A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex (original) (raw)
. Author manuscript; available in PMC: 2015 Feb 25.
Published in final edited form as: Nature. 2008 Dec 18;456(7224):976–979. doi: 10.1038/nature07422
Summary
Most transmembrane proteins are selected as transport vesicle cargo through the recognition of short, linear amino acid motifs in their cytoplasmic portions by vesicle coat proteins. In the case of clathrin-coated vesicles (CCVs) the motifs are recognised by clathrin adaptors. The AP2 adaptor complex (subunits α,β2,μ2,σ2) recognises both major endocytic motifs: YxxΦ motifs 1 and [DE]xxxL[LI] acidic dileucine motifs. Here we describe the binding of AP2 to the endocytic dileucine motif from CD4 2. The major recognition events are the two leucine residues binding in hydrophobic pockets on σ2. The hydrophilic residue four residues upstream from the first leucine sits on a positively charged patch made from residues on σ2 and α subunits. Mutations in key residues inhibit the binding of AP2 to ‘acidic dileucine’ motifs displayed in liposomes containing PtdIns4,5P2, but do not affect binding to YxxΦ motifs via μ2. In the ‘inactive’ AP2 core structure 3, both motif binding sites are blocked by different parts of the β2 subunit. To allow a dileucine motif to bind, the β2 N-terminus is displaced and becomes disordered; however, in this structure the YxxΦ binding site on μ2 remains blocked.
Keywords: clathrin, receptor, sorting signal, membrane protein
In CCVs, cargo is mainly selected through the recognition of one of three types of signal by clathrin adaptors: the short, linear, transplantable peptide motifs that are widely found on general cargo; covalently attached ubiquitin molecules (reviewed in 4); and the folded determinants on SNARE proteins 5,6. The two major classes of transplantable motifs used in CCVs are the YxxΦ and acidic dileucine motifs. The latter can be divided into two further classes: the [ED]xxxL[LI] motifs, which, along with YxxΦ motifs, are recognised by the heterotetrameric AP clathrin adaptor complexes on numerous different pathways (reviewed in 7 and references therein); and the DxxLL motifs, recognised by GGAs for TGN-to-endosome transport 8,9. AP2 plays a pivotal role in regulating the formation of endocytic CCVs destined for early endosomes. Whilst the mechanism of YxxΦ motif binding by APs has been characterised at the molecular level, there is no mechanistic insight into [DE]xxxL[LI] binding, although several recent studies indicate that the binding site resides on the α/σ2 heterodimer 10,11. Eight different ‘acidic dileucine’ peptides were tried in co-crystallisation experiments with the 200kDa AP2 core and several produced poorly-diffracting crystals. However, co-crystallisation with the peptide RM(phosphoS)QIKRLLSE (Q peptide) from the T-cell cell-surface antigen protein CD4, and a variant of this peptide (RM(phosphoS)EIKRLLSE) (E peptide), resulted in crystals that diffracted to resolutions of 3.0Å and 3.4Å respectively. The structure of the Q peptide complex was solved by molecular replacement using models derived from the closed structure 3 see Figure 1. Rebuilding of the α and β2 helical solenoids was required due to alterations in their overall curvature (see later), but even in the initial electron density maps the ‘dileucine peptide’ could be seen clearly. The identity and orientation of the peptide was confirmed as described in online Materials and Methods.
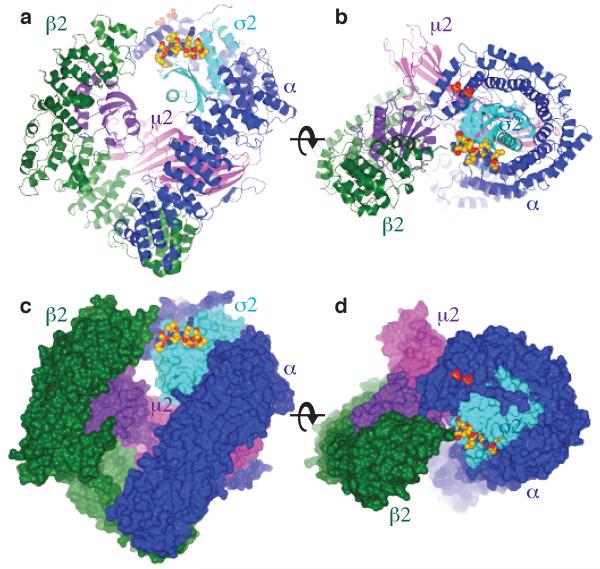
Figure 1. Structure of the AP2 adaptor core in complex with the dileucine peptide from CD4.
