Segmental Duplications in Euchromatic Regions of Human Chromosome 5: A Source of Evolutionary Instability and Transcriptional Innovation (original) (raw)
Abstract
Recent analyses of the structure of pericentromeric and subtelomeric regions have revealed that these particular regions of human chromosomes are often composed of blocks of duplicated genomic segments that have been associated with rapid evolutionary turnover among the genomes of closely related primates. In the present study, we show that euchromatic regions of human chromosome 5—5p14, 5p13, 5q13, 5q15–5q21—also display such an accumulation of segmental duplications. The structure, organization and evolution of those primate-specific sequences were studied in detail by combining in silico and comparative FISH analyses on human, chimpanzee, gorilla, orangutang, macaca, and capuchin chromosomes. Our results lend support to a two-step model of transposition duplication in the euchromatic regions, with a founder insertional event at the time of divergence between Platyrrhini and Catarrhini (25–35 million years ago) and an apparent burst of inter- and intrachromosomal duplications in the Hominidae lineage. Furthermore, phylogenetic analysis suggests that the chronology and, likely, molecular mechanisms, differ regarding the region of primary insertion—euchromatic versus pericentromeric regions. Lastly, we show that as their counterparts located near the heterochromatic region, the euchromatic segmental duplications have consistently reshaped their region of insertion during primate evolution, creating putative mosaic genes, and they are obvious candidates for causing ectopic rearrangements that have contributed to evolutionary/genomic instability.
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: D. Le Paslier, A. McKenzie, J. Melki, C. Sargent, J. Scharf and S. Selig.]
There is compelling evidence that gene duplication and transposition events have played essential roles during vertebrate evolution (Brosius 1999a,b; Patthy 1999; Long 2001; Friedman and Hughes 2001a; McLysaght et al. 2002). However, the importance of such evolutionary mechanisms is not restricted to ancient times. The initial sequencing and analysis of the human genome, in combination with previously published reports, have revealed that our genome is constituted of a remarkably complex pattern of both ancient and recent duplications (Lander et al. 2001; Bailey et al. 2002a). It is now estimated that ∼5% (probably more) of our genetic material is composed of duplicated genomic segments that have emerged during the past 35 million years of primate evolution. These blocks (termed segmental duplications) range in size from a few kilobases to hundreds of kilobases, and share a high degree of sequence identity (>90%). In contrast to whole-genome polyploidization events, segmental duplications have originated from the duplicative transpositions of small portions of chromosomal material, often containing intron-exon structure of known genes, and tend to be localized in pericentromeric and subtelomeric regions (for review, see Eichler 2001; Samonte and Eichler 2002). Preliminary analyses indicate that these particular regions of the genome have experienced extraordinary rates of evolutionary turnover, which result in considerable structural change and rapid gene innovation in the genomes of man and great apes (for review, see Nahon 2002; Samonte and Eichler 2002).
In earliest studies (Viale et al. 1998, 2000; Courseaux and Nahon 2001), we have seen that recently duplicated segments of the human chromosome 5 (HSA5) were harboring two novel chimeric genes PMCHL1 and PMCHL2, whose mRNA or protein have been exapted (Gould and Vrba 1982) into a new functional role during the last stages of primate evolution. These genes were shown to have been created by a complex mechanism of exon shuffling through retrotransposition of an antisense MCH-messenger RNA (AROM mRNA) coupled to de novo creation of splice sites, followed by a last event of duplication involving a large chromosomal region (Courseaux and Nahon 2001). In the course of the characterization of the PMCHL genes, we established that PMCHL2 was located on chromosome band 5q13, proximal to the predisposition locus of the spinal muscular atrophy (SMA)—an autosomal recessively inherited neurodegenerative disorder with variable clinical severity (Munsat et al. 1990; Viale et al. 2000). Interestingly, this region of the human genome was shown to harbor a class of transcribed pseudogenes/gene-derived sequences that are similar in their organization to the PMCHL genes (Sargent et al. 1994; Theodosiou et al. 1994; Selig et al. 1995). These sequences are not typical processed pseudogenes, but rather contain introns and exons with related copies on both the q and p arms of human chromosome 5, and show at least 90% nucleic acid sequence identity to functional genes located elsewhere in the genome. A preliminary PCR analysis of the DNA of different primate species revealed that those sequences have emerged during the last 35 million years (A. Viale and C. Ortola, unpubl.). Such a clustering of recent transcribed pseudogenes in duplicated genomic regions is reminiscent of the characteristics of primate-specific segmental duplications (Bailey et al. 2002b; Crosier et al. 2002) and raise questions about when these sequences have arisen, how they were maintained, and the role that they may have played in chromosome and genome evolution.
To address these questions, we first focus on three types of gene-derived sequences evidenced by different laboratories during their attempts to identify potential candidate genes for SMA disease as follows: (1) Psdex4 and c41-cad, two sequences derived from the CDH12 gene, a neuronal cadherin gene residing on 5p14.3. They were shown to be almost identical to CDH12 exons 4 and 6 and surrounding intronic sequences, respectively (Selig et al. 1995). (2) Glu5–10, a sequence that exhibits high-sequence similarity to exons 5, 9, 10, and adjacent intronic sequences of the β-glucuronidase gene (GUSB), which is located on chromosome band 7q11.21 (Sargent et al. 1994; Theodosiou et al. 1994). By combining FISH and in silico analyses, we reveal the series of transposition and inter- and intrachromosomal duplication events that have led to the creation and spreading of the GUSB and _CDH12_-derived sequences. Many of the recently duplicated segments demonstrated to date, were shown to be associated with recurrent chromosomal structural rearrangements and generation of gene diversity (Samonte and Eichler 2002). We thus investigate the potential of human chromosome 5 euchromatic segmental duplications (1) to be involved in the chromosomal/evolutionary instability associated with the predisposition locus of the SMA disease (Lewin 1995), and (2) to evolve novel transcripts by examining the duplicated regions for evidence that a transposition and/or duplication event has generated a new transcript.
RESULTS
Clustering of Gene-Derived Sequences in Euchromatic Regions of HSA5
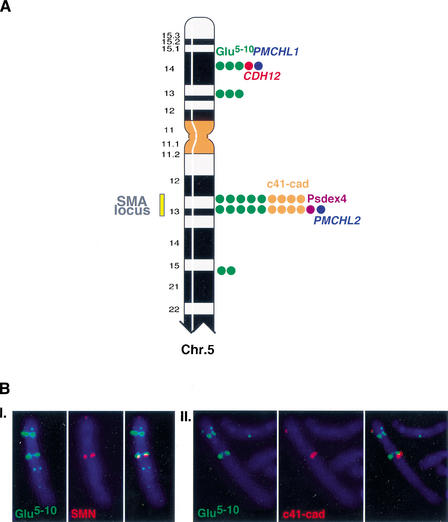
To first determine the precise chromosomal localization of all the paralogous copies of the gene segments, we used a combination of mono- and dual-color FISH experiments on human metaphase chromosomes. This analysis revealed that the _CDH12_- and _GUSB_-derived sequences were present in several copies on different loci of the human chromosome 5, and that they were colocalized with PMCHL1 and PMCHL2 on both 5p14 and 5q13 (Fig. 1A,B). Interestingly, the hybridization signals obtained with c41-cad and Glu 5–10 on chromosome 5 were shown to vary between individuals (data not shown). These observations suggest that the copy number of the duplicated fragments on chromosome 5 is highly polymorphic, as proposed previously (Sargent et al. 1994; Theodosiou et al. 1994).
Figure 1.
(A) Colocalization of the gene-derived sequences on human chromosome 5. The FISH hybrization signals observed on human chromosome 5 with gene segment-specific probes are represented by colored dots. (Blue) PMCHL genes; (green) Glu 5–10; (red) Br-cadherin gene (CDH12); (orange) c41-cad; (purple) Psdex4. The number of dots is proportional to the signal intensity, which was scored on a scale of from 0–5. The yellow bar delimits the predisposition locus to the SMA disease. (B) Illustration of some dual-color FISH experiments. (I.) Glu 5–10 biotin-labeled (5F10R probe)/SMN digoxigenin-labeled (132SE23 probe, specific to the SMN gene in the SMA locus), (II.) Glu 5–10 biotin-labeled (5F10R probe)/c41-cad digoxigenin-labeled (Φ-98-1 probe). The hybridization signals observed on 5p with the c41-cad probe are due to cross-hybridization with the genuine CDH12 gene.
