Rational design of a split-Cas9 enzyme complex (original) (raw)
Significance
Bacteria have evolved clustered regularly interspaced short palindromic repeats (CRISPRs) together with CRISPR-associated (Cas) proteins to defend themselves against viral infection. RNAs derived from the CRISPR locus assemble with Cas proteins into programmable DNA-targeting complexes that destroy DNA molecules complementary to the guide RNA. In type II CRISPR-Cas systems, the Cas9 protein binds and cleaves target DNA sequences at sites complementary to a 20-nt guide RNA sequence. This activity has been harnessed for a wide range of genome-engineering applications. This study explores the structural features that enable Cas9 to bind and cleave target DNAs, and the results suggest a way of regulating Cas9 by physical separation of the catalytic domains from the rest of the protein scaffold.
Keywords: CRISPR-Cas9, genome engineering, split enzyme
Abstract
Cas9, an RNA-guided DNA endonuclease found in clustered regularly interspaced short palindromic repeats (CRISPR) bacterial immune systems, is a versatile tool for genome editing, transcriptional regulation, and cellular imaging applications. Structures of Streptococcus pyogenes Cas9 alone or bound to single-guide RNA (sgRNA) and target DNA revealed a bilobed protein architecture that undergoes major conformational changes upon guide RNA and DNA binding. To investigate the molecular determinants and relevance of the interlobe rearrangement for target recognition and cleavage, we designed a split-Cas9 enzyme in which the nuclease lobe and α-helical lobe are expressed as separate polypeptides. Although the lobes do not interact on their own, the sgRNA recruits them into a ternary complex that recapitulates the activity of full-length Cas9 and catalyzes site-specific DNA cleavage. The use of a modified sgRNA abrogates split-Cas9 activity by preventing dimerization, allowing for the development of an inducible dimerization system. We propose that split-Cas9 can act as a highly regulatable platform for genome-engineering applications.
Bacteria use RNA-guided adaptive immune systems encoded by clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated (Cas) genomic loci to defend against invasive DNA (1, 2). In type II CRISPR-Cas systems, a single enzyme called Cas9 is responsible for targeting and cleavage of foreign DNA (3). The ability to program Cas9 for DNA cleavage at sites defined by engineered single-guide RNAs (sgRNAs) (4) has led to its adoption as a robust and versatile platform for genome engineering (for recent reviews, see refs. 5–7).
Cas9 contains two nuclease active sites that function together to generate DNA double-strand breaks (DSBs) at sites complementary to the 20-nt guide RNA sequence and adjacent to a protospacer adjacent motif (PAM). Structural studies of the Streptococcus pyogenes Cas9 showed that the protein exhibits a bilobed architecture comprising the catalytic nuclease lobe and the α-helical lobe of the enzyme (8). Electron microscopy (EM) studies and comparisons with X-ray crystal structures with and without a bound guide RNA and target DNA revealed a large-scale conformational rearrangement of the two lobes relative to each other upon nucleic acid binding (8, 9). Strikingly, RNA binding induces the nuclease lobe to rotate ∼100° relative to the α-helical lobe, generating a nucleic-acid binding cleft that can accommodate DNA, and interactions between the two lobes seem to be mediated primarily through contacts with the bound nucleic acid rather than direct protein–protein contacts (8, 9). These observations suggested that the two structural lobes of Cas9 might be separable into independent polypeptides that retain the ability to assemble into an active enzyme complex. Such a system would enable analysis of the functionally distinct properties of each Cas9 structural region and might offer a unique mechanism for controlling active protein assembly.
Here, we show that two distinct Cas9 polypeptides encompassing the α-helical and nuclease lobes can be stably expressed and purified. Filter binding and negative-stain EM experiments demonstrate that the split-Cas9 assembles with sgRNA into a ternary complex resembling that of full-length Cas9 RNA. Furthermore, DNA cleavage assays reveal that the enzymatic activity of split-Cas9 closely mimics that of WT Cas9. Split-Cas9 is functional for genome editing in human cells with full-length sgRNAs but can be inactivated with shortened sgRNAs that give rise to destabilized complexes. Together, these data show how the Cas9 protein can be reengineered as a split enzyme whose assembly and function is regulatable by the sgRNA, providing a platform for controlled use of Cas9 for genome-engineering applications in cells.
