Large-Scale Analysis of the Meningococcus Genome by Gene Disruption: Resistance to Complement-Mediated Lysis (original) (raw)
Abstract
The biologic role of a majority of the Neisseria meningitidis 2100 predicted coding regions is still to be assigned or experimentally confirmed. Determining the phenotypic effect of gene disruption being a fundamental approach to understanding gene function, we used high-density signature-tagged transposon mutagenesis, followed by a large-scale sequencing of the transposon insertion sites, to construct a genome-wide collection of mutants. The sequencing results for the first half of the 4548 mutants composing the library suggested that we have mutations in 80%–90% of N. meningitidis nonessential genes. This was confirmed by a whole-genome identification of the genes required for resistance to complement-mediated lysis, a key to meningococcal virulence. We show that all the genes we identified, including four previously uncharacterized, were important for the synthesis of the polysialic acid capsule or the lipooligosaccharide (LOS), suggesting that these are likely to be the only meningococcal attributes necessary for serum resistance. Our work provides a valuable and lasting resource that may lead to a global map of gene function in N. meningitidis.
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: E. Carbonnelle, M. Joussemet, D. Caugant, M.-J. Quentin-Millet.]
The ongoing accumulation of complete genome sequences in the public databases made it clear that a genome sequencing project is a small step in the understanding of the biology of an organism. This can be illustrated by the example of Neisseria meningitidis, a human pathogen that is one of the leading causes of fatal sepsis and meningitis worldwide (Tikhomirov et al. 1997). The two complete meningococcal sequences contain an average of 2100 predicted coding regions, of which half were assigned no biologic role, whereas most of the remaining half were assigned roles that await experimental validation (Parkhill et al. 2000; Tettelin et al. 2000). The conversion of these sequence data into meaningful biologic information, by performing whole-genome experimental investigations, is therefore an important scientific challenge. Significant functional information can be acquired on a genome scale by analyzing, among other things, gene expression using DNA microarrays (Schena et al. 1996), protein expression using proteome chips (Zhu et al. 2001), protein subcellular localization using reporter genes (Whiteley et al. 1999), or protein–protein interactions using two-hybrid technologies (Rain et al. 2001). However, as shown in the model organism Saccharomyces cerevisiae, arguably the most direct approach for functional genomics is the phenotypic analysis of a collection of mutants containing mutations in each nonessential gene (Winzeler et al. 1999). Moreover, this approach helps in defining the minimal set of genes necessary to support cellular life as described in Mycoplasma, the sequenced bacterium with the smallest genomic content (Hutchison et al. 1999).
Unfortunately, the construction of an exhaustive library of mutants is an extremely laborious process that has severely hindered the general use of this approach. Indeed, a PCR-mediated gene-by-gene deletion strategy is probably beyond the scope of most individual laboratories, and is practical only when used by international consortia as demonstrated in S. cerevisiae (Winzeler et al. 1999). Saturating transposon mutagenesis is an alternative approach for obtaining a genome-wide collection of mutants. However, large libraries including numerous mutants with transposon insertions in noncoding regions are to be created, and the precise number of genes that are mutagenized remains unknown. Determining the transposon insertion sites in such a large number of mutants is usually impractical, and represents the limiting step in this approach for the rational creation of a complete library of mutants.
Taking advantage of its small genome and tractable genetics, we constructed an ordered library of 4548 signature-tagged mutants of N. meningitidis using in vitro Himar1 mariner mutagenesis (Pelicic et al. 2000). We subsequently demonstrated by a large-scale determination of the insertion sites of the transposon in half of the mutants that our library is 80%–90% complete. In addition, this was confirmed by a functional analysis of the N. meningitidis ability to resist complement-mediated lysis, a well-defined phenotypic trait particularly important for its virulence (Figueroa and Densen 1991). These results validated our library as a valuable resource for large-scale analysis of the meningococcus genome.
RESULTS
Creating a Genome-Wide Collection of N. meningitidis Transposition Mutants
Bearing in mind that the overall coding density of the N. meningitidis genome is 83% (Parkhill et al. 2000; Tettelin et al. 2000), and assuming that the selected transposon shows no preference for coding or noncoding regions, by applying the formula P = 1 − e_−_a/b (Walbot, 2000), where P is the probability that a gene is mutated, a is the number of mutants in genes, and b is the number of nonessential genes, we can estimate that a library of 4500 transposon mutants may be large enough to contain insertions within 92% of meningococcus nonessential genes. Indeed, based on recent whole-genome analyses of essential genes in Haemophilus influenzae (Akerley et al. 2002) and Staphylococcus aureus (Forsyth et al. 2002), we can assume that 70% of N. meningitidis genes, 1470 out of a total of 2100, are nonessential and can therefore be mutagenized. It should, however, be pointed out that this prediction does not take into account the length of the genes, which obviously influences their probability of being mutated.
