Diversity of Methane-Cycling Archaea in Hydrothermal Sediment Investigated by General and Group-Specific PCR Primers (original) (raw)
Abstract
The zonation of anaerobic methane-cycling Archaea in hydrothermal sediment of Guaymas Basin was studied by general primer pairs (mcrI, ME1/ME2, mcrIRD) targeting the alpha subunit of methyl coenzyme M reductase gene (mcrA) and by new group-specific mcrA and 16S rRNA gene primer pairs. The mcrIRD primer pair outperformed the other general mcrA primer pairs in detection sensitivity and phylogenetic coverage. Methanotrophic ANME-1 Archaea were the only group detected with group-specific primers only. The detection of 14 mcrA lineages surpasses the diversity previously found in this location. Most phylotypes have high sequence similarities to hydrogenotrophs, methylotrophs, and anaerobic methanotrophs previously detected at Guaymas Basin or at hydrothermal vents, cold seeps, and oil reservoirs worldwide. Additionally, five mcrA phylotypes belonging to newly defined lineages are detected. Two of these belong to deeply branching new orders, while the others are new species or genera of Methanopyraceae and Methermicoccaceae. Downcore diversity decreases from all groups detected in the upper 6 cm (∼2 to 40°C, sulfate measurable to 4 cm) to only two groups below 6 cm (>40°C). Despite the presence of hyperthermophilic genera (Methanopyrus, Methanocaldococcus) in cooler surface strata, no genes were detected below 10 cm (≥60°C). While _mcrA_-based and 16S rRNA gene-based community compositions are generally congruent, the deeply branching mcrA cannot be assigned to specific 16S rRNA gene lineages. Our study indicates that even among well-studied metabolic groups and in previously characterized model environments, major evolutionary branches are overlooked. Detecting these groups by improved molecular biological methods is a crucial first step toward understanding their roles in nature.
INTRODUCTION
Hydrothermal surface sediments in Guaymas Basin support exceptionally high microbial activity and microbial diversity (1–4). The highly productive overlying water column, combined with terrestrial runoff, leads to sedimentation rates exceeding 1 mm yr−1 (5) and organic carbon contents of 2 to 4% by weight (6). In addition, the upward flow of hydrothermal fluids supplies hydrocarbons (methane, petroleum) and volatile fatty acids produced by thermal degradation of buried organic matter in deeper sediment layers to surface sediments and their microbial communities (1, 7–9).
Methanogenesis and sulfate-dependent methane oxidation in Guaymas Basin are carried out by diverse microbial lineages. 16S rRNA gene-based surveys have detected known methane-cycling groups (Methanococcoides, Methanocorpusculum, Methanoculleus, Methermicoccus, ANME-1, ANME-2) (4, 10−12), a new lineage of ANME-1, ANME-1Guaymas (12), and unknown deeply branching euryarchaeotal groups within the phylogenetic vicinity of known methane-cycling Archaea (4, 10). Phylogenetic analyses of mcrA, a gene diagnostic of methanogenic and anaerobic methanotrophic Archaea (13, 14), indicate an even higher diversity of methane-cycling Archaea than those detected by 16S rRNA gene surveys (Methanococcoides, Methanohalophilus, Methanosaeta, Methanoculleus, Methanocorpusculum, Methanocaldococcus, Methermicoccus, group e, ANME-1, ANME-2 [10, 12]) and, using a revised general mcrA primer pair (15), a novel deeply branching mcrA cluster (12). Perhaps surprisingly, the anaerobic methanotrophic ANME-1 cluster, a dominant group in clone libraries of archaeal 16S rRNA genes (4), was initially absent from mcrA clone libraries (10). This inconsistency has since been resolved by using an ANME-1-specific mcrA primer pair (15), with which high diversity and widespread occurrence of ANME-1 in Guaymas Basin sediment have been shown (12).
The fact that revised general and new ANME-1-specific mcrA primers detected a wider phylogenetic range of mcrA gene diversity than that obtained in previous studies on Guaymas Basin sediments (10, 11) raises several questions. (i) How do published general mcrA primer pairs compare in diversity coverage and detection sensitivity? (ii) To what extent do these general mcrA primer pairs cover the diversity of mcrA genes present in Guaymas Basin sediment? (iii) How do different sites within the Guaymas Basin compare in terms of methanogen and anaerobic methanotroph diversity? (iv) How do Guaymas Basin methanogenic and anaerobic methanotrophic communities compare to methanogenic and anaerobic methanotrophic communities elsewhere on Earth?
We examine these questions in surficial sediments of the Everest Mound area in the southern Guaymas vent field, home to diverse lineages of methanogens and methanotrophs (4, 10). We first compare the detection sensitivity and diversity coverage of three degenerate, general mcrA PCR primer pairs, designed by Springer et al. (13), Hales et al. (16), and more recently Lever (15). We then evaluate detection sensitivity and phylogenetic biases by comparing clone libraries based on these general mcrA primer pairs to ones obtained using 27 nondegenerate, group-specific mcrA primer pairs (15). As a further check for the phylogenetic range of general and group-specific mcrA primers, we compare the total community detected based on mcrA sequences to that detected with 17 new methanogen- and anaerobic methanotroph-targeted 16S rRNA gene primer pairs. We conclude with an analysis of methane-cycling archaeal zonation within the steep thermal and sulfate gradients of the site, investigate possible metabolic pathways of phylotypes detected based on closest relatives with known metabolisms, and compare the overall phylogenetic range and community structure to those found in previous Guaymas Basin sediment studies and other habitats worldwide.
MATERIALS AND METHODS
Sampling and site characteristics.
The sediment core used in this study was obtained during Alvin Dive 3204 from the “Everest Mound” area in the southern Guaymas Trench. Temperature gradients were determined in situ using Alvin's high-temperature probe. The pore water sulfate concentration profile was determined using standard ion chromatographic methods (12). An extended site description is provided in the supplemental material.
DNA extraction and primer design.
DNA was extracted according to Lever et al. (17; see also the supplemental material). All primer information is shown in the supplemental material and in Table 1. The general mcrIRD primer pair and the group-specific mcrA primer pairs as well as all new archaeal 16S rRNA gene primers were designed based on DNA sequence alignments in the ARB software (http://www.arb-home.de/ [18]) and tested with the DNA extracts. Annealing temperatures were based on calculated melting temperatures. For each primer pair, the lower of the two melting temperatures was used as annealing temperature.
TABLE 1.
Names, target groups, nucleotide sequences, primer lengths, amplicon lengths, annealing temperatures, and amplification success of mcrA and archaeal 16S rRNA gene primers with DNA extracts from Everest Mounda
| Name | Target groups | Primer sequences | No. of nucleotides | Length (bp) | Tanneal (°C) | Amplificationb | Specificityc |
|---|---|---|---|---|---|---|---|
| mcrA gene primers | |||||||
| mcrI | General | F, 5′-TAYGAYCARATHTGGYT | 17; 17 | ∼490 | 51 | + | + |
| R, 5′-ACRTTCATNGCRTARTT | |||||||
| ME1/ME2 | General | F, 5′-CMATGCARATHGGWATGTC | 20; 21 | ∼750 | 58 | + | + |
| R, 5′-TCATKGCRTAGTTDGGRTAGT | |||||||
| mcrIRD | General except ANME-1 | F, 5′-TWYGACCARATMTGGYT | 17; 17 | ∼490 | 55 | + | + |
| R, 5′-ACRTTCATBGCRTARTT | |||||||
| ANME-1-mcrI | ANME-1, ANME-1-related group | F, 5′-GACCAGTTGTGGTTCGGAAC | 20; 20 | ∼480 | 63 | + | + |
| R, 5′-ATCTCGAATGGCATTCCCTC | |||||||
| mcrMS | Methanosarcina | F, 5′-GACCAGATCTGGCTCGGATC | 20; 20 | 425 | 66 | + | − |
| R, 5′-TCGCCCTGGTAGGACAGAAC | |||||||
| mcrANME-2 | ANME-2 | F, 5′-GGATTCACGCAGTACGCAAC | 20; 20 | 155 | 64 | + | + |
| R, 5′-CAAGAAGCGTTGGGTAGTCC | |||||||
| mcrMsaeta | Methanosaeta | F, 5′-TACACCAACGATGTCCTGGA | 20; 20 | 305 | 64 | + | − |
| R, 5′-CACTGATCCTGCAGGTCGTA | |||||||
| mcrURFS | Unidentified Rice Field Soil McrA/Zoige cluster | F, 5′-TATGCAACACCAGCATACACC/GTATGCCACAGCAGCATACAC | 22/21; 17 | 385 | 64 | + | − |
| R, 5′-CACCGCACTGATCCTGC | |||||||
| mcrAM-3 I | ANME-3 | F, 5′-GATATCATTCAGACAAGCCG | 20; 20 | 525 | 60 | − | NA |
| R, 5′-AGTTCAAGAGGCTCTCCTTC | |||||||
| mcrAM-3 II | ANME-3 | F, 5′-CCTTGAGGTAGTCGGTGCAG | 20; 22 | 480 | 64 | − | NA |
| R, 5′-AGTTCAAGAGGCTCTCCTTCGT | |||||||
| mcrAM-3etal | ANME-3, Methanococcoides, Methanomethylovorans, Methanolobus, Methanohalophilus | F, 5′-GATATCATTCAGACAAGCCGTR, 5′-CACCACACTGGTCCTGC | 21; 17 | 480 | 60 | + | − |
| mcrRCI/FC | Rice Cluster I, Fen cluster | F, 5′-TACAAGATGTGCGCCGGT | 18; 20 | 560 | 64 | − | NA |
| R, 5′-CATGCTTCCTTGTGCAGGTA | |||||||
| mcrFCI | Fen cluster | F, 5′-AGCCAGGTGGCATCAAGTT | 19; 20 | 510 | 64 | − | NA |
| R, 5′-ACTGGTCCTGCAGGTCGTAG | |||||||
| mcrFCII | Fen cluster | F, 5′-AGCCAGGTGGCATCAAGTT | 19; 19 | 445 | 64 | + | − |
| R, 5′-GACAGGTACCAGCCGTTCA | |||||||