Emergence and Spreading of Gene-Derived Sequences During Primate Evolution
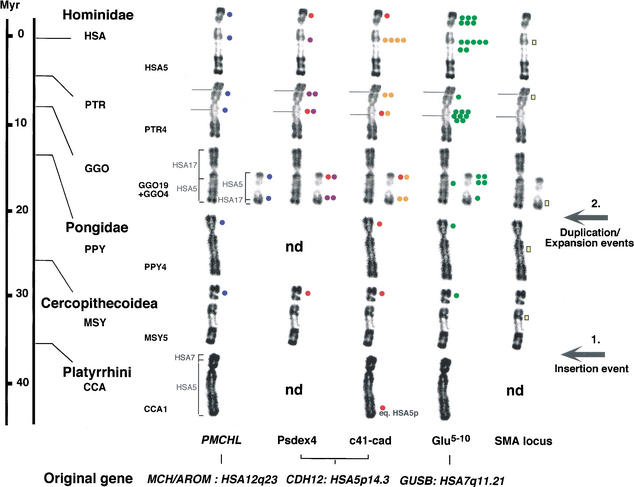
To provide further insights into the evolutionary mechanisms that have generated such a genomic diversity and gene-segment clustering, we extended our FISH analysis to other primates. The metaphase chromosomes of six species were compared (Homo sapiens [HSA], Pan troglodytes [PTR], Gorilla gorilla [GGO], Pongo pygmaeus [PPY], Macaca sylvana [MSY], and Cebus capucinus [CCA]). The FISH hybridization signals suggested that the mechanism responsible for the emergence and spreading of the gene-variant sequences comprised at least two distinct steps (Fig. 2). First, transposition events from the ancestral chromosomal regions equivalent to HSA12q23 and HSA7q21.11 have led to the insertion of PMCHL1 and Glu 5–10, respectively, on the ancestral chromosomal segment equivalent to the short arm of HSA5, 25–35 million years ago (Mya) before the divergence of Catarrhini. This was followed by a burst of duplication events, which operated in the Hominidae lineage within the last 5–10 million years, and which led to their distribution/expansion onto various loci of the ancestral chromosome equivalent to HSA5. The history of the spreading of the _CDH12_-derived sequences is quite different, as the founder event was more likely to be operated only at the time of divergence of humans and African great apes, about 10 Mya. Although we could not firmly exclude the presence of copies of Psdex4 and c41-cad on the equivalent of HSA5p in PPY, MSY, and CCA, we hypothesized that the hybridization signals observed were due to cross-hybridization with the homologous functional CDH12 gene that is present on the band equivalent to HSA5p14 (Selig et al. 1997).
Figure 2.
Summary of the in situ hybridization results of the gene-derived sequences/SMA genes on human and other primate chromosomes equivalent to human chromosome 5 (HSA5). The scale indicates the generally accepted times of divergence of Anthropoids from the human lineage (Hacia 2001). On the basis of the phylogenetic classification of Goodman (1999), we placed Pongo in the family Pongidae and grouped Gorilla, Pan, and Homo in the family Hominidae. The same color code as in Figure 1 was used. The number of dots indicates the average signal intensity observed at a given location with a particular clone. Red dots on Psdex4 and c41-cad hybridization patterns indicate cross-hybridization with the genuine CDH12 gene. The names and chromosomal localizations of the original genes, whose gene segments derived from are indicated at bottom. The yellow bar indicates hybrization signals obtained with two genes (SMN and NAIP) specific to the SMA locus. The position of the homologs to human SMA locus is indicated as a chromosomal index of the region equivalent to HSA 5q13. Brackets indicate the homology between HSA and other primate chromosomes. For Cebus capucinus (CCA), CCA1 is formed by the equivalent of the whole HSA5, plus a small segment of HSA7 (Richard et al. 1996). For Macaca sylvana (MSY) and Pongo pygmaeus (PPY), the equivalent to HSA5 (MSY5 and PPY4, respectively) have a banding pattern very similar to HSA5 (Dutrillaux 1979). For Gorilla gorilla (GGO), GGO4 and GGO19 are the products of a reciprocal translocation between ancestral chromosomes equivalent to HSA5 and HSA17, respectively (Dutrillaux et al. 1973). Evolutionary breakpoints occurred in regions equivalent to HSA5q13.3 and HSA17p12 (Stankiewicz et al. 2001). Brackets indicate the homology between GGO and HSA chromosomes. For Pan troglodytes (PTR), PTR4 differs from HSA5 by a pericentromeric inversion, inv(5)(p14.3;q13.3) (Yunis and Prakash 1982; Marzella et al. 1997). Grey lines on PTR chromosomes indicate the pericentromeric inversion breakpoints.
To further address the question of the original emergence of the _CDH12_-derived sequences, we performed an in silico analysis of the genomic structure of the human CDH12 gene. A BLAST search against the HTGS and nr databases of GenBank allowed us to identify several clones in HTGS phase encompassing the CDH12 locus. The analysis of the draft sequence revealed that the neuronal cadherin gene spanned a larger region than suspected previously (>1 Mb) (Selig et al. 1997). Interestingly, both PMCHL1 and a _GUSB_-derived sequence were detected inside _CDH12_-intronic sequences in the vicinity of corresponding exons 4 and 6, respectively. PMCHL and _CDH12_-exon 4-derived sequences linkage was established by both (1) PCR amplification from BAC clones bearing either the PMCHL1 locus on 5p14 (CIT-HSP283L20) or the PMCHL2 locus on 5q13 (CIT-HSP811M22, CIT-HSP 484D2), which we analyzed previously (Courseaux and Nahon 2001), and (2) sequence comparison between clones AC091956/AC092358 and AC108106 in HTGS phase-specific 5p and 5q loci, respectively. This analysis led us to conclude that both PMCHL2 and Psdex4 have arisen, 5–10 Mya (Fig. 2) through an intra-chromosomal duplication event that involved a large chromosomal region equivalent to HSA5p14, encompassing more than 100 kilobases. Similarly, the link between a _GUSB_-derived sequence and the exon 6 of CDH12 strongly suggested that c41-cad have arisen through the duplication of a large genomic region equivalent to HSA5p14. The additional copies of Psdex4 and c41 detected, particularly on the equivalent of HSA5p in PTR and GGO, were probably generated by further rearrangements after the divergence between African great apes and human lineages.
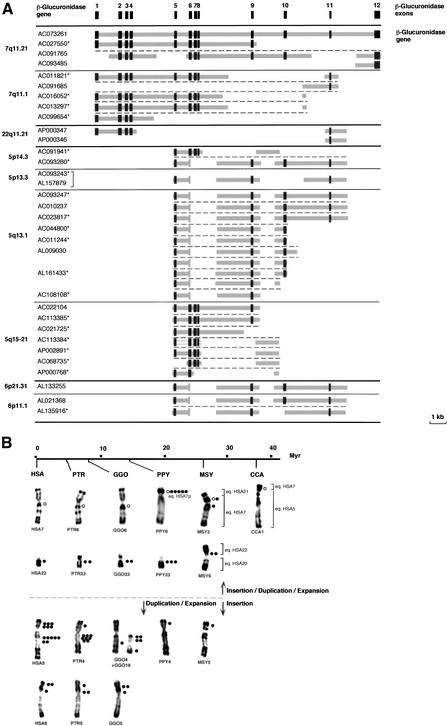
To refine the evolutionary scenario responsible for the appearance and spreading of Glu 5–10, structural analysis of _GUSB_-derived paralogous sequences was performed by an in silico analysis of the human genome draft sequence. This study revealed the existence of several classes of unprocessed pseudogenes that share a high degree (90%–95%) of identity with different parts of the GUSB genomic locus (Fig. 3A). The genomic distribution of those sequences was in agreement with our previous FISH analysis. The Glu 5–10 probe strongly hybridized on chromosome 5 (5p14, 5p13, 5q13, 5q15) (Figs. 2 and 3B). Additional hybridizing loci were found at 6p11 and 6p21, and further weak signals were also detected on 22q11 and 7q11 (Fig. 3B). The chronology of emergence and spreading of the _GUSB_-derived sequences was evidenced from the chromosomal distribution of the related sequences found in the different primate species. As illustrated in Figure 3B, the hybridization signals on primate chromosomes suggested that after Platyrrhini divergence—about 35 Mya—the ancestral _GUSB_-genomic locus served as a donor hot spot for transposition to the ancestral chromosomal regions homologous to HSA5p and to pericentromeric regions of HSA7 and HSA22. According to the present genomic organization of the _GUSB_-paralogous sequences in human, we propose that initial transposition event(s) may have involved large portion(s) of the β-glucuronidase gene, parts of which subsequently became deleted. Once the initial GUSB autosomal copy had been transposed into a pericentromeric region, it was rapidly used as a seed for further transposition/duplication in nonhuman primates. On the ancestral region homologous to HSA5p, the initial GUSB copy has taken a different evolutionary route. The first event of insertion was followed by a period of apparent paucity in terms of duplication of ∼20 million years. In contrast, the multiple copies on the equivalents of HSA5 and HSA6 in the Hominidae genomes (Fig. 3B) seem to be the result of a burst of intra- and inter-chromosomal duplication events that occurred after the separation from the ancestral Pongidae, 14 Mya.