As recently demonstrated (Pelicic et al. 2000), in vitro transposition of Himar1 mariner on meningococcal chromosomal DNA is an efficient random mutagenesis technique. We adapted the mini-transposon for signature-tagged mutagenesis (STM) (Hensel et al. 1995), an effective high-throughput screening method, by labeling it with unique identifying DNA signature tags. This allows the presence or the absence of individual mutants within a pool of mutants to be detected by PCR amplification and detection of their signature tags (Hensel et al. 1995). We used this methodology in N. meningitidis 8013, a well-characterized clinical isolate, to construct an ordered library of 4548 signature-tagged mutants that can be analyzed in 96 pools of 44–48 mutants. Southern blot analysis of several mutants (data not shown) demonstrated that they harbored single transposon insertions that were different, in agreement with a random transposition of Himar1. To enable the rational creation of a genome-wide library of mutants, we prepared and ordered on microtitre plates the chromosomal DNAs for each of the 4548 mutants of the library and started a systematic sequencing of the transposon insertion sites. For the characterization of the genomic DNA sequences flanking the transposon, we used a simple ligation-mediated PCR (LMPCR) (Prod'hom et al. 1998). Here we present the sequencing results for the first half of the library corresponding to 2281 mutants.
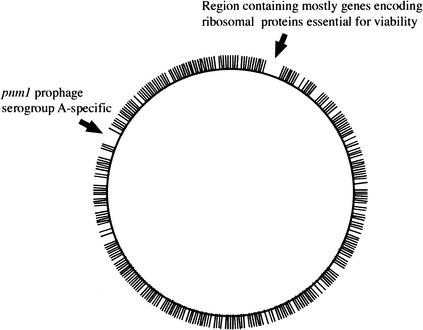
We succeeded in sequencing the transposon insertion sites in 1886 mutants or 83% of the analyzed clones (Table 1), a success rate in accord with what was previously described for LMPCR, demonstrating that this approach is feasible on a large scale, in a reasonable time period. We mapped 1557 insertion sites (Table 1) using the genome sequence of N. meningitidis Z2491 (Parkhill et al. 2000) and N. meningitidis MC58 (Tettelin et al. 2000). The 329 insertion sites that were not mapped, as much as 17% of the total, were mostly in regions that are specific to our target strain and will, therefore, be mapped once its genome sequence will be available. This tentative mapping demonstrated that transposon insertions were scattered all around the genome (Fig. 1), including 613 different genes, except for two large genomic regions of the Z2491 genome, which were devoid of inserted transposons. The first one is a 38-kb region, starting at position 100000 of the Z2491 sequence, which contains mostly genes encoding ribosomal proteins, and is therefore almost certainly essential for bacterial viability. The second is a 40-kb region, starting at position 1768000 of the Z2491 sequence, which corresponds to the prophage pnm1 known to be specific to serogroup A strains such as Z2491 (Klee et al. 2000), and therefore absent in strain 8013. Importantly, 71% of the insertions were in genes (Table 1), a rate substantially different from the coding density of the N. meningitidis genome, that is, 83%. This is consistent with what was previously reported in a similar approach in the model organism Mycoplasma (Hutchison et al. 1999), and strengthens the idea that intergenic regions are less critical for viability. This bias lowers somewhat the initial statistical prediction to 88% of meningococcus nonessential genes being mutated in our library, which is, nevertheless, expected to be useful for large-scale analysis of the N. meningitidis genome.
Table 1.
Summary of Sequenced Transposon Insertion Sites
| Insertion sites sequenced | 1886 |
|---|---|
| Insertions mapped | 1557 |
| Insertions in intergenic regions | 451 |
| Insertions in genes | 1106 |
Figure 1.
Mapping of 826 sequenced transposon insertions on the genome sequence of N. meningitidis Z2491.
Genome-Wide Identification of Serum-Sensitive Mutants
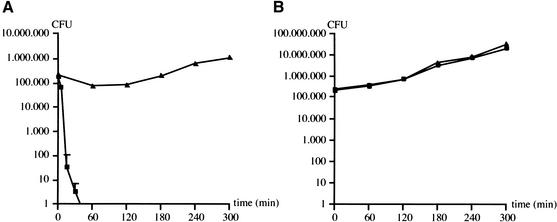
To appreciate the usefulness of our resource, we identified the genes required for resistance to complement-mediated lysis, a key virulence attribute of N. meningitidis. We chose this phenotype because the polysialic acid capsule and LOS are known to be implicated in meningococcal serum resistance (Mackinnon et al. 1993; Kahler et al. 1998), which was expected to help in the validation of our resource. To ensure optimal comparability between the results obtained in different bactericidal assays, we used a unique serum throughout this work. This serum did not harm the wild-type strain, but killed within minutes an unencapsulated siaA isogenic mutant known to be serum-sensitive (Fig. 2A). This bactericidal activity was exclusively due to the action of the complement, because mutant and wild-type strains grew equally well in heat-inactivated serum (Fig. 2B).
Figure 2.
Survival of N. meningitidis 8013 (triangles) and an isogenic unencapsulated mutant (squares) in native (A) and heat-inactivated serum (B). Error bars indicate standard errors of the means (n = 4).