| mcrMcorp | Methanocorpusculum | F, 5′-TGTCATCAACATGGCCCAC | 19; 20 | 550 | 64 | + | − |
| R, 5′-TCGTAGCCGAAGAAACCAAG | |||||||
| mcrMspir | Methanospirillum | F, 5′-GATGAGTTCACCTACTATGGTATR, 5′-CTGACAGAGAGTGAGTTGGT | 23; 20 | 335 | 56 | + | − |
| mcrMmicrob | Methanomicrobiales | F, 5′-CACCTACTACGGTATGGACTA | 21; 20 | 310 | 61 | + | (−/+) |
| R, 5′-GAGTTTGCTGAACCACACTG | |||||||
| mcrMcul | Methanoculleus and close relatives | F, 5′-GGTATGGACTACATCAAGGACAAR, 5′-ACTGGTCCTGGAGGTCGTA | 23; 19 | 290 | 62 | + | (−/+) |
| mcrGMcul | Guaymas Basin Methanomicrobiales mcrA cluster | F, 5′-CACCTACTACGGTATGGACTA | 21; 18 | 355 | 61 | + | + |
| R, 5′-AGCTCTCCGAGCAGACCT | |||||||
| mcrMbacA | Methanobacterium aarhusense group | F, 5′-GCAAAACACGCAGAAGTTGT | 20; 22 | 515 | 63 | + | − |
| R, 5′-GTCTGGAGTGCTGTTCTTTGTG | |||||||
| mcrMbac | Methanobacteriales except M. aarhusense and Methanothermobacter | F, 5′-GGTTAGGTTCTTACATGTCTGGTGR, 5′-GCACCACATTGATCTTGTAAATC/TGCTCCACACTGGTCCTG/CACCACACTGGTCCTGGA | 24; 23 | 365 | 63 | + | (−/+) |
| mcrMcoc | Methanococcaceae | F, 5′-AAGAAGAGCAAGAGGTCCAAA | 21; 23 | 490 | 61 | + | −/+ |
| R, 5′-TCGTATCCGTAGAATCCTAATCT | |||||||
| mcrMtb | Methanothermobacter | F, 5′-AGCCTACACAGACAACATCCTCR, 5′-CACCACACTGGTCCTGGA | 22; 18 | 315 | 63 | + | − |
| mcrMpK | Methanopyrus kandleri | F, 5′-CTAGGATCCTACATGTCAGGAGGR, 5′-CCTCACGCTCAGCGAGTT | 23; 18 | 385 | 64 | + | + |
| mcrDBGrII | Deeply Branching mcrA group II | F, 5′-GGGAGTAGGATTCACGCAGTA | 21; 22 | 410 | 64 | + | + |
| R, 5′-GATAGTTTGGACCACGCAGTTC | |||||||
| mcrMlas | Clone mlas, clone DEBITS and relatives | F, 5′-ACGACTTCTGCTACTACGGTGCR, 5′-CCTGCCCATCTCCTCCTT | 22; 18 | 255 | 64 | + | − |
| mcrDBGrIII | Deeply Branching mcrA group III | F, 5′-GCAGTATGCAACCGCTGTT | 18; 19 | 395 | 64 | + | + |
| R, 5′-GTCTGCACCTCTGAGCTCAAG | |||||||
| mcrMpyralesI&II | Methanopyrus mcrA subclusters I and II | F, 5′-GTGTACACGGACAACATCCTGGR, 5′-ACGCTCAGCGAGTTGGC | 22; 17 | 330 | 64 | + | − |
| Archaeal 16S rRNA gene primers | |||||||
| MS 183F/MS 1138R | Methanosarcina | F, 5′-TGCTGGAATGCTTTATGCGT | 20; 19 | 955 | 65 | − | − |
| R, 5′-CCGGAGGACATGCTGGTAA | |||||||
| MS 184F/MS 1009R | Methanosarcina | F, 5′-GCTGGAATGCTTTATGCGT | 19; 20 | 825 | 62 | + | − |
| R, 5′-TGGCCTACATATTGCTGTCG | |||||||
| MCC 221F/MCC&Mlob 1155R | Methanococcoides, Methanolobales, Methanohalophilus | F, 5′-CCTAAGGATGGATCTGCGG | 19; 21 | 934 | 64 | + | −/+ |
| R, 5′-CCACAGAGTACCCATCATCCC | |||||||
| RCI 549F/RCI 1014R | Rice Cluster I | F, 5′-TGGTGGCCGATATTATTGAGTC | 22; 20 | 465 | 64 | + | (−/+) |
| R, 5′-TCAGCCTGGCCTTCATACAA | |||||||
| ANME-2 244F/ANME-2 836R | ANME-2 | F, 5′-TCAGGTTGTAGTGGGTGTAA | 20; 20 | 592 | 60 | − | NA |
| R, 5′-CTGACACATAGCGAGCATCG | |||||||
| Msta 268F/Msta 927R | Methanosaeta | F, 5′-CCTACTAGCCTACGACGGGT | 20; 21 | 659 | 62 | + | (−/+) |
| R, 5′-CCCGCCAATTCCTTTAAGTTT | |||||||
| Msta 571F/Msta 927R | Methanosaeta | F, 5′-TAAAGGGTCTGTAGCCGGCC | 20; 21 | 356 | 65 | + | (−/+) |
| R, 5′-CCCGCCAATTCCTTTAAGTTT | |||||||
| MM 48F/MM 1171R | Methanomicrobiales | F, 5′-TTAAGCCATGCGAGTCGAGA | 20; 19 | 1123 | 65 | + | + |
| R, 5′-TTTAGCAGAGGCGGTCCCA | |||||||
| ANME-1 42F/ANME-1 898R | ANME-1 | F, 5′-GAGTTCGATTAAGCCATGTTAGTR, 5′-CGACCGTACTCCCCAGAT | 23; 18 | 856 | 60 | + | + |
| ANME-1 Deep 35F/ANME-1 Deep 1038R | Deeply Branching ANME-1 | F, 5′-GCTATCAGCGTCCGACTAAGC | 21; 19 | 1003 | 65 | + | (−/+) |
| R, 5′-TAATCCGGCAGGGTCTTCA | |||||||
| ANME-1 Deep 176F/ANME-1 Deep 1038R | Deeply Branching ANME-1 | F, 5′-CGGATAGGCCTCTGATACCTG | 21; 19 | 852 | 64 | − | NA |
| R, 5′-TAATCCGGCAGGGTCTTCA | |||||||
| MB 136F/MB 873R | Methanobacteriales | F, 5′-CCTTAGGACTGGGATAACCC | 20; 19 | 737 | 62 | − | NA |
| R, 5′-TTAACAGCTTCCCTTCGGC | |||||||
| MB 137F/MB 873R | Methanobacteriales | F, 5′-CTTGGGACCGGGATAACC | 18; 19 | 736 | 64 | − | NA |
| R, 5′-TTAACAGCTTCCCTTCGGC | |||||||
| MB 310F/MB 873R | Methanobacteriales | F, 5′-CGGAGATGGAACCTGAGACA | 20; 19 | 563 | 64 | − | NA |
| R, 5′-TTAACAGCTTCCCTTCGGC | |||||||
| MC 266F/MC 910R | Methanococcales | F, 5′-GCCCACCAAGCCTACGATC | 19; 22 | 644 | 65 | − | NA |
| R, 5′-TTTCAGTCTTGCGACCGTACTC | |||||||
| MP&MT 235F/ | Methanopyrales and Methanothermales | F, 5′-TGCGGCCGATTAGGTAGTT | 19; 20 | 786 | 64 | + | (−/+) |
| MT 1021R | R, 5′-AAGGTCATCAACCTGGCCAT | ||||||
| MP&MT 235F/ | Methanopyrales | F, 5′-TGCGGCCGATTAGGTAGTT | 19; 20 | 748 | 64 | + | (−/+) |
| MP 983 R | R, 5′-TAAGGTTTCCGGCGTTGAAT |
PCR protocol.
PCR assays were performed with the TaKaRa SpeedSTAR HS DNA polymerase kit (TaKaRa Bio USA, Madison, WI) using the manufacturer's recommended reaction mixture, except that bovine serum albumin was added to a final concentration of 1 μg μl−1. The PCR protocol was as follows: (i) one 2-min denaturation (98°C), (ii) 40 cycles of 10-s denaturation (98°C), 30-s annealing (Table 1 for temperatures), and 1-min extension (72°C), and (iii) one 5-min extension (72°C). For assays with general mcrA primers, 10 μl of DNA extract was used. For all other assays, 1 μl of extract was used. Where necessary, 1 μl of PCR product from the first PCR was transferred to tubes containing fresh PCR reagents and reamplified for a second PCR of 40 cycles.
Cloning and sequencing.
PCR products were purified in a 2.5% low-melting-point agarose gel using Tris acetate-EDTA (TAE) buffer. Gel slices containing the correct PCR fragment length were excised and purified using the SN.A.P. minikit (Invitrogen, Carlsbad, USA) and cloned into electrocompetent Escherichia coli using the Topo TA kit (Invitrogen, Carlsbad, CA, USA). Plasmid extraction, purification, and cycle sequencing were performed at the Josephine Bay Paul Center at the Marine Biological Laboratory (Woods Hole, MA).
Phylogenetic trees.
Sequences were BLAST analyzed (www.ncbi.nlm.nih.gov/blast). Chimeras were identified by visual alignment checks in ARB and using the online software Database Enabled Code for Ideal Probe Hybridization Employing R (DECIPHER; http://decipher.cee.wisc.edu/index.html). Phylogenetic trees were created using ARB neighbor joining with Jukes-Cantor correction and bootstrap analyses with 1,000 replicates. The taxonomic identification and classification of novel mcrA phylotypes were checked and substantiated with nucleotide sequence similarity matrixes, as specified in the supplemental material.
Nucleotide sequence accession numbers.
Nucleotide sequences were deposited to the GenBank archive under the following accession numbers: for mcrA, KM370762 to KM370786; for archaeal 16S rRNA genes, KM370787 to KM370815).
RESULTS
General results.
DNA extractions were successful to 10 cm below seafloor (cmbsf), at which the sediment temperature was ∼60°C (Fig. 1A). Sulfate concentrations decreased steeply in the upper cmbsf and were below the detection limit (∼0.1 mM) below a 4-cm depth, indicating active sulfate reduction within the surface layers and sulfate limitation below (Fig. 1B). The absence of sulfate in the deeper sediment layers indicates minimal core disturbance and seawater in-mixing during sampling and retrieval.
FIG 1.
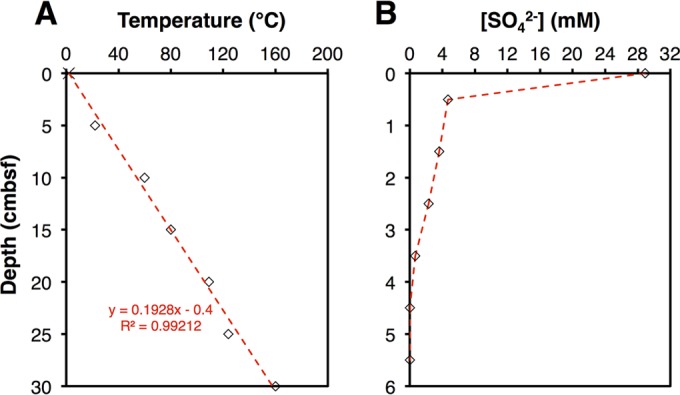
(A) Measured temperature data for the sediment interval examined by Weber & Jørgensen (5 to 30 cmbsf). The dashed red line indicates the best-fit line, assuming a temperature of 2°C at the sediment surface. The slope of this line was used to model temperatures at higher depth resolution throughout the sediment interval examined in this study. (B) Sulfate concentration profile. Sulfate could not be detected below 4 cmbsf.
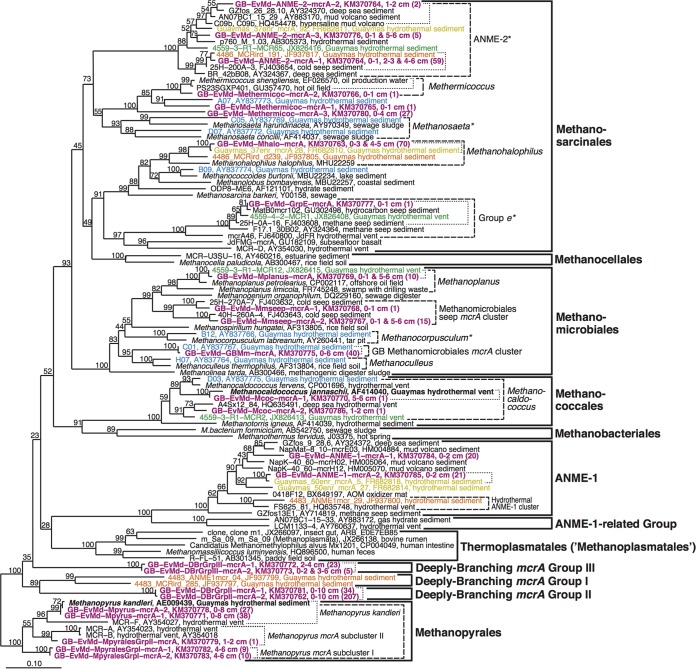
The combined mcrA and archaeal 16S rRNA gene surveys identified a total of 22 phylotypes of methane-cycling Archaea. Here, distinct phylotypes were defined as having <97% sequence similarity to other sequences detected in this study; the same 97% cutoff is used consistently for mcrA and 16S rRNA genes. Similarly, all mcrA sequences from this and previous studies that shared ≥97% sequence similarity were classified as belonging to the same phylotype. A total of 21 phylotypes were detected among the mcrA sequences (Fig. 2), and only one additional phylotype, belonging to the candidate order “Methanoplasmatales,” was found among the 16S rRNA gene sequences (see Fig. S4 in the supplemental material). Several additional 16S rRNA phylotypes belonging to euryarchaeotal groups without known physiologies might, however, be involved in methane cycling. Of the phylotypes that could be linked to methane cycling based on mcrA detection or based on 16S rRNA gene sequences that are monophyletic with known methane-cycling Archaea, 20 fall into groups with known energy substrates. The remaining two mcrA phylotypes (termed “Deeply Branching mcrA groups II and III”) lack close cultured relatives or in fact any environmental sequences with high sequence similarity (Fig. 2; Table 1). The numbers of phylotypes had a bimodal distribution, with the highest numbers in cool, sulfate-containing surface samples (0 to 2 cm, ∼2 to 12°C) and in a warm, sulfate-depleted layer (5 to 6 cm; ∼20 to 30°C) (Fig. 2; Tables 2 and 3; see also Fig. S4 in the supplemental material). Only two phylotypes were detected below 6 cmbsf.