Figure 3.
(A) Schematic representation of the genomic structure of the β-glucuronidase gene (GUSB) and _GUSB_-derived paralogous sequences. The genomic structure of the GUSB gene was established according to sequence comparison and alignment between the β-glucuronidase mRNA complete sequence (M15182) and genomic sequence of the BAC clone RP11-252P18 (AC073261). Dark gray boxes indicate the exons, and thick light-gray lines indicate intervening intronic sequences. For each _GUSB_-derived sequence found in the draft sequence of the human genome, the chromosomal localization and the GeneBank ID of the clone(s) they were derived from are indicated. (*) Clones in HTGS phase in GenBank—according to the June 2002 freeze of the genome draft sequence. Horizontal black bold lines illustrate chromosomal separations, horizontal black plain lines, subchromosomal separations, and horizontal black broken lines, gene-copy separations. The bracket indicates that the same clone was sequenced twice. (B) Summary of the in situ hybridization results obtained with a Glu 5–10 cosmid probe on human (HSA) and other primate chromosomes. A molecular timescale for primate evolution is indicated. The number of dots indicates the average signal intensity observed at a given location, resulting from the number of loci and the hybridization efficiency. The brackets illustrate the homologies between Macaca or Cebus and human chromosomes. In the presumed ancestral karyotype of placental mammals, the HSA7 homolog was composed of two parts. These two components fused before the separation of Cercopithecoidea and Hominidae (∼25 Mya), and then pericentric and paracentric inversions occurred in the Hominidae lineage. Thus, HSA7 is a fairly recent chromosome shared by HSA and PTR only. In Cebus capucinus, the smallest part homolog to HSA7 is on CCA1 associated with the homolog to the whole HSA5 (Richard et al. 1996). In Macaca sylvana, MSY2 is formed by the equivalents of the whole HSA7 and HSA21, and MSY9 by the equivalents of HSA20 and HSA22 (Muleris et al. 1984). HSA22/PTR23/GGO23/PPY23 on one hand, and HSA6/PTR5/GGO5 on the other hand, differ mostly only by heterochromatic variations (Yunis and Prakash 1982). The differences in the chronology of expansion/spreading of the GUSB paralogous sequences is depicted by a horizontal broken line, top, primo-insertion in pericentromeric regions on ancestral HSA7 and HSA22, bottom, primo-insertion in the ancestral HSA5p euchromatic region.
Spreading of the Gene-Derived Sequences Through a Process of Segmental Genomic Duplication
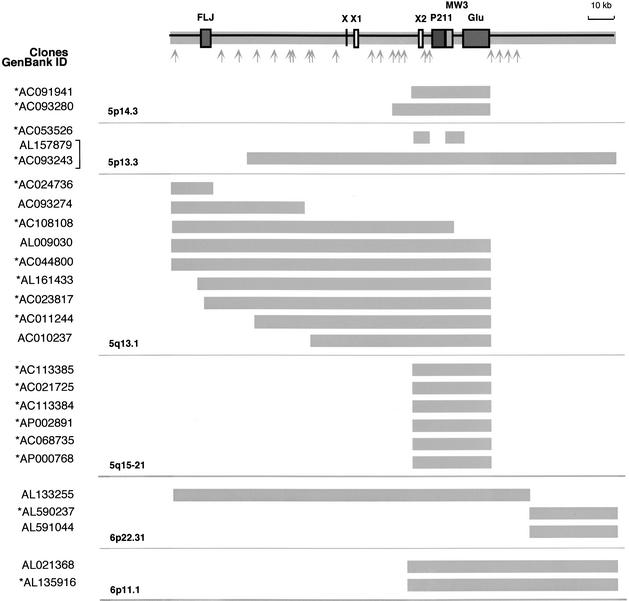
As proposed previously (Courseaux and Nahon 2001) and in this study, the last events of duplication likely involved large genomic segments. Sequence homology was thus expected to extend to flanking sequences that lie between the genes-derived sequences. To confirm this hypothesis, we expanded our comparative in silico analysis of the genomic structure to the flanking regions of GUSB pseudogenes residing on HSA5 and HSA6. This allowed us to identify several BAC/PAC clones that share strong sequence similarity (>90%) over large regions. A schematic representation of the homology extent between the different loci is given in Figure 4. Interestingly, the series of database searches used to characterize these segmental duplications has led to the identification of three other paralogous sequence variants (PSVs), MW3, P211, and FLJ. MW3 shows a high degree of identity to the THE-1 (transposable human element) retrotransposon gene family (Francis et al. 1995). Further FISH analysis on human and primate chromosomes suggested that MW3 was distributed onto Hominidae chromosomes equivalent to HSA5 (represented graphically in supplemental information) and HSA6 at the same time and place as Glu 5–10. FLJ and P211 were named on the basis of their strong similarity to cDNAs FLJ23032 fis and DKFZp434P211 (85%–92%), respectively. Both appeared highly amplified throughout the human genome. Other FLJ-related sequences (80%–95% nucleic acid identity) have been found in clones specific to the HSA2q14–HSA2q21, HSA13q32, and Xq21 genomic regions, and numerous P211-related sequences were also identified on HSA22q11 and HSA20p11 (Table 1). The respective genomic organization and chromosomal localization of FLJ and P211-related sequences suggest that those sequences have originated primarily on the ancestral chromosome homologous to HSA5 through the transposition of segmental parts of genes located elsewhere in the genome, as PMCHL and Glu 5–10 have done. Sequences between and outside of the PSVs are composed of a mixture of highly repetitive elements (∼50%, mainly LINE1, Alu repeats, and LTR elements) and putative transcripts—sequences that display strong sequence similarity (>95%) with ESTs and Unigene clusters as illustrated in Figure 4. The molecular structure of the paralogous regions proved to be amazingly complex, with components being present, duplicated, or absent among different representative copies. When located adjacent to one another, the duplicated elements are arranged tandemly. The molecular basis of the variation in the extent of paralogy is unclear. Although it is possible that these differences reflect the size variation of the genomic segments that were further distributed, they may also be the result of secondary events that rearranged these large blocks after duplication. The net effect of these duplication events is the generation of large (30 to >100 kb) blocks of paralogy among multiple nonhomologous chromosomal loci, each of which is a mosaic of smaller duplicated segments.
Figure 4.
Paralogy map and sequence content of the HSA5/HSA6 duplicon. The genomic organization of the duplicated region was deduced from the AL157879 and AL133255 clones that were chosen as reference seed sequence. The thick gray lines illustrate the homology extent between paralogous loci. The chromosomal localization and the GeneBank ID of the clones are indicated. The P211, FLJ, and GUSB gene-derived sequences are boxed in dark gray. For exon-intron organization of the _GUSB_-derived sequences, see Figure 3A. The THE-1 transposable element MW3 is boxed in light gray. X, X1, and X2 are sequences paralogous to exons of the chimeric genes described in Figure 6. Gray arrows indicate the localization of sequences highly homologous (>95%) to ESTs and human UNIGENE clusters as follows: Hs.186379, Hs.312136, Hs.224604, Hs.145839, Hs.274528, Hs.297663, Hs.50454, Hs.202243, Hs.183256, Hs.132586, AW963166, BE780832, Hs.317160, Hs.326016, Hs.121081, Hs.294040, Hs.166361, Hs.321499, Hs.324135, Hs.7569, Hs.131950, and Hs.186180. The brackets indicate that the same clone was sequenced twice. (*) Clones in HTGS phase in GenBank—according to the June, 2002 freeze of the genome draft sequence.
Table 1.
