A New Muscle Glycogen Storage Disease Associated with Glycogenin-1 Deficiency (original) (raw)
. Author manuscript; available in PMC: 2015 Mar 4.
Published in final edited form as: Ann Neurol. 2014 Oct 31;76(6):891–898. doi: 10.1002/ana.24284
Abstract
We describe a slowly progressive myopathy in 7 unrelated adult patients with storage of polyglucosan in muscle fibers. Genetic investigation revealed homozygous or compound heterozygous deleterious variants in the glycogenin-1 gene (GYG1). Most patients showed depletion of glycogenin-1 in skeletal muscle, whereas 1 showed presence of glycogenin-1 lacking the C-terminal that normally binds glycogen synthase. Our results indicate that either depletion of glycogenin-1 or impaired interaction with glycogen synthase underlies this new form of glycogen storage disease that differs from a previously reported patient with GYG1 mutations who showed profound glycogen depletion in skeletal muscle and accumulation of glycogenin-1.
Skeletal muscle relies on glycogen storage for contraction and relaxation. Defects in glycogen synthesis or degradation pathways provoke glycogen accumulation and/or depletion, leading to energy deficiency.1
The 3 main enzymes involved in biosynthesis of glycogen are glycogenin-1, glycogen synthase, and branching enzyme.2 Glycogenin-1 is a glycosyltransferase that forms a short glucose polymer of approximately 10 glucose residues by autoglucosylation. Glycogen synthase and branching enzyme allow further elongation and branching of the glucose polymer primer.2
Defects in various enzymes involved in glycogen metabolism cause glycogen storage diseases. Some of these are characterized by accumulation of polyglucosan, which is variably resistant to digestion by alpha-amylase, and contains abnormally long and poorly branched glucosyl chains. Such diseases include branching enzyme (GBE1) deficiency, phosphofructokinase deficiency, aden-osine monophosphate–activated protein kinase deficiency, and Lafora disease.1,3,4 Deficiency of RanBP-type and C3HC4-type zinc finger–containing 1 (RBCK1) has recently been shown to cause a polyglucosan storage disease with cardiomyopathy and skeletal myopathy.5
A specific mutation in the glycogenin-1 gene (GYG1) has been shown to cause an impaired priming of glycogen synthesis in 1 patient with cardiomyopathy and depletion of glycogen in skeletal muscle.6,7
We describe a new polyglucosan body myopathy due to pathogenic variants in GYG1.
Patients and Methods
Patients
Seven adult patients with polyglucosan body myopathy from 7 unrelated families of various ethnic backgrounds were included in the present study. These patients belonged to a cohort of some 20 patients with polyglucosan body myopathy and/or cardiomyopathy negative for GBE1 and RBCK1 disease-causing variants explaining the phenotype. After informed consent, the patients underwent open biopsy of the deltoid or quadriceps femoris muscle for morphologic and histochemical analyses of fresh-frozen muscle tissue. Patients P5 and P6 have been previously reported.8
Molecular Genetic Analysis
Genomic DNA was extracted from blood or frozen skeletal muscle by standard methods, and Sanger sequencing was used to screen for GYG1 (NM_004130) variants. Total RNA was isolated from frozen skeletal muscle using the RNeasy Fibrous Tissue Mini Kit (Qiagen, Valencia, CA). RNA was reverse-transcribed with the QuantiTect reverse transcription kit (Qiagen), and GYG1 cDNA was analyzed by Sanger sequencing.
Morphological Analysis
Cryostat sections of fresh-frozen deltoid or quadriceps femoris muscle tissue were histochemically analyzed by standard techniques. For immunohistochemistry the following antibodies were used: SQSTM1 (D-3; Santa Cruz Biotechnology, Santa Cruz, CA; 1:1,000), Anti-Ubiquitin (P4D1-A11; Merck Millipore, Billerica, MA; 1:100), and Anti-Human Desmin (D33; Dako Cytomation, Carpinteria, CA; 1:50).
