calls into question molecular-clock estimates for the date of the _C. elegans_-C. briggsae divergence. Genetic divergence data have been used to calculate that C. briggsae and C. elegans diverged 80-110 million years ago (2, 32). This estimation assumed (i) an arthropod-nematode divergence date of 800-1,000 million years ago, (ii) the existence of an arthropod-nematode clade called Ecdysozoa (36), and (iii) a universal molecular clock. However, there are problems with all of these assumptions: (i) there is no nematode fossil record to calibrate an accurate molecular clock for divergence times within nematodes and especially within Caenorhabditis; (ii) support for an Ecdysozoa clade is controversial (37-39); and (iii) genes have evolved at very different rates in different lineages of animals and even within rhabditids (K.K., N.P.G., and D.H.A.F., unpublished data). Considering all of these caveats, estimates for the date of divergence between C. elegans and C. briggsae are very unreliable. Thus, the best measure of taxonomic difference in this group is simply the relative amount of sequence difference (as in Fig. 2_B_), not the date of divergence.
By providing a fully resolved phylogeny for the closest available relatives of C. elegans, we now have a powerful new tool for informed selection of taxa for comparative genome projects. In fact, our phylogeny has been used as the basis for selecting C. remanei, Caenorhabditis sp. CB5161, and C. japonica for comparative genome sequencing projects (P. Sternberg, personal communication). The phylogeny will also be useful for uncovering proximate mechanisms underlying the origin of traits, such as hermaphroditism, and analyzing patterns of gene evolution. As more Caenorhabditis genome sequences become available, augmenting our comparative analytical tool kit in the context of a fully resolved phylogeny, the molecular basis for nematode evolution should become ever clearer.
Note Added in Proof. By using the genes fog-1, fog-3, cpb-1, cpb-2, and cpb-3, Cho et al. (40) have inferred similar phylogenetic relationships with respect to the Elegans group species, C. japonica, and Caenorhabditis sp. PS1010; they also suggest that frequent intron loss has occurred.
Supplementary Material
Supporting Information
Acknowledgments
We thank Claude Desplan, Morris Goodman, E. Jane Albert Hubbard, Stephen Small, and Walter Sudhaus for comments on the manuscript, stimulating discussion, and/or strains. We also thank four anonymous reviewers whose thoughtful comments led to a much improved manuscript. This work was supported by grants to D.H.A.F. from the National Science Foundation and the Human Frontier Science Program. N.P.G., Y.R., and C.R. are undergraduates in the College of Arts and Sciences of New York University, which also provided fellowship support.
Abbreviations: SSU, small subunit; LSU, large subunit; rDNA, rRNA-encoding DNA; RNAP2, largest subunit of the RNA polymerase II gene; Ti/Tv, transition/transversion.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY601770-AY601780, AY602167-AY602189, and AY604469-AY604482).
References
- 1.Riddle, D. L. (1997) C. elegans II (Cold Spring Harbor Lab. Press, Plainview, NY).
- 2.Stein, L. D., Bao, Z., Blasiar, D., Blumenthal, T., Brent, M. R., Chen, N., Chinwalla, A., Clarke, L., Clee, C., Coghlan, A., et al. (2003) PLoS Biol. 1**,** E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. (2003) Nature 423**,** 241-254. [DOI] [PubMed] [Google Scholar]
- 4.Eisen, J. A. (1998) Genome Res. 8**,** 163-167. [DOI] [PubMed] [Google Scholar]
- 5.Sudhaus, W. & Kiontke, K. (1996) J. Zool. Syst. Evol. Res. 34**,** 217-233. [Google Scholar]
- 6.Fitch, D. H., Bugaj-Gaweda, B. & Emmons, S. W. (1995) Mol. Biol. Evol. 12**,** 346-358. [DOI] [PubMed] [Google Scholar]
- 7.Haag, E. S. & Kimble, J. (2000) Genetics 155**,** 105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haag, E. S., Wang, S. & Kimble, J. (2002) Curr. Biol. 12**,** 2035-2041. [DOI] [PubMed] [Google Scholar]
- 9.Rudel, D. & Kimble, J. (2001) Genetics 157**,** 639-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudel, D. & Kimble, J. (2002) Evol. Dev. 4**,** 319-333. [DOI] [PubMed] [Google Scholar]
- 11.Sudhaus, W. & Fitch, D. (2001) J. Nematol. 33**,** 1-70. [PMC free article] [PubMed] [Google Scholar]
- 12.Sulston, J. & Hodgkin, J. (1988) in The Nematode Caenorhabditis elegans, ed. Wood, W. B. (Cold Spring Harbor Lab. Press, Cold Spring Harbor, NY), pp. 587-606.