Localization of the Human Paralogous Sequences
| PSVs | Chromosome | Chromosomal band | Clones GenBank ID |
|---|---|---|---|
| MW3 | HSA5 | 5p14 | AC093280 |
| 5p13 | AL157879, AC093243, AC053526 | ||
| 5q13 | AC108108, AC023817, AL161433, AC044800, AC011244, AL009030, AC010237 | ||
| 5q15-21 | AP002891, AP000768, AC068735, AC021725 | ||
| HSA6 | 6p11 | AL021368, AL135916 | |
| 6p21 | AL133255 | ||
| FLJ | HSA2 | 2q14-q21 | AC069195, AC012306 |
| ?? | AC010981 | ||
| HSA5 | 5q13 | AC024736, AC093274, AC108108, AL161433, AC044800, AL009030, AC023817 | |
| HSA6 | 6p21 | AL133255 | |
| HSA13 | 13q32 | AL356098, AL356011 | |
| HSAX | Xq21 | AL592563 | |
| P211 | HSA5 | 5p14 | AC093280 |
| 5p13 | AL157879, AC093243 | ||
| 5q13 | AC108108, AC023817, AL161433, AC044800, AC011244, AL009030, AC010237 | ||
| 5q15-21 | AP002891, AP000768, AC068735, AC021725 | ||
| HSA6 | 6p11 | AL021368, AL135916 | |
| 6p21 | AL133255 | ||
| HSA20 | 20p11 | AL133466 | |
| ?? | AL389896 | ||
| HSA22 | 22q11 | AC000069, AP000550, AC008103, AC008132, AC007050, AC000095, AC002522, AP000552, AP000356, AP000354 | |
| ?? | AC011896, AC009288, AC007981, AC007324, AC023491, AC002051, AC012330, AC002054, AC007708, AC008018, AC002308, AC012331, AC000051, AC007664, AC007325 |
Organization of Segmental Duplications at the SMA Locus on 5q13
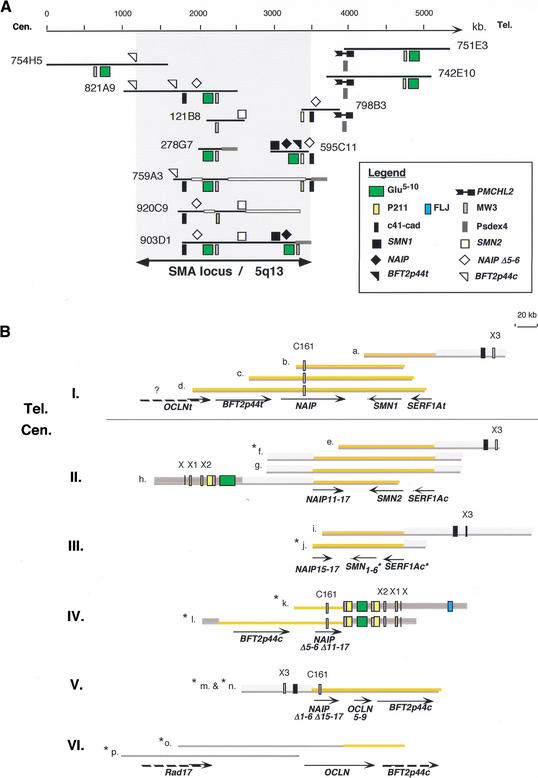
To study the impact of segmental duplications on the genomic organization of the SMA region (Lewin 1995), we first analyzed the distribution of the PSVs through a PCR analysis on a series of overlapping YAC clones spanning the region of predispostion to the disease (Fig. 5A). To further specify the genomic structure of this locus, a series of BLAST similarity searches against the nr and HTGS databases of GenBank were performed to reveal all of the clones sequenced as part of the Human Genome Project that contain one or more genes specific to the SMA region (SMN, SERF1A, NAIP, or BFT2p44) (Lefebvre et al. 1995; Burglen et al. 1997; Carter et al. 1997; Scharf et al. 1998). The searches identified 15 clones issued from 6 different genomic libraries that were selected for subsequent sequence comparisons to provide the best alignment (Fig. 5B). Such an analysis showed that what was considered primarily to be an inverted duplication of some several hundreds of kilobases (Lewin 1995), was far more complex than expected previously. The telomeric part of the 5q duplicon that contains the functional copies of the SMA locus genes (BTF2p44t, NAIP, SMN1, SERF1At) was shown to display a compact gene organization of ∼200 kb in length. In contrast, the centromeric duplicon appeared to be composed of a complex patchwork of recently duplicated genomic segments (Fig. 5B). A combination of PCR and FISH analysis with PAC clones described previously (Roy et al. 1995) was used to confirm the chromosomal distribution and the cytogenetic band positions of the duplicated segments (data not shown).
Figure 5.
Genomic organization of the SMA locus on 5q13. (A) Consensus YAC contig spanning the SMA region. Genes of the locus (SMN, NAIP, and BFTp44) are symbolized on the black line representing the extent of the clones, and the PSVs underneath. Legend at right. Thick gray and white lines symbolize YAC chimerism and instability, respectively. (B) Genomic organization of the SMA locus established through sequence analysis of PAC/BAC clones (a–p). Despite the fact that critical pieces of information about this chromosomal region are still missing or unclear, we succeeded in reconstructing the sequence organization of six nonoverlapping genomic areas encompassing the disease locus (I. to VI.). Tel. and Cen. refer, respectively, to the telomeric and centromeric parts of the duplication as described in the literature. Sequences specific to this duplicon are highlighted in orange. Thick gray lines represent duplicons whose original locus is on 5p14; (1) thick light-gray lines represent areas paralogous to the 5p14 region, and (2) thick dark-gray lines represent paralogous sequences distributed among several loci on human chromosomes 5 and 6 (see Fig. 4). Arrows indicate the extent and the 5′–3′ orientation of the genes. The NAIPΔ represent deleted forms of the NAIP gene. SMN1-6, NAIP11-17, and OCLN5-9 are deleted forms of the SMN, NAIP, and OCLN genes, respectively. (C161) CATT1-G1/C161 dinucleotide repeat marker (Burghes et al. 1994). (X, X1, X2, X3, C161) Sequences paralogous to exons of the chimeric genes depicted in Figure 6. Gene symbols and PSVs are denoted according to the symbol used in Figures 4 and 5A. (a) AC016554/CTC-340H12; (b) AC005031/GSP13996; (c) U80017/125D9; (d) AC044797/RP11-195E2; (e) AC004999/D215P15; (f) AC022119/CTC-566F17; (g) AC010272/CTC-492P2; (h) AC010237/CTC-348J20; (i) AC010217/CTC-202F24; (j) AC093202/CTC-249C14; (k) AC044800/RP11-34J8; (l) AC011244/RP11-497H16; (m) AC012369/RP11-551B22; (n) AC110009/RP11-508M8; (o) AC010355/CTD-2027K22; (p) AC015470/RP11-2H18. (*) Clones in HTGS phase in GenBank—according to the June, 2002 freeze of the genome draft sequence.
Segmental Duplications of HSA5: A Source of Gene Diversity?
To determine whether the recently duplicated regions of HSA5 could contain functional genes specific to the primate lineage, we have looked for transcripts that were likely to have originated recently during primate evolution as the PMCHL genes did (Courseaux and Nahon 2001). By systematic searches in databases and careful examination of the literature, we discovered nine chimeric-processed transcripts whose genomic organization is suggestive of a recent origin (Fig. 6). Their creation could not predate the duplication events that have led to the juxtaposition of exonic sequences of which they are constituted. The first class of transcripts (SMA3, SMA4, SMA5, NIH_MGC_19) contains exons homologous to exons 5, 9, and 10 of the GUSB gene associated with exonic sequences, which were shown to be present only in the duplicons specific to HSA5 and HSA6 (Fig. 4). These mosaic transcripts could lead to the fusion of new protein modules as illustrated in Figure 6 (ORF, a–j). Another transcript, 5CMP1, was shown to display a mosaic structure. It is composed of (1) four exons homologous to the GUSB gene (exons 5, 9, 10, and 11), (2) an exon X3 present in the duplicon paralogous to the HSA5p14 (see Fig. 5B), and (3) two exons, N7 and C161, homologous to the functional NAIP gene. N7 is paralogous to exon 7 of NAIP, and C161 was named on the basis of its strong sequence identity to the CATT-G1/C161 multiple subloci polymorphic marker, whose one copy lies into the intron between exons 7 and 8 of the NAIP gene. The mapping of CATT1 suggests that the 5CMP1 gene may be transcribed from one subloci localized on HSA5q13.1 (Burghes et al. 1994). Regarding the last class of transcripts (CE3, CD19, CD23, CC14.1), only partial information was available. No sequence has been deposited in GenBank, and consequently, the sequence of the 3′ part was obtained from the literature (Roy et al. 1995). Although fragmentary, these data were sufficient to establish that the four transcripts resulted from the fusion of NAIP and _OCLN_-derived exons associated with an exon X4, whose genomic origin remains undetermined.