Protein and Functional Analyses
Autoglucosylation was analyzed in vitro applying a cell-free protein expression system. The cloned sequence of glycogenin-1 (NM_001184720) was inserted downstream of the 6xHis tag in the pEXP5-NT/TOPO vector (Invitrogen Life Technologies, Carlsbad, CA). Variants identified in the patients were introduced using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) with the primers listed in the Supplementary Table. The recombinant glycogenin-1 was expressed using the FastLane Escherichia coli protein synthesis kit (RiNA, Berlin, Germany) for 1 hour at 37° C and allowed to autoglucosylate in N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid (50mM, pH7.5), MnCl2 (5mM); dithiothreitol (0.5mM), and 0.1M uridine diphosphate (UDP) glucose (Calbiochem, San Diego, CA) for 1 hour at 30° C.7,9
Western blot analysis was performed on protein extracted from skeletal muscle tissue as well as recombinant protein. Cryostat sections from biopsies were treated with 10μg/ml human alpha amylase (Sigma-Aldrich, St Louis, MO) in a total volume of 10μl phosphate-buffered saline, pH 6.5 for 1 hour at 37° C. Protein extracts were loaded and separated on 10% Bis-Tris gel or 3 to 8% Tris-acetate Gel (Novex; Life Technologies, Grand Island, NY) followed by electroblotting. The membranes were incubated with primary antihuman glycogenin-1 N-terminal antibody M07 clone 3B5 (Abnova, Taipei, Taiwan; 1:500) or C-terminal anti-GYG1 (Atlas Antibodies, Stockholm, Sweden; 1:500) or anti-GBE1 (Atlas Antibodies; 1:500) or anti-GYS1 (Atlas Antibodies). Western Breeze (Invitrogen) was used for antibody detection.
Results
Clinical Findings
A clinical summary is provided in the Table. Age at onset ranged from childhood to adult life. Muscle weakness was progressive in 1 patient and slowly progressive in the other patients. Four patients developed symmetrical proximodistal muscle weakness with variable involvement of hip and shoulder girdle muscles, lower legs, and hand muscles. Two patients showed isolated proximal muscle weakness. One patient showed only hand and finger involvement starting in adulthood. Facial and ocular muscles were spared. Serum creatine kinase was elevated in 1 and normal in the other patients. Electromyography was invariably myopathic, but 1 patient showed in addition neurogenic involvement. Electrocardiography and cardiac ultrasound were normal in all patients. Cardiac magnetic resonance imaging was normal in P4. Myocardial biopsy was not performed.
TABLE.
Clinical, Laboratory, and Genetic Findings
| Patient | |||||||
|---|---|---|---|---|---|---|---|
| Characteristic | PI | P2 | P3 | P4 | P5 | P6 | P7 |
| Gender | M | F | M | F | M | F | F |
| Age at onset, yr | 17 | Probably childhood | 39 | 65 | 62 | 61 | 49 |
| Initial symptoms | Difficulty running | Trouble rising from squatting since childhood; progressive weakness after age 39 years | Difficulty walking | Difficulty climbing stairs and walking | Difficulty climbing stairs and walking | Loss of grip strength in both hands | Difficulty climbing stairs and loss of grip strength in right hand |
| Age at examination, yr | 26 | 50 | 43 | 72 | 72 | 66 | 57 |
| Clinical features | Hip girdle muscle weakness, atrophy, and fatigability | Shoulder, hip girdle, and distal anterior leg muscle weakness | Hip girdle, tibial, and peroneal muscle weakness | Shoulder and hip girdle muscle weakness | Hip girdle and thigh muscle weakness and atrophy; milder asymmetric weakness of shoulder girdle and upper arm muscles | Intrinsic hand muscle weakness and atrophy; mild extrinsic finger extensor muscle weakness | Shoulder and hip girdle muscle weakness; wrist and finger extensor, interosseus, foot and toe extensor paralysis |