FIG 2.
mcrA gene phylogeny. Representative phylotypes from this study are shown in bold magenta type font. The depth interval of origin is shown, along with the number of clone sequences in parentheses. Phylotypes from other studies on the Guaymas Basin have the following color codes: bold black, pure culture isolates; blue, Dhillon et al. (10); yellow, Holler et al. (11); orange, Biddle et al. (12); green, Y. He and F. Wang, unpublished data. Bootstrap values of ≥50% are shown at branch nodes. Based on sequence similarity calculations, we made the following phylogenetic distinctions: sequences likely to belong to the same species are marked by dotted lines, dashed lines indicate members of the same genus, asterisks (*) indicate genus level mcrA branches that also represent families, and thick solid lines indicate sequences that belong to the same order (for more information on these calculations, see the supplemental material, as well as Discussion).
TABLE 2.
Overview of phylotypes from within Guaymas Basin, and outside Guaymas Basin, with highest BLAST sequence similarity to ones detected at Everest Mounda
| OTU name | Closest BLAST hit from Guaymas Basin (accession no.) | Habitat | % similarity | Reference | Closest BLAST hit outside Guaymas Basin (accession no.) | Origin | % similarity | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| mcrA genes | ||||||||
| GB-EvMd-Mhalo-mcrA | Guaymas_37enr_mcrA 28 (FR682810) | Hydrothermal sediment | 99 | 11 | Methanohalophilus halophilus (AB703633) | Shark Bay hypersaline microbial mat | 86 | 21 |
| GB-EvMd-GrpE-mcrA | 4559–4-2-MCR1 (JX826408) | Hydrothermal vent | 98 | Y. He and F. Wang, unpublished | 25H-0A-16 (FJ403608) | Sea of Ohkotsk cold seeps | 99 | 22 |
| GB-EvMd-ANME-2-mcrA-2 | 4559-3-R1-MCR65 (JX826416) | Hydrothermal vent | 93 | Y. He and F. Wang, unpublished | C09b (HQ454478) | Napoli mud volcano | 97 | 23 |
| GB-EvMd-ANME-2-mcrA-1 | 4486_MCRird_191 (JF937817) | Hydrothermal sediment | 99 | 12 | 25H-200A-3 (FJ403654) | Sea of Ohkotsk cold seeps | 99 | Zhang et al., unpublished |
| GB-EvMd-ANME-2-mcrA-3 | Guaymas_37enr_mcrA 92 (FR682811) | Hydrothermal sediment | 93 | 11 | p760_M_1.03 (AB305373) | Okinawa Trough hydrothermal sediment | 95 | 24 |
| GB-EvMd-Methermicoc-mcrA-2 | A07 (AY837773) | Hydrothermal sediment | 82 | 10 | PS23SGXP401 (GU357470) | Hot Alaskan oil field | 96 | 25 |
| GB-EvMd-Methermicoc-mcrA-1 | A07 (AY837773) | Hydrothermal sediment | 85 | 10 | FS448_53 (HQ635543) | Mariana Arc, hydrothermal vent fluid | 94 | 26, 27 |
| GB-EvMd-Methermicoc-mcrA-3 | A07 (AY837773) | Hydrothermal sediment | 77 | 10 | PS23SGXP401 (GU357470) | Hot Alaskan oil field | 83 | 25 |
| GB-EvMd-Mmseep-mcrA-2 | 4559-3-R1-MCR12 (JX826415) | Hydrothermal vent | 82 | Y. He and F. Wang, unpublished | 40H-260A-4 (FJ403643) | Sea of Ohkotsk cold seep | 98 | Zhang et al., unpublished |
| GB-EvMd-Mmseep-mcrA-1 | None | 25H-270A-7 (FJ403632) | Sea of Ohkotsk cold seep | 96 | Zhang et al., unpublished | |||
| GB-EvMd-Mplanus-mcrA | 4559-3-R1-MCR12 (JX826415) | Hydrothermal vent | 99 | Y. He and F. Wang, unpublished | LW-6 (JX430031) | Min River estuary | 92 | C.-X. She and C. Tong, unpublished |
| GB-EvMd-Mcull-mcrA | C01 (AY837767) | Hydrothermal sediment | 99 | 10 | ATB-EN-10393-M138 (FJ226731) | Agricultural biogas plant | 87 | 28 |
| GB-EvMd-Mcoc-mcrA-1 | Methanocaldococcus jannaschii (L77117) | Hydrothermal sediment | 99 | 19 | FS625_56 (HQ635721) | Juan de Fuca Ridge hydrothermal vent fluid | 99 | 26 |
| GB-EvMd-Mcoc-mcrA-2 | 4559-3-R1-MCR2 (JX826413) | Hydrothermal sediment | 96 | Y. He and F. Wang, unpublished | A4Sx12_84 (HQ635491) | Juan de Fuca Ridge hydrothermal vent fluid | 95 | 26 |
| GB-EvMd-ANME-1-mcrA-1 | None | NapMat-8_10-mcrE03 (HM004884) | Napoli mud volcano hypersaline sediment | 92 | 23 | |||
| GB-EvMd-ANME-1-mcrA-2 | Guaymas_50enr_mcrA 63 (FR682818) | Hydrothermal sediment | 98 | 11 | NapK-40_60-mcrH12 (HM005070) | Napoli mud volcano hypersaline sediment | 92 | 29 |
| GB-EvMd-DbrGrpII-mcrA-1 | 4483_MCRird_285 (JF937797) | Hydrothermal sediment | 72 | 12 | M_mcrA-11 (JQ406852) | Huabei oil field production water | 72 | 30 |
| GB-EvMd-DbrGrpII-mcrA-2 | 4483_MCRird_285 (JF937797) | Hydrothermal sediment | 71 | 12 | M_mcrA-11 (JQ406852) | Huabei oil field production water | 72 | 30 |
| GB-EvMd-DbrGrpIII-mcrA-1 | Methanopyrus kandleri | Hydrothermal sediment | 71 | 20 | FS625_63 (HQ635729) | Juan de Fuca Ridge hydrothermal vent fluid | 74 | 26 |
| GB-EvMd-DbrGrpIII-mcrA-2 | Methanopyrus kandleri | Hydrothermal sediment | 71 | 20 | FS625_63 (HQ635729) | Juan de Fuca Ridge hydrothermal vent fluid | 75 | 26 |
| GB-EvMd-MpyralesGrpI-mcrA | Methanopyrus kandleri | Hydrothermal sediment | 88 | 20 | MCR-A (AY354023) | EPR diffuse vent covered by mats | 94 | 31 |
| GB-EvMd-MpyralesGrpII-mcrA-1 | Methanopyrus kandleri | Hydrothermal sediment | 86 | 20 | MCR-B (AY354018) | EPR diffuse vent covered by mats | 89 | 31 |
| GB-EvMd-MpyralesGrpII-mcrA-2 | Methanopyrus kandleri | Hydrothermal sediment | 86 | 20 | MCR-B (AY354018) | EPR diffuse vent covered by mats | 89 | 31 |
| GB-EvMd-Mpyrus-mcrA-2 | Methanopyrus kandleri | Hydrothermal sediment | 99 | 20 | 1crD2T36 (FN650318) | Rainbow ultramafic hydrothermal vent fluid | 89 | 32 |
| GB-EvMd-Mpyrus-mcrA-1 | Methanopyrus kandleri | Hydrothermal sediment | 99 | 20 | 1crD2T36 (FN650318) | Rainbow ultramafic hydrothermal vent fluid | 90 | 32 |
| Archaeal 16S rRNA genes | ||||||||
| GB-EvMd-Methermicoc-A | 4A08 (AY835419) | Hydrothermal sediment | 97 | 10 | Methermicoccus shengliensis (NR_043960) | Shengli oil field, production water | 99 | 44 |
| GB-EvMd-Methermicoc-C | 4A08 (AY835419) | Hydrothermal sediment | 100 | 10 | Methermicoccus shengliensis (NR_043960) | Shengli oil field, production water | 99 | 44 |
| GB-EvMd-Halo-C | GUAY_37enr_Arch72 (FR682485) | Hydrothermal sediment | 99 | 51 | SS_WC_06 (FJ656258) | Salton Sea, hypersaline lake sediment | 99 | 56, 57 |
| GB-EvMd-Mcull-A | 7H12 (AY835412) | Hydrothermal sediment | 99 | 10 | QHYA-25 (JF741950) | Oil reservoir | 96 | Tang et al., unpublished |
| GB-EvMd-Mcull-B | 7H12 (AY835412) | Hydrothermal sediment | 100 | 10 | Methanoculleus sp. clone A3 (AJ133793) | Freshwater sediment enriched with hexadecane | 96 | 58 |
| GB-EvMd-MBGD-A | 4559-4-C2-82 (JX507259) | Hydrothermal vent | 98 | Y. He and F. Wang, unpublished | KSTwh-C1-7-A-028 (JQ611039) | Kueishan Is shallow hydrothermal field | 90 | Lin et al., unpublished |
| GB-EvMd-MBGD-B | 4559-4-C2-82 (JX507259) | Hydrothermal vent | 98 | Y. He and F. Wang, unpublished | IAN1-62 (AB175594) | Oknawa Trough, hydrothermal vent chimney | 92 | 59 |
| GB-EvMd-Mplasmatales | 7C08 (AY835423) | Hydrothermal sediment | 99 | 10 | 0DA-79 (JQ772440) | High-temperature oil reservoir | 99 | Z. Fang, unpublished |
| GB-EvMd-DHVE5-A | None | a87R29 (DQ417481) | EPR, seafloor basaltic flanks | 97 | 60 | |||
| GB-EvMd-DHVE5-B | None | ARCHDER07_1A1 (FN598017) | Almeria, Spain, seawater-processed activated sludge | 88 | 61 | |||
| GB-EvMd-ANME-1a-B | GUAY_50enr_Arch41 (FR682490) | Hydrothermal sediment | 99 | 11 | MC118_26C2 (HM600925) | Gulf of Mexico hydrocarbon seep sediment | 99 | Twing et al., unpublished |
| GB-EvMd-ANME-1a-C | Arch4483_112 (JF937751) | Hydrothermal sediment | 99 | 12 | MC118_26C2 (HM600925) | Gulf of Mexico hydrocarbon seep sediment | 99 | Twing et al., unpublished |
| GB-EvMd-ANME-1b-A | BG410(1)9 (JQ740749) | Hydrothermal vent fluid | 99 | 27 | FS725(3)73 (JQ740760) | Endeavor Segment hydrothermal fluid | 97 | 27 |
| GB-EvMd-ANME-1b-C | GBa2r032 (AF419632) | Hydrothermal sediment | 99 | 4 | FS625(3)16 (JQ740759) | Endeavor Segment hydrothermal fluid | 99 | 27 |
| GB-EvMd-GBEury-G | Arch4486_075 (JF937738) | Hydrothermal sediment | 99 | 12 | KZNMV-25-A37 (FJ712390) | Kazan Mud Volcano, hydrate sediment | 100 | 62 |
| GB-EvMd-GBEury-E | Arch4486_075 (JF937738) | Hydrothermal sediment | 96 | 12 | Arch-Q12 (JQ241421) | Huabei oil field | 98 | 63 |
| GB-EvMd-GBEury-K | 4E09 (AY835427) | Hydrothermal sediment | 98 | 10 | KZNMV-25-A37 (FJ712390) | Kazan Mud Volcano, hydrate sediment | 95 | 62 |
| GB-EvMd-ThCoc-D | Thermococcus sp. GB18 (FJ862790) | Hydrothermal sediment | 99 | 52 | Thermococcus alcaliphilus (NR040870) | Vulcano submarine solfataric field | 99 | 64 |
| GB-EvMd-ThCoc-E | T. mexicalis (AY099181) | Hydrothermal sediment | 99 | 53 | HibPWCl-ARC3 (JF789485) | Hibernia oil field, production water | 99 | Yeung et al., unpublished |
| GB-EvMd-ThCoc-F | T.coccus guaymasensis (JQ346762) | Hydrothermal sediment | 98 | 53 | Thermococcus sp. Ax00-27 (AY559130) | Axial seamount, diffuse hydrothermal vent | 97 | J. A. Huber and J. A. Baross, unpublished |
| GB-EvMd-ThCoc-G | G26_C48 (AF356631) | Sulfide chimney | 95 | K. Longnecker and A.-L. Reysenbach, unpublished | Q2-a15.seq (FR852947) | Hydrothermal vent chimney | 94 | J. Li, unpublished |
| GB-EvMd-Archaeoglobales | Archaeoglobus profundus (AF297529) | Hydrothermal sediment | 93 | 54 | PW15.7A (EU573156) | North Sea chalk petroleum reservoir | 98 | 65 |