Figure 6.
Schematic representation of chimeric transcripts and potentially functional ORFs. Exonic sequences are boxed and were deduced from comparative analyses between the DNA sequences of the transcripts and genomic clones. Sequences highly similar (86%–100% identity) to other known gene-exonic sequences are colored as follows: green, GUSB derived; blue, OCLN derived; orange, NAIP derived. These exons are numbered according to the exonic sequences they are derived from, that is, N7, sequence paralogous to NAIP exon 7. _GUSB_-derived chimeric transcripts are drawn to scale. Regarding _NAIP-OCLN_-derived transcripts, only partial information was available. The 3′ part of these cDNAs was assigned previously as NAIP exon 17 (Roy et al. 1995). However, further in silico analysis revealed that the 3′ ends actually consisted of four exons homologous to OCLN exons 5–9, and were associated with the exonic sequence X4, whose genomic origin remains undetermined. (Rp) Repeated sequence as follows: (Rp.1) LTR/pTR5; (Rp.2) LINE/HAL1-SINE/Alu Sx; (Rp.3) MER3-SINE/Alu Y; (Rp.4) LTR/pTR5-LINE/L1. (C161) Exonic sequence containing the CATT1-G1/C161 dinucleotide repeat marker. Gray brackets numbered a–l indicate the extent of potentially functional ORFs initiated from an ATG codon in a reasonable initiation context (Kozak 1984). For genomic localizations of sequences paralogous to exons C161 and X3, see Figure 5B, and for exons X, X1, and X2, see Figures 4 and 5B.
DISCUSSION
The recent data concerning the organization of segmental duplications in primate genomes revealed that these unusual genomic regions represent an extraordinary source of information about several biological processes (Eichler 2001; Samonte and Eichler 2002). They constitute a rich palaeontological record, holding crucial clues about evolutionary events and forces. As passive markers, they shed light on chromosome structure and dynamics; as active agents, they have reshaped the genome by creating entirely new genes and causing ectopic rearrangements that may further have led to inherited pathology.
On the basis of previous analysis (Christian et al. 1999; Jackson et al. 1999; Shaikh et al. 2000; Courseaux and Nahon 2001; Horvath et al. 2001) and the present findings, a plausible scenario for the emergence of paralogous segments in the human lineage can be drawn. The multiplicity of segmental gene copies in the human genome might be explained by an intense activity of gene duplication—through retrotransposition for PMCHL1, but yet unknown mechanisms for the other gene models—that occurred in the ancestral genome at the time of divergence between Catarrhini and _Plathyrrhini_—about 25–35 Mya for PMCHL1 and Glu 5–10, predating Catarrhini divergence for the other gene models. Some ancestral gene loci appeared to have acted as hot-spot donors of sequences (e.g., the ancestral MCH/AROM and GUSB loci). Whereas other regions of the genome appeared to have served as a safe haven for transposons/duplicons that have escaped inactivation or elimination—for example, ancestral HSA5p14 or pericentromeric or subtelomeric regions of chromosomes (Horvath et al. 2000; Bailey et al. 2001). The further expansion/spreading of the segmental gene copies occurred then by subsequent inter- and intra-chromosomal duplication events involving large chromosomal regions, whose chronology and mechanism should be different regarding the region of primary insertion. We have shown that, on the ancestral region homologous to HSA5p, a period of ∼20 million years, has apparently separated the first event of insertion and the following massive duplication events (Fig. 3B). Conversely, once transposed into a pericentromeric region, DNA segments were shown to have been rapidly used as a seed for further transposition/duplication. The quantitative differences in the distribution of the _GUSB_-derived sequences, as we observed among representative members of the Hominidae (Fig. 3B), are consistent with the rapid evolutionary turnover that has been found in the pericentromeric region of human and other primate chromosomes (Eichler et al. 1999). Although it is possible that these regions simply have a great tolerance for large insertions, the possibility of specific mechanisms for recent insertion of segmental genomic duplication has been suggested on the basis of unusual GC-rich DNA (Eichler et al. 1999) and satellite sequences (Regnier et al. 1997; Guy et al. 2000) flanking the insertions. The authors postulated that these elements, in addition to serving as transposition integration signals, may also represent focal points for the transfer of genomic material among pericentromeric regions. The quantitative and qualitative differences in the distribution of these duplicons shown among representative members of human and apes, suggests that the dynamic spread of these sequences may be a still-ongoing evolutionary process in the Hominoid genomes.
In the last few years, intrachromosomal duplications have been implicated in many recurrent chromosomal rearrangements associated with microdeletion and microduplication syndromes (Lupski 1998). It is noteworthy that many of the regions associated with intrachromosomal rearrangements and disease have also been active for segmental duplication events, suggesting that the processes of duplication and genomic instability are closely intertwined and contribute to disease etiology and genomic structural variations (for review, see Ji et al. 2000; Emanuel and Shaikh 2001). Here, we address this question in the context of the etiology of the SMA pathology by analyzing the impact of segmental duplications on the genomic organization of the predisposition locus. This region of the human genome has proven to have an highly unstable and intricate genomic structure (Lewin 1995) and to show a marked heterogeneity in the normal population (Burghes 1997; Campbell et al. 1997). Differences in the structure of the SMA region among human individuals are well documented. Markers and genes that lie within this locus are represented multiple times but also may vary in copy number even in a highly inbred population (Burghes 1997; Campbell et al. 1997). Sequence homology between the recently duplicated DNA segments, which we show here (Fig. 5), does provide a chance for misalignment during meiosis, leading to unequal exchange and chromosome rearrangements that result in the multiplication of all or parts of genes/PSVs observed within this region. This unusual form of polymorphism may also explain why this region is purported to be particularly recalcitrant to YAC subcloning and the lack of correlation between the different physical maps that have been published previously (Lewin 1995). Human genomic libraries consist of clones from at least two different haplotypes, making a consistent map of such a region from mixed haplotype libraries present even more of a challenge. Finally, it is obvious that the complex, highly duplicated nature of these regions is not amenable to high-throughput assembly methods—that is, restriction fingerprinting methods for determining clone overlap—(Bailey et al. 2001). As a consequence, the current assembly of the SMA locus was revealed to be erroneous and not able to recapitulate the organization published in the literature. The most common error was the merging of nearly identical sequence-duplicated segments into a single contig. To circumvent all of the problems inherent in physical mapping, we have used the sensitivity of sequence data to further specify the potential implication of PSVs in mediating the high degree of instability observed within the SMA region. Such an analysis showed that the insertion of PSVs associated with the burst of duplication events that occurred in the Hominidae lineage has fully reshaped the SMA locus during primate evolution. Although a direct connection is yet to be established, it is particularly tempting to speculate that there is a link between the dynamic duplicative nature of genomic duplicons and the rearrangements mediating the SMA disease (e.g., deletions and gene conversion events). The definite proof will come from the isolation and characterization of breakpoints from multiple patients.