We screened the 96 pools of mutants in native serum in duplicate and identified 553 serum-sensitive candidates. Such a large number of clones, already seen in another STM analysis of N. meningitidis (Sun et al. 2000), was not unexpected because meningococcus displays a vast repertoire of genes, often involved in virulence, which expression is controlled by phase variation mediated through reversible changes in the length of simple DNA repeats (Snyder et al. 2001). The phenotypes observed can, therefore, be due to phase variation events in genes unlinked to transposon insertions. To exclude this possibility, we transformed into the wild-type strain 8013 each of the 553 mutations that were selected, assembled new pools of mutants, and tested them as above. In this second-round of screening, we identified 159 candidates that presented reduced hybridization signals when their respective signature tags were amplified from the bacteria recovered after passage in human serum. However, due to the inherent limitations of the procedure used to detect the signature tags, already pointed out in other STM studies (Camacho et al. 1999), some of these mutants were expected to correspond to false positives, which prompted us to test them individually in native and heat-inactivated serum. One hundred nine mutants as well as the wild type resisted in native serum and clearly represented false positives. Thirteen mutants, which harbored transposon insertions in various housekeeping genes and genes necessary for cell wall biosynthesis, were also discarded because they grew poorly in heat-inactivated serum. It is likely that these mutants were indeed underrepresented after passage in human serum because they are less fit for growth and are therefore out-competed when grown in pools with other mutants. Finally, 37 mutants presented a reduced survival in native serum but were unaffected for growth in heat-inactivated serum, and were thus considered as serum-sensitive (Table 2). They contained transposon insertions in 18 genes or their promoters, and two intergenic regions. For both these latter mutants, the serum-sensitive phenotype is likely to be due to the effect of the inserted transposon on one of the previous 18 genes. However, the affected gene could not be unambiguously identified as the transposon insertion in the first mutant was in the _siaA_-ctrA intergenic region, whereas it was downstream the lgtE gene in the second one. Finally, we selected one mutant in each of the 18 genes and determined their survival indexes, ranging from 10−1 to 10−7 (Table 2).
Table 2.
Characteristics of the Serum-Sensitive Mutants Identified in this Study
| NMB | NMA | Number of mutants | Gene | Predicted function in databases | S.I. |
|---|---|---|---|---|---|
| Capsule biosynthesis genes | |||||
| 0069 | 1 | siaB | sialic acid biosynthesis protein | 9.20 10−2 | |
| 0070 | 3 | siaA | sialic acid biosynthesis protein | <2.72 10−7 | |
| 0071 | 0198 | 1 | ctrA | capsule export outer-membrane protein | 9.36 10−3 |
| 0072a | 0197 | 1 | ctrB | capsule export inner-membrane protein | 3.47 10−4 |
| 0073 | 0196 | 1 | ctrC | capsule export inner-membrane protein | <2.72 10−7 |
| 0074 | 0195 | 2 | ctrD | capsule export ATP-binding protein | 8.34 10−3 |
| 0082 | 0186 | 2 | lipA | capsule phospholipid modification protein | <2.72 10−7 |
| 0083 | 0185 | 5 | lipB | capsule phospholipid modification protein | <2.72 10−7 |
| — | 1 | — | siaA-ctrA intergenic region | 3.66 10−4 | |
| LOS biosynthesis genes | |||||
| 0790 | 1001 | 3 | pgm | phosphoglucomutase | 1.06 10−3 |
| 0825 | 1034 | 1 | rfaE | ADP-heptose synthetase | 2.57 10−2 |
| 0828 | 1037 | 2 | rfaD | ADP-L-_glycero_-d-_manno_-heptose epimerase | 1.39 10−1 |
| 1704 | 1958 | 3 | lgtF | β-1,4-glucosyl transferase | 8.43 10−3 |
| 1926 | 0527 | 1 | IgtE | latco-_N_-neotetraose biosynthesis glycosyl transferase | 9.00 10−3 |
| 1929 | 0524 | 4 | lgtA | lacto-_N_-neotetraose biosynthesis glycosyl transferase | 3.07 10−2 |
| —b | 1 | — | region downstream lgtE | 1.93 10−1 | |
| New genes | |||||
| 0065 | 2 | hypothetical protein | <2.72 10−7 | ||
| 0352a | 2135 | 1 | possible sugar isomerase | 4.44 10−3 | |
| 0638 | 0848 | 1 | galU | probable UTP-glucose-1-phosphate uridylyltransferase | 1.65 10−2 |
| 2076 | 0356 | 1 | possible transferase (aut family) | 8.99 10−3 |
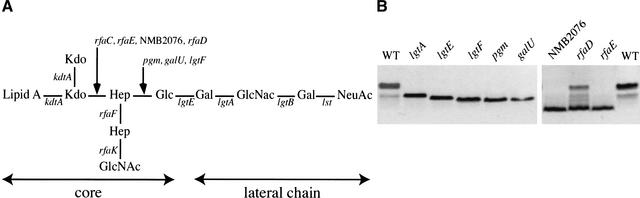
As mentionned above, meningococcal polysialic acid capsule and LOS are important for serum resistance (Mackinnon et al. 1993; Kahler et al. 1998). We were therefore not surprised to identify mutants harboring insertions in as much as 14 genes necessary for the biosynthesis of these two surface molecules (Table 2). A major result of this screen was the identification of insertions in 8 out of 10 known capsule synthesis genes that are present in the cps gene cluster and are sufficient to promote capsule expression on the Escherichia coli surface (Frosch et al. 1989). Besides underlining the efficiency of our screening procedure, this offered a functional confirmation of the completeness of our library. Regarding the LOS, the number of genes that can be identified is uncertain because some mutants, for example a mutant in the _kdt_A gene (Fig. 3A), are expected to be affected for growth and therefore probably absent from the library, whereas other mutants may be serum-resistant. This latter hypothesis was confirmed by the sequencing program that led to the identification of two additional LOS mutants, with insertions in lst and rfaF genes, which we did not identify during the screening. Individual analysis confirmed that these genes are dispensable for serum-resistance (data not shown). For the lst gene, which is necessary for LOS sialylation, this is in agreement with a previous report (Vogel et al. 1999), whereas it is surprising for the rfaF gene, which is supposed to be required for the synthesis of the LOS (Fig. 3A) (Jennings et al. 1995).