Results
Design and Functional Validation of Split-Cas9.
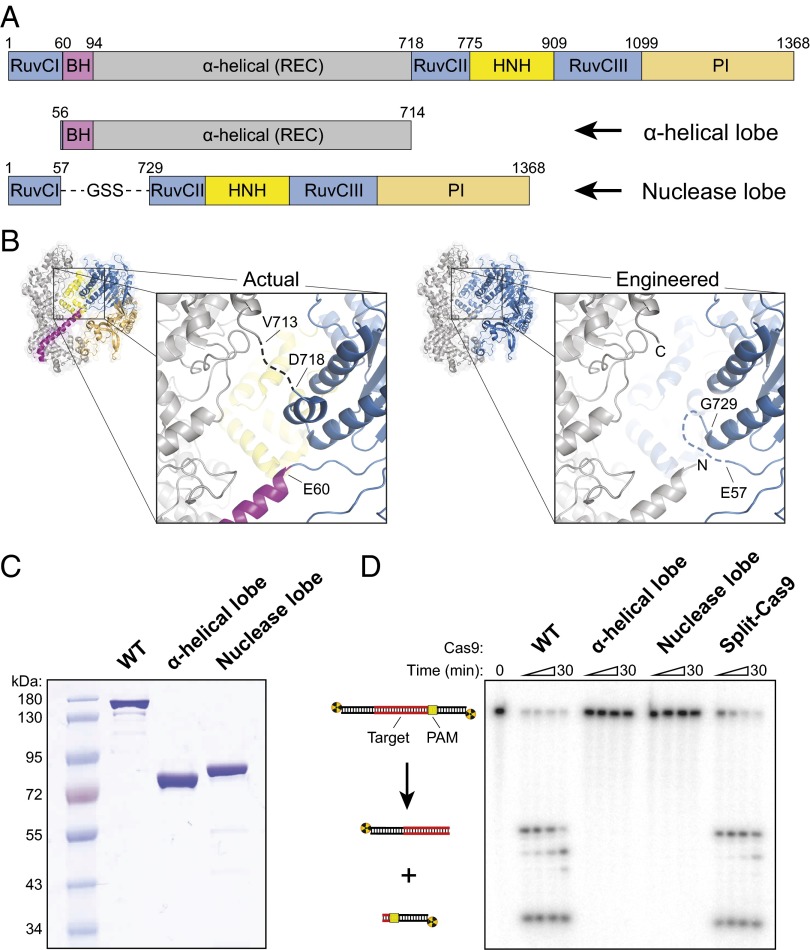
The nuclease lobe of Cas9 includes the RuvC and HNH nuclease domains, as well as a C-terminal domain that is involved in PAM recognition (Fig. 1_A_) (8–10). The RuvC domain comprises three distinct motifs: Motifs II and III are interrupted by the HNH domain, and motifs I and II are interrupted by a large lobe composed entirely of α-helices. This α-helical lobe, also referred to as the recognition (REC) lobe (9), forms a broad cleft that makes extensive contacts with the sgRNA and target DNA. We previously showed that the α-helical lobe undergoes a large rotation relative to the nuclease lobe upon guide RNA binding to create a central channel where target DNA is bound (8).
Fig. 1.