Despite its possible deleterious effects, gene duplication remains one of the primary forces of evolutionary change. An evolutionary benefit of such genomic plasticity may be to juxtapose different cassettes from diverse genes to create a reservoir of gene/protein modules with a potentially new function. In the present study, we revealed that the evolutionary rearrangements that have participated in the origin of the two chimeric functional genes PMCHL1 and PMCHL2 during primate evolution (Courseaux and Nahon 2001) were also involved in the emergence and dispersal of various gene segments containing duplicated regions throughout several loci of human chromosomes 5 and 6. It was therefore crucial to determine whether such an abundant recently duplicated material could contain other functional genes specific to the primate lineage. By in silico screening, we identified numerous chimeric transcripts. A first class (SMA3, SMA4, SMA5, NIH_MGC_19) consisted of _GUSB_-derived exons associated with different combinations of exonic sequences that were shown to be present only in the duplicon described in Figure 4 and to display no similarity to any other expressed sequence of the GenBank databases. This observation rendered very unlikely the hypothesis that those new sequences might be part or duplicates of previously existing genes. Rather, those exons should have originated from unique genomic sequences that fortuitously evolved as standard intron-exon structures, as we found previously for the PMCHL1 gene (Courseaux and Nahon 2001). This led us to propose that after its insertion, about 35 Mya, the transposed GUSB gene segment has recruited through subsequent mutations events, intronic and exonic components into transcription unit(s). Furthermore, sequence analysis revealed that these mRNAs did not result from alternative splicing events, but rather suggested that they were the product of genes located on different paralogous segments. Several ORFs were deduced from the sequences of the SMA3, SMA4, SMA5, and NIH_MGC_19 transcripts (bracketed in Fig. 6). The analogy here, with the PMCHL genes, was particularly striking, as these ORFs overlapped exons originating from distinct paralogous loci and led to putative new fused proteins domains. Another class of transcripts was found to result from the juxtaposition of NAIP and _OCLN_-derived exons (Fig. 6). In the human genome, as many as six copies of truncated and/or internally deleted NAIP loci have been detected previously (Rajcan-Separovic et al. 1996). Our in silico analysis of the SMA locus allowed us to analyze the genomic environment of three of them (Fig. 5B). Interestingly, the three loci were localized at the boundaries between different duplicons. In particular, one of them, the internally deleted form NAIP_Δ_1–6_Δ_15–17 (Fig. 5B), was adjacent to a 5′ truncated version of the OCLN gene (OCLN5-9). This strongly suggested that CE3, CD19, CD23, and CC14.1 are the transcriptional products of at least one chimeric gene created 5–10 Mya, as the result of the rearrangements at the SMA locus. These have led to the clustering of new emerging deleted forms of the NAIP and OCLN genes, thus supporting the concept of exon shuffling (Gilbert 1978).
Recently, several fusion transcripts involving endogenous genes and exonic portions of segmental duplications have been described in the human genome. Although their biological significance is unknown, it is notable that the transcription of all of these recent transcripts is found mainly in germ-line, fetal, or cancerous tissues (Fig. 6; Viale et al. 2000; Courseaux and Nahon 2001; Bailey et al. 2002b). The vast majority of such evolutionary experiments are probably failures at a functional level, and thus require further experimental confirmation. However, according to the sheer abundance of transcripts that have been created or modified through transposition and/or duplication events during the last 35 million years—an estimate of 500–1100 (Bailey et al. 2002b)—we can expect functional genes to arise from the hodgepodge of segmental duplications. The identification within recently duplicated segments of members of the PMCHL and morpheus gene families that were shown to exhibit, respectively, indications of exaption and of positive selection among Hominoids, strongly support this hypothesis (Courseaux and Nahon 2001; Johnson et al. 2001). Taken together, these results reveal one of the most unexpected recurrent roles of chromosomal duplication during primate evolution and support the idea that processes of duplicon clustering joined to positional polymorphism could be a nursery for generating gene diversity and a source of phenotypic variation among humans as proposed previously by B. Trask, E. Eichler, and coworkers (Linardopoulou et al. 2001; Samonte and Eichler 2002). The generation of human-specific genes may also be related to the major quantitative changes in gene/protein expression in the human brain by comparison with other mammalian species, including chimpanzees, as reported recently by S. Pääbo and coworkers (Enard et al. 2002). It is tempting to speculate that newly created genes in Hominoids may contribute as master genes in the control of the species- and tissues-specific enhancement of gene activity (Nahon 2003).
Finally, whether the process of segmental duplication as described here is a true attribute of primate genome evolution, remains to be determined. The contribution of large genomic duplications to generate new gene families is firmly established in nonvertebrate species, such as Caenorhabditis elegans and yeast (Friedman and Huges 2001a). However, human and its closest relatives are the only ones to demonstrate duplication of unrelated gene cassettes within nonrandom regions of chromosomes. Primary analyses of the human genome draft sequence revealed that such a high proportion of large duplications containing mosaic genes clearly distinguishes the human genome from other genomes sequenced to date (Lander et al. 2001). A firm answer will probably come from the complete sequences of other vertebrates, such as the mouse and rat. In a more general context, there is a highly controversial debate regarding the overt contribution (or not) of ancient whole-genome duplication (WGD) to shape vertebrate genomes (Friedman and Huges 2001b; McLysaght et al. 2002). Recently, a global search in the draft human-genome sequence for groups of parologs (parologons) and a molecular clock analysis of orthologs in fly and nematodes provided some support for the WGD hypothesis (McLysaght et al. 2002). However, if massive transposition of large genomic duplicons, as exemplified here during Hominidae evolution, was an active process during early vertebrate evolution, this might also explain recurrent duplications in the genomes of vertebrates. Lastly, it is worth noting that transposable elements and retrotransposons have played a major role in shaping the vertebrate genomes (Brosius 1999b; Kidwell and Lisch 2001). Recent studies have revealed that apart from an exceptional burst of activity of Alu repeats peaking around 40 Mya, a fairly steady decline in transposon activity has occurred in the Hominid lineage over the past 35–50 million years, whereas no similar decline was detected in the mouse genome (Lander et al. 2001). Thus, it would be interesting to investigate whether the occurence of segmental duplication and transposon activity may be inter-related at the molecular level in vertebrates, particularly in the Hominids.
METHODS
FISH Analysis
On human chromosomes, FISH was performed (as described previously) (Courseaux and Nahon 2001) on metaphase chromosomes from peripheral blood lymphocytes of seven normal unrelated individuals. For polymorphism estimation, at least 100 metaphases per individual and per probe were analyzed. Fluorescent images were captured using high-resolution cooled charge-coupled device (CCD) camera C4880 (Hamamatsu). Image acquisition, processing, and analysis were performed using the Vysis software package (Quips SmartCapture FISH).
Nonhuman metaphase spreads were obtained from cell cultures of fibroblasts conserved in liquid nitrogen in the tissue bank of the Institut Curie (Paris, France). Probes were labeled with biotin-11-dUTP by nick translation (kit Mix enzyme NTL, Roche Molecular Biochemicals). Fluorescence in situ hybridization was performed essentially as described previously (Viale et al. 1998). Observations were performed under an epifluorescent microscope (Microphot-FXA, Nikon) and images were captured using a cooled CCD camera (Photometrics), and a capture software (Quips-Smart, Vysis).
A complete list of the clones used as FISH probe and the combination of probes used in dual-color FISH experiments are detailed in Supplemental Table 1.
In Silico Modeling
The in silico analysis and detection of segmental duplications were conducted through BLAST searches of GenBank against many databases in the Web site of the National Center for Biotechnology Information of the National Institutes of Health (www.ncbi.nlm.nih.gov/). Multiple alignments of the DNA sequences were done using the flat query-anchored with identities alignment view option of the BLAST program. The extent of the homology of paralogous regions, analysis of HTGS, extension, and completion of contigs were further performed by using the NIX tool (www.hgmp.mrc.ac.uk/), which combines many DNA analysis programs (GRAIL, Fex, Hexon, MZEF, Genemark, Genefinder, Fgene, BLAST [against many databases], Polyah, RepeatMasker, and tRNAscan).
The genomic organization of the SMA locus was established by a method in which public sequences (GenBank databases) were analyzed at the clone level. SMA genes and PSVs were first used as anchor markers for sequence comparison to show overlapping genomic areas encompassing the disease locus (100% sequence identity). The extent of the overlaps was further refined through adjacent sequence comparison.
The translation of DNA sequences to protein sequences was conducted on the Web site of National Cancer BI of the National Institutes of Health (www.NCBI.nlm.nih.gov/). Chromosomal localizations were established according to both ENSEMBL (http://www.ensembl.org/) and the Mapview resource (http://www.ncbi.nlm.nih.gov/) and/or sequence comparisons.
YACs Contig Construction
The degree of overlap between YACs is based on sequence content and data of the literature (Roy et al. 1995). Sequence content of the YAC clones were established by PCR experiments. Primers and PCR conditions are described in Supplemental Table 2.
WEB SITE REFERENCES
http://www.ensembl.org/; Ensembl, a joint project between EMBL-EBI and the Sanger Institute that produces automatic annotation on eukaryotic genomes.
http://www.ncbi.nlm.nih.gov/; National Center for Biotechnology Information (NCBI).
www.hgmp.mrc.ac.uk/; UK Human Genome Mapping Project Resource Centre (HGMP-RC).