| GB-EvMd-DHVEG | 90-PY-10 (FR692155) | Hydrothermal sediment | 99 | 55 | pCIRA-L (AB095122) | Central Indian Ridge hydrothermal field | 96 | 66 |
| GB-EvMd-AcidProf-A | G26_C56 (AF356635) | Sulfide chimney | 98 | K. Longnecker and A.-L. Reysenbach, unpublished | HTM1039Pn-A31 (AB611429) | Okinawa Trough, polychaete nest, hydroth. vent | 99 | 67 |
| GB-EvMd-AcidProf-B | G26_C56 (AF356635) | Sulfide chimney | 98 | K. Longnecker and A.-L. Reysenbach, unpublished | Aciduliprofundum isolate EPR07-39 (FR865186) | EPR hydrothermal vent | 99 | 68 |
| GB-EvMd-AcidProf-F | G26_C56 (AF356635) | Sulfide chimney | 95 | K. Longnecker and A.-L. Reysenbach, unpublished | Aciduliprofundum isolate EPR07-39 (FR865186) | EPR hydrothermal vent | 96 | 68 |
| GB-EvMd-MCG-A | 4559-4-C2-11 (JX507250) | Hydrothermal vent | 99 | Y. He and F. Wang, unpublished | PNG_Kap4_A49 (JF935159) | Alkaline hot spring | 99 | Amend et al., unpublished |
| GB-EvMd-MCG-B | 4559-4-C2-9-5 (JX507244) | Hydrothermal vent | 97 | Y. He and F. Wang, unpublished | A257-49 (FN554070) | Logatchev ultramafic hydrothermal vent field | 98 | 69 |
| GB-EvMd-Thermoproteales-1 | 4559-4-C2-61 (JX507245) | Hydrothermal vent | 100 | Y. He and F. Wang, unpublished | HTM1036Pn-A14 (AB611436) | Okinawa Trough, polychaete nest, hydrothermal vent | 96 | 67 |
TABLE 3.
Depth intervals in which various mcrA clusters were detected using general and group-specific primersa
| Primer | Depth (cmbsf) | No. of mcrA clusters (of 14) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methanoplanus | Methanomicrobiales Seep mcrA cluster | GB Methanomicrobiales mcrA cluster | Methanocaldococcus | Methanopyrus kandleri | Methanohalophilus | Methermicoccusb | ANME-1 | ANME-2 | Group e | DB Guaymas group II | DB Guaymas group III | Methanopyrus mcrA subcluster I | Methanopyrus mcrA subcluster II |
| ME1/ME2 | 0–1, 5–6 | 0–1, 5–6 | 0–1, 4–5 | 4–5 | 0–2, 4–5 | 0–1 | 0–2, 4–6 | 7 | |||||
| mcrI | 0–1 | 0–1 | 0–1, 2–3, 4–5 | 5–6 | 2–3, 4–6 | 0–1 | 0–1, 2–3 | 0–1 | 0–1, 2–3 | 2–3 | 10 | ||
| mcrIRD | 0–1 | 0–1, 5–6 | 0–3 | 1–2 | 0–1, 2–8 | 0–3 | 0–4 | 0–1 | 0–1 | 0–10 | 4–6 | 1–2 | 12 |
| _mcr_ANME-1 | 0–2 | 1 | |||||||||||
| _mcr_ANME-2 | 0–3 | 1 | |||||||||||
| _mcr_FCI | 1–2 | 1 | |||||||||||
| _mcr_Mmicrob | 0–4 | 1 | |||||||||||
| _mcr_Mcul | 2–3 | 1 | |||||||||||
| _mcr_GMcul | 0–3, 5–6 | 1 | |||||||||||
| _mcr_Mbac | 0–1 | 0–2 | 0–1 | 3 | |||||||||
| _mcr_Mcoc | 1–2, 3–4 | 2–4 | 1 | ||||||||||
| _mcr_MpK | 0–8 | 1 | |||||||||||
| _mcr_DBGrII | 0–10 | 1 | |||||||||||
| _mcr_DBGrIII | 0–6 | 2 |
Among the closest BLAST hits to the 21 mcrA phylotypes, 8 were from Guaymas Basin sediment, 1 was from a Guaymas Basin hydrothermal vent, and 12 were from outside Guaymas Basin (Table 2). These closest BLAST hits included the pure-culture isolates Methanocaldococcus jannaschii and Methanopyrus kandleri, both of which were first isolated from a hydrothermal vent and a hydrothermal sediment in Guaymas Basin, respectively (19, 20).
mcrA diversity.
We designed a sequence similarity matrix for taxonomic classification of mcrA phylotypes that is analogous to sequence similarity cutoffs that have been employed to classify 16S rRNA gene sequences for decades (33, 34). To discuss mcrA diversity in consistent terms, we use the term “group” to refer to sequences classified as forming their own family, order, or class, the term “cluster” to refer to sequences classified as forming their own genus, and the term “subcluster” to refer to sequences classified as belonging to the same phylotype. In the following, we discuss mcrA diversity at Everest Mound based on this classification scheme. For uncultured clusters, these designations are necessarily qualified, for example, by referring to “genus level lineages.”
Within the Methanosarcinales, we detected 3 phylotypes in the ANME-2 family (Fig. 2), of which one is nearly identical to a previously detected sequence from Guaymas Basin. Three other phylotypes are monophyletic with Methermicoccus shengliensis and related environmental sequences. One of these phylotypes belongs to the same genus as M. shengliensis. The other two phylotypes are considerably divergent and likely belong to two additional genera within the family Methermicoccaceae (see Table S4 in the supplemental material). Phylotypes of Methanohalophilus and group e are virtually identical to previously detected sequences from Guaymas Basin sediment and a hydrothermal vent, respectively. Group e forms a sister family to the family Unidentified Rice Field Soil mcrA group/Zoige cluster I (represented by the sequence with accession number GU182109), which in turn forms a sister family to its neighboring branch (represented by the sequence with the accession number AY354030).
Within the Methanomicrobiales, a Methanoplanus phylotype is nearly identical to a previously detected sequence from a Guaymas Basin hydrothermal vent and closely related to Methanoplanus petrolearius, which was isolated from an oil field (35). Sequences of the genus level Methanomicrobiales seep mcrA cluster appear for the first time in Guaymas Basin. We also detect mcrA sequences of the same phylotype as a sequence previously detected in Guaymas Basin sediment (AY837767); this phylotype clusters separately from other genera of Methanomicrobiales and has only low DNA sequence similarity to the closest genera, Methanoculleus and Methanofollis (83.0% ± 2.3% and 84.1%, respectively; Fig. 2 [Methanofollis not shown]; see also Table S4 in the supplemental material). We classify this phylotype, which so far lacks mcrA sequences with high sequence similarity outside Guaymas Basin, as a member of the new genus level “Guaymas Basin Methanomicrobiales mcrA cluster.”
Within the Methanococcales, one sequence is nearly identical to Methanocaldococcus jannaschii; a second sequence forms a distinct phylotype with high sequence similarity to other Methanocaldococcus phylotypes from hydrothermal vents, including one from Guaymas Basin (Fig. 2).
Two phylotypes fall into the ANME-1 Archaea, which forms its own order and even a separate class along with the neighboring order level ANME-1-related group (Fig. 2; see also Table S4 in the supplemental material). One of these phylotypes clusters with sequences from cold deep sea and mud volcano sediments. The other is nearly identical to thermophilic ANME-1 previously enriched by Holler et al. (11). All ANME-1 archaea detected here fall into the same genus level lineage. The JF937800 phylotype, which is the likely mcrA equivalent of the ANME-1Guaymas 16S rRNA phylotype (12), forms a separate genus level lineage together with a phylotype from hydrothermal fluid of the Endeavor Segment (HQ635748; 24). We named this lineage “Hydrothermal ANME-1 cluster.”
Within the Methanopyrales, we detected three different branches, all identified as members of the genus Methanopyrus. Besides a phylotype that is nearly identical to Methanopyrus kandleri, we detected a new phylotype, which we called “Methanopyrus mcrA subcluster I,” and one phylotype that clusters with sequences from hydrothermal vents, which we termed “Methanopyrus mcrA subcluster II” (Fig. 2).
In addition to the previously known groups, we detected novel deep branches on the mcrA phylogenetic tree, which each consist of one phylotype. We call these “Deeply Branching mcrA groups II and III.” Interestingly, these two groups appear equidistant to each other and to Deeply Branching mcrA group I: Deeply Branching mcrA group II has sequence similarities of 67.5 and 70.6%, and Deeply Branching mcrA group III has sequence similarities of 63.0 and 68.8% to the two members of Deeply Branching mcrA group I (see Table S4 in the supplemental material). Sequence similarities of groups II and III to each other are also low (65.7%). Based on our taxonomic classifier, Deeply Branching mcrA groups I, II, and III represent three distinct orders, with the two phylotypes in group I being equivalent to separate families within the same order. Our classification thus suggests that the methane-cycling Archaea consist of at least 10 rather than the currently recognized 7 orders (36).
Inferred energy substrates of mcrA phylotypes.
Based on published information on the closest relatives within the Methanomicrobiales, Methanococcales, and Methanopyrales, which consist almost exclusively of hydrogenotrophic methanogens (19−20, 35, 37−42), H2 and/or formate is a likely energy source of 9 of 21 mcrA phylotypes detected. In addition, we detected one phylotype of the genus Methanohalophilus and three of the family Methermicoccaceae, lineages that catabolize methanol and methylamines (43, 44). Aceticlastic groups (Methanosaeta, Methanosarcina, Zoige cluster I) (45−46) were not detected. ANME-1 and ANME-2 Archaea have been linked to anaerobic methane oxidation (47–50). Nothing is known about the substrates of group e or the two deeply branching groups.
Comparison of general mcrA primers.