Figure 3.
Functional analysis of LOS mutants. (A) Representation of N. meningitidis 8013 LOS. (B) Silver-stained SDS-PAGE of LOS preparations with wild-type (WT) included as a control.
Functional Characterization of the Serum-Sensitive Mutants with Insertions in New Genes
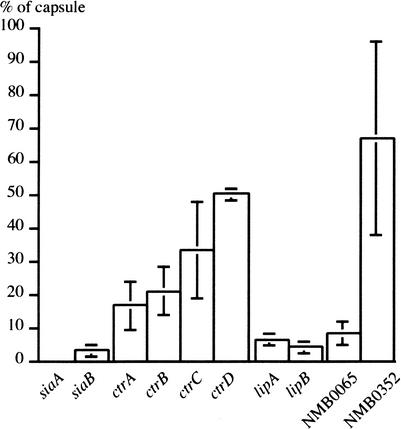
The most interesting category of mutants contained insertions within four previously uncharacterized genes of highly probable (NMB0638), possible (NMB0352 and NMB2076), or unknown function (NMB0065) (Table 2). We investigated the possibility that these genes too were involved in the synthesis of capsule and/or LOS by respectively quantifying the amount of polysialic acid they produced (Fig. 4) using a whole-cell ELISA assay (Abdillahi and Poolman 1987), and by visualizing their LOS on SDS-PAGE (Fig. 3B).
Figure 4.
Functional analysis of capsule mutants. The amount of surface-bound capsular polysaccharide relative to the wild-type was determined by whole-cell ELISA quantitation. Error bars indicate standard errors of the means (n = 3).
Mutants with transposon insertions in NMB0065 and NMB0352 presented a reduced capsule production (Fig. 4), which suggested that these genes were previously unidentified determinants of polysialic acid capsule synthesis. The NMB0065 mutant had a dramatically reduced capsule production, and can therefore be considered as unencapsulated, which was confirmed by immunofluorescence microscopy (data not shown). Interestingly, in the MC58 genome (Tettelin et al. 2000), this gene is localized within the cps gene cluster required for capsule production (Frosch et al. 1989), and is not found in the Z2491 genome, a strain that does not produce a polysialic acid capsule. A detailed PSI-BLAST analysis showed that NMB0065, which was annotated as an hypothetical protein (Tettelin et al. 2000), has homologies with neuE, a gene essential for encapsulation of E. coli K1 (Steenbergen et al. 1992). In contrast, the NMB0352 mutant had a slightly reduced capsule production (Fig. 4), which nevertheless makes the corresponding gene the first one involved in capsule biosynthesis that is not localized in the cps cluster (Frosch et al. 1989). This gene was predicted to encode a protein with a domain found in many phosphosugar isomerases and phosphosugar binding proteins, one of which is interestingly KpsF, a protein of unknown function conserved among the E. coli group II capsule gene clusters (Simpson et al. 1996).
The two remaining mutants, with transposon insertions in NMB0638 and NMB2076, presented a truncated LOS (Fig. 3B), which demonstrated that these genes are previously unidentified determinants of LOS biosynthesis. On the basis of strong sequence homologies, NMB0638 was predicted in the databases to encode GalU, an enzyme involved in the formation of UDP-glucose, which is a precursor of the biosynthesis of LOS (Kahler and Stephens 1998) (Fig. 3A). Accordingly, the LOS produced by this mutant (Fig. 3B) was as truncated as the LOS produced by a pgm mutant (Fig. 3B), which is also unable to synthesize UDP-glucose (Kahler and Stephens 1998). This is an indirect but strong indication that NMB0638 encodes GalU. On the other hand, a BLASTP analysis demonstrated that NMB2076, which was annotated as a possible transferase from the aut family, is homologous to the domain II of E. coli RfaE. In E. coli, RfaE is a bifunctional enzyme involved in the biosynthesis of ADP-L-_glycero_-D-_manno_-heptose (Valvano et al. 2000), another precursor of LOS. This observation was validated by the fact that the LOS produced by this mutant is deeply truncated as in other rfa mutants also affected in the synthesis of the core (Fig. 3B).
DISCUSSION
The release of two genomic sequences (Parkhill et al. 2000; Tettelin et al. 2000) ushered N. meningitidis into the era of postgenomics where sequence needs to be turned into function. A major challenge is to assign, or experimentally confirm, a biologic role to a majority of the 2100 meningococcal predicted coding regions. A powerful method for determining function being the phenotypic analysis of genome-wide collections of mutants, we aimed at creating such a library in N. meningitidis. Unfortunately, the two sequenced meningococcus strains are either not (Z2491) or poorly (MC58) transformable, and cannot be used for such an approach. Therefore, we chose N. meningitidis 8013, a serogroup C strain (C:22:NST:L3,7,9) initially isolated in 1989 from the blood of a patient, because it is genetically tractable and has been widely used to study meningococcal biology (Nassif et al. 1993; Deghmane et al. 2000). We describe here means for the construction of a genome-wide library in a reasonable time period, which relies, in the first place, on the use of high-density transposon mutagenesis to create an ordered library of signature-tagged mutants. We performed in vitro transposition of Himar1 mariner on N. meningitidis 8013 chromosomal DNA to construct a library of 4548 viable mutants, organized in pools of 44–48 mutants that can be assayed simultaneously. This step is followed, in a second phase, by the sequencing of the transposon insertion sites in all the mutants. Importantly, we show that LMPCR (Prod'hom et al. 1998) is a practical means for this otherwise often limiting step. We present the results of the sequencing analysis of the first half of the library, corresponding to the mutants of the pools 1 to 48. One thousand eight hundred eighty-six transposon insertion sites, in 83% of the analyzed mutants, were successfully sequenced. We can therefore estimate that approximately 3761 insertion sites could ultimately be sequenced using this reasonably practical method. If necessary, the sequencing of the remaining insertion sites may be done using conventional methods, usually more labor-intensive, such as the construction of libraries containing the junctions between the transposon and the chromosome, as demonstrated in Mycoplasma (Hutchison et al. 1999).