Acknowledgments
We thank M.C. Potier (ESPCI, CNRS UMR7637, Paris, France) and P. Vernier (Institut A. Fessard, CNRS UPR 2197, Gif-sur-Yvette, France) for critical reading of the manuscript. We thank the following individuals for providing reagents or samples: D. Le Paslier (UMR CNRS 8030, Evry, France), A. McKenzie (Molecular Genetics Laboratory Children's Hospital, Ottawa, Canada), J. Melki (INSERM EMI-9913, Universite d'Evry, France.), C. Sargent (University of Cambridge, UK), J. Scharf (Howard Hughes Medical Institute, Boston, USA) and S. Selig (Department of Nephrology, Rambam Medical Center, Haifa, Israel). A.V. was supported by the ADER-PACA (CAR9312/2679; 1994–1996) and the Association Française contre les Myopathies (AFM) (1997). A.C. was a recipient of post-doctoral fellowships from AFM (1997) and from Association pour la Recherche contre le Cancer (ARC) (1998–2000). We thank F. Cuzin (U470 INSERM, Nice) for the financial support (Hoechst Marion Roussel) attributed to A.C. (2001). Supported by grants from the AFM (ASI 1996–1998) and the Centre National de la Recherche Scientifique (CNRS) (Programme OHLL, 2002).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Present addresses:
E-MAIL nahonjl@ipmc.cnrs.fr; FAX 33-493-957-708.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.490303\. Article published online before print in February 2003.
REFERENCES
- 1.Bailey J.A., Yavor, A.M., Massa, H.F., Trask, B.J., and Eichler, E.E. 2001. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 11**:** 1005-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002a. Recent segmental duplications in the human genome. Science 297**:** 1003-1007. [DOI] [PubMed] [Google Scholar]
- 3.Bailey J.A., Yavor, A.M., Viggiano, L., Misceo, D., Horvath, J.E., Archidiacono, N., Schwartz, S., Rocchi, M., and Eichler, E.E. 2002b. Human-specific duplication and mosaic transcripts: The recent paralogous structure of chromosome 22. Am. J. Hum. Genet. 70**:** 83-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosius J. 1999a. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene 238**:** 115-134. [DOI] [PubMed] [Google Scholar]
- 5.___, 1999b. Vertebrate genomes were forged by massive bombardments with retroelements and retrosequences. Genetica 107**:** 209-238. [PubMed] [Google Scholar]
- 6.Burghes A.H. 1997. When is a deletion not a deletion? When it is converted. Am. J. Hum. Genet. 61**:** 9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burghes A.H., Ingram, S.E., Kote-Jari, Z., Rosenfeld, S., Herta, N., Nadkarni, N., DiNato, C.J., Carpten, J., Hurko, O., Florence, J., et al. 1994. A multicopy dinucleotide marker that maps close to the spinal muscular atrophy gene. Genomics 21**:** 394-402. [DOI] [PubMed] [Google Scholar]
- 8.Burglen L., Seroz, T., Miniou, P., Lefebvre, S., Burlet, P., Munnich, A., Pequignot, E.V., Egly, J.M., and Melki, J. 1997. The gene encoding p44, a subunit of the transcription factor TFIIH, is involved in large-scale deletions associated with Werdnig-Hoffmann disease. Am. J. Hum. Genet. 60**:** 72-79. [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell L., Potter, A., Ignatius, J., Dubowitz, V., and Davies, K. 1997. Genomic variation and gene conversion in spinal muscular atrophy: Implications for disease process and clinical phenotype. Am. J. Hum. Genet. 61**:** 40-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter T.A., Bonnemann, C.G., Wang, C.H., Obici, S., Parano, E., DeFatima Bonaldo, M., Ross, B.M., Penchaszadeh, G.K., MacKenzie, A., et al. 1997. A multicopy transcription-repair gene, BTF2p44, maps to the SMA region and demonstrates SMA associated deletions. Hum. Mol. Genet. 6**:** 229-236. [DOI] [PubMed] [Google Scholar]
- 11.Christian S.L., Fantes, J.A., Mewborn, S.K., Huang, B., and Ledbetter, D.H. 1999. Large genomic duplicons map to sites of instability in the prader-willi/Angelman syndrome chromosome region (15q11-q13). Hum. Mol. Genet. 8**:** 1025-1037. [DOI] [PubMed] [Google Scholar]
- 12.Courseaux A. and Nahon, J.L. 2001. Birth of two chimeric genes in the Hominidae lineage. Science 291**:** 1293-1297. [DOI] [PubMed] [Google Scholar]
- 13.Crosier M., Viggiano, L., Guy, J., Misceo, D., Stones, R., Wei, W., Hearn, T., Ventura, M., Archidiacono, N., Rocchi, M., et al. 2002. Human paralogs of KIAA0187 were created through independent pericentromeric-directed and chromosome-specific duplication mechanisms. Genome Res. 12**:** 67-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutrillaux B. 1979. Chromosomal evolution in primates: Tentative phylogeny from Microcebus murinus (Prosimian) to man. Hum. Genet. 48**:** 251-314. [DOI] [PubMed] [Google Scholar]
- 15.Dutrillaux B., Rethore, M.O., Prieur, M., and Lejeune, J. 1973. Analysis of the structure of chromatids of Gorilla gorilla. Comparison with Homo sapiens and Pan troglodytes. Humangenetik 20**:** 343-354. [PubMed] [Google Scholar]
- 16.Eichler E.E. 2001. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 17**:** 661-669. [DOI] [PubMed] [Google Scholar]
- 17.Eichler E.E., Archidiacono, N., and Rocchi, M. 1999. CAGGG repeats and the pericentromeric duplication of the hominoid genome. Genome Res. 9**:** 1048-1058. [DOI] [PubMed] [Google Scholar]
- 18.Emanuel B.S. and Shaikh, T.H. 2001. Segmental duplications: An ‘expanding’ role in genomic instability and disease. Nat. Rev. Genet. 2**:** 791-800. [DOI] [PubMed] [Google Scholar]
- 19.Enard W., Khaitovich, P., Klose, J., Zollner, S., Heissig, F., Giavalisco, P., Nieselt-Struwe, K., Muchmore, E., Varki, A., Ravid, R., et al. 2002. Intra- and interspecific variation in primate gene expression patterns. Science 296**:** 340-343. [DOI] [PubMed] [Google Scholar]
- 20.Francis M.J., Nesbit, M.A., Theodosiou, A.M., Rodrigues, N.R., Campbell, L., Christodoulou, Z., Qureshi, S.J., Porteous, D.J., Brooks, A.J., and Davis, K.E. 1995. Mapping of retrotransposon sequences in the unstable region surrounding the spinal muscular atrophy locus in 5q13. Genomics 27**:** 366-369. [DOI] [PubMed] [Google Scholar]
- 21.Friedman R. and Hughes, A.L. 2001a. Gene duplication and the structure of eukaryotic genomes. Genome Res. 11**:** 373-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.___, 2001b. Pattern and timing of gene duplication in animal genomes. Genome Res. 11**:** 1842-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert W. 1978. Why genes in pieces? Nature 271**:** 501. [DOI] [PubMed] [Google Scholar]
- 24.Goodman M. 1999. The genomic record of Humankind's evolutionary roots. Am. J. Hum. Genet. 64**:** 31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gould S.J. and Vrba, E.S. 1982. Exaptation—a missing term in the science of form. Paleobiology 8**:** 4-15. [Google Scholar]
- 26.Guy J., Spalluto, C., McMurray, A., Hearn, T., Crosier, M., Viggiano, L., Miolla, V., Archidiacono, N., Rocchi, M., Scott, C., et al. 2000. Genomic sequence and transcriptional profile of the boundary between pericentromeric satellites and genes on human chromosome arm 10q. Hum. Mol. Genet. 9**:** 2029-2042. [DOI] [PubMed] [Google Scholar]
- 27.Hacia J.G. 2001. Genome of the apes. Trends Genet. 17**:** 637-645. [DOI] [PubMed] [Google Scholar]
- 28.Horvath J.E., Schwartz, S., and Eichler, E.E. 2000. The mosaic structure of human pericentromeric DNA: A strategy for characterizing complex regions of the human genome. Genome Res. 10**:** 839-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvath J.E., Bailey, J.A., Locke, D.P., and Eichler, E.E. 2001. Lessons from the human genome: Transitions between euchromatin and heterochromatin. Hum. Mol. Genet. 10**:** 2215-2223. [DOI] [PubMed] [Google Scholar]