Orthogonal views of the dileucine motif liganded AP2 complex in ribbon (a and b) and in molecular surface (c and d) representations. In a and c the membrane is parallel to the upper face of the complex and in b and d the complex is viewed through the membrane with the membrane interacting surface facing up. The α subunit is coloured dark blue, β2 green, σ2 pale blue, N-μ2 purple and C-μ2 mauve. The dileucine motif peptide, shown as spheres with carbons coloured gold. The two sulphate groups bound to the α subunit in the PtdInsP2 site are shown.
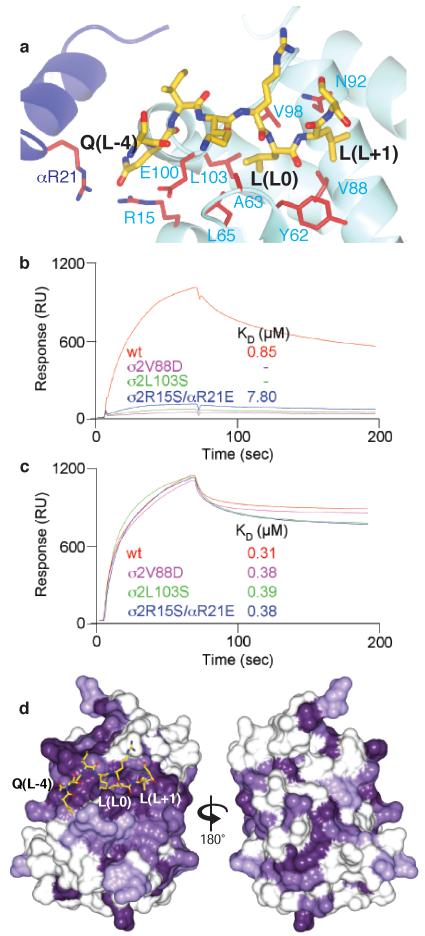
The structure shows that the peptide binds in an extended conformation near the α subunit binding site for PtdIns4,5P2, which is replaced here by two sulphate ions (see Figures 1 & 4 and Figure S3). The LL moiety at positions L0 and L+1 (for nomenclature see 7 and Figure S1b) binds in two adjacent hydrophobic pockets on the small σ2 subunit that are lined by a number of hydrophobic residues (see Figure 2), most of which are conserved in all σ subunits from APs 1-4 in species from yeast to mammals (Figure 3 and Supplementary Information). Mutation of a number of these to hydrophilic residues (σ2L65S, σ2V88D, σ2V98S or σ2L103S), or filling in the pocket by replacement of σ2A63 or σ2N92 with tryptophan, strongly inhibited binding of recombinant AP2 core complexes to different dileucine motifs displayed on PtdIns4,5P2 - containing liposomes (Table S1, online Materials and Methods and 12). The KD for wild-type AP2 binding to the CD4 dileucine motif was 0.85 μM, whilst the σ2V88 and σ2L103 mutations reduced binding to below detectable levels. A double mutant in σ2R15 and αR21 decreased binding to the dileucine motif by around an order of magnitude, with the KD for σ2R15S/σ2R21E reduced to 7.8 μM. In contrast, the binding of wild-type and mutant AP2 cores to PtdIns4,5P2 and to YxxΦ was unaffected, with KDs for binding to the TGN38 YxxΦ motif between 0.31μM and 0.38 μM, indicating that all recombinant core variants were correctly folded and functional (Figure 3).
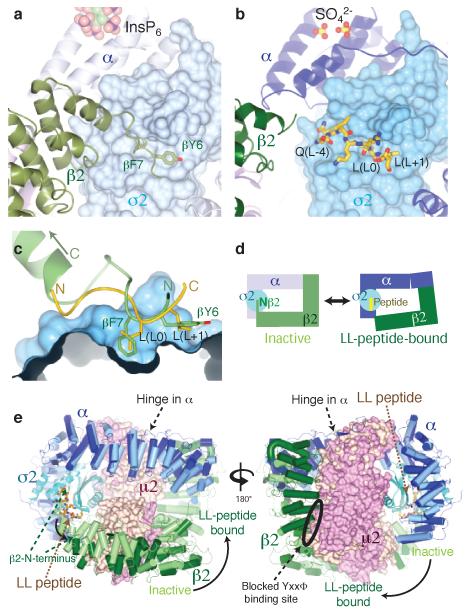
Figure 4. A conformational change in AP2 is required for dileucine peptide binding.