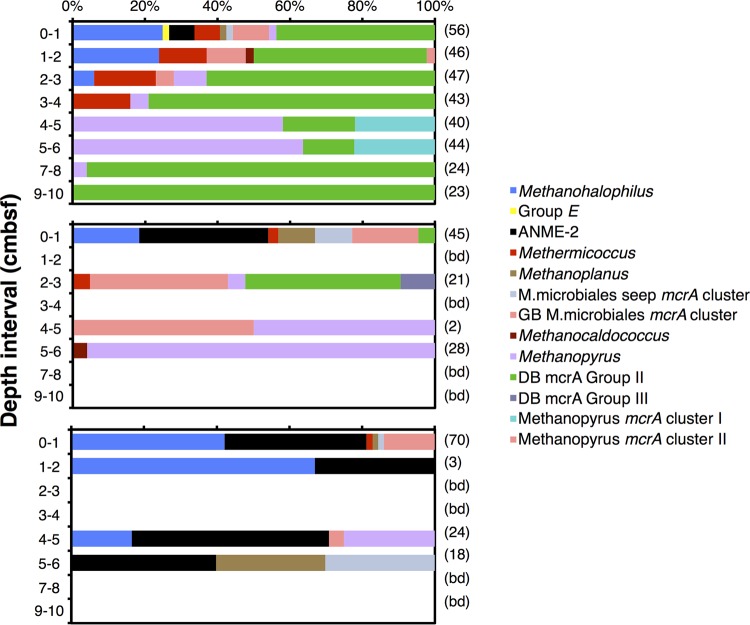
The detection sensitivity and detected number of mcrA clusters varied considerably between the three general primer pairs (Fig. 3). With the ME1/ME2 and mcrI primer pairs, we detected mcrA genes in four horizons, with only weak PCR amplification and cloning success in one of these horizons (for mcrI, 4 to 5 cmbsf; for ME1/ME2, 1 to 2 cmbsf). In contrast, the mcrIRD primer pair produced suitable PCR products for cloning throughout the upper eight horizons. The number of mcrA clusters was also higher; with the mcrIRD primer pair, 12 clusters were detected, compared to 10 and 7 clusters detected with the mcrI and ME1/ME2 primer pairs, respectively.
FIG 3.
Comparison of primer performance versus depth using the three general mcrA primer pairs. (Top panel) mcrIRD primer pair; (middle panel) mcrI primer pair; (bottom panel) ME1/ME2 primer pair. After these initial clone libraries, which led to the use of the mcrIRD primer pair from then on, additional sequencing on new PCR products was done with the mcrIRD primer pair, resulting in detection of further clusters (Table 3). We omit these results here to make primer performances more comparable.
Each primer pair produced a different community profile. Not surprisingly, profiles obtained with the mcrIRD and mcrI primer pairs, which target the same loci and are similar in primer sequence (Table 1), are more similar to one another than to the community profile obtained with the ME1/ME2 primer pair. The biggest difference is the lack of detection of group e, both deeply branching mcrA groups, and both Methanopyrales subclusters with the ME1/ME2 primer pair. Three of these five groups were detected with the mcrI primer pair and four with the mcrIRD primer pair; mcrIRD clone libraries were even dominated by Deeply Branching group II in six of eight sediment horizons (Fig. 3).
Comparison of group-specific mcrA primers.
Eleven of the 27 group-specific primer pairs resulted in successful mcrA amplifications. Five of these primer pairs exclusively amplified their target groups (Table 2; see also Table S1A in the supplemental material), which are ANME-1 Archaea, ANME-2 Archaea, Methanopyrus kandleri, and Deeply Branching mcrA groups II and III.
In contrast to our results with the general mcrA primers, we detected only one new phylotype (ANME-1) using group-specific primers (Table 3). Moreover, certain phylotypes were detected in depth intervals, where they had not been detected with general primer pairs, e.g., Methanopyrus kandleri at 1 to 2 cm, ANME-2 at 2 to 3 cm, and most strikingly, Deeply Branching group III, which was shown to be present from 0 to 6 cmbsf rather than only from 2 to 3 cmbsf. Interestingly, with the notable exception of ANME-1, primer pairs that were designed to target previously undetected groups, e.g., _mcr_Msaeta for Methanosaeta or _mcr_Mbac for Methanobacteriales, still did not detect these groups. Instead, they amplified nontarget mcrA clusters already detected with general primers (_mcr_Mbac) (Table 3) or non-mcrA genes (_mcr_Msaeta) (Table 1A). Moreover, certain clusters detected with general primers, e.g., Methanoplanus, Methanocaldococcus, or Methanopyrus mcrA subclusters I and II were not detected with group-specific primers. These primers amplified (i) not all their target groups (_Mcr_Mmicrob in the case of Methanoplanus), (ii) nontarget mcrA (Methanocaldococcus), or (iii) non-mcrA genes (Methanopyrus mcrA subclusters I and II) (Table 2; see also Table S1A in the supplemental material).
Archaeal 16S rRNA gene diversity.
The archaeal 16S rRNA gene survey yielded a total of 14 lineages and 29 phylotypes (Table 2; see also Fig. S4 in the supplemental material). Five euryarchaeotal lineages belong to known methane-cycling groups: the methyl-disproportionating genera Methanohalophilus and Methermicoccus within the Methanosarcinales, the methylotrophic order Methanoplasmatales, the members of which gain energy by methanol reduction with H2 (71, 72), and the likely hydrogenotrophic sister lineage of Methanoculleus within the hydrogenotrophic Methanomicrobiales (the plausible 16S rRNA gene equivalent of the Guaymas Basin Methanomicrobiales mcrA cluster), as well as the ANME-1 methane oxidizers. Phylotypes from four groups with cultured members—the Thermococcales, Archaeoglobales, Thermoproteales, and the Deep-Sea Hydrothermal Vent Euryarchaeotal group 2 (DHVE2) (73)—are not linked to methane cycling. The remaining phylotypes are from uncultured lineages: the Guaymas Euryarchaeotal group (10), the Marine Benthic group D (74), the Deep-Sea Hydrothermal Vent Euryarchaeotal group 5 (DHVE5) (73), the Deep-Sea Hydrothermal Vent Euryarchaeotal group/Rice Cluster V (DHVEG/RC-V) (66, 75), and the Miscellaneous Crenarchaeotal group (MCG) (76, 77). Though primer pairs were designed to target groups of Euryarchaeota, they also amplified phylotypes of crenarchaeotal MCG and Thermoproteales. The archaeal 16S rRNA gene lineages are further discussed in the supplemental material.
All 16S rRNA gene lineages were previously detected in sediments or hydrothermal vents of the Guaymas Basin, with the exception of the DHVE5 (Table 2; see also Fig. S4 in the supplemental material). Of the phylotypes detected, approximately one-half (14) had the closest BLAST hits from within the Guaymas Basin, including hydrothermal vents and hydrothermal sediments in roughly equal proportions. The other phylotypes (15) had the closest BLAST hits from outside the Guaymas Basin, mainly from hydrothermal vents, oil reservoirs, cold seeps, and mud volcanoes.
DISCUSSION
Methane cycling archaeal lineages.
Our study demonstrates a diversity hot spot for methane-cycling archaea in Guaymas Basin sediments. Collectively, the mcrA and 16S rRNA gene data include at least 22 phylotypes, of which 21 were among sequenced mcrA genes. The only known methane-cycling archaea detected with 16S rRNA gene primers but missed with the _mcrA_-targeted approach were the Methanoplasmatales (Fig. 2; see also Fig. S4 in the supplemental material).
The mcrA diversity exceeds the cumulative diversity detected in previous mcrA gene surveys in Guaymas sediments (10, 12; see Table 5). As did previous studies, we detected phylotypes of the methylotrophic Methanosarcinales, hydrogenotrophic Methanomicrobiales and Methanococcales, and anaerobic methanotrophic ANME-1 and ANME-2 Archaea. Seven of these share the same phylotype (≥97% sequence similarity) with mcrA sequences that were previously detected in Guaymas Basin (Fig. 2). The key differences from past studies are the detection of two novel deeply branching mcrA clusters and the first detection of Methanopyrus in a genetic survey of sediment. Our sequence similarity analyses also suggest two new genus level lineages in the family Methermicoccaceae, one of which was previously detected but not phylogenetically assigned (AY837773) (10), as well as the presence of the genus Methermicoccus.
TABLE 5.
Comparison of mcrA community profiles across three environmental surveys in the Guaymas Basin
| Characteristic (unit) | Everest Mound, Alvin dive 3205 (Dhillon et al. [10]) | Biddle et al. (12) | Everest Mound, Alvin dive 3204 (this study) | ||
|---|---|---|---|---|---|
| Mat Mound, Alvin dive 4483 | Megamat, Alvin Dive 4486 | UNC Mat, Alvin Dive 4489 | |||
| Location | Lat 27.0148, long −111.4122 | Lat 27.0065, long −111.4093 | Lat 27.0077, long −111.4085 | Lat 27.0074, long −111.4088 | Lat 27.0149, long −111.4105 |
| Sediment depth (cmbsf) | 0–15 | 8–10 | 6–8 | 8–14 | 0–10 |
| Temp (°C) | 3–94 | 15–20 | 30–35 | 60–95 | ∼2–54 |
| Sulfate concn (mM) | 0–19 | ∼8–25 | ∼24–28 | ∼22–29 | 0–4 |
| Other descriptors | No mat, hydrocarbon rich | White and orange Beggiatoa mat | Edge of white Beggiatoa mat, hydrocarbon rich | Orange Beggiatoa mat | Patchy mat, hydrocarbon rich |
| mcrA phylotypes detected | |||||
| Methylotrophic | |||||
| Methanococcoides | + | + | |||
| Methanohalophilus | + | + | |||
| Methermicoccaceae | + | + | |||
| Aceticlastic | |||||
| Methanosaeta | + | ||||
| Hydrogenotrophic | |||||
| Methanoplanus | + | ||||
| GB Methanomicrobiales mcrA cluster | + | + | |||
| Methanomicrobiales Seep mcrA cluster | + | ||||
| Methanocorpusculum | + | + | |||
| Methanocaldococcus | + | + | |||
| Methanopyrus | + | ||||
| Methanotrophic | |||||
| ANME-1 | + | + | + | + | |
| ANME-2 | + | + | + | ||
| Unknown | |||||
| Group e | + | + | |||
| Deeply Branching mcrA group I | + | ||||
| Deeply Branching mcrA group II | + | ||||
| Deeply Branching mcrA group III | + |
We confirm that previous failures to detect ANME-1 Archaea using general mcrA primers are due to primer bias of general mcrA primers against this group (12, 15). This problem is overcome by running PCR assays with ANME-1-specific mcrA primers complementarily to ones with general mcrA primers.
Inferred energy substrates.
(i) Hydrogenotrophic methanogens. Five of the mcrA branches detected have high sequence similarity to cultivated, obligate H2- and formate-utilizing methanogens (Fig. 2; Table 4); thus, H2 and/or formate are likely to be important in situ methanogenic substrates. This finding implies that microbial sulfate reduction in the upper centimeters (0 to 4 cmbsf; Fig. 1B), is not drawing H2 concentrations below the threshold required by methanogens, unlike in coastal sediments (78). Fluctuating redox conditions, caused by temporal shifts in advection of highly reduced hydrothermal fluids and oxygenated bottom seawater through surficial sediment (79), may prevent anaerobic microbes from exerting a thermodynamic control over H2 concentrations. Interestingly, while two clusters were detected throughout (Guaymas Basin Methanomicrobiales mcrA cluster, Methanopyrus), the three remaining clusters were detected only in surface sediments (0 to 2 cmbsf) and in one deeper layer (5 to 6 cmbsf; Methanomicrobiales mcrA seep cluster, Methanoplanus, Methanocaldococcus) (Table 4). This bimodal distribution may reflect local peaks in hydrogenotrophic activity and different H2 sources, for example microbial diagenesis near the surface and thermal degradation of organic matter in deeper layers.
TABLE 4.