Seventeen percent of the sequenced insertion sites could not be mapped using the available heterologous genome sequences. This is consistent with the fact that as much as 17% of N. meningitidis gene content may be strain-specific (Parkhill et al. 2000; Tettelin et al. 2000). This makes it clear that the genome sequence of the target strain is indispensable for an exhaustive exploitation and precise interpretation of the sequencing results, which prompted us to start sequencing the genome of N. meningitidis 8013 (unpublished data). Nevertheless, although it clearly leads to an imperfect estimation of the efficiency of our approach, the mapping of the insertion sites on the available sequences demonstrated that transposition was random. However, it should be pointed out that there was a substantial bias towards transposition in intergenic regions, a phenomenom already observed in a similar global transposon mutagenesis approach performed in Mycoplasma (Hutchison et al. 1999), which should therefore be taken into account in the initial calculation. Therefore, when a library of 4500 transposition mutants is available for an organism with a genome size and coding density similar to N. meningitidis, it can be estimated that LMPCR may lead to the sequencing of 3721 transposon insertion sites, of which 2642 may be in genes. This number is high enough to provide an 83% saturation in nonessential genes (Walbot 2000), which is supported both by our sequencing data and our functional analysis. Therefore, the modest increase in saturation expected to result from the sequencing of the remaining insertion sites, which would require the use of laborious methods, may not justify such a profusion of efforts. Instead, it may be preferable to construct directly the missing mutants, corresponding probably to small genes that were less likely to be mutagenized.
We illustrated the validity of this resource for functional genomics, by identifying the genes required for serum-resistance, a phenotype central to meningococcal virulence never assayed before by random mutagenesis. Although the functional analysis of virulence phenotypes in N. meningitidis is complicated by the fact that the expression of numerous virulence factors can be switched on and off through changes in the length of simple DNA repeats (Snyder et al. 2001), the identification of 37 serum-sensitive mutants confirmed that this approach is valid. These mutants were characterized for the synthesis of the polysialic acid capsule and LOS, which demonstrated that the 18 genes that were disrupted were all involved in the synthesis of these two molecules. Besides underlining the efficiency of our screening procedure, the identification of 8 of 10 genes that are necessary for the production of the capsule (Frosch et al. 1989) is a functional confirmation that our library contains mutants in approximately 80% of meningococcus nonessential genes. It is therefore very likely that capsule and LOS are the only meningococcal attributes necessary for serum resistance. Furthermore, our analysis assigned a role in LOS or capsule biosynthesis, the biosynthesis of which has been extensively studied in the past 13 years, to four previously uncharacterized genes. For example, we show that NMB2076 is necessary for the synthesis of the LOS and presents homologies to domain II of RfaE, the E. coli ADP-heptose synthetase. Surprisingly, NMB0825, which was also identified during this analysis, was annotated as rfaE in the databases (Parkhill et al. 2000; Tettelin et al. 2000). This apparent discrepancy was explained by a detailed BLASTP analysis that demonstrated that these two proteins are indeeed homologous to the bifunctional E. coli RfaE enzyme (Valvano et al. 2000), but to the first and second domain, respectively. This not only shows that the two domains are genetically separated in N. meningitidis, but also suggests that the actual ADP-heptose synthetase function, catalyzed by the second domain of the E. coli RfaE protein, is not encoded by NMB0825 but rather by NMB2076.
In summary, a large-scale analysis of the N. meningitidis genome allowed us to revisit serum-resistance, an “old” phenotype essential for pathogenicity, and helped us to demonstrate that we have an extensive ordered collection of meningococcal mutants that is a good starting point for the creation of a collection containing mutants with insertions in each nonessential gene. We expect this resource to be valuable for the production of a global map of gene function, and the definition of the minimal set of genes necessary for meningococcal life.
METHODS
Bacterial Strains and Growth Conditions
N. meningitidis 8013, a serogroup C strain (C:22:NST:L3,7,9), was used in this study. As a serum-sensitive control, we used an unencapsulated siaA isogenic mutant that we generated by allelic exchange as described (Edwards et al. 1994). N. meningitidis was grown on GCB medium and E. coli was grown on Luria Bertani medium. When required, kanamycin was added at 20 and 100 μg/mL for E. coli and N. meningitidis, respectively. Transformation of N. meningitidis was done as previously described (Pelicic et al. 2000).