In the IP6-liganded inactive conformation (a) the N-terminus of β2 is held in place by β2F7 and β2Y6 sitting in the L0 and L+1 pockets. For dileucine motifs to bind the N-terminus of β2 is displaced from the surface of σ2 (b). (c) close-up view of the dileucine binding site: note that the LL peptide (gold) runs in the opposite direction to the N-terminus of β2 (green). (d) Schematic representation of the conformational change: a 20° hinge movement in α moves the N-terminus of β2 out of the LL-binding site, allowing the motif to bind. The μ2 subunit has been omitted for clarity. (e) front & back views of the conformational change: the IP6-liganded inactive conformation is shown in pale colours, the unlocked dileucine-peptide bound conformation in dark colours. The YxxΦ site on μ2 remains blocked, and is remote from the dileucine binding site.
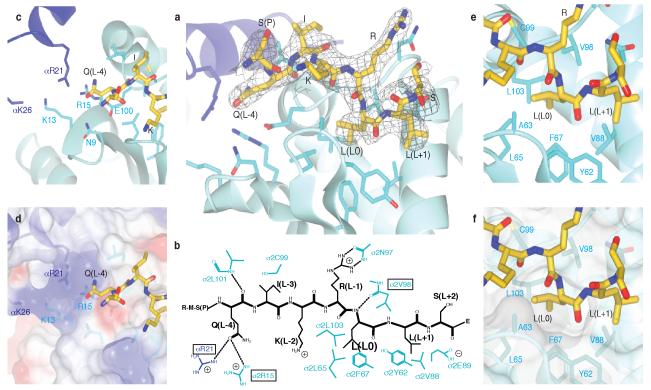
Figure 2. Details of binding of the CD4 dileucine signal by the σ2 and α subunits of AP2.
a The dileucine peptide is bound mainly by the pale blue σ2 subunit near its interface with the α subunit (dark blue). The peptide is shown in its final 2mFo-dFc electron density (cropped around the peptide and contoured at 0.11 e/Å3).
b Schematic representation of the dileucine peptide with the principal side chains involved in its binding. Mutants in the boxed residues were kinetically analysed.
c and d details of the polar Q(L-4) binding pocket: d with semi transparent electrostatic surface representation (coloured from red −0.5V to blue +0.5V), showing the positive charge principally due to αR21, σ2K13 and σ2R15.
e and f: the deep and shallow pockets on σ2 involved in recognising the leucine residues at position L0 and L+1. Labelled side chains are in the σ2 subunit unless otherwise indicated.
Figure 3. Confirmation of location and conservation amongst different σ subunits of the dileucine motif binding site.
a The dileucine peptide binding site with residues whose mutation strongly inhibit dileucine peptide binding whilst not affecting YxxΦ motif binding are coloured red (See Supplementary Table S1 and Figure S8).
b and c Sensorgrams and KD values for binding of wild type AP2 core and three mutants thereof that strongly inhibit binding of AP2 to PtdIns4,5P2 containing liposomes displaying the CD4 Q peptide motif (b) but do not effect binding to PtdIns4,5P2 containing liposomes displaying the TGN38 YxxΦ motif (c).
d Mammalian σ2, σ1a, σ3 and σ4 were aligned using ClustalW (Figure S7) and the residue conservation plotted from dark purple (absolute conservation) to white (no conservation) onto the surface of mammalian σ2 in two views related by a rotation of 180°. The binding site for the dileucine motif peptide is the outstanding feature of surface residue conservation.
The L-4 position of AP2-binding dileucine motifs is most commonly occupied by the acidic residues glutamate or aspartate, but other residues are possible, such as glutamine (as in the human CD4 dileucine motif used here), histidine (in feline CD4) or arginine (in GLUT4) (reviewed in 7). The binding of the residue in the L-4 position occurs on a hydrophilic patch with an overall positive charge. The electron density for its side chain is not as strong as that for the two leucine residues or the peptide backbone, indicating some degree of flexibility in binding. Indeed, simultaneous mutation of two basic residues in the patch (αR21 and σ2R15) were necessary to reduce dileucine motif binding significantly (Figure 3). In the E peptide where the L-4 position is the more commonly found glutamate 7, the electron density is similar to that for the corresponding glutamine in the Q peptide complex (see supplementary information). The use of general electrostatic complementarity rather than a specific side chain/side chain interaction is supported by the fact that exchange of a motif’s L-4 glutamate to a shorter aspartate has little effect on binding of dileucine motifs to AP2 11. This general electrostatic interaction probably assists in correctly orienting the peptide, a proposal supported by the observations that mutation of the L-4 acidic residue to an alanine significantly weakens but does not abolish binding, whereas mutation to a positively charged arginine abrogates binding altogether in the context of the sequence EDEPLL 11. Consistent with this we observed a slightly weaker binding of AP2 to the Q peptide compared to the E peptide (see data in Table S2). Intriguingly, however, in GLUT4 transporters (RRTPSLL), the L-4 and L-5 positions of the internalisation motifs are arginine residues (reviewed in 7); suggesting that AP2 might possess alternate modes of dileucine motif binding that would likely utilise the same dileucine moiety binding pocket but could place the remainder of the peptide in a different location. Taken together, these structural and mutagenesis data explain the inhibition of the endocytosis of dileucine motif-containing cargoes when the key L[LI] and [ED] residues are mutated 11,13,14.