Substrate use by closest relatives of all 14 mcrA clusters, depth intervals and temperature ranges in which they were detected, and total number of mcrA clusters detected per depth layera
| Depth (cmbsf) | Temp (°C) | H2/HCO3−, formate | MeOH, TMA | Methane | Unknownb | No. of clusters (of 12) per depth layer | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methanoplanusc | Methanomicrobiales mcrA seep cluster | GB Methanomicrobiales mcrA cluster | Methanocaldococcusc | Methanopyrusc,d | Methanohalophilusc | Methermicoccusc,e | ANME-1f | ANME-2f | Group e | DB Guaymas group II | DB Guaymas group III | |||
| 0–1 | 2–7 | + | + | + | − | + | + | + | + | + | + | + | + | 11 |
| 1–2 | 8–12 | − | + | + | + | + | + | + | + | + | − | + | + | 11 |
| 2–3 | 13–17 | − | − | + | − | + | + | + | − | + | − | + | + | 7 |
| 3–4 | 18–23 | − | − | + | − | + | − | + | − | − | − | + | + | 5 |
| 4–5 | 24–28 | − | − | + | − | + | + | − | − | + | − | + | + | 7 |
| 5–6 | 29–33 | + | + | + | + | + | − | − | − | + | − | + | + | 10 |
| 7–8 | 39–43 | − | − | − | − | + | − | − | − | − | − | + | − | 2 |
| 9–10 | 49–54 | − | − | − | − | − | − | − | − | − | − | + | − | 1 |
| 12–13 | 64–69 | − | − | − | − | − | − | − | − | − | − | − | − | 0 |
| 15–16 | 85–89 | − | − | − | − | − | − | − | − | − | − | − | − | 0 |
| 19–20 | 100–105 | − | − | − | − | − | − | − | − | − | − | − | − | 0 |
| No. of depth intervals | NA | 3 | 3 | 6 | 2 | 7 | 4 | 4 | 2 | 5 | 1 | 8 | 6 | NA |
(ii) Methylotrophic methanogens.
The detection of mcrA and 16S rRNA genes with high sequence similarity to known methylotrophic methanogens (Fig. 2; see also Fig. S4 in the supplemental material) is consistent with past studies on Guaymas Basin sediments (Table 5) (10, 12). These phylotypes were detected throughout the upper 5 cm across a range of temperatures (2 to 33°C) and sulfate concentrations (0 to 4 mM). Yet, if clone libraries obtained with general mcrA primers are an indication (Fig. 3), the proportion of methylotrophs of the total methane-cycling community decreases downward. In line with this interpretation, methylotrophy at our site is probably supported mainly by the biological production of methylamines and/or methanol from photosynthesis-derived organic matter.
(iii) Anaerobic methanotrophs.
The presence of mcrA genes of ANME-1 and ANME-2 Archaea suggests that anaerobic methanotrophy (AOM) is concurrent with methanogenesis. If anaerobic methanotrophy is a reversal of methanogenesis (80) and coupled to sulfate reduction via syntrophic activity by sulfate-reducing bacteria (49), then the coexistence of methanogenesis and methanotrophy in the same samples is thermodynamically possible under two scenarios: the presence of chemically distinct microenvironments or pulsating hydrothermal and bottom seawater flow resulting in redox conditions that are alternatingly favorable for methanogenesis and AOM. While the existence of microniches has not been determined, there is evidence in support of the latter scenario (79, 81). Alternatively, anaerobic methanotrophy may not be a reversal of methanogenesis and thus readily cooccur with methanogenesis. Recent studies on ANME-2 Archaea suggest that these methanotrophs can intracellularly couple the oxidation of methane to the reduction of elemental S0 (50).
(iv) Acetoclastic methanogens.
The absence of identifiable acetoclastic methanogens in this survey may seem puzzling, given that acetate is a key intermediate in the anaerobic microbial breakdown of organic matter (82) and produced by thermal degradation or organic matter (83). Acetate is also known to occur at locally high pore water concentrations in Guaymas Basin sediments (8, 10), and mcrA sequences of the acetoclastic genus Methanosaeta were previously detected in Guaymas Basin sediments (10), and in oil- and hydrocarbon-rich environments elsewhere (62, 84–86). Plausible explanations are as follows: (i) mcrA phylotypes detected are unrecognized acetoclastic methanogens; the primary suspects are the uncultivated group e and Deeply Branching mcrA groups II and III, and especially group e, which belongs to the Methanosarcinales, an order that includes the acetoclastic Methanosarcina, Unidentified Rice Field Soil McrA/Zoige cluster I (represented by the sequence with accession number GU182109), and Methanosaeta; (ii) acetate is consumed by other metabolic guilds; the genetic potential for acetate oxidation is present based on 16S rRNA gene sequences of archaeal groups that oxidize acetate with sulfate, S(0), Fe(III), nitrate, or nitrite as electron acceptors (see the supplemental material); and (iii) failure of our PCR primers to detect acetoclastic methanogens. The last scenario is the least likely, given that the mcrIRD primer pair PCR amplifies all three known groups of acetoclastic methanogens in samples from other locations (15, 17; M. A. Lever, unpublished data) and that group-specific primers also did not result in their detection (Table 1).
(v) The unknowns.
The energy substrates used by the five uncultivated lineages are unknown, and phylogenetic affiliation can suggest only working hypotheses. The two phylotypes clustering with the methanol-disproportionating Methermicoccus shengliensis belong to the same family, the Methermicoccaceae, and could equally represent methylotrophic methanogens. Group e is a candidate for generalistic methanogens, due to its clustering with the Unidentified Rice Field Soil mcrA/Zoige cluster I group, members of which use H2, acetate, methanol, and methylamines as energy substrates (46). Such inferences are impossible for Deeply Branching mcrA groups II and III. The mcrA sequence similarity of both groups to the closest cultured relative, Methanopyrus kandleri, is low (∼67%) (see Table S4 in the supplemental material), so low that our taxonomic classifier indicates Deeply Branching mcrA groups II and III to belong to a different order or even class from Methanopyrus kandleri (see Fig. S3 in the supplemental material).
Biogeographical implications.
The high DNA sequence similarity of many phylotypes detected in this study to ones detected at other Guaymas Basin sites as well as in hydrothermal environments, oil reservoirs, and hydrocarbon seeps worldwide is striking—as is the near absence of highly similar DNA sequences from other well-studied methanogenic environments, such as sewage digesters, bovine intestines, rice fields, freshwater, and estuarine and marine sediments (Table 2; see also Table S5 in the supplemental material). This indicates that the global distribution of methane-cycling Archaea follows biogeographic patterns. Methane-cycling Archaea may populate Guaymas Basin sediments in exceptional phylogenetic diversity because these sediments combine key characteristics of hydrothermal environments, oil reservoirs, and hydrocarbon seeps. These characteristics are as follows: (i) hydrothermal fluid flow and circulation, which produce a dynamic environment with profound physicochemical changes over short distances and time scales, (ii) abundance of petroleum compounds and hydrocarbons produced thermogenically from relict organic carbon, and (iii) high supply of microbial energy substrates by thermal degradation of relict organic carbon, petroleum compounds, and hydrocarbons and by microbial fermentation of photosynthesis-derived organic matter, petroleum compounds, and hydrocarbons. Thus, the exceptional diversity of methane-cycling Archaea in sediments of the Guaymas Basin could reflect the fact that these sediments host the cumulative diversity of ecological niches that are otherwise found in three physically separated habitat types.
In addition to high diversity, the discovery of three deeply branching groups, each likely to represent a new order of methane-cycling Archaea, raises the questions as to why these groups were first detected here and which characteristics of Guaymas Basin sediment select for their presence. Surface sediments of Guaymas Basin are a distinctive environment because they combine extreme fluctuations in temperature, fluid flow, and redox conditions with exceedingly high energy fluxes. We propose that high energy availability combined with high physiological stress and mortality due to frequent physicochemical disturbances prevents the establishment of microbial climax communities and enables disturbance-resilient groups that are competitively excluded in more stable environments to thrive here.
Primer recommendation.
Our study underscores the importance of using suitable general and lineage-specific PCR primers to detect and accurately map the phylogenetic diversity of (unknown) microorganisms. Comparisons of primer performance and phylogenetic coverage (discussed in detail in the supplemental material) suggest that the mcrIRD primer pair, when combined with the ANME-1-mcrA primer pair, covers a wide diversity of mcrA genes. Though we cannot preclude that certain divergent sequences are missed, our results show that the mcrIRD primers perform well beyond the range of known mcrA sequences and even amplify novel, deeply branching groups. The mcrIRD primer pair thus combines wide phylogenetic breadth with a lower detection limit than the mcrI and ME1/ME2 primer pairs and illustrates how reduced primer degeneracy and hence higher amplification efficiency do not necessarily compromise the breadth of phylotypes targeted. The new phylotypes and mcrA gene lineages detected show that the methanogenic and methane-oxidizing archaea—a metabolic guild generally thought of as well explored—are considerably more diverse than expected and contain previously uncharacterized “microbial dark matter” (87) that needs genomic and physiological investigation.
Supplementary Material
Supplemental material
ACKNOWLEDGMENTS
Sediment sampling in Guaymas Basin in 1998 was made possible by NSF grant OCE 9714195 (Life in Extreme Environments) to A.P.T. Sequencing was supported by the NASA Astrobiology Institute “From Early Biospheric Metabolisms to the Evolution of complex systems” and performed at the Josephine Bay Paul Center for Comparative Molecular Biology and Evolution at the Marine Biological Laboratory, Woods Hole, MA. M.A.L. was supported by a Schlanger Ocean Drilling Fellowship, a University of North Carolina Dissertation Completion Fellowship, a Marie-Curie Intra-European Fellowship (number 255135), and funding provided to Bo Barker Jørgensen by the Danish National Research Foundation and Max Planck Society.
The Hanse Wissenschaftskolleg provided a supportive writing environment to A.P.T. during the complex genesis of the manuscript.
We declare that we have no conflicts of interests.