Construction of the Library of Mutants
We previously described the _mariner_-based in vitro transposition system for N. meningitidis (Pelicic et al. 2000). For this study, the mini-transposon, which consists of Himar1 inverted repeats flanking a kanamycin resistance cassette and a neisserial uptake sequence, was adapted for STM (Hensel et al. 1995) by cloning 80-bp oligonucleotide signature tags, with _Sal_I restriction sites at their extremities, within a _Sal_I site internal to the transposon. Forty-eight tagged transposons, each labeled with a different signature tag, were selected and used to produce 48 mutant libraries. Ninety-six mutants of each library were organized on microtiter plates that were immediately frozen at −80°C in 20% glycerol. This was done in such a way that the same wells from different microtiter plates contained different mutants with the same signature tag, whereas mutants from different wells within the same microtiter plate harbored different signature tags. This original library was replicated, and the replica, consisting of 4548 viable mutants, was used throughout this study.
Characterization of Transposon Insertion Sites
The identification of genomic DNA sequences flanking the inserted transposons was done by LMPCR (Prod'hom et al. 1998) as described (Pelicic et al. 2000). We first prepared chromosomal DNAs for each of the 4548 viable mutants of the library and ordered them on microtiter plates in the same order as the corresponding mutants of the original library. To facilitate the precise identification of the transposon insertion site, the only change to the published method consisted in the sequencing with outward-reading primers internal to the mini-transposon, ISR (5′-CGCTCTTGAAGGGAACTATGT TGA-3′) or ISL (5′-AATCATTTGAAGGTTGGTACTATA-3′), instead of the LMP1 primer located in the linker that was previously used (Pelicic et al. 2000). Sequence homology searches were performed using BLASTN against the meningococcal sequences present in the databases at TIGR (http://tigrblast.tigr.org/cmr-blast/) and the Sanger Center (http://www.sanger.ac.uk/Projects/N_meningitidis/blast_server.shtml). When searches were positive, there was in general 90%–100% identity with sequences in the databases, especially in the immediate vicinity of the insertion. Negative searches did not return any homologous sequences, indicating that the insertions occurred in regions specific to our target strain. Where indicated, the corresponding proteins were compared, using BLASTP and PSI-BLAST programs, with the protein sequences in the NCBI (http://www.ncbi.nlm.nih.gov/BLAST/) and PRODOM (http://prodes.toulouse.inra.fr/prodom/2002.1/ html/home.php) databases.
Serum Bactericidal Assays
Blood from several individuals with hereditary haemochromatosis, which are treated by frequent phlebotomies (Provan and Weatherall 2000), was obtained on a regular basis from Prof. M. Joussemet (Centre de Transfusion Sanguine des Armées). The serum from this blood that is otherwise discarded, was purified by three rounds of centrifugation at 3000 × g for 15 min at 4°C, divided into aliquots, and immediately stored at −80°C. Laboratory tests showed that serum composition was normal, except for a ferritin concentration of approximately 2,000 μg/L. Mutants to be tested were grown on GCB agar plates overnight and colonies were inoculated in 5 mL of RPMI and grown for 2 h at 37°C. Optical density (OD) was read at 600 nm and adjusted at 0.1 in RPMI. Serum, RPMI and bacteria were mixed in four-well NUNC plates (POLYLABO) so that the final 400 μL suspension contained 2 × 105 colony-forming units (CFU) in 25% serum (vol/vol). A control reaction was performed in the same conditions with serum decomplemented by heating at 56°C for 30 min. The reactions, performed at least in triplicate, were incubated at 37°C in an atmosphere containing 5% CO2. Twenty-microliter samples were taken from the reaction mixtures at different time points, and serial dilutions were plated on GCB. Colony counts were performed after overnight incubation of the plates and the number of CFU was calculated at each sampling time. Data were expressed as a survival index, defined as the ratio of mutant-to-wild-type CFU at a defined time, corrected by the initial ratio of mutant-to-wild-type CFU.
When pools of mutants were assayed, clones were resuspended individually in RPMI on microtitre plates, OD600 was measured on a microtiter plate reader, and equivalent amounts of each mutant were pooled. After 2 h of growth at 37°C, the OD600 was measured and adjusted to 0.5 in RPMI. Serum, RPMI, and bacteria were mixed in four-well NUNC plates so that the final 400-μL suspension contained 106 CFU in 25% serum (vol/vol), or approximately 2 × 104 CFU of each mutant. The reactions, performed in duplicate, were incubated at 37°C for 5 h in an atmosphere containing 5% CO2. We chose a 5-h incubation time to avoid a bias towards the most serum resistant meningococci being selected, which is much more likely to happen at shorter incubation times. More than 104 CFU were collected after overnight incubation of serial dilutions plated on GCB and subjected to signature tag amplification.
Signature Tag Manipulation and Hybridization
Chromosomal DNA was prepared from isolated or pooled meningococci using the Wizard Genomic DNA Purification kit (Promega) following the manufacturer's recommendations. Signature tag amplification, labeling, and hybridization were done as described (Camacho et al. 1999).
Capsule and LOS Analysis
Serogroup C capsular polysaccharide was detected using the Pastorex Meningitis 61160 kit (Biorad). It was quantified by whole-cell ELISA as described (Abdillahi and Poolman 1987), using a serogroup C capsular polysaccharide-specific mouse serum that was generously donated by M.-J. Quentin-Millet (Aventis Pasteur). LOS was prepared from standard numbers of CFU by a proteinase K treatment (Hitchcock and Brown 1983), resolved on 16% tricine SDS-PAGE gels (Lesse et al. 1990), and visualized by silver staining (Tsai and Frasch 1982).