| Serum CK | 977U/1 | Normal | Normal | Normal | Normal | Normal | Normal |
| Cardiac examination | Normal | Normal | Normal | Normal | Normal | Normal | Normal |
| EMC | Myopathic | Myopathic with fibrillation potentials and brief myotonic discharges | Myopathic | Myopathic | Myopathic and neurogenic, few positive sharp waves and fibrillation potentials | Myopathic, positive sharp waves and fibrillation potentials | Myopathic, polyphasic motor unit action potentials with early recruitment |
| Muscle pathology by light microscopy | Partially x-amylase–resistant, PAS-positive inclusions, desmin, SQSTM1, and ubiquitin positive | Partially α-amylase–resistant, PAS-positive inclusions | Partially α-amylase–resistant, PAS-positive inclusions; moderate interstitial fibrous and fat tissue | Partially α-amylase–resistant, PAS-positive inclusions, desmin, SQSTM1, and ubiquitin positive | Partially α-amylase–resistant, PAS-positive inclusions, desmin, SQSTM1, and ubiquitin positive; moderate interstitial fibrous and fat tissue | Partially α-amylase–resistant, PAS-positive inclusions, desmin, SQSTM1, and ubiquitin positive | Partially α-amylase–resistant, PAS-positive inclusions, desmin, SQSTM1, and ubiquitin positive; prominent interstitial fibrous and fat tissue |
| Muscle pathology by electron microscopy | Polyglucosan bodies | Polyglucosan bodies | Polyglucosan bodies | Polyglucosan bodies | Polyglucosan bodies | Polyglucosan bodies | Polyglucosan bodies |
| DNA analysis | Homozygous c.143 + 3G>C | Homozygous c.143 + 3G>C | Compound heterozygous C.304>C, c.749G>A | Homozygous c.46G>C | Compound heterozygous c.143 + 3G>C, c.7G>C | Homozygous c.484delG | Compound heterozygous c.143 + 3G>C c.970C>T |
| RNA analysis | r.8_l43del | r.8_l43del | ND | r.46g>c | r.8_l43del r.O | r.O | r.8_l43del r.970c>u |
| Protein change | p.Asp3Glufs*4 | p.Asp3Glufs*4 | p.(Aspl02His) p.(Trp250*) | p.Alal6Pro | p.Asp3Glufs*4 p.spl | p.Thrl63Aspfs*5 | p.Asp3Glufs*4 p.Arg324* |
Morphological Analysis
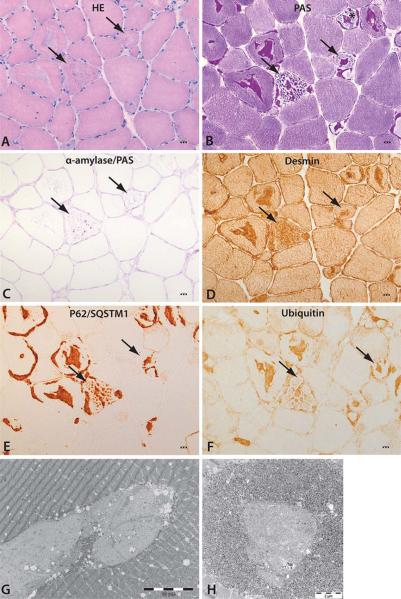
Morphological investigations by light- and electron-microscopy of muscle revealed characteristic alterations. Thirty to forty percent of the fibers, frequently in clusters, showed inclusions in the center and subsarcolemmal regions of the fibers (Fig 1). Periodic acid–Schiff (PAS)-positive material formed the main content of the inclusions. Some fibers presented inclusions surrounded by normal glycogen content, whereas other fibers showed depletion of normal glycogen around the inclusions. Alpha-amylase treatment showed a variable degree of digestion of the PAS-positive material. Inclusions were positively immunostained for desmin, ubiquitin-binding protein sequestosome-1 (p62/SQSTM1), and ubiquitin. Electron microscopy demonstrated presence of storage material in the subsarcolemmal and central regions of the fibers. The inclusions frequently formed a lobulated grape structure composed of oval subunits with the same electron density. A rim of normal glycogen particles and mitochondria surrounded the storage material. Smaller inclusions often contained central areas composed of less electron-dense filamentous material. Necrotic or regenerating fibers were not identified. Increase of interstitial connective tissue and fat was noted in P3 and P5, and more pronounced in P7.