Footnotes
REFERENCES
- 1.Bazylinski DA, Wirsen CO, Jannasch HW. 1989. Microbial utilization of naturally occurring hydrocarbons at the Guaymas Basin hydrothermal vent site. Appl Environ Microbiol 55:2832–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson DC, Wirsen CO, Jannasch HW. 1989. Characterization of large, autotrophic Beggiatioa spp. abundant at hydrothermal vents of the Guaymas Basin. Appl Environ Microbiol 55:2909–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elsgaard L, Isaksen MF, Jørgensen BB, Alayse A-M, Jannasch HW. 1994. Microbial sulfate reduction in deep-sea sediments at the Guaymas Basin hydrothermal vent area: influence of temperature and substrates. Appl Environ Microbiol 58:3335–3343. [Google Scholar]
- 4.Teske A, Hinrichs K-U, Edgcomb V, Gomez AD, Kysela D, Sylva SP, Sogin ML, Jannasch HW. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl Environ Microbiol 68:1994–2007. doi: 10.1128/AEM.68.4.1994-2007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curray JR, Moore DG, Smith SM, Chase TE. 1982. Underway geophysical data from Deep Sea Drilling Project Leg 64: navigation, bathymetry, magnetics, and seismic profiles, p 505–507. _In_Blakeslee J, Platt LW, Stout LN (ed), Deep Sea Drilling Initial Reports, vol 64 U.S. Government Printing Office, Washington, DC. [Google Scholar]
- 6.Simoneit BRT, Bode GW. 1982. Carbon/carbonate and nitrogen analyses, Leg 64, Gulf of California, p 1303–1307. _In_Blakeslee J, Platt LW, Stout LN (ed), Deep Sea Drilling Initial Reports, vol 64 U.S. Government Printing Office, Washington, DC. [Google Scholar]
- 7.Bazylinski DA, Farrington JW, Jannasch HW. 1988. Hydrocarbons in surface sediments from a Guaymas Basin hydrothermal vent site. Org Geochem 12:547–559. doi: 10.1016/0146-6380(88)90146-5. [DOI] [Google Scholar]
- 8.Martens CS. 1990. Generation of short chain organic acid anions in hydrothermally altered sediments of the Guaymas Basin, Gulf of California. Appl Geochem 5:71–76. doi: 10.1016/0883-2927(90)90037-6. [DOI] [Google Scholar]
- 9.Pearson A, Seewald JS, Eglinton TI. 2005. Bacterial incorporation of relict carbon in the hydrothermal environment of Guaymas Basin. Geochim Cosmochim Acta 69:5477–5486. doi: 10.1016/j.gca.2005.07.007. [DOI] [Google Scholar]
- 10.Dhillon A, Lever M, Lloyd KG, Albert DB, Sogin ML, Teske A. 2005. Methanogen diversity evidenced by molecular characterization of methyl coenzyme M reductase A (mcrA) genes in hydrothermal sediments of the Guaymas Basin. Appl Environ Microbiol 71:4592–4601. doi: 10.1128/AEM.71.8.4592-4601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holler T, Widdel F, Knittel K, Amann R, Kellermann MY, Hinrichs K-U, Teske A, Boetius A, Wegener G. 2011. Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J 5:1946–1956. doi: 10.1038/ismej.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biddle JF, Cardman Z, Mendlovitz H, Albert DB, Lloyd KG, Boetius A, Teske A. 2012. Anaerobic oxidation of methane at different temperature regimes in Guaymas Basin hydrothermal sediments. ISME J 6:1018–1031. doi: 10.1038/ismej.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Springer E, Sachs MS, Woese CR, Boone DR. 1995. Partial gene sequences for the α subunit of methyl-coenzyme M reductase (mcrI) as a phylogenetic tool for the family Methanosarcinaceae. Int J Syst Bacteriol 45:554–559. doi: 10.1099/00207713-45-3-554. [DOI] [PubMed] [Google Scholar]
- 14.Hallam SJ, Girguis PR, Preston CM, Richardson PM, DeLong EF. 2003. Identification of methyl coenzyme M reductase A (mcrA). genes associated with methane-oxidizing Archaea. Appl Environ Microbiol 69:5483–5549. doi: 10.1128/AEM.69.9.5483-5491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lever MA. 2008. Anaerobic carbon cycling pathways in the subseafloor investigated via functional genes, chemical gradients, stable carbon isotopes, and thermodynamic calculations. Ph.D. thesis University of North Carolina at Chapel Hill, Chapel Hill, NC. [Google Scholar]
- 16.Hales BA, Edwards C, Ritchie DA, Hall G, Pickup RW, Saunders JR. 1996. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl Environ Microbiol 62:668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lever MA, Rouxel OJ, Alt J, Shimizu N, Ono S, Coggon RM, Shans WC III, Lapham L, Elvert M, Prieto-Mollar X, Hinrichs K-U, Inagaki F, Teske A. 2013. Evidence for microbial carbon and sulfur cycling in deeply buried ridge flank basalt. Science 339:1305–1308. doi: 10.1126/science.1229240. [DOI] [PubMed] [Google Scholar]
- 18.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Bucher A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüssmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H. 2004. ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones WJ, Leigh JA, Mayer F, Woese CR, Wolfe RS. 1983. Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch Microbiol 136:254–261. doi: 10.1007/BF00425213. [DOI] [Google Scholar]
- 20.Kurr M, Huber R, König H, Jannasch HW, Fricke H, Trincone Kristjansson JK, Stetter KO. 1991. Methanopyrus kandleri, gen. and sp. nov. represents a novel group of hyperthermophilic methanogens, growing at 110°C. Arch Microbiol 156:239–247. doi: 10.1007/BF00262992. [DOI] [Google Scholar]
- 21.Zhilina TN. 1983. New obligate halophilic methane-producing bacterium. Microbiology (Engl Transl) 52:290–297. [Google Scholar]
- 22.Dang H, Luan X, Zhao J, Li J. 2009. Diverse and novel nifH and nifH-like gene sequences in the deep-sea methane seep sediments of the Okhotsk Sea. Appl Environ Microbiol 75:2238–2245. doi: 10.1128/AEM.02556-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazar CS, L'Haridon S, Pignet P, Toffin L. 2011. Archaeal populations in hypersaline sediments underlying orange microbial mats in the Napoli Mud Volcano. Appl Environ Microbiol 77:3120–3131. doi: 10.1128/AEM.01296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nunoura T, Oida H, Nakaseama M, Kosaka A, Ohkubo SB, Kikuchi T, Kazama H, Hosoi-Tanabe S, Nakamura K, Kinoshita M, Hirayama H, Inagaki F, Tsunogai U, Ishibashi J, Takai K. 2010. Archaeal diversity and distribution along thermal and geochemical gradients in hydrothermal sediments at the Yonaguni Knoll IV hydrothermal field in the Southern Okinawa Trough. Appl Environ Microbiol 76:1198–1211. doi: 10.1128/AEM.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gieg LM, Davidova IA, Duncan KE, Suflita JM. 2010. Methanogenesis, sulfate reduction and crude oil biodegradation in hot Alaskan oilfields. Environ Microbiol 12:3074–3086. doi: 10.1111/j.1462-2920.2010.02282.x. [DOI] [PubMed] [Google Scholar]
- 26.Ver Eecke HC, Butterfield DA, Huber JA, Lilley MD, Olson EJ, Roe KK, Evans LJ, Merkel AY, Cantin HV, Holden JF. 2012. Hydrogen-limited growth of hyperthermophilic methanogens at deep-sea hydrothermal vents. Proc Natl Acad Sci U S A 109:13674–13679. doi: 10.1073/pnas.1206632109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merkel AY, Huber JA, Chernyh NA, Bonch-Osmolovskaya EA, Lebedinsky AV. 2013. Detection of putatively thermophilic anaerobic methanotrophs in diffuse hydrothermal vent fluids. Appl Environ Microbiol 79:915–923. doi: 10.1128/AEM.03034-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nettmann E, Bergmann I, Pramschüter S, Mundt K, Plogsties V, Herrmann C, Klocke M. 2010. Polyphasic analyses of methanogenic archaeal communities in agricultural biogas plants. Appl Environ Microbiol 76:2540–2548. doi: 10.1128/AEM.01423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lazar CS, Parkes RJ, Cragg BA, L'Haridon S, Toffin L. 2011. Methanogenic diversity and activity in hypersaline sediments of the Napoli mud volcano, Eastern Mediterranean Sea. Environ Microbiol 13:2078–2091. doi: 10.1111/j.1462-2920.2011.02425.x. [DOI] [PubMed] [Google Scholar]
- 30.Li D, Midgley DJ, Ross JP, Oytam Y, Abell GCJ, Volk H, Daud WAW, Hendry P. 2012. Microbial biodiversity in a Malaysian oil field and a systematic comparison with oil reservoirs worldwide. Arch Microbiol 194:513–523. doi: 10.1007/s00203-012-0788-z. [DOI] [PubMed] [Google Scholar]
- 31.Nercessian O, Bienvenu N, Moreira D, Prieur D, Jeanthon C. 2005. Diversity of functional genes of methanogens, methanotrophs and sulfate reducers in deep-sea hydrothermal environments. Environ Microbiol 7:118–132. doi: 10.1111/j.1462-2920.2004.00672.x. [DOI] [PubMed] [Google Scholar]
- 32.Roussel EG, Konn C, Charlou J-L, Donval J-P, Fouquet Y, Querellou J, Prieur D, Cambon Bonavita M-A. 2011. Comparison of microbial communities associated with three Atlantic ultramafic hydrothermal systems. FEMS Microbiol Ecol 77:647–665. doi: 10.1111/j.1574-6941.2011.01161.x. [DOI] [PubMed] [Google Scholar]
- 33.Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849. doi: 10.1099/00207713-44-4-846. [DOI] [Google Scholar]
- 34.Stackebrandt E. 2009. Phylogeny based on 16S rRNA/DNA. Encyclopedia of Life Sciences. Wiley Online Library, Hoboken, NJ. doi: 10.1002/9780470015902.a0000462.pub2. [DOI] [Google Scholar]
- 35.Ollivier B, Cayol J-L, Patel BKC, Magot M, Fardeau M-L, Garcia J-L. 1997. Methanoplanus petrolearius sp. nov., a novel methanogenic bacterium from an oil-producing well. FEMS Microbiol Lett 147:51–56. doi: 10.1111/j.1574-6968.1997.tb10219.x. [DOI] [PubMed] [Google Scholar]
- 36.Borrel G, O'Toole PW, Harris HMB, Peyret P, Brugère J-F, Gribaldo S. 2013. Phylogenomic data support a seventh order of methylotrophic methanogens and provide insights into the evolution of methanogenesis. Genome Biol Evol 5:1769–1780. doi: 10.1093/gbe/evt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romesser JA, Wolfe RS, Mayer F, Spiess E, Walther-Mauruschat AW. 1979. Methanogenium, a new genus of marine methanogenic bacteria, and characterization of Methanogenium cariaci sp. nov. and Methanogenium marisnigri sp. nov. Arch Microbiol 121:147–153. doi: 10.1007/BF00689979. [DOI] [Google Scholar]
- 38.Wildgruber G, Thomm M, König H, Ober K, Ricchiuto T, Stetter KO. 1982. Methanoplanus limicola, a plate-shaped methanogen representing a novel family, the Methanoplanaceae. Arch Microbiol 132:31–36. doi: 10.1007/BF00690813. [DOI] [Google Scholar]
- 39.L'Haridon S, Reysenbach A-L, Banta A, Messner P, Schumann P, Stackebrandt E, Jeanthon C. 2003. Methanocaldococcus indicus sp. nov., a novel hyperthermophilic methanogen isolated from the Central Indian Ridge. Int J Syst Evol Microbiol 53:1931–1935. doi: 10.1099/ijs.0.02700-0. [DOI] [PubMed] [Google Scholar]
- 40.Mikucki JA, Liu Y, Delwiche M, Colwell FS, Boone DR. 2003. Isolation of a methanogen from deep marine sediments that contain methane hydrates, and description of Methanoculleus submarinus sp. nov. Appl Environ Microbiol 69:3311–3316. doi: 10.1128/AEM.69.6.3311-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kendall MM, Liu Y, Sieprawska-Lupa M, Stetter KO, Whitman WB, Boone DR. 2006. Methanococcus aeolicus sp. nov., a mesophilic, methanogenic archaeon from shallow and deep marine sediments. Int J Syst Evol Microbiol 56:1525–1529. doi: 10.1099/ijs.0.64216-0. [DOI] [PubMed] [Google Scholar]
- 42.Cheng L, Qiu T-L, Li X, Wang W-D, Deng Y, Yin X-B, Zhang H. 2008. Isolation and characterization of Methanoculleus receptaculi sp. nov. from Shengli oil field, China. FEMS Microbiol Lett 285:65–71. doi: 10.1111/j.1574-6968.2008.01212.x. [DOI] [PubMed] [Google Scholar]
- 43.Wilharm T, Zhilina TN, Hummel P. 1991. DNA-DNA hybridization of methylotrophic halophilic methanogenic bacteria and transfer of Methanococcus halophilus to the genus Methanohalophilus as Methanohalophilus halophilus comb. nov. Int J Syst Bacteriol 41:558–562. doi: 10.1099/00207713-41-4-558. [DOI] [Google Scholar]