WEB SITE REFERENCES
http://tigrblast.tigr.org/cmr-blast; BLAST on TIGR comprehensive microbial resource.
http://www.sanger.ac.uk/Projects/N_meningitidis/blast_server.shtml; BLAST on N. meningitidis genome sequences at the Sanger Institute.
http://www.ncbi.nlm.nih.gov/BLAST/; BLAST on the NCBI databases.
http://prodes.toulouse.inra.fr/prodom/2002.1/html/home.php; ProDom database of protein domain families.
Acknowledgments
We thank E. Carbonnelle for delivery of blood generously donated by Prof. M. Joussemet (Centre de Transfusion Sanguine des Armées, France), D. Caugant (National Institute of Public Health, Norway) for serologic typing of N. meningitidis 8013 strain, and M.-J. Quentin-Millet (Aventis Pasteur, France) for the gracious gift of serogroup C capsular polysaccharide-specific serum. We are grateful to L.R. Camacho for helpful discussions, J.-M. Reyrat for help with protein homology searches, and S. Nair, C. Recchi, and P. Trieu-Cuot for critical reading of the manuscript. This work was funded by INSERM and Université Paris V.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E MAIL pelicic@necker.fr; FAX 33 1 40 61 55 92.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.664303\. Article published online before print in February 2003.
REFERENCES
- 1.Abdillahi H. and Poolman, J.T. 1987. Whole-cell ELISA for typing Neisseria meningitidis with monoclonal antibodies. FEMS Microbiol. Lett. 48**:** 367-371. [PubMed] [Google Scholar]
- 2.Akerley B.J., Rubin, E.J., Novick, V.L., Amaya, K., Judson, N., and Mekalanos, J.J. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. 99**:** 966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camacho L.R., Ensergueix, D., Perez, E., Gicquel, B., and Guilhot, C. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34**:** 257-267. [DOI] [PubMed] [Google Scholar]
- 4.Deghmane A.E., Petit, S., Topilko, A., Pereira, Y., Giorgini, D., Larribe, M., and Taha, M.K. 2000. Intimate adhesion of Neisseria meningitidis to human epithelial cells is under the control of the crgA gene, a novel LysR-type transcriptional regulator. EMBO J. 19**:** 1068-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards U., Müller, A., Hammerschmidt, S., Gerardy-Schahn, R., and Frosch, M. 1994. Molecular analysis of the biosynthesis pathway of the α-2,8 polysialic acid capsule by Neisseria meningitidis serogroup B. Mol. Microbiol. 14**:** 141-149. [DOI] [PubMed] [Google Scholar]
- 6.Figueroa J.E. and Densen, P. 1991. Infectious diseases associated with complement deficiencies. Clin. Microbiol. Rev. 4**:** 359-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forsyth R.A., Haselbeck, R.J., Ohlsen, K.L., Yamamoto, R.T., Xu, H., Trawick, J.D., Wall, D., Wang, L., Brown-Driver, V., Froelich, J.M., et al. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43**:** 1387-1400. [DOI] [PubMed] [Google Scholar]
- 8.Frosch M., Weisgerber, C., and Meyer, T.F. 1989. Molecular characterization and expression in Escherichia coli of the gene complex encoding the polysaccharide capsule of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. 86**:** 1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hensel M., Shea, J.E., Gleeson, C., Jones, M.D., Dalton, E., and Holden, D.W. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269**:** 400-403. [DOI] [PubMed] [Google Scholar]
- 10.Hitchcock P.J. and Brown, T.M. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154**:** 269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutchison C.A., Peterson, S.N., Gill, S.R., Cline, R.T., White, O., Fraser, C.M., Smith, H.O., and Venter, J.C. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286**:** 2165-2169. [DOI] [PubMed] [Google Scholar]
- 12.Jennings M.P., Bisercic, M., Dunn, K.L., Virji, M., Martin, A., Wilks, K.E., Richards, J.C., and Moxon, E.R. 1995. Cloning and molecular analysis of the lsi1 (rfaF) gene of Neisseria meningitidis which encodes a heptosyl-2-transferase involved in LPS biosynthesis: Evaluation of surface exposed carbohydrates in LPS mediated toxicity for human endothelial cells. Microb. Pathog. 19**:** 391-407. [DOI] [PubMed] [Google Scholar]
- 13.Kahler C.M. and Stephens, D.S. 1998. Genetic basis for biosynthesis, structure, and function of meningococcal lipooligosaccharide (endotoxin). Crit. Rev. Microbiol. 24**:** 281-334. [DOI] [PubMed] [Google Scholar]
- 14.Kahler C.M., Martin, L.E., Shih, G.C., Rahman, M.M., Carlson, R.W., and Stephens, D.S. 1998. The (α2→8)-linked polysialic acid capsule and lipooligosaccharide structure both contribute to the ability of serogroup B Neisseria meningitidis to resist the bactericidal activity of normal human serum. Infect. Immun. 66**:** 5939-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klee S.R., Nassif, X., Kusecek, B., Merker, P., Beretti, J.L., Achtman, M., and Tinsley, C.R. 2000. Molecular and biological analysis of eight genetic islands that distinguish Neisseria meningitidis from the closely related pathogen Neisseria gonorrhoeae. Infect. Immun. 68**:** 2082-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lesse A.J., Campagnari, A.A., Bittner, W.E., and Apicella, M.A. 1990. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Immunol. Methods 126**:** 109-117. [DOI] [PubMed] [Google Scholar]