- 13.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25**,** 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maddison, D. R. & Maddison, W. P. (2001) macclade, Version 4.05 (Sinauer, Sunderland, MA).
- 15.Swofford, D. L. (2002) paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10 (Sinauer, Sunderland, MA).
- 16.Graur, D. & Li, W.-H. (2000) Fundamentals of Molecular Evolution (Sinauer, Sunderland, MA), p. 38.
- 17.Hillis, D. M., Huelsenbeck, J. P. & Cunningham, C. W. (1994) Science 264**,** 671-677. [DOI] [PubMed] [Google Scholar]
- 18.Brusca, R. C. & Brusca, G. J. (2003) Invertebrates (Sinauer, Sunderland, MA), 2nd. Ed.
- 19.Llopart, A. & Aguade, M. (1999) Genetics 152**,** 269-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farris, J. S., Källersjö, M., Kluge, A. G. & Bult, C. (1995) Cladistics 10**,** 315-319. [Google Scholar]
- 21.Rokas, A., Williams, B. L., King, N. & Carroll, S. B. (2003) Nature 425**,** 798-804. [DOI] [PubMed] [Google Scholar]
- 22.Kiontke, K., Mantaro, H. & Sudhaus, W. (2002) Nematology 4**,** 933-941. [Google Scholar]
- 23.Brenner, S. (1988) in The Nematode Caenorhabditis elegans, ed. Wood, W. B. (Cold Spring Harbor Lab. Press, Cold Spring Harbor, NY), pp. ix-xiii.
- 24.Stewart, A. D. & Phillips, P. C. (2002) Genetics 160**,** 975-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobsen, A. G. & Sater, A. K. (1988) Development (Cambridge, U.K.) 104**,** 341-359. [DOI] [PubMed] [Google Scholar]
- 26.Logsdon, J. M., Jr., Stoltzfus, A. & Doolittle, W. F. (1998) Curr. Biol. 8**,** R560-R563. [DOI] [PubMed] [Google Scholar]
- 27.Tarrio, R., Rodriguez-Trelles, F. & Ayala, F. J. (2003) Proc. Natl. Acad. Sci. USA 100**,** 6580-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson, H. M. (1998) Genome Res. 8**,** 449-463. [DOI] [PubMed] [Google Scholar]
- 29.Robertson, H. M. (2000) Genome Res. 10**,** 192-203. [DOI] [PubMed] [Google Scholar]
- 30.Rogozin, I. B., Wolf, Y. I., Sorokin, A. V., Mirkin, B. G. & Koonin, E. V. (2003) Curr. Biol. 13**,** 1512-1517. [DOI] [PubMed] [Google Scholar]
- 31.Kent, W. J. & Zahler, A. M. (2000) Genome Res. 10**,** 1115-1125. [DOI] [PubMed] [Google Scholar]
- 32.Coghlan, A. & Wolfe, K. H. (2002) Genome Res. 12**,** 857-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, X. & Chamberlin, H. M. (2002) Genes Dev. 16**,** 2345-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King, M. C. & Wilson, A. C. (1975) Science 188**,** 107-116. [DOI] [PubMed] [Google Scholar]
- 35.Mushegian, A. R., Garey, J. R., Martin, J. & Liu, L. X. (1998) Genome Res. 8**,** 590-598. [DOI] [PubMed] [Google Scholar]
- 36.Aguinaldo, A. M., Turbeville, J. M., Linford, L. S., Rivera, M. C., Garey, J. R., Raff, R. A. & Lake, J. A. (1997) Nature 387**,** 489-493. [DOI] [PubMed] [Google Scholar]
- 37.Blair, J. E., Ikeo, K., Gojobori, T. & Hedges, S. B. (2002) BMC Evol. Biol. 2**,** 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giribet, G. (2003) Zoology 106**,** 303-326. [DOI] [PubMed] [Google Scholar]
- 39.Scholtz, G. (2002) Org. Divers. Evol. 2**,** 197-215. [Google Scholar]
- 40.Cho, S., Jin, S.-W., Cohen, A. & Ellis, R. E. (2004) Genome Res., in press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information