- 44.Cheng L, Qiu T-L, Yin X-B, Wu X-L, Hu G-Q, Deng Y, Zhang H. 2007. Methermicoccus shengliensis gen. nov., sp. nov., a thermophilic, methylotrophic methanogen isolated from oil-production water, and proposal of Methermicoccaceae fam. nov. Int J Syst Evol Microbiol 57:2964–2969. doi: 10.1099/ijs.0.65049-0. [DOI] [PubMed] [Google Scholar]
- 45.Whitman WB, Bowen TL, Boone DR. 2006. The methanogenic bacteria, p 165–207. _In_Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer, New York, NY. [Google Scholar]
- 46.Zhang G, Tian J, Jiang N, Guo X, Wang Y, Dong X. 2008. Methanogen community in Zoige wetland of Tibetan plateau and phenotypic characterization of a dominant uncultured methanogen cluster ZC-I. Environ Microbiol 10:1850–1860. doi: 10.1111/j.1462-2920.2008.01606.x. [DOI] [PubMed] [Google Scholar]
- 47.Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jørgensen BB, Witte U, Pfannkuche O. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–625. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 48.Orphan VJ, House CH, Hinrichs K-U, McKeegan KD, DeLong EF. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc Natl Acad Sci U S A 99:7663–7668. doi: 10.1073/pnas.072210299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knittel K, Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol 63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 50.Milucka J, Ferdelman TG, Polerecky L, Franzke D, Wegener G, Schmid M, Lieberwirth I, Wagner M, Widdel F, Kuypers MMM. 2012. Zero-valent sulphur is a key intermediate in marine methane oxidation. Nature 491:541–546. doi: 10.1038/nature11656. [DOI] [PubMed] [Google Scholar]
- 51.Kellermann MY, Wegener G, Elvert M, Yoshinaga MY, Lin YS, Holler T, Prieto Mollar X, Knittel K, Hinrichs K-U. 2012. Autotrophy as a predominant mode of carbon fixation in anaerobic methane-oxidizing microbial communities. Proc Natl Acad Sci U S A 109:19321–19326. doi: 10.1073/pnas.1208795109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teske A, Edgcomb V, Rivers AR, Thompson JR, de Vera Gomez A, Molyneaux SJ, Wirsen CO. 2009. A molecular and physiological survey of a diverse collection of hydrothermal vent Thermococcus and Pyrococcus isolates. Extremophiles 13:905–915. doi: 10.1007/s00792-009-0278-7. [DOI] [PubMed] [Google Scholar]
- 53.Canganella F, Jones WJ, Gambacorta A, Antranikian G. 1998. Thermococcus guaymasensis sp. nov. and Thermococcus aggregans sp. nov., two novel thermophilic archaea isolated from the Guaymas Basin hydrothermal vent site. Int J Syst Bacteriol 48:1181–1185. doi: 10.1099/00207713-48-4-1181. [DOI] [PubMed] [Google Scholar]
- 54.Burggraf S, Jannasch HW, Nicolaus B, Stetter KO. 1990. Archaeoglobus profundus sp. nov., represents a new species within the sulfate-reducing archaebacteria. Syst Appl Microbiol 13:24–28. doi: 10.1016/S0723-2020(11)80176-1. [DOI] [Google Scholar]
- 55.Parkes RJ, Linnane CD, Webster G, Sass H, Weightman AJ, Hornibrook ERC, Horsfield B. 2011. Prokaryotes stimulate mineral H2 formation for the deep biosphere and subsequent thermogenic activity. Geology 39:219–222. doi: 10.1130/G31598.1. [DOI] [Google Scholar]
- 56.Swan BK, Ehrhardt CJ, Reifel KM, Moreno LI, Valentine DL. 2010. Archaeal and bacterial communities respond differently to environmental gradients in anoxic sediments of a California hypersaline lake, the Salton Sea. Appl Environ Microbiol 76:757–768. doi: 10.1128/AEM.02409-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watts JM, Swan BK, Tiffany MA, Hurlbert SH. 2001. Thermal, mixing, and oxygen regimes of the Salton Sea, California, 1997-1999. Hydrobiologia 466:159–176. doi: 10.1023/A:1014599719989. [DOI] [Google Scholar]
- 58.Zengler K, Richnow HH, Rosselló-Mora R, Michaelis W, Widdel F. 1999. Methane formation from long-chain alkanes by anaerobic microorganisms. Nature 401:266–269. doi: 10.1038/45777. [DOI] [PubMed] [Google Scholar]
- 59.Nakagawa S, Takai K, Inagaki F, Chiba H, Ishibashi J, Kataoka S, Hirayama H, Nunoura T, Horikoshi K, Sako Y. 2005. Variability in microbial community and venting chemistry in a sediment-hosted backarc hydrothermal system: impacts of subseafloor phase-separation. FEMS Microbiol Ecol 54:141–155. doi: 10.1016/j.femsec.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 60.Ehrhardt CJ, Haymon RM, Lamontagne MG, Holden PA. 2007. Evidence for hydrothermal Archaea within the basaltic flanks of the East Pacific Rise. Environ Microbiol 9:900–912. doi: 10.1111/j.1462-2920.2006.01211.x. [DOI] [PubMed] [Google Scholar]
- 61.Sánchez O, Garrido L, Forn I, Massana R, Maldonado MI, Mas J. 2011. Molecular characterization of activated sludge from a seawater-processing wastewater treatment plant. Microb Biotechnol 4:628–642. doi: 10.1111/j.1751-7915.2011.00256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pachiadaki MG, Lykousis V, Stefanou EG, Kormas KA. 2010. Prokaryotic community structure and diversity in the sediments of an active submarine volcano (Kazan mud volcano, East Mediterranean Sea). FEMS Microbiol Ecol 72:429–444. doi: 10.1111/j.1574-6941.2010.00857.x. [DOI] [PubMed] [Google Scholar]
- 63.Zhao L, Ma T, Gao M, Gao P, Cao M, Zhu X, Li G. 2012. Characterization of microbial diversity and community in water flooding oil reservoirs in China. World J Microbiol Biotechnol 28:3039–3052. doi: 10.1007/s11274-012-1114-2. [DOI] [PubMed] [Google Scholar]
- 64.Keller M, Braun F-J, Dirmeier R, Hafenbradl D, Burggraf S, Rachel R, Stetter KO. 1995. Thermococcus alcaliphilus sp. nov., a new hyperthermophilic archaeum growing on polysulfide at alkaline pH. Arch Microbiol 164:390–395. doi: 10.1007/BF02529736. [DOI] [PubMed] [Google Scholar]
- 65.Kaster KM, Bonaunet K, Berland H, Kjeilen-Eilertsen G, Brakstad OG. 2009. Characterisation of culture-independent and –dependent microbial communities in a high-temperature offshore chalk petroleum reservoir. Antonie Van Leeuwenhoek 96:423–439. doi: 10.1007/s10482-009-9356-1. [DOI] [PubMed] [Google Scholar]
- 66.Takai K, Gamo T, Tsunogai U, Nakayama N, Hirayama H, Nealson KH, Horikoshi K. 2004. Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles 8:269–282. doi: 10.1007/s00792-004-0386-3. [DOI] [PubMed] [Google Scholar]
- 67.Yoshida-Takashima Y, Nunoura T, Kazama H, Noguchi T, Inoue K, Akashi H, Yamanaka T, Toki T, Yamamoto M, Furushima Y, Ueno Y, Yamamoto H, Takai K. 2012. Spatial distribution of viruses associated with planktonic and attached microbial communities in hydrothermal environments. Appl Environ Microbiol 78:1311–1320. doi: 10.1128/AEM.06491-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flores GE, Wagner ID, Liu Y, Reysenbach A-L. 2012. Distribution, abundance, and diversity patterns of the thermoacidophilic “deep-sea hydrothermal vent euryarchaeota 2.” Frontiers Microbiol 3:1–17. doi: 10.3389/fmicb.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schauer R, Røy H, Augustin N, Gennerich H-H, Peters M, Wenzhoefer F, Amann R, Meyerdierks A. 2011. Bacterial sulfur cycling shapes microbial communities in surface sediments of an ultramafic hydrothermal vent field. Environ Microbiol 13:2633–2648. doi: 10.1111/j.1462-2920.2011.02530.x. [DOI] [PubMed] [Google Scholar]
- 70.Weber A, Jørgensen BB. 2002. Bacterial sulfate reduction in hydrothermal sediments of the Guaymas Basin, Gulf of California, Mexico. Deep Sea Res I 49:827–841. doi: 10.1016/S0967-0637(01)00079-6. [DOI] [Google Scholar]
- 71.Dridi B, Fardeau M-L, Ollivier B, Raoult D, Drancourt M. 2012. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol 62:1902–1907. doi: 10.1099/ijs.0.033712-0. [DOI] [PubMed] [Google Scholar]
- 72.Paul K, Nonoh JO, Mikulski L, Brune A. 2012. “Methanoplasmatales,” Thermoplasmatales-related Archaea in termite guts and other environments, are the seventh order of methanogens. Appl Environ Microbiol 78:8245–8253. doi: 10.1128/AEM.02193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takai K, Horikoshi K. 1999. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics 152:1285–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vetriani C, Jannasch HW, MacGregor BJ, Stahl DA, Reysenbach A-L. 1999. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl Environ Microbiol 65:4375–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Großkopf R, Stubner S, Liesack W. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl Environ Microbiol 64:4983–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Inagaki F, Suzuki M, Takai K, Oida H, Sakamoto T, Aoki K, Nealson KH, Horikoshi K. 2003. Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl Environ Microbiol 69:7724–7735. doi: 10.1128/AEM.69.12.7224-7235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kubo K, Lloyd KG, Biddle JF, Amann R, Teske A, Knittel K. 2012. Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. ISME J 6:1949–1965. doi: 10.1038/ismej.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoehler TM, Alperin MJ, Albert DB, Martens CS. 1998. Thermodynamic control on hydrogen concentrations in anoxic sediments. Geochim Cosmochim Acta 62:1745–1756. doi: 10.1016/S0016-7037(98)00106-9. [DOI] [Google Scholar]
- 79.Gundersen JK, Jørgensen BB, Larsen E, Jannasch HW. 1992. Mats of giant sulphur bacteria on deep-sea sediments due to fluctuating hydrothermal flow. Nature 360:454–456. doi: 10.1038/360454a0. [DOI] [Google Scholar]
- 80.Thauer RK. 2011. Anaerobic oxidation of methane with sulfate: on the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr Opin Microbiol 14:292–299. doi: 10.1016/j.mib.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 81.McKay L. 2014. Microbial ecology of a manmade oil spill in the Gulf of Mexico and a natural hydrothermal oil seep in the Gulf of California. Ph.D. thesis University of North Carolina at Chapel Hill, Chapel Hill, NC. [Google Scholar]
- 82.Zehnder AJB, Ingvorsen K, Marti T. 1982. Microbiology of methane bacteria, p 45–68. _In_Hughes DE, Stafford DA, Wheatley BI, Baader W, Lettinga G, Nyns EJ, Verstraete W (ed), Anaerobic digestion. Elsevier Biomedical Press, Amsterdam, Netherlands. [Google Scholar]
- 83.Wellsbury P, Goodman K, Barth T, Cragg BA, Barnes SP, Parkes RJ. 1997. A deep bacterial biosphere fuelled by low-temperature acetate generation during burial. Nature 388:573–576. doi: 10.1038/41544. [DOI] [Google Scholar]
- 84.Mochimaru H, Uchiyama H, Yoshioka H, Imachi H, Hoaki T, Tamaki H, Nakamura K, Sekiguchi Y, Kamagata Y. 2007. Methanogen diversity in deep subsurface gas-associated water at the Minami-Kanto gas field in Japan. Geomicrobiol J 24:93–100. doi: 10.1080/01490450701266571. [DOI] [Google Scholar]
- 85.Pham VD, Hnatow LL, Zhang S, Fallon RD, Jackson SC, Tomb JF, DeLong EF, Keeler SJ. 2009. Characterizing microbial diversity in production water from an Alaskan mesothermic petroleum reservoir with two independent molecular methods. Environ Microbiol 11:176–187. doi: 10.1111/j.1462-2920.2008.01751.x. [DOI] [PubMed] [Google Scholar]
- 86.Orcutt BN, Joye SB, Kleindienst S, Knittel K, Ramette A, Reitz A, Samarkin V, Treude T, Boetius A. 2010. Impact of natural oil and higher hydrocarbons on microbial diversity, distribution, and activity in Gulf of Mexico cold-seep sediments. Deep Sea Res II 57:2008–2012. doi: 10.1016/j.dsr2.2010.05.014. [DOI] [Google Scholar]
- 87.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund HP, Tsiamis G, Sievert S, Liu W-T, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material