- 17.Mackinnon F.G., Borrow, R., Gorringe, A.R., Fox, A.J., Jones, D.M., and Robinson, A. 1993. Demonstration of lipooligosaccharide immunotype and capsule as virulence factors for Neisseria meningitidis using an infant mouse intranasal infection model. Microb. Pathog. 15**:** 359-366. [DOI] [PubMed] [Google Scholar]
- 18.Nassif X., Lowy, J., Stenberg, P., O'Gaora, P., Ganji, A., and So, M. 1993. Antigenic variation of pilin regulates adhesion of Neisseria meningitidis to human epithelial cells. Mol. Microbiol. 8**:** 719-725. [DOI] [PubMed] [Google Scholar]
- 19.Parkhill J., Achtman, M., James, K.D., Bentley, S.D., Churcher, C., Klee, S.R., Morelli, G., Basham, D., Brown, D., Chillingworth, T., et al. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404**:** 502-506. [DOI] [PubMed] [Google Scholar]
- 20.Pelicic V., Morelle, S., Lampe, D., and Nassif, X. 2000. Mutagenesis of Neisseria meningitidis by in vitro transposition of Himar1 mariner. J. Bacteriol. 182**:** 5391-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prod'hom G., Lagier, B., Pelicic, V., Hance, A.J., Gicquel, B., and Guilhot, C. 1998. A reliable amplification technique for the characterization of genomic DNA sequences flanking insertion sequences. FEMS Microbiol. Lett. 158**:** 75-81. [DOI] [PubMed] [Google Scholar]
- 22.Provan D. and Weatherall, D. 2000. Red cells II: Acquired anaemias and polycythaemia. Lancet 355**:** 1260-1268. [DOI] [PubMed] [Google Scholar]
- 23.Rain J., Selig, L., De Reuse, H., Battaglia, V., Reverdy, C., Simon, S., Lenzen, G., Petel, F., Wojcik, J., Schachter, V., et al. 2001. The protein–protein interaction map of Helicobacter pylori. Nature 409**:** 211-215. [DOI] [PubMed] [Google Scholar]
- 24.Schena M., Shalon, D., Heller, R., Chai, A., Brown, P.O., and Davis, R.W. 1996. Parallel human genome analysis: Microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. 93**:** 10614-10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson D.A., Hammarton, T.C., and Roberts, I.S. 1996. Transcriptional organization and regulation of expression of region 1 of the Escherichia coli K5 capsule gene cluster. J. Bacteriol. 178**:** 6466-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder L.A.S., Butcher, S.A., and Saunders, N.J. 2001. Comparative whole-genome analyses reveal over 100 putative phase-variable genes in the pathogenic Neisseria spp. Microbiology 147**:** 2321-2332. [DOI] [PubMed] [Google Scholar]
- 27.Steenbergen S.M., Wrona, T.J., and Vimr, E.R. 1992. Functional analysis of the sialyltransferase complexes in Escherichia coli K1 and K92. J. Bacteriol. 174**:** 1099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y.H., Bakshi, S., Chalmers, R., and Tang, C.M. 2000. Functional genomics of Neisseria meningitidis pathogenesis. Nat. Med. 6**:** 1269-1273. [DOI] [PubMed] [Google Scholar]
- 29.Tettelin H., Saunders, N.J., Heidelberg, J., Jeffries, A.C., Nelson, K.E., Eisen, J.A., Ketchum, K.A., Hood, D.W., Peden, J.F., Dodson, R.J., et al. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287**:** 1809-1815. [DOI] [PubMed] [Google Scholar]
- 30.Tikhomirov E., Santamaria, M., and Esteves, K. 1997. Meningococcal disease: Public health burden and control. World Health Stat. Q. 50**:** 170-177. [PubMed] [Google Scholar]
- 31.Tsai C.M. and Frasch, C.E. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119**:** 115-119. [DOI] [PubMed] [Google Scholar]
- 32.Valvano M.A., Marolda, C.L., Bittner, M., Glaskin-Clay, M., Simon, T.L., and Klena, J.D. 2000. The rfaE gene from Escherichia coli encodes a bifunctional protein involved in biosynthesis of the lipopolysaccharide core precursor ADP-L-_glycero_-D-_manno_-heptose. J. Bacteriol. 182**:** 488-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogel U., Claus, H., Heinze, G., and Frosch, M. 1999. Role of lipopolysaccharide sialylation in serum resistance of serogroup B and C meningococcal disease isolates. Infect. Immun. 67**:** 954-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walbot V. 2000. Saturation mutagenesis using maize transposons. Curr. Opin. Plant Biol. 3**:** 103-107. [DOI] [PubMed] [Google Scholar]
- 35.Whiteley M., Lee, K.M., and Greenberg, E.P. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 96**:** 13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winzeler E.A., Shoemaker, D.D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J.D., Bussey, H., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285**:** 901-906. [DOI] [PubMed] [Google Scholar]
- 37.Zhu H., Bilgin, M., Bangham, R., Hall, D., Casamayor, A., Bertone, P., Lan, N., Jansen, R., Bidlingmaier, S., Houfek, T., et al. 2001. Global analysis of protein activities using proteome chips. Science 293**:** 2101-2105. [DOI] [PubMed] [Google Scholar]