FIGURE 1.
Characteristic morphological alterations of skeletal muscle. (A) Hematoxylin–eosin (HE) staining showing bluish inclusions in muscle fibers (arrows). (B) The inclusions contain periodic acid–Schiff (PAS)-positive material (arrows), and some fibers lack normal intermyofibrillar glycogen (asterisk). (C) The storage material is partially resistant to alpha-amylase treatment (arrows). (D–F) The storage material (arrows) is stained with antibodies for desmin, P62/SQSTM1, and ubiquitin. Scale bars in A–F correspond to 5μm. (G, H) Electron microscopy demonstrating abnormal glycogen presenting with ovoid structures composed of partly filamentous material surrounded by a rim of more dense glycogen granules and mitochondria.
Molecular Genetic Analysis
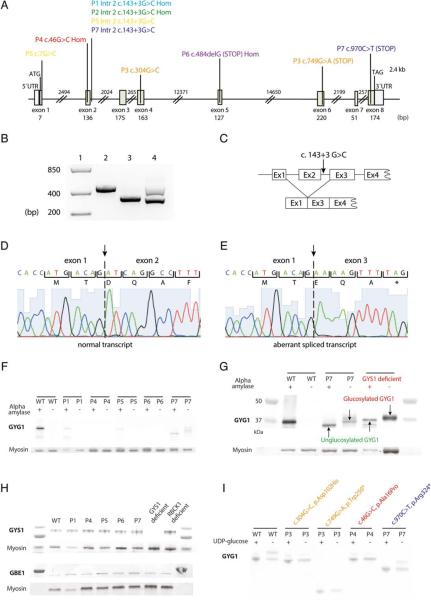
The identified GYG1 pathogenic variants are summarized in the Table and Figure 2A. The most common variant was a single nucleotide substitution at the donor splice site in intron 2 (c.143+3G>C), either homozygous or compound heterozygous. Analysis of GYG1 cDNA with this variant revealed aberrant splicing with skipping of exon 2 (r.8_143del) creating a frameshift p.Asp3Glufs*4 (see Fig 2B–E). P5 was compound heterozygous for the common splice site variant and a c.7G>C variant affecting the last nucleotide in exon 1, within the donor splice site. The corresponding transcript was not detected in cDNA, suggesting that this allele was not expressed. P7 was compound heterozygous for the common splice site variant and a C-terminal nonsense (c.970C>T, p.Arg324*) variant. P3 was compound heterozygous for a missense (c.304G>C, p.Asp102His) and a nonsense (c.749G>A, p.Trp250*) variant. P4 was homozygous for a N-terminal missense variant (c.46G>C, p.Ala16Pro) that was confirmed at the RNA level. P6 was homozygous for a single nucleotide deletion (c.484delG), and no transcript was detected in cDNA.
FIGURE 2.
