Alternative Lengthening of Telomeres Renders Cancer Cells Hypersensitive to ATR Inhibitors (original) (raw)
. Author manuscript; available in PMC: 2016 Jan 16.
Published in final edited form as: Science. 2015 Jan 16;347(6219):273–277. doi: 10.1126/science.1257216
Abstract
Cancer cells rely on telomerase or the alternative lengthening of telomeres (ALT) pathway to overcome replicative mortality. ALT is mediated by recombination and is prevalent in a subset of human cancers, yet whether it can be exploited therapeutically remains unknown. Loss of the chromatin remodeling protein ATRX associates with ALT in cancers. Here, we show that ATRX loss compromises cell-cycle regulation of the telomeric non-coding RNA TERRA and leads to persistent association of replication protein A (RPA) with telomeres after DNA replication, creating a recombinogenic nucleoprotein structure. Inhibition of the protein kinase ATR, a critical regulator of recombination recruited by RPA, disrupts ALT and triggers chromosome fragmentation and apoptosis in ALT cells. Importantly, the cell death induced by ATR inhibitors is highly selective for cancer cells that rely on ALT, , suggesting that such inhibitors may be useful for treatment of ALT-positive cancers.
Cancer cells overcome replicative senescence by activating telomerase or the alternative lengthening of telomeres (ALT) pathway (1–3). ALT is used in ~5–15% of all human cancers and is prevalent in specific cancer types, including osteosarcoma and glioblastoma (4). Currently, there are no therapies specifically targeting ALT. ALT relies on recombination to elongate telomeres (3), but how the recombinogenic state of ALT telomeres is established remains elusive. In contrast to cancer cells defective for homologous recombination (HR) and susceptible to Poly(ADP-ribose) polymerase (PARP) inhibition (5, 6), ALT-positive cells are HR-proficient (7). Thus, the reliance of ALT on recombination raises an important question as to whether recombination can be exploited in ALT-positive cancers as a means for targeted therapy.
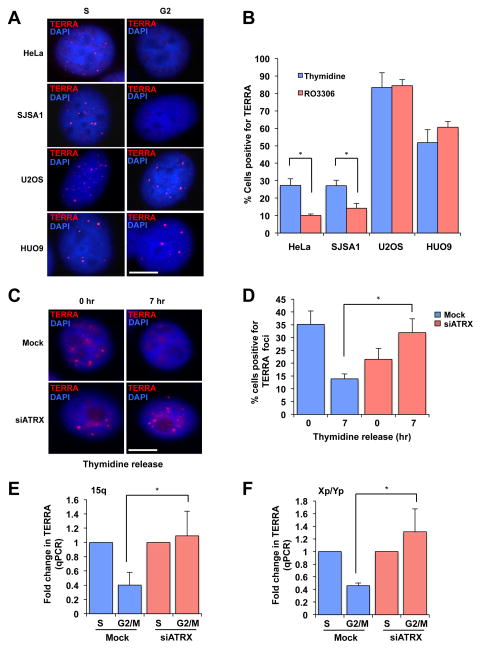
Single-stranded DNA (ssDNA) coated by replication protein A (RPA) is a key intermediate in both DNA replication and HR (8). RPA transiently associates with telomeres during DNA replication, but is released from telomeres after S phase (9, 10). The release of RPA may be an important mechanism to suppress HR at telomeres. The association of RPA with telomeres in S phase is facilitated by TERRA, the telomere repeat-containing RNA, which is also present at telomeres during this period (9, 11–13). To investigate how ALT is established, we determined whether the association of TERRA with telomeres is altered in ALT cells. TERRA colocalized with the telomere-binding protein TRF2 in telomerase-positive HeLa cervical cancer cells (fig. S1) (9). However, in both HeLa and telomerase-positive SJSA1 osteosarcoma cells (fig. S24B), the number of TERRA foci declined from S phase to G2 (Fig. 1A–B) (fig.S2) (9, 12). Although in ALT-positive U2OS osteosarcoma cells TERRA also colocalized with the telomere marker TRF2 (fig. S3A–B), neither the levels of TERRA, nor the colocalization of TERRA and TRF2, declined from S to G2 (fig. S2, S3B–C, S4A–B). Furthermore, in ALT-positive U2OS and HUO9 osteosarcoma cells (Fig. 3D) (fig. S25A–B), the number of TERRA foci increased significantly in S phase and remained high into G2 (Fig. 1A–B) (fig. S2). Thus, in contrast to telomerase-positive cells, ALT cells are defective in the cell-cycle regulation of TERRA.
Fig. 1. Loss of ATRX compromises the cell-cycle regulation of TERRA.
(A) RNA fluorescence in situ hybridization (FISH) analyses of TERRA in HeLa, SJSA1, U2OS and HUO9 cells during the cell cycle. TERRA foci colocalized with TRF2 at telomeres (Fig. S1, S3A–B). To enrich cells in S phase, cells were treated with thymidine alone. To enrich cells in G2, cells were first arrested in S phase with thymidine and then released into medium containing the CDK1 inhibitor RO3306 (Fig. S2). Scale bar: 10 μm. (B) The percentage of cells positive for TERRA foci (> 5 foci) was graphed as the mean with error bars representing standard deviation (n=2). (C) HeLa cells were mock treated or treated with ATRX siRNA, and RNA FISH analysis of TERRA was performed following thymidine release. The knockdown of ATRX was confirmed by Western blot (Fig. S5A). Cells were enriched in late S and G2 phases 7 h after thymidine release (Fig. S5B). Scale bar: 10 μm. (D) The percentage of cells positive for TERRA foci was graphed as the mean with error bars representing the standard deviation (n=3). (E–F) HeLa cells were mock treated or treated with ATRX siRNA, and were enriched in S or M phase with thymidine and nocodazole, respectively (Fig. S5B). TERRA was analyzed by RT-qPCR using the subtelomeric/telomeric primers of chromosome 15q or Xp/Yp. The results are graphed as the mean fold-change with error bars representing the standard deviation (15q n=3, Xp/Yp n=4). *: P-Value < 0.05.
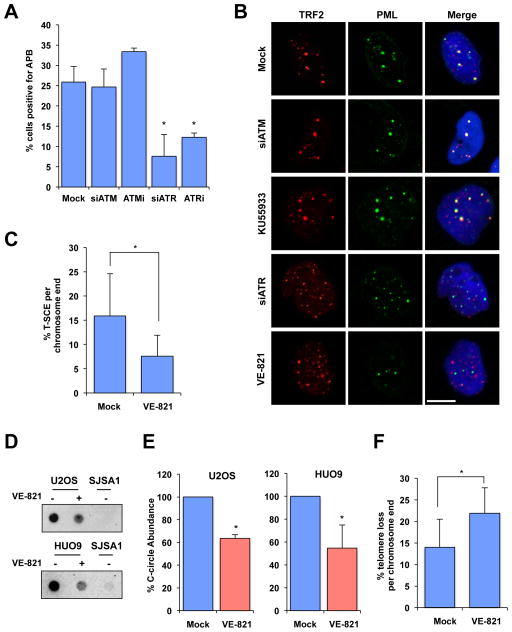
Fig. 3. ATR inhibitor disrupts ALT activity.
(A) U2OS cells were mock treated, treated with 5 μM VE-821, 5 μM KU-55933, or treated with siRNA for ATR, or ATM, and then immunostained for TRF2 and PML. The percentage of cells positive for APBs were graphed as the mean with error bars representing the standard deviation, experiment performed in triplicate (n=3). P-Value < 0.02. (B) Representative images from cells quantified in (A). Scale bar: 10 μm. (C) U2OS cells were mock treated or treated with 2.5 μM VE-821 for 4 days and analyzed for T-SCE events using G-rich (green) and C-rich (red) PNA probes. The fraction of chromosome ends with T-SCE was quantified and graphed as the mean with error bars representing the standard deviation (Mock n=1032, VE-821 n=1556). *: P-Value < 0.01. (DE) HUO9 and U2OS cells were mock treated or treated with 5 μM VE-821 for 24 and 48 h, respectively. C-circle amplification products were detected by dot blot in (D). The levels of C-circles were graphed in (E) as the mean with error bars representing standard deviation (n=2). Telomerase-positive SJSA1 cells were used as a negative control. P-Value < 0.02 (F) The fraction of chromosome ends with telomere loss was quantified and graphed as the mean with error bars representing the standard deviation (Mock n=1032, VE-821 n=1556). *: P-Value < 0.01.
We next explored why TERRA persistently associates with telomeres in ALT cells. Recent cancer genome studies have revealed a correlation of ALT with mutations in the ATRX gene and loss of the chromatin remodeling protein ATRX in cancer (14–17). ATRX was detected in HeLa but not U2OS cells (Fig. S5A, see Fig. S25C) (14), prompting us to investigate whether the dysregulation of TERRA in ALT cells is a result of ATRX loss. Indeed, knockdown of ATRX in HeLa cells resulted in persistent TERRA foci, and elevated TERRA levels in G2/M (Fig. 1C–D, S5, S6). Furthermore, the levels of TERRA derived from individual telomeres (15q and Xp/Yp) declined from S phase to mitosis in control HeLa cells but not in ATRX knockdown cells (Fig. 1E–F). These results suggest that TERRA is repressed by ATRX in G2/M.
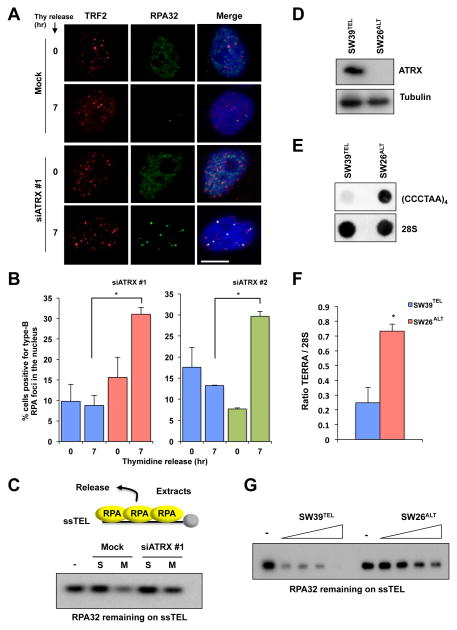
Considering that RPA is released from telomeres in G2/M when TERRA is repressed by ATRX (9), we examined whether ATRX is required for the release of RPA. In HeLa cells, numerous small replication-associated RPA foci (type-A RPA foci) were detected in S phase (Fig. S7). As cells progressed from S to G2, type-A RPA foci became largely undetectable (Fig. 2A). However, upon ATRX knockdown, bright damage-associated RPA foci (type-B RPA foci) were detected at telomeres in a fraction of G2 cells (Fig. 2A, S7, S8). Knockdown of ATRX with two independent siRNAs led to a significant increase of type-B RPA foci in G2 cells (Fig. 2B). To examine the release of RPA from telomeric ssDNA biochemically, we followed this process in cell extracts using an in vitro assay that we previously established (9). A biotinylated ssDNA oligo of telomeric repeats (ssTEL) was coated with recombinant RPA and incubated in extracts from S-phase or mitotic HeLa cells. Consistent with the release of RPA from telomeres in G2/M, RPA was released from ssTEL more efficiently in mitotic extracts than in S-phase extracts (Fig. 2C) (9). Knockdown of ATRX reduced the release of RPA from ssTEL in mitotic extracts (Fig. 2C), demonstrating that ATRX contributes to the RPA release in G2/M. To test if the loss of ATRX in ALT cells affects RPA release, we analyzed IMR90 myofibroblast-derived SW39TEL (telomerase-positive) and SW26ALT (ALT-positive) cells (Fig. S9) (7). ATRX was detected in SW39TEL but not SW26ALT (Fig. 2D). Moreover, the loss of ATRX in SW26ALT associated with a 4-fold increase of TERRA compared with SW39TEL (Fig. 2E–F). Importantly, RPA was released from ssTEL more efficiently in SW39TEL cell extracts than in SW26ALT cell extracts (Fig. 2G), showing that ALT cells lacking ATRX indeed have a reduced ability to release RPA from telomeric ssDNA.
Fig. 2. Loss of ATRX compromises RPA release from telomeres.
(A) HeLa cells were mock treated or treated with ATRX siRNA, and RPA and TRF2 foci were analyzed in S and G2 phases as in Fig. 1C. Scale bar: 10 μm. (B) The percentage of cells positive for RPA foci was graphed as the mean with error bars representing the standard deviation (n=2) *: P-Value = 0.008 (left) and P-Value = 0.002 (right). (C) HeLa cells were either mock treated or treated with ATRX siRNA, and whole-cell extracts (WCE) were generated from cells in S or M phase. Biotinylated ssTEL was coated with RPA and incubated with the WCE. After the incubation, ssTEL was retrieved and the remaining RPA32 on ssTEL was analyzed by Western blot. (D) SW39TEL and SW26ALT cells were analyzed for ATRX protein expression by Western blot. (E) SW39TEL and SW26ALT cells were analyzed for TERRA transcript by dot blot using digoxigenin (DIG)-labeled anti-TERRA or 28S RNA probes. (F) Quantification of dot blots for TERRA transcript in SW39TEL and SW26ALT cells. TERRA signal was normalized to 28S signal and ratios were graphed as the mean with error bars representing the standard deviation (n=2) *: P-Value = 0.001. (G) RPA-ssTEL was incubated in WCE from SW39TEL or SW26ALT cells. The RPA32 remaining on ssTEL was analyzed by Western blot.
Given that RPA-ssDNA is a key HR intermediate, we asked if ATRX loss induces ALT. Knockdown of ATRX in HeLa cells did not inactivate telomerase, nor did it induce telomere lengthening (Fig. S10A–B). These results are consistent with a previous study (14) and suggest that loss of ATRX is insufficient to establish ALT. Nevertheless, ATRX knockdown in HeLa cells promoted some features of ALT, such as the persistent association of TERRA and RPA with telomeres. Interestingly, a recent study showed that loss of the histone chaperone ASF1 led to the acquisition of several ALT phenotypes, including accumulation of RPA at telomeres (18). We postulate that ALT is established via a multistep process in which loss of ATRX poises telomeres for ALT, but additional genetic/epigenetic changes are needed to fully activate the ALT pathway (see Fig. S29).
RPA-ssDNA is not only an HR intermediate, but also the nucleoprotein structure that recruits ATR, a protein kinase that is a key regulator of HR (19, 20). The defective RPA release from telomeres in ATRX knockdown cells and ALT cells suggests that ATR may be recruited to telomeres during the establishment of ALT. Consistent with our hypothesis, ATR colocalizes with promyelocytic leukemia protein (PML) in U2OS cells but not in HeLa cells (21), suggesting its presence in ALT-associated PML bodies (APBs) (22). Furthermore, ATRIP, the regulatory partner of ATR, associates with telomeres in ALT-positive WI38-VA13 cells but not in HeLa cells (23). These findings prompted us to test if ATR is functionally required for ALT. The ATR inhibitor VE-821 (24) and ATR siRNA significantly reduced APBs in U2OS and SW26ALT cells (Fig. 3A–B, S11A–B, S12A). VE-821 also disrupted APBs in U2OS cells synchronized in G2 (Fig. S12B) (25), ruling out cell-cycle changes as the cause of APB dispersal. In marked contrast, the ATM inhibitor KU-55933 and ATM siRNA did not affect APBs in U2OS cells (Fig. 3A–B, S12B–C), highlighting the role for ATR, but not ATM, in the maintenance of APBs in ALT cells.
To determine whether VE-821 affects the recombinogenic state of ALT telomeres, we analyzed telomere sister-chromatid exchange (T-SCE) and extrachromosomal telomeric C-rich DNA (C-circles) in ALT cells. VE-821 not only decreased T-SCE in U2OS cells (Fig. 3C, S13A), but also reduced C-circle levels in U2OS and HUO9 cells (Fig. 3D–E), showing that ALT is indeed inhibited. Furthermore, VE-821 elevated the frequency of telomere loss in U2OS cells (Fig. 3F, S13B), suggesting that the stability of ALT telomeres is compromised. Consistent with the idea that TERRA acts upstream of ATR to promote RPA retention at ALT telomeres, VE-821 did not affect TERRA levels and telomere association in U2OS cells (Fig. S14A–B).
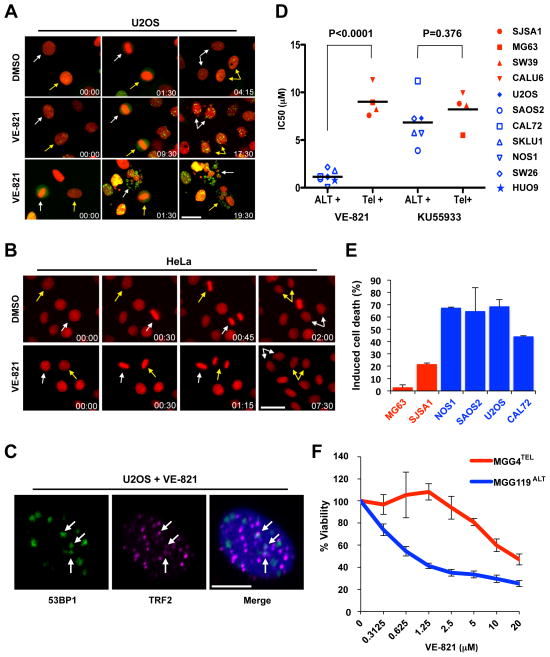
The effects of VE-821 on ALT telomeres prompted us to investigate whether VE-821 selectively kills ALT cells. SW26ALT was indeed more sensitive to VE-821 than SW39TEL (Fig. S15). Importantly, SW26ALT and SW39TEL were similarly sensitive to a panel of DNA-damaging agents (Fig. S16), demonstrating that the effects of VE-821 are unique to ATR inhibition but not a result of general genotoxicity. Moreover, VE-821 induced accumulation of the phosphorylated form of histone H2AX (γH2AX) more efficiently in SW26ALT than in SW39TEL cells (Fig. S17), suggesting that it inflicts more DNA damage in ALT cells. At a concentration that kills? U2OS cells, VE-821 only modestly reduced the proliferation of untransformed RPE-1 retinal pigment epithelial cells (Fig. S18). Using H2B-mRFP and live-cell imaging, we followed the chromosome segregation in U2OS, HeLa, and RPE-1 cells after VE-821 treatment. Furthermore, we used 53BP1-GFP to visualize DNA double-stranded breaks (DSBs) in U2OS cells. VE-821 induced dramatic errors in anaphase chromosome segregation in U2OS but not HeLa or RPE-1 cells (Fig. 4A–B, S19; Movie S1). In the following interphase, U2OS cells displayed increased micronucleation compared to HeLa or RPE-1 cells (Fig. S20A; Movie S1). Moreover, U2OS cells exhibited numerous 53BP1 foci (Fig. 4A, 4C; Movie S1). A fraction of the 53BP1 foci in U2OS cells colocalized with telomeres (Fig. 4C, S20B), although only a minority of telomeres associated with 53BP1. The colocalization of 53BP1 with telomeres but not centromeres was significantly induced by VE-821 (Fig. S21A-B), suggesting that ALT telomeres are particularly fragile upon ATR inhibition. Interestingly, knockdown of ATRX in HeLa cells and BJ fibroblasts did not increase the induction of γH2AX by VE-821 or VE-821 sensitivity (Fig. S22A–C), suggesting that while ATRX loss may prime cells for ALT, it is not directly responsible for the vulnerability of ALT cells to ATR inhibition.
Figure 4. Selective killing of ALT cells by ATR inhibitor.
(A–B) Stills from time-lapse live-cell imaging experiments of (A) U2OS cells stably expressing H2B-mRFP and 53BP1-GFP or (B) HeLa cells expressing H2B-mRFP following treatment with either 5 μM VE-821 or vehicle control (DMSO). Colored arrows mark individual cells as they progress through mitosis. Time scale: hr:min. Scale bar: 30 μm. At least 150 cells were scored for each condition over two independent experiments. (C) U2OS cells were treated with VE-821 as in (A), and analyzed by immunofluorescence using 53BP1 and TRF2 antibodies. Scale bar: 10 μm. (D) A panel of osteosarcoma cell lines were mock treated, treated with VE-821, or KU-55933 for 4–6 days. Cell viability was determined using CellTiter Glo. Dots represent IC50s calculated from experiments preformed in triplicate (n=3). (E) The osteosarcoma cell lines were treated with 3 μM VE-821 for 6 days, and cell death was quantified by Annexin V staining. Induced cell death is graphed as the mean with error bars representing standard deviation (n=2). (E) MGG4TEL and MGG119ALT cells were treated with increasing concentrations of VE-821 for 6 days. Cell viability was determined using CellTiter Glo. Error bars represent standard deviation, experiment performed in duplicate (n=2).
Given the prevalence of ALT in osteosarcoma (26), we tested the effects of VE-821 on a panel of osteosarcoma cell lines. These cell lines clearly clustered into two groups (Fig. 4D). The mean IC50 of VE-821 for one group (U2OS, SAOS2, CAL72, NOS1, and HUO9) was ~0.8 μM, whereas the mean IC50 for the other group (MG63 and SJSA1) was ~9 μM (Fig. 4D, S23A). Among these lines, U2OS and SAOS2 are known ALT lines without detectable ATRX protein (Fig. S24A) (14). CAL72, NOS1, and HUO9 lacked detectable telomerase activity, ATRX protein, and displayed APBs (Fig. S24A-C, S25A-C), suggesting that they are also ALT-positive. In contrast, MG63 and SJSA1 were positive for telomerase activity, ATRX protein, and negative for APBs (Fig. S24A–C). In this panel of cell lines, VE-821 induced substantially higher levels of apoptosis in the ALT lines than in the telomerase-positive lines (Fig. 4E). The hypersensitivity of ALT cells to ATR inhibition was confirmed with a second ATR inhibitor, AZ20 (Fig. S23B). In contrast to ATR inhibitors, neither the ATM inhibitor KU-55933 nor the DNA replication inhibitor gemcitabine showed significant selectivity toward ALT cells (Fig. 4D, S23C–D). Notably, several ATRX-expressing ALT lines were also hypersensitive to VE-821 (Fig. S25A–D, S26A–B) (14), again suggesting that the state of ALT telomeres but not ATRX loss per se renders cells hypersensitive to ATR inhibitors.
ALT is prevalent not only in osteosarcoma but also in pediatric glioblastoma (27). MGG119, a newly developed glioma stem cell (GSC) line (28), lacked detectable telomerase activity and ATRX protein, but expressed high levels of TERRA and displayed APBs (Fig. S27A–D), suggesting that it is ALT-positive. In contrast, the GSC line MGG4 was positive for telomerase activity and ATRX protein, but expressed low levels of TERRA and lacked APBs (Fig. S27A–D) (29). Importantly, although MGG119ALT and MGG4TEL were similarly sensitive to a panel of DNA-damaging agents (Fig. S28A–C), MGG119ALT was significantly more sensitive to VE-821 than MGG4TEL (Fig. 4F), suggesting that VE-821 is uniquely effective in killing ALT GSCs.
The HR defects of specific cancers have offered an opportunity for targeted therapy using PARP inhibitors (5, 6). However, in contrast to HR-defective cancers, ALT-positive cancers actively use recombination to sustain immortality. We show that ATR inhibitors disrupt ALT (Fig. S29) and selectively kill ALT cells in vitro, suggesting a rational strategy for the treatment of ALT-positive cancers. Several ATR inhibitors are entering clinical trials for cancer treatment (24, 30–33). Our findings suggest that cancers reliant on recombination, which include but are not limited to ALT-positive cancers, are hypersensitive to ATR inhibitors, offering an unexplored direction for future preclinical and clinical studies.
Supplementary Material
Sup Info and Figs
Acknowledgments
We thank S. Artandi, W. Wright, H. Yu, M.P. Junier, and Q. Yang for reagents; A. Manning, A. Jimenez, D. Ramirez, and T. Kortulewski for technical assistance; and members of the Zou lab for helpful discussions. C.H.B. is supported by a grant from the Wellcome Trust (102696). M.J. has a fellowship from Irtelis CEA PhD program. F.B. has a grant from INCA ‘TetraTips’ (PLBIO10-030). R.L.F. is supported by the NIH K99/R00 Award (CA166729), the Karin Grunebaum Cancer Research Foundation and the Foster Foundation. L.Z. is a Jim and Ann Orr Massachusetts General Hospital Research Scholar, a Senior Scholar of the Ellison Medical Foundation, and supported by the NIH grant GM076388.
Footnotes
Materials and Methods
References (34–35)
References
- 1.Shay JW, Wright WE. Role of telomeres and telomerase in cancer. Semin Cancer Biol. 2011;21:349. doi: 10.1016/j.semcancer.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 4.Heaphy CM, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol. 2011;179:1608. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 6.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 7.Bechter OE, Zou Y, Shay JW, Wright WE. Homologous recombination in human telomerase-positive and ALT cells occurs with the same frequency. EMBO Rep. 2003;4:1138. doi: 10.1038/sj.embor.7400027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 9.Flynn RL, et al. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471:532. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schramke V, et al. RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat Genet. 2004;36:46. doi: 10.1038/ng1284. [DOI] [PubMed] [Google Scholar]
- 11.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 12.Porro A, Feuerhahn S, Reichenbach P, Lingner J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol Cell Biol. 2010;30:4808. doi: 10.1128/MCB.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 14.Lovejoy CA, et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bower K, et al. Loss of wild-type ATRX expression in somatic cell hybrids segregates with activation of Alternative Lengthening of Telomeres. PLoS One. 2012;7:e50062. doi: 10.1371/journal.pone.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heaphy CM, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 18.O’Sullivan RJ, et al. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat Struct Mol Biol. 2014;21:167. doi: 10.1038/nsmb.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Powell SN, Iliakis G, Wang Y. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res. 2004;64:7139. doi: 10.1158/0008-5472.CAN-04-1289. [DOI] [PubMed] [Google Scholar]
- 20.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 21.Barr SM, Leung CG, Chang EE, Cimprich KA. ATR kinase activity regulates the intranuclear translocation of ATR and RPA following ionizing radiation. Curr Biol. 2003;13:1047. doi: 10.1016/s0960-9822(03)00376-2. [DOI] [PubMed] [Google Scholar]
- 22.Yeager TR, et al. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175. [PubMed] [Google Scholar]
- 23.Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reaper PM, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 25.Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol. 2007;14:581. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- 26.Scheel C, et al. Alternative lengthening of telomeres is associated with chromosomal instability in osteosarcomas. Oncogene. 2001;20:3835. doi: 10.1038/sj.onc.1204493. [DOI] [PubMed] [Google Scholar]
- 27.Hakin-Smith V, et al. Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet. 2003;361:836. doi: 10.1016/s0140-6736(03)12681-5. [DOI] [PubMed] [Google Scholar]
- 28.Wakimoto H, et al. Targetable Signaling Pathway Mutations Are Associated with Malignant Phenotype in IDH-Mutant Gliomas. Clin Cancer Res. 2014 doi: 10.1158/1078-0432.CCR-13-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakimoto H, et al. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009;69:3472. doi: 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fokas E, et al. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis. 2012;3:e441. doi: 10.1038/cddis.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charrier JD, et al. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J Med Chem. 2011;54:2320. doi: 10.1021/jm101488z. [DOI] [PubMed] [Google Scholar]
- 32.Foote KM, et al. Discovery of 4-{4-[(3R)-3-Methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-y l}-1H-indole (AZ20): a potent and selective inhibitor of ATR protein kinase with monotherapy in vivo antitumor activity. J Med Chem. 2013;56:2125. doi: 10.1021/jm301859s. [DOI] [PubMed] [Google Scholar]
- 33.Toledo LI, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18:721. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sup Info and Figs