- 30.Jackson M.S., Rocchi, M., Thompson, G., Hearn, T., Crosier, M., Guy, J., Kirk, D., Mulligan, L., Ricco, A., Piccininni, S., et al. 1999. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications and unstable sequences with homologies to telomeric and other centromeric locations. Hum. Mol. Genet. 8**:** 205-215. [DOI] [PubMed] [Google Scholar]
- 31.Ji Y., Eichler, E.E., Schwartz, S., and Nicholls, R.D. 2000. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res. 10**:** 597-610. [DOI] [PubMed] [Google Scholar]
- 32.Johnson M.E., Viggiano, L., Bailey, J.A., Abdul-Rauf, M., Goodwin, G., Rocchi, M., and Eichler, E.E. 2001. Positive selection of a gene family during the emergence of humans and African apes. Nature 413**:** 514-519. [DOI] [PubMed] [Google Scholar]
- 33.Kidwell M.G. and Lisch, D.R. 2001. Perspective: Transposable elements, parasitic DNA, and genome evolution. Evolution 55**:** 1-24. [DOI] [PubMed] [Google Scholar]
- 34.Kozak M. 1984. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 12**:** 857-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lander E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Deron, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409**:** 860-921. [DOI] [PubMed] [Google Scholar]
- 36.Lefebvre S., Burglen, L., Reboullet, S., Clermont, O., Burlet, P., Viollet, L., Benichou, B., Cruaud, C., Millasseau, P., Zeviani, M., et al. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80**:** 155-165. [DOI] [PubMed] [Google Scholar]
- 37.Lewin B. 1995. Genes for SMA: Multum in parvo. Cell 80**:** 1-5. [DOI] [PubMed] [Google Scholar]
- 38.Linardopoulou E., Mefford, H.C., Nguyen, O., Friedman, C., van den Engh, G., Farwell, D.G., Coltrera, M., and Trask, B.J. 2001. Transcriptional activity of multiple copies of a subtelomerically located olfactory receptor gene that is polymorphic in number and location. Hum. Mol. Genet. 10**:** 2373-2383. [DOI] [PubMed] [Google Scholar]
- 39.Long M. 2001. Evolution of novel genes. Curr. Opin. Genet. Dev. 11**:** 673-680. [DOI] [PubMed] [Google Scholar]
- 40.Lupski J.R. 1998. Genomic disorders: Structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 14**:** 417-422. [DOI] [PubMed] [Google Scholar]
- 41.Marzella R., Viggiano, L., Ricco, A.S., Tanzariello, A., Fratello, A., Archidiacono, N., and Rocchi, M. 1997. A panel of radiation hybrids and YAC clones specific for human chromosome 5. Cytogenet. Cell Genet. 77**:** 232-237. [DOI] [PubMed] [Google Scholar]
- 42.McLysaght A., Hokamp, K., and Wolfe, K.H. 2002. Extensive genomic duplication during early chordate evolution. Nat .Genet. 31**:** 200-204. [DOI] [PubMed] [Google Scholar]
- 43.Muleris M., Paravatou-Petsota, M., and Dutrillaux, B. 1984. Diagrammatic representation for chromosomal mutagenesis studies. II. Radiation-induced rearrangements in Macaca fascicularis. Mutat. Res. 126**:** 93-103. [DOI] [PubMed] [Google Scholar]
- 44.Munsat T.L., Skerry, L., Korf, B., Paber, B., Schapira, Y., Gascon, G.G., al-Rajeh, S.M., Dubowitz, V., Davies, K., Brzustowicz, L.M., et al. 1990. Phenotypic heterogeneity of spinal muscular atrophy mapping to chromosome 5q11.2–13.3 (SMA 5q). Neurology 40**:** 1831-1836. [DOI] [PubMed] [Google Scholar]
- 45.Nahon, J.L. 2003. Birth of “human-specific” genes during primate evolution. Genetica (in press). [DOI] [PubMed]
- 46.Patthy L. 1999. Genome evolution and the evolution of exon-shuffling—a review. Gene 238**:** 103-114. [DOI] [PubMed] [Google Scholar]
- 47.Rajcan-Separovic E., Mahadevan, M.S., Lefebvre, C., Besner-Johnston, A., Ikeda, J.E., Korneluk, R.G., and MacKenzie, A. 1996. FISH detection of chromosome polymorphism and deletions in the spinal muscular atrophy (SMA) region of 5q13. Cytogenet. Cell Genet. 75**:** 243-247. [DOI] [PubMed] [Google Scholar]
- 48.Regnier V., Meddeb, M., Lecointre, G., Richard, F., Duverger, A., Nguyen, V.C., Dutrillaux, B., Bernheim, A., and Danglot, G. 1997. Emergence and scattering of multiple neurofibromatosis (NF1)-related sequences during hominoid evolution suggest a process of pericentromeric interchromosomal transposition Hum. Mol. Genet. 6**:** 9-16. [DOI] [PubMed] [Google Scholar]
- 49.Richard F., Lombard, M., and Dutrillaux, B. 1996. ZOO-FISH suggests a complete homology between human and capuchin monkey (Platyrrhini) euchromatin. Genomics 36**:** 417-423. [DOI] [PubMed] [Google Scholar]
- 50.Roy N., Mahadevan, M.S., McLean, M., Shutler, G., Yaraghi, Z., Farahani, R., Baird, S., Besner-Johnston, A., Lefebvre, C., Kang, X., et al. 1995. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell 80**:** 167-178. [DOI] [PubMed] [Google Scholar]
- 51.Samonte R.V. and Eichler, E.E. 2002. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3**:** 65-72. [DOI] [PubMed] [Google Scholar]
- 52.Sargent C.A., Chalmers, I.J., Leversha, M., and Affara, N.A. 1994. A rearrangement on chromosome 5 of an expressed human β-glucuronidase pseudogene. Mamm. Genome 5**:** 791-796. [DOI] [PubMed] [Google Scholar]
- 53.Scharf J.M., Endrizzi, M.G., Wetter, A., Huang, S., Thompson, T.G., Zerres, K., Dietrich, W.F., Wirth, B., and Kunkel, L.M. 1998. Identification of a candidate modifying gene for spinal muscular atrophy by comparative genomics. Nat. Genet. 20**:** 83-86. [DOI] [PubMed] [Google Scholar]
- 54.Selig S., Bruno, S., Scharf, J.M., Wang, C.H., Vitale, E., Gilliam, T.C., and Kunkel, L.M. 1995. Expressed cadherin pseudogenes are localized to the critical region of the spinal muscular atrophy gene. Proc. Natl. Acad. Sci. 92**:** 3702-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selig S., Lidov, H.G., Bruno, S.A., Segal, M.M, and Kunkel, L.M. 1997. Molecular characterization of Br-cadherin, a developmentally regulated, brain-specific cadherin. Proc. Natl. Acad. Sci. 94**:** 2398-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaikh T.H., Kurahashi, H., Saitta, S.C., O'Hare, A.M., Hu, P., Roe, B.A., Driscoll, D.A., McDonald-McGinn, D.M., Zackai, E.H., Budarf, M.L., et al. 2000. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: Genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 9**:** 489-501. [DOI] [PubMed] [Google Scholar]
- 57.Stankiewicz P., Park, S.S., Inoue, K., and Lupski, J.R. 2001. The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res. 11**:** 1205-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Theodosiou A.M., Morrison, K.E., Nesbit, A.M., Daniels, R.J., Campbell, L., Francis, M.J., Christodoulou, Z., and Davies, K.E. 1994. Complex repetitive arrangements of gene sequence in the candidate region of the spinal muscular atrophy gene in 5q13. Am. J. Hum. Genet. 55**:** 1209-1217. [PMC free article] [PubMed] [Google Scholar]
- 59.Viale A., Ortola, C., Richard, F., Vernier, P., Presse, F., Schilling, S., Dutrillaux, B., and Nahon, J.L. 1998. Emergence of a brain-expressed variant melanin-concentrating hormone gene during higher primate evolution: A gene “in search of a function”. Mol. Biol. Evol. 15**:** 196-214. [DOI] [PubMed] [Google Scholar]
- 60.Viale A., Courseaux, A., Presse, F., Ortola, C., Breton, C., Jordan, D., and Nahon, J.L. 2000. Structure and expression of the variant melanin-concentrating hormone genes: Only PMCHL1 is transcribed in the developing human brain and encodes a putative protein. Mol. Biol. Evol. 17**:** 1626-1640. [DOI] [PubMed] [Google Scholar]
- 61.Yunis J.J. and Prakash, O. 1982. The origin of man: A chromosomal pictorial legacy. Science 215**:** 1525-1530. [DOI] [PubMed] [Google Scholar]