Genetic and protein analyses. (A) Schematic illustration of the coding and noncoding region of the gene GYG1 (NM_004130). Detected variants are marked in different colors for each patient (P1, light blue; P2, green; P3, orange; P4, red; P5, yellow; P6, purple; P7, dark blue). (B) Reverse transcriptase polymerase chain reaction products covering exons 1 to 4. Lane 1: size ladder; lane 2: control sample with wild-type sequence; lane 3: patient homozygous for the GYG1 c.14313G>C variant resulting in transcripts with skipping of exon 2; lane 4: patient heterozygous for the GYG1 c.14313G>C variant, showing 2 bands, 1 of normal size and 1 with aberrant splicing with exon 2 skipping (lower band). (C) Schematic illustration of the consequences of incorrect splicing. The GYG1 c.14313G>C variant results in skipping of exon 2. (D) Normally spliced transcript. (E) Aberrantly spliced transcript with exon 2 skipping. (F) Results of Western blot analysis of glycogenin-1 in skeletal muscle from a normal control (WT) and patients (P1, P4, P5, P6, and P7) performed with (1) and without (2) alpha-amylase treatment. After alpha-amylase treatment, glycogenin-1 was detected in P1, P5, and P7, demonstrating that these patients produce some residual glycogenin-1. Without alpha-amylase treatment, free glycogenin-1 was detected in P7. (G) Western blot analysis of a normal control, P7, and a patient with glycogen synthase deficiency. The glycogenin-1 detected in P7 as well as in the glycogen synthase– deficient patient revealed a gel shift corresponding to approximately 1kDa, indicating that it was autoglucosylated. Glycogenin-1 in P7 is reduced in size due to the truncating variant. (H) Western blot analyses of glycogen synthase (GYS1) and branching enzyme (GBE1) in P1, P4, P5, P6, P7, 1 patient with GYS1 deficiency, and 1 with RBCK1 deficiency. No obvious upregulation or downregulation of these enzymes was evident in patients with pathogenic GYG1 variants. (I) In vitro autoglucosylation demonstrated that glycogenin-1 in P3 and P4 was unable to autoglucosylate when uridine diphosphate (UDP) glucose was added (1). The truncated glycogenin-1 in P7 increased in size when UDP glucose was added, showing that it was autoglucosylated.
Protein and Functional Analyses
Expression of glycogenin-1 in skeletal muscle tissue was studied by Western blot analysis either without or with alpha-amylase treatment, to cleave off the glucose in the glycogen particles (see Fig 2F). Free autoglucosylated glycogenin-1 weighs approximately 1kDa more than the unglucosylated protein. In normal individuals, there is no free glycogenin-1. A weak band corresponding to glycogenin-1 was identified in P1 and P5 only after alpha-amylase treatment, demonstrating presence of some residual functional glycogenin-1. P4 and P6 did not show any glycogenin-1. Glycogenin-1 was detected both with and without alpha-amylase treatment in P7. Glycogenin-1 had a reduced molecular weight in P7, corresponding to a shorter protein produced by the truncating variant c.970C>T p.Arg324*. The gel shift induced by alpha-amylase treatment revealed that the protein was functionally active with regard to autoglucosylation. Material from P2 and P3 was not available for Western blot analysis.
A gel shift after alpha-amylase treatment was identified only in P7, lacking the C-terminal of glycogenin-1 (see Fig 2F), suggesting defective elongation of the glucose polymer primer. To further explore this hypothesis, an additional Western blot experiment of glycogenin-1 was performed. Samples from P7 and a patient lacking glycogen synthase due to GYS1 nonsense variants were analyzed (see Fig 2G). In both cases, a gel shift of similar size after alpha-amylase treatment was demonstrated. These results suggest that elongation of the glycogen polymer, normally catalyzed by glycogen synthase, was impaired in P7 by loss of the glycogenin-1 C-terminal. Western blot analyses for glycogen synthase and branching enzyme showed no changes in the expression in the patients (see Fig 2H).
Assay of autoglucosylation in vitro of the GYG1 variants resulting in p.Trp250*, p.Asp102His, and p.Ala16Pro did not show any gel shift after the addition of UDP glucose, suggesting nonfunctional glycogenin-1 (see Fig 2I). Proteins with the missense variants were detected with double bands even in the absence of UDP glucose, suggesting that the shift was due to a conformational change and not incorporation of glucose. The C-terminal nonsense variant p.Arg324* resulted in truncated protein with reduced molecular weight but preserved autoglucosylation, which was in accordance with the results from Western blot analysis of glycogenin-1 in patients’ muscle tissue (see Fig 2F).