Point mutation and motif-swapping indicates that the identity of the residues in the L-1 to L-3 positions have some effect on motif binding 10,11. In the structure presented here the arginine in the L-1 position interacts with σ2N97. A proline, often found at this position, would increase the strength of binding by favouring the conformation of the peptide backbone observed in the bound peptide 14. In this AP2:CD4 peptide complex the −2 and −3 position residue side chains point into solvent away from the AP2 surface. In vivo, however, the conformational context of the motif may be affected by these and other surrounding residues, as has been proposed for the L-5 position phosphorylated serine residue, which has no visible electron density for the phosphate in the structures presented here, despite its presence as confirmed by mass spectrometry (data not shown). The apparent lack of sequence specificity for residues other than the L[LI] moiety which is manifest in the increased tolerance of AP2 for alterations in residues adjacent to the L[LI] as compared with AP1 and AP3 11, is reflected in the lack of strong contacts between non L[LI] residues of the motif and AP2 displayed here. Such comparatively broad specificity, as is shown by the similar binding affinities displayed by AP2 for a number of different ‘acidic dileucine’ motifs (see Supplementary Table S2), may ensure the efficient internalisation of dileucine motif-containing proteins that arrive at or mis-sort to the cell surface. The ability of the other AP complexes to discriminate between different L[LI] signals may be necessary to ‘fine tune’ the directing of cargoes to different intracellular compartments.
In the closed and therefore inactive conformation of AP2 3, the hydrophobic L[LI] binding pockets are occupied by side chains from residues in the N-terminal extension to the first helix of the β2 subunit. β2F7 occupies the deep L pocket whilst β2Y6 lies in the broader and shallower L+1 L[LI] pocket (Figure 4 and Figure S8c). Phosphorylation of β2Y6 by EGFR 15 would inhibit the binding of β2Y6 in the L+1 pocket and so greatly weaken the interaction between the N-terminus of β2 and σ2. In the overall structure of the heterotetrameric complex has the helical solenoids of the α and β2 chains form a puckered ring around σ2 and the N-terminal domain of μ2 (N-μ2), which together make a bowl for the C-terminal domain of μ2 (C-μ2). Comparing this dileucine motif-liganded, unlocked structure with the ‘inactive’ conformation, the α/β2 ring splits approximately as two rigid bodies, α(1-400)/σ2 and α(401-end)/β2/N-μ2, with a relative rotation of ~20° about a point near residue α400. This widens the opening of the ring at the N-terminus of β2, and the N-terminal extension swings away into solvent, thus exposing the peptide binding site. C-μ2 forms a third rigid body, bridging between α and β2, and roughly preserving its contacts with both α and β2, such that the YxxΦ binding site on μ2 remains blocked by β2V365 and β2Y405. In the case of AP1, and very likely therefore also in AP2, it has been proposed that binding of either an acidic dileucine or YxxΦ motif drives a conformational change that favours the binding of the other motif 16. Therefore, a further conformational change must occur in order to produce a fully active conformation that is able to bind both types of motif and PtdIns4,5P2 simultaneously. The failure to attain a fully active ‘open’ conformation is driven by a combination of factors including crystal packing, the lack of a YxxΦ peptide to compete out the autoinhibitory binding of β2 to μ2 and the high concentration of ammonium sulphate in the crystallisation conditions which will increase the apparent strength of the interaction between β2V365 and the C-μ2 Φ residue binding pocket and which also helps to stabilise the interaction of β2 residues 350 and 500 with C-μ2.