Cas9 can be split into two separate polypeptides that retain the ability to catalyze RNA-guided dsDNA cleavage. (A) Domain organization of WT Cas9 (Top) and split-Cas9 (Bottom), composed of the α-helical lobe and nuclease lobe. Domain junctions are numbered according to Nishimasu et al. (9). BH, bridge helix; PI, PAM-interacting; REC, recognition lobe. The PI domain can be further subdivided into Topo-homology and C-terminal domains (8). (B) Crystal structures of sgRNA/DNA-bound Cas9 (PDB ID code 4OO8) (9) colored according to domain (Left) or by lobe (Right), with the α-helical and nuclease lobes depicted in gray and blue, respectively. Nucleic acids are omitted for clarity. In the observed interface between the lobes (Inset, Left), the dashed line represents a disordered linker spanning residues V713–D718. In the engineered interface (Inset, Right), the dashed line represents a GGS linker connecting E57 to G729, and new N and C termini of the α-helical lobe are shown. (C) SDS polyacrylamide gel electrophoresis (SDS/PAGE) analysis of purified WT Cas9 (159 kDa), the α-helical lobe (77 kDa), and the nuclease lobe (81 kDa). The gel was stained with Coomassie Brilliant Blue. (D) DNA cleavage assay with the indicated Cas9 construct, analyzed by denaturing PAGE. Reactions contained ∼1 nM radiolabeled dsDNA and 100 nM protein–sgRNA complex; split-Cas9 contained a twofold molar excess of the α-helical lobe. Quantified data and kinetic analysis can be found in Fig. S2 and Table S1.
Using available crystal structures as a guide, we designed a split-Cas9 in which the native structure of both lobes was kept as intact as possible (Fig. 1_A_). In particular, rather than simply split the full-length Cas9 sequence internally at a single junction, we constructed the nuclease lobe by directly linking the N-terminal RuvCI motif to the remainder of the nuclease lobe located ∼650 amino acids away in primary sequence, with the intervening polypeptide comprising the α-helical lobe. Two crossover points between the lobes occur at residues ∼56 and ∼720 (Fig. 1_B_): the C-terminal connection is disordered in both apo-Cas9 and sgRNA/DNA-bound structures, and the N-terminal connection occurs between the RuvCI motif and the bridge helix. We connected residue E57 from RuvCI with residue G729 from RuvCII using a three-amino acid linker and removed a short, poorly conserved α-helix from the RuvCII motif that does not seem to play an important structural role in the sgRNA/DNA-bound state (Fig. 1_B_). The α-helical lobe spans residues G56–S714, with the N terminus encompassing the entirety of the bridge helix.
To determine whether the lobes could function as separate polypeptides, we separately overexpressed both lobes in Escherichia coli and purified them by affinity and size-exclusion chromatography (Fig. 1_C_ and Fig. S1). We investigated whether split-Cas9 (α-helical lobe plus nuclease lobe) would recapitulate the activity of WT Cas9 using a standard cleavage assay with sgRNA and a radiolabeled double-stranded DNA (dsDNA) target (Fig. 1_D_). No cleavage was observed with either lobe individually, but the reconstituted split-Cas9 enzyme complex exhibited robust target DNA cleavage (Fig. 1_D_ and Fig. S2). Split-Cas9 maintained the same site and pattern of cleavage as WT Cas9, including the “trimming” of the nontarget strand that we observed previously (4), and functioned equally well with a dual-guide RNA composed of CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) (Fig. S2). In addition, we confirmed that split-Cas9 activity was dependent on complementarity between the sgRNA and target DNA as well as the presence of a 5′-NGG-3′ PAM (Fig. S2).
When we investigated the kinetics of DNA cleavage under pseudo first-order conditions using excess enzyme, we found that split-Cas9 was ∼10-fold slower than WT although it reached the same endpoint after 5 min (Fig. S2 and Table S1). This reduced rate may result from slower kinetics of protein–RNA complex formation, a reduced rate of dsDNA recognition and unwinding, or a minor defect in nuclease domain activation. DNA-binding experiments using nuclease-inactive split dCas9 (D10A/H840A mutations) revealed a significantly weaker affinity of split-Cas9 for target DNA than WT Cas9 (Fig. S3), suggesting that slower kinetics of dsDNA binding likely limit the observed rate of cleavage. Collectively, these results demonstrate that the enzymatic activity of WT Cas9 does not require a direct linkage between the α-helical and nuclease lobes although their physical connection within RNA–protein complexes increases the affinity for the target DNA substrate. Remarkably, whereas previous work shows that the RNA-induced large-scale rearrangement of both lobes is necessary for WT Cas9 to achieve an active conformation (8), our experiments revealed that the sgRNA is entirely sufficient to recruit and dimerize the separate lobes into an active enzyme complex. Furthermore, communication through the sgRNA enables PAM recognition, dsDNA unwinding, and DNA cleavage, despite the absence of extensive protein–protein interactions between the lobes.
sgRNA Motifs Recruit both Cas9 Lobes to Form a Ternary Complex.
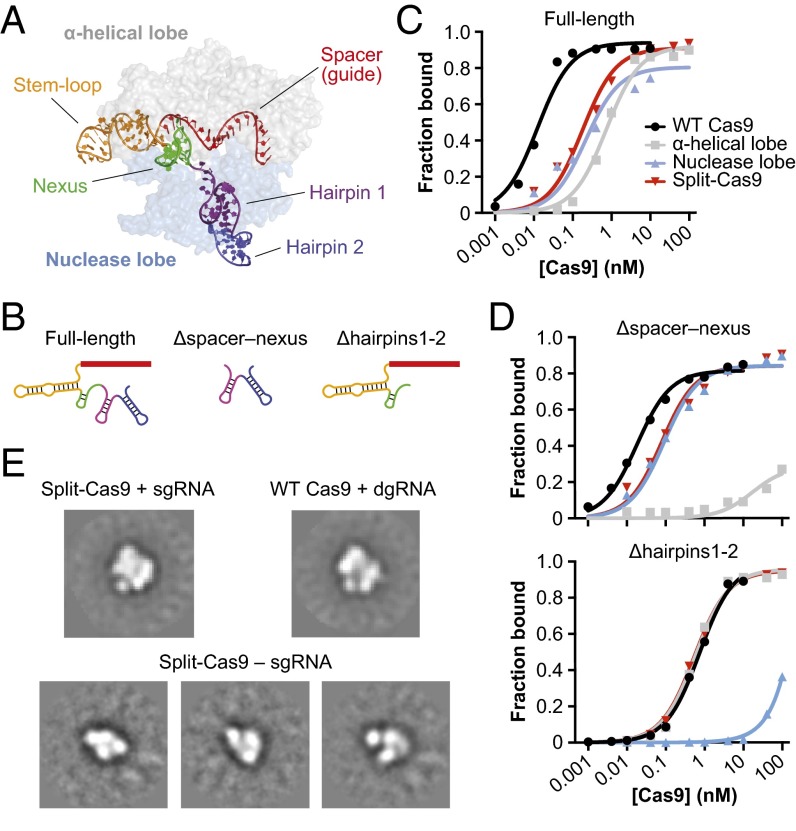
We next wanted to investigate RNA molecular determinants that promote heterodimerization of the α-helical and nuclease lobes. Crystal structures of sgRNA/DNA-bound Cas9 show that the spacer (guide) and stem-loop motifs at the 5′ end of the sgRNA primarily contact the α-helical lobe whereas two hairpins at the 3′ end bind the outside face of the nuclease lobe (Fig. 2_A_). The nexus motif, recently shown to be critical for activity (11), occupies a central position between the lobes and forms extensive interactions with the bridge helix. Based on this interaction profile, we generated a full-length sgRNA and two shorter sgRNA constructs that were selectively truncated from either the 5′ or 3′ end (Fig. 2_B_ and Table S2) and determined their affinities for WT Cas9, the individual α-helical and nuclease lobes, and split-Cas9 using a filter binding assay.
Fig. 2.
Split-Cas9 assembly requires the sgRNA. (A) Crystal structure of sgRNA/DNA-bound Cas9 (PDB ID code 4OO8) (9). Cas9 is colored by lobe and shown as a transparent surface, and the sgRNA is colored by motif according to Briner et al. (11), and the DNA is omitted for clarity. (B) Diagrams of full-length and truncated sgRNA variants used in binding experiments; specific motifs of the sgRNA are colored as in A. (C and D) Results from binding experiments using full-length sgRNA (C), and Δhairpins1-2 and Δspacer–nexus sgRNA truncations (D). Radiolabeled RNAs were incubated with increasing concentrations of WT Cas9, individual α-helical and nuclease lobes, or split-Cas9, and the fraction of protein-bound RNA was determined by nitrocellulose filter binding. Equilibrium dissociation constants (_K_d) determined from three independent experiments are shown in Table 1. (E) Reference-free class averages from negative-stain EM images of split-Cas9 reconstituted with single-guide RNA (Top Left), WT Cas9 reconstituted with dual-guide RNA (Top Right), and split-Cas9 in the absence of guide RNA (Bottom). For split-Cas9 without sgRNA, several class averages are shown. The width of the boxes corresponds to ∼336 Å. Data with WT Cas9 are adapted from Jinek et al. (8).
The full-length sgRNA is bound by WT Cas9 with an equilibrium dissociation constant (_K_d) of 10 ± 2 pM whereas the lobes individually and together have _K_d values in the range of 0.2–0.8 nM (Fig. 2_C_ and Table 1). The difference between WT and split-Cas9 likely reflects the increased entropic cost required to assemble a ternary versus binary complex. Interestingly, WT Cas9 bound a truncated sgRNA comprising only the 3′ hairpins (Δspacer–nexus) with an affinity that was indistinguishable from the full-length sgRNA (Fig. 2_D_ and Table 1), indicating that these hairpins provide the major source of binding energy for the WT protein–RNA complex (12). Consistent with the crystal structure, the nuclease lobe still bound the 5′-truncated sgRNA as tightly as the full-length sgRNA whereas the affinity of the α-helical lobe was reduced by over three orders of magnitude (_K_d > 100 nM).
Table 1.
Equilibrium dissociation constants for protein–sgRNA interactions
| Protein | Equilibrium dissociation constant (_K_d) for indicated sgRNA* | ||
|---|---|---|---|
| Full-length | ΔHairpins1-2 | ΔSpacer–nexus | |
| WT Cas9 | 10 ± 2 pM | 0.86 ± 0.12 nM | 16 ± 2 pM |
| α-Helical lobe | 0.75 ± 0.12 nM | 0.70 ± 0.13 nM | >100 nM |
| Nuclease lobe | 0.30 ± 0.07 nM | >100 nM | 0.17 ± 0.06 nM |
| Split-Cas9 | 0.23 ± 0.04 nM | 1.05 ± 0.05 nM | 0.17 ± 0.07 nM |
We similarly reasoned that removing the two hairpins from the 3′ end of the sgRNA (Δhairpins1-2) would selectively perturb interactions with the nuclease lobe. Indeed, the affinity of the 3′-truncated sgRNA for the nuclease lobe decreased by over three orders of magnitude relative to full-length sgRNA (_K_d >100 nM) whereas the affinity of the α-helical lobe was unchanged (Fig. 2_D_ and Table 1). Our results demonstrate that sgRNA truncations specifically destabilize binding to only one of the two lobes and that the affinity of split-Cas9 is limited by the highest affinity interaction with either lobe. These findings highlight the multiple, independent molecular contacts formed between the sgRNA and the two lobes of Cas9.
Based on our binding data and on the minimal contacts observed between the two Cas9 lobes in available structures, we hypothesized that the sgRNA would be required for heterodimerization of the α-helical and nuclease lobes. To test this idea, we performed analytical negative-stain electron microscopy with each lobe alone and together in the presence and absence of sgRNA. Raw micrographs of a sample containing both polypeptides and the sgRNA revealed bilobed densities that had dimensions consistent with our earlier reconstructions of the Cas9–RNA complex (Fig. S4) (8), and the resulting class averages were indistinguishable from those obtained using WT Cas9 (Fig. 2_E_). In contrast, we observed smaller particles that had dimensions more consistent with single lobes when the polypeptides were mixed together in the absence of sgRNA (Fig. 2_E_ and Fig. S4). These results indicate that a full-length sgRNA acts as a molecular scaffold in dimerizing the two lobes.
Split-Cas9 Functions in Mammalian Cells for Genome Editing.
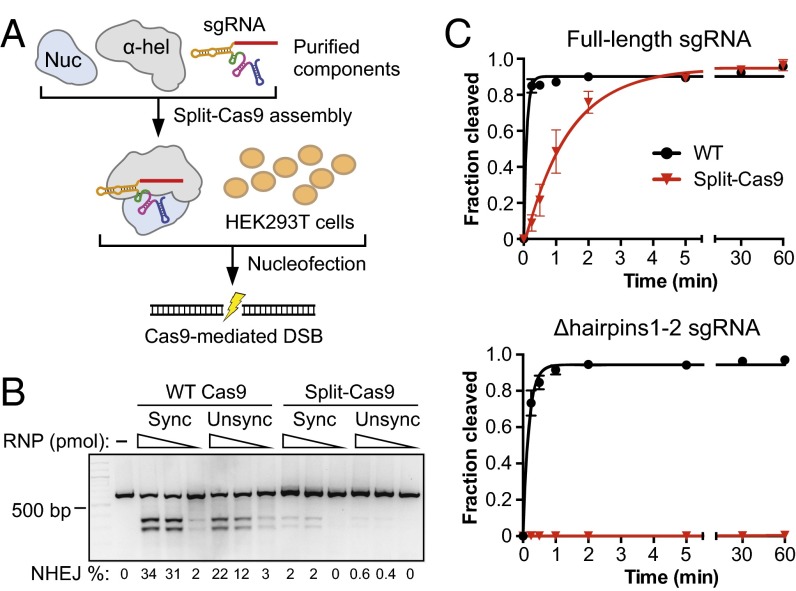
To determine whether split-Cas9 would retain the ability to generate site-specific genomic edits in vivo, we targeted the EMX1 locus in HEK293T cells by nucleofection using reconstituted Cas9–sgRNA ribonucleoprotein (RNP) complexes (Fig. 3_A_) (13). Split-Cas9 generated indels with efficiencies of up to 0.6% and 2% in cells that were unsynchronized or nocodazole-synchronized, respectively, compared with 22% and 34% with WT Cas9 (Fig. 3_B_). The reduced levels of editing may be due in part to disruption of the ternary complex during dilution and nucleofection because the complex is limited by the affinity of the α-helical lobe for sgRNA, or to slower kinetics of DNA cleavage in cells. Additionally, because each copy of sgRNA must recruit both lobes to form an active complex, we suspect that the activity in cells may be particularly sensitive to the stoichiometry between the sgRNA and either lobe. In agreement with this hypothesis, we found that the in vitro DNA cleavage activity of split-Cas9 decreased as the sgRNA concentration was increased above that of both lobes (Fig. S5), suggesting that excess sgRNA likely titrates the lobes apart from each other. Although our results leave room for optimization of split-Cas9 activity in cells, they demonstrate that the intrinsic genome-editing capabilities are retained when Cas9 comprises two individual polypeptides.
Fig. 3.
Genomic editing function and selective inactivation of split-Cas9. (A) Schematic of the split-Cas9 RNP nucleofection assay using a full-length _EMX1_-targeting sgRNA. Illustration and protocol adapted from Lin et al. (13). (B) Analysis of editing efficiencies by nonhomologous end joining (NHEJ) using a T7 endonuclease I assay and agarose-gel electrophoresis. Cells were nucleofected with 100, 30, or 10 pmol of WT or split-Cas9 ribonucleoprotein (RNP) complexes after arrest at mitosis with nocodazole (Sync) or during normal growth (Unsync). Editing efficiencies are shown at the bottom. (C) DNA cleavage time courses using WT and split-Cas9 with either a full-length sgRNA (Top) or the Δhairpins1-2 sgRNA (Bottom). Values were averaged from three independent experiments, and error bars represent the SD. Rate constants can be found in Table S1.
Engineered sgRNAs Selectively Preclude Split-Cas9 but