Discussion
In the present study, we report a new recessive muscle glycogen storage disorder caused by GYG1 pathogenic variants. All patients presented a skeletal myopathy without cardiac involvement. Most patients manifested slowly progressive, adulthood onset, hip girdle, shoulder girdle, and/or hand and leg muscle weakness. One patient showed childhood onset, and 1 presented a peculiar phenotype characterized by late onset weakness of hands and fingers.
In the 7 patients, we identified different missense, nonsense, or frameshift GYG1 pathogenic variants, distributed all over the gene. There was either reduced or complete absence of glyogenin-1 protein, in accordance with the deleterious effects of the variants. The most frequent variant was c.143+3G>C, identified in 4 patients from different ethnic backgrounds. This common splice site variant caused a complete or nearly complete alternative splicing, with profound reduction of wild-type glycogenin-1.
PAS-positive storage material was found in around 30 to 40% of muscle fibers. This picture had some similarities with polyglucosan body myopathies of other causes, especially branching enzyme or RBCK1 deficiency.5,10,11 However, the inclusions found in glycogenin-1–deficient patients appeared to be less alpha-amylase resistant. These were also more frequently found in fibers with apparently normal glycogen content compared to myopathy associated with GBE1 and RBCK1. These findings suggest morphological heterogeneity in polyglucosan body myopathies and probably reflect the differences in primary gene defects. Polyglucosan body formation has in some cases been suggested to be caused by an imbalance between the activities of glycogen synthase and branching enzyme.12,13 We did not show any apparent upregulation or downregulation of glycogen synthase or branching enzyme in our glycogenin-1–deficient patients. However, analysis of glycogen synthase and branching enzyme activities were not available for our patients.
Unlike our patients, a previously reported patient with pathogenic GYG1 variants showed cardiomyopathy with storage of abnormal glycogen in cardiomyocytes and glycogen depletion in skeletal muscle.6 This subject presented a different phenotype compared to our patients. He showed a cardiomyopathy at young age, whereas our patients developed late onset skeletal myopathy. His muscle biopsy demonstrated profound glycogen depletion, whereas our patients showed polyglucosan body myopathy. Glycogenin-1 was accumulated in the previous patients but depleted in ours. The type of pathogenic variant and other possible factors might explain these differences. Further studies may demonstrate a large variability with regard to cardiac and skeletal muscle involvement in patients with GYG1 pathogenic variants.
Various amounts of normal glycogen were found in all patients despite depletion or absence of glycogenin-1. This observation raises the question how this glycogen is formed. Apparently, glycogenin-1 is not mandatory for glycogen formation in muscle, which may explain the relatively late onset and the slow progression of the disease. Whether this is due to upregulation of another autoglucosylating protein, or alternative primers for glycogen synthase, remains to be investigated.
In P7, with glycogenin-1 lacking the C-terminal, we demonstrated free autoglucosylated glycogenin-1. This is similar to what is found in patients with glycogen synthase deficiency.14 Because glycogen synthase is known to bind the glycogenin-1 C-terminal,15 our results strongly indicate that this binding is essential for glycogen synthase function.
In summary, we have described a new muscle glycogen storage disorder characterized by polyglucosan bodies that is due to deficiency of glycogenin-1, and have shown that the C-terminal of glycogenin-1 is essential for glycogen synthase activity.
Supplementary Material
Supplemental materials
Acknowledgment
Supported by grants from the Swedish Research Council (7122; A.O.), the NIH (NINDS, NS6277; A.G.E.), the Keith B. Hayes Foundation (S.D. and H.O.A.) and the Adult Polyglucosan Body Disease Research Foundation (S.D and H.O.A.).
We thank Dr D. Fischer for the clinical investigations of the patients; G. Brochier, G. Alm en, L. Seifi, and A.-C. Ericsson for technical assistance; and the patient's families for their support.