The binding site on σ2 used by AP2 to bind acidic dileucine motifs is not the same as that used by other proteins with σ2 or ‘longin domain’ folds to bind their cognate ligands: these include the SNAREs Sec22b 17 and Vamp7 5 and the signal recognition particle SRα subunit 18. Intriguingly, however, the hydrophobic L[LI] binding pocket, but not the −4 position acidic residue binding patch, is largely conserved in the ζCOP subunit of the β, γ, δ, ζ subcomplex of COPI that is homologous in structure to AP complexes (PDB 2HF6 and supplementary information), pointing to a possible conservation of function in the binding of a di-hydrophobic motif, candidates for which would include the FF motif of the COPI cargoes, the p24 family and ERGIC-53 19 and the N-terminus of β-COP.
The mechanism of the binding of a dileucine motif by the AP2 clathrin adaptor complex conforms to the established paradigm of a CCV/cargo interaction. The peptide is bound in an extended conformation by a folded adaptor domain, with specificity arising from a few residue side chains fitting into compatible pockets. The mode of binding is therefore superficially similar to the binding of DxxLL motifs by the VHS domains of GGAs 8,9 but the design of the LL and acidic residue recognition pockets, their spacing, and the structures underlying them are completely different (see figure S9). The interaction between the two isolated components, a dileucine motif-containing peptide molecule and a single AP2 complex, is dynamic with a KD in the micromolar range (1-3 μM) 12 and buries around 1100Å2 of solvent accessible surface area, which are both similar to the values demonstrated for other cargo/clathrin adaptor interactions 6. When the signal is presented in, and so co-recognised with, a PtdIns4,5P2 -containing membrane, the apparent strength of the interaction with AP2 is increased with the KD dropping into the high nM range as shown here and it is this coincidence detection of PtdIns4,5P2 and acidic dileucine motif that allows the signal to outcompete the N-terminus of β2 for binding to σ2. It is also the requirement both for co-recognition with PtdIns4,5P2 and a conformational change that prevents inappropriate recognition of dileucine and also YxxΦ sequences in the cytoplasm.
Online Materials and Methods
Expression, crystallisation & structure solution
The four subunits of the AP2 core (1-621 mouse α-adaptin; 1-591 human β2-adaptin; 1-435 rat μ2-adaptin, 1-143 mouse σ2-adaptin) were co-expressed in E. coli from two bicistronic plasmids of different (ampicillin and kanamycin) antibiotic resistances as in 3 in E. coli BL21DE3 pLysS cells at 22°C. Protein expression was allowed to continue for 14 hours, the cells harvested, lysed using a Cell Disrupter (Constant Cell Disruption Systems) and insoluble material removed by centrifugation. AP2 was purified throughout in 10mM Tris pH 8.7, 250mMNaCl, 1mM DTT. The complex was bound to glutathione sepharose (Pharmacia), the column extensively washed and the complex eluted by overnight cleavage at 20°C with bovine thrombin (Sigma). The resulting AP2 complex was bound to NiNTA agarose (Quiagen), the column extensively washed and the complex eluted with buffer containing 0.3M imidazole. The complex was then purified by gel filtration on Superdex S200 (Pharmacia) and finally passed back down a glutathione sepharose column to remove any remaining uncleaved GST-AP2 complex. The yield was 1-2 mg of purified AP2 per litre of culture.
Crystals of the CD4 peptide/AP2 core complexes were grown over a period of three weeks by hanging drop vapour diffusion against a reservoir containing 1.7-2.2M ammonium sulphate, 100mM sodium citrate pH6.5 and 5mM DTT from a mixture of 10mg/ml AP2 core and 7mg/ml peptide. The best crystals had dimensions of around 200⌠m and diffracted to a best resolution of 3.0Å. Crystals were cryoprotected with 1.8-2.3M ammonium sulphate, 100mM sodium citrate pH 6.5, 17% glycerol and 7mg/ml CD4 dileucine peptide. Data were collected at 100K on Diamond beamline I03, integrated using Mosflm 21, scaled with Scala 22. The crystals belong to space group P43212 with two molecules in the asymmetric unit. The structure was solved by molecular replacement using Phaser 20, with models derived from the IP6-liganded ‘inactive’ AP2 core structure (3 PDB code 2vgl). Extra electron density representing the peptide bound to the σ2 subunit was clear from the first maps after molecular replacement (figure S1). The final model refined at 3.0Å resolution comprises residues 3-623 of α (missing 1-2 and including a final 2 residues from the cloning linker), 12-582 of β2 (missing 1-11 and 583-591), 1-142 of σ2 (missing none), and residues 1-141 and 159-435 of μ2 (missing 142-158, as well as the loop 223-231 which are also missing in the structure of isolated C-μ2), the dileucine peptide, sulphates and waters. For statistics relating to the data collection and structure solution see Table 1 and for a sample of typical electron density see FigureS2.
Table 1. Data collection & refinement statistics.
| Dataset | Q peptide | E peptide |
|---|---|---|
| Space group, unit cell (a, c) | P43212, 169.9, 321.7 Å | P43212, 171.2,324.3 Å |
| Wavelength | 0.976 Å | 0.976 Å |
| Rotation range (nearest axis to spindle) | 112° (c*) | 114° (a*) |
| Resolution range Å (high resolution) | 51–3.0 (3.16-3.0) | 47-3.4 (3.58-3.40) |
| Rmerge | 0.198 (1.17) | 0.195 (0.96) |
| Rmerge in top intensity bin | 0.050 | 0.058 |
| Rmeas | 0.223 (1.32) | 0.212 (1.06) |
| Rpim | 0.102 | 0.079 (0.42) |
| Number of observations (unique) | 853264 (96683) | 410234 (64153) |
| < <Ih> / ⌠(<Ih>) > | 8.6 (1.8) | 7.8 (1.9) |
| Completeness | 100.0% (100.0%) | 95.8% (89.1%) |
| Multiplicity | 8.8 (9.0) | 6.4 (5.0) |
| Wilson | 60Å2 | 58 Å2 |
| Refinement | ||
| Rwork | 0.204 (0.33) | 0.24 (0.33) |
| Rfree | 0.261 (0.37) | 0.267 (0.38)) |
| Number of reflections (test set) | 183747 (9168) | 116095 (5902) |
| RMSD bonds (angles) | 0.007Å (1.1°) | 0.009Å (1.3°) |
| Number of TLS groups | 28 | 28 |
| <Biso> (peptides) | 79Å2 (84Å2) | 86Å2 (108Å2) |
| Number of atoms | 28125 | 28125 |
| Number of sulphate ions (waters) | 34 (26) | 34 (26) |
The orientation and registration of the RMS(P)QIKRLLSE peptide in this density was decided as follows. (1) There were clearly two pockets for hydrophobic residues, one deep and one shallow. Electron density consistent with leucine side chains could be seen in each pocket, but the orientation of the backbone connecting them could be in either direction. (2) The electron density for the peptide extends for about 4 or 5 residues on one side of the deep pocket, but not more than 1 or 2 on the other side of the shallow pocket: since the LL sequence is only two residues from the C-terminus of the peptide, the longer part of the density must be N-terminal to the LL sequence. This places the invariant first L in the deep pocket, and the conserved second L (which can also be I or M) in the shallower pocket. With this model of the peptide, the structure refined well, showing good density for the residues SQIKRLLS (see Figure 2), but no density for the phosphate moiety of the phophoserine (S(P)) side chain, which was therefore modelled as a serine, although mass spectrometry of a sample of peptide incubated in crystallisation conditions for 2 weeks confirmed that the serine residue remained phosphorylated. The lack of density for the phosphate is likely to be caused by static disorder of the phosphate group. The structure was refined using Phenix.refine 23 with default tight NCS restraints between the two molecules, and rebuilt with Coot 24. Large positive difference density peaks close to Arg or Lys side chains were interpreted as SO42− ions, given the crystallisation conditions in 2M (NH4)2SO4, and a few well resolved waters were added to the model (Figure S3). TLS group assignment used the TLSMD server (http://skuld.bmsc.washington.edu/~tlsmd/) 25 splitting the α chain into 6 groups, the β2 chain into 5, the μ2 chain into 2 groups and the σ2 chain as 1 group. Refinement without TLS gave slightly higher Rfactors (Rwork 0.222, Rfree 0.266) but little difference in the maps (map correlation 0.99, 0.93 at 3Å resolution). Ramachandran analysis of the final model showed 86% of residues in the most favoured region, 96% in “allowed” regions.
The structure of a complex between the AP2 core and a version of the CD4 acidic dileucine motif peptide termed “E-peptide” in which the L-4 Q residue was replaced with an E (ie RMSpEIKRLLSE) was also solved. The “E-peptide” dataset was of lower quality than the best Q peptide data set. The crystal used to collect the Q-peptide dataset gave better diffraction than the many others which were tried, and also had the serendipitous advantage of rotating approximately around the long c axis, which reduced the spot overlap compared to rotation around an axis roughly perpendicular to the long axis, as for the E-peptide crystal. The final model from the Q-peptide after the Q to E substitution was refined against the E-peptide data, with & without the peptide: as expected, the map quality was poorer than for the Q-peptide, but there was no significant difference in the structure. The electron density for the peptide shows no evidence of any difference in conformation (figure S4), nor are the residues contacting the E side chain noticeably different: in particular, the side chain density for αR21 is still very weak. The lack of crystallisability of other AP2 core:dileucine motif peptide complexes is likely be explained by the packing interaction that occur in the crystal between the L-1 position arginine in the peptide and the side chain of β2Glu157 and the backbone amide of β2Q154
Peptides sequences, conjugation of peptides to lipid and liposome and analyte preparation
The following peptides were coupled to lipid for incorporation into liposomes, which were then used in the SPR-based binding experiments:
| TGN 38* | CKVTRRPKASDYQRL, |
|---|---|
| Phosphorylated CD4 Q peptide | CHRRRQAERM(S_P)_QIKRLLSEK |
| Phosphorylated CD4 E peptide | CHRRRQAERM(S_P)_EIKRLLSEK |
| Limp-II | CRGQGSTDEGTADERAPLIRT |
| Tyrosinase* | CKKQPQEERQPLLMDKDDYHSLLYQSHL |
| TRP1* | CRSRSTKNEANQPLLTDHYQRYAED |
| LRP* | CKVTRRPKASEDEPLLS |
Peptides with and asterisk were also synthesized with mutations in their critical sorting relevant amino acid residues (Y to A in TGN 38 and pairs of alanine residues substituting for the two leucine residues in dileucine containing peptides) and served as negative controls.
Coupling of peptides to MPB-PE and the subsequent incorporation into liposomes were performed as described in 12. In brief, peptides were covalently linked via their amino-terminal cysteine to 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidophenyl) butyramide], Avanti, USA. The lipid-linked peptide was subsequently incorporated into 100nm liposomes that were composed of 60% PC, 20%PE, 10% PtdIns4,52 and 10% peptido-lipid. The efficiency of peptide coupling was determined by mass spectrometry and the integrity of the liposomes was controlled by dynamic light scattering. The average variation of the PtdIns4,5P2 content between different liposome preparations was less than 3% as determined by binding of the ENTH domain of Epsin1 (See figure S5).
Peptido-liposomes were used to generate a stable “membrane mimic” on a L1sensorsurface of a BIAcore 3000 (BIAcore AB, Sweden) SPR biosensor. The system was first equilibrated in 10mM Tris pH 8.7, 250mM NaCl 1mM DTT, which was used as the running buffer followed by priming with two injections of 20mM CHAPS for 1min (flow rate 10μl/min) and injection of the liposomes (0.25 mM final concentration) for 4min at 5μl/min. Loosely bound liposomes were removed by two pulse injections of 50mM sodium hydroxide for 30sec at 30μl/min. This procedure resulted in an increase of the baseline by app. 9500 RU with less than 4% variation between the four flow-cells. After liposome capture, wild type and mutant AP2 cores were injected at concentrations ranging from 50nM to 1μM at a flow rate of 30μl/min for one min (association) followed by buffer flow for 4 min (dissociation). All protein that did not dissociate within this period from the membrane was stripped off by a 20sec pulse injection of 50mM NaOH. In a typical experiment the sensor surface was derivatized with basic liposomes (PC/PE) in flow-cell 1, PC/PE+ PtdIns4,5P2 in flow cell 2, PC/PE+ PtdIns4,5P2 +TGN38 peptide in flow-cell 3 and with PC/PE+ PtdIns4,5P2 +phosphoCD4 peptide in flow-cell 4. This set up allowed to differentiate between background binding to PC/PE liposomes, binding to PtdIns4,5P2 and PtdIns4,5P2 together with a sorting signal containing peptide. In other experiments TGN38 and LRP dileucine sorting signal containing peptido-liposomes were compared with liposomes containing mutant versions of both peptides in which the critical tyrosine or both leucine residues were substituted for alanine. This setup was used to control binding of AP2 to a mutated sorting signal in a PtdIns4,5P2 containing membrane (not shown). The kinetic analysis of binding was carried out exactly as described 26 and rate constants were calculated using the Evaluation software supplied by the manufacturer (BIAcore evaluation software 27). All GST-AP2 complexes used in the SPR binding assay expressed to similar levels and were checked for intactness and folding by SDS PAGE and circular dichroism prior to use. A typical Coomassie Blue stained SDS PAGE gel of AP2 cores used in the SPR-based assay is shown in figure S5. The samples are actually those used in the determination of KDs shown in Figure 3 in the main text.
Supplementary Material
1
2
Acknowledgements
We thank the protein crystallography beamline staff at Diamond, especially Liz Duke, Katherine McAuley and Ralf Flaig for their support and assistance. DJO, BTK and SEM are funded by a Wellcome Trust Senior Research Fellowship to DJO. SH and KS are supported by grands from the Deutsche Forschungsgemeinschaft (SFB635 and SFB670). Atomic coordinates and structure factors have been deposited with the Protein Data Bank, accession numbers 2jkr and r2jkrsf for the Q peptide complex and 2jkt and r2jktsf for the E peptide complex respectively
References
- 1.Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282:1327–32. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pitcher C, Honing S, Fingerhut A, Bowers K, Marsh M. Cluster of differentiation antigen 4 (CD4) endocytosis and adaptor complex binding require activation of the CD4 endocytosis signal by serine phosphorylation. Mol Biol Cell. 1999;10:677–91. doi: 10.1091/mbc.10.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–35. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- 4.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–72. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pryor PR, et al. Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell. 2008 doi: 10.1016/j.cell.2008.07.023. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller SE, Collins BM, McCoy AJ, Robinson MS, Owen DJ. A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles. Nature. 2007;450:570–4. doi: 10.1038/nature06353. [DOI] [PubMed] [Google Scholar]
- 7.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 8.Shiba T, et al. Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature. 2002;415:937–41. doi: 10.1038/415937a. [DOI] [PubMed] [Google Scholar]
- 9.Misra S, Puertollano R, Kato Y, Bonifacino JS, Hurley JH. Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature. 2002;415:933–7. doi: 10.1038/415933a. [DOI] [PubMed] [Google Scholar]
- 10.Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, Bonifacino JS. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol. 2007;81:3877–90. doi: 10.1128/JVI.02725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doray B, Lee I, Knisely J, Bu G, Kornfeld S. The gamma/sigma1 and alpha/sigma2 hemicomplexes of clathrin adaptors AP-1 and AP-2 harbor the dileucine recognition site. Mol Biol Cell. 2007;18:1887–96. doi: 10.1091/mbc.E07-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honing S, et al. Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2. Mol Cell. 2005;18:519–31. doi: 10.1016/j.molcel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 13.Letourneur F, Klausner RD. A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell. 1992;69:1143–57. doi: 10.1016/0092-8674(92)90636-q. [DOI] [PubMed] [Google Scholar]
- 14.Doray B, Knisely JM, Wartman L, Bu G, Kornfeld S. Identification of Acidic Dileucine Signals in LRP9 that Interact with Both GGAs and AP-1/AP-2. Traffic. 2008 doi: 10.1111/j.1600-0854.2008.00786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang F, Jiang X, Sorkin A. Tyrosine phosphorylation of the beta2 subunit of clathrin adaptor complex AP-2 reveals the role of a di-leucine motif in the epidermal growth factor receptor trafficking. J Biol Chem. 2003;278:43411–7. doi: 10.1074/jbc.M306072200. [DOI] [PubMed] [Google Scholar]
- 16.Lee I, Doray B, Govero J, Kornfeld S. Binding of cargo sorting signals to AP-1 enhances its association with ADP ribosylation factor 1-GTP. J Cell Biol. 2008;180:467–72. doi: 10.1083/jcb.200709037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mancias JD, Goldberg J. The transport signal on Sec22 for packaging into COPII-coated vesicles is a conformational epitope. Mol Cell. 2007;26:403–14. doi: 10.1016/j.molcel.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz T, Blobel G. Structural basis for the function of the beta subunit of the eukaryotic signal recognition particle receptor. Cell. 2003;112:793–803. doi: 10.1016/s0092-8674(03)00161-2. [DOI] [PubMed] [Google Scholar]
- 19.Bethune J, et al. Coatomer, the coat protein of COPI transport vesicles, discriminates endoplasmic reticulum residents from p24 proteins. Mol Cell Biol. 2006;26:8011–21. doi: 10.1128/MCB.01055-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Adams PD, Read RJ. PHASER crystallographic software. J. Appl. Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional References for online Materials and Methods
- 21.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 22.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 23.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallographica Section D - Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–50. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 26.Ricotta D, Conner SD, Schmid SL, von Figura K, Honing S. Phosphorylation of the AP2 mu subunit by AAK1 mediates high affinity binding to membrane protein sorting signals. J Cell Biol. 2002;156:791–5. doi: 10.1083/jcb.200111068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonsson U, et al. Real-time biospecific interaction analysis using surface plasmon resonance and a sensor chip technology. Biotechniques. 1991;11:620–7. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2