Footnotes
Additional supporting information can be found in the online version of this article.
Potential Conflicts of Interest
None.
References
- 1.Oldfors A, DiMauro S. New insights in the field of muscle glycogenoses. Curr Opin Neurol. 2013;26:544–553. doi: 10.1097/WCO.0b013e328364dbdc. [DOI] [PubMed] [Google Scholar]
- 2.Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS. Glycogen and its metabolism: some new developments and old themes. Biochem J. 2012;441:763–787. doi: 10.1042/BJ20111416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiMauro S, Spiegel R. Progress and problems in muscle glycoge-noses. Acta Myol. 2011;30:96–102. [PMC free article] [PubMed] [Google Scholar]
- 4.Malfatti E, Birouk N, Romero NB, et al. Juvenile-onset permanent weakness in muscle phosphofructokinase deficiency. J Neurol Sci. 2012;316:173–177. doi: 10.1016/j.jns.2012.01.027. [DOI] [PubMed] [Google Scholar]
- 5.Nilsson J, Schoser B, Laforet P, et al. Polyglucosan body myopathy caused by defective ubiquitin ligase RBCK1. Ann Neurol. 2013;74:914–919. doi: 10.1002/ana.23963. [DOI] [PubMed] [Google Scholar]
- 6.Moslemi AR, Lindberg C, Nilsson J, et al. Glycogenin-1 deficiency and inactivated priming of glycogen synthesis. N Engl J Med. 2010;362:1203–1210. doi: 10.1056/NEJMoa0900661. [DOI] [PubMed] [Google Scholar]
- 7.Nilsson J, Halim A, Moslemi AR, et al. Molecular pathogenesis of a new glycogenosis caused by a glycogenin-1 mutation. Biochim Biophys Acta. 2012;1822:493–499. doi: 10.1016/j.bbadis.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 8.Jeub M, Kappes-Horn K, Kornblum C, Fischer D. Polyglycosan body myopathy [in German]. Nervenarzt. 2006;77:1487–1491. doi: 10.1007/s00115-006-2184-x. [DOI] [PubMed] [Google Scholar]
- 9.Nilsson J, Halim A, Larsson E, et al. LC-MS/MS characterization of combined glycogenin-1 and glycogenin-2 enzymatic activities reveals their self-glucosylation preferences. Biochim Biophys Acta. 2014;1844:398–405. doi: 10.1016/j.bbapap.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Boisson B, Laplantine E, Prando C, et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat Immunol. 2012;13:1178–1186. doi: 10.1038/ni.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, Kim C, Bradfield J, et al. Whole-genome DNA/RNA sequencing identifies truncating mutations in RBCK1 in a novel Mendelian disease with neuromuscular and cardiac involvement. Genome Med. 2013;5:67. doi: 10.1186/gm471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCue ME, Valberg SJ, Lucio M, Mickelson JR. Glycogen synthase 1 (GYS1) mutation in diverse breeds with polysaccharide storage myopathy. J Vet Intern Med. 2008;22:1228–1233. doi: 10.1111/j.1939-1676.2008.0167.x. [DOI] [PubMed] [Google Scholar]
- 13.Raben N, Danon M, Lu N, et al. Surprises of genetic engineering: a possible model of polyglucosan body disease. Neurology. 2001;56:1739–1745. doi: 10.1212/wnl.56.12.1739. [DOI] [PubMed] [Google Scholar]
- 14.Kollberg G, Tulinius M, Gilljam T, et al. Cardiomyopathy and exercise intolerance in muscle glycogen storage disease 0. N Engl J Med. 2007;357:1507–1514. doi: 10.1056/NEJMoa066691. [DOI] [PubMed] [Google Scholar]
- 15.Skurat AV, Dietrich AD, Roach PJ. Interaction between glycogenin and glycogen synthase. Arch Biochem Biophys. 2006;456:93–97. doi: 10.1016/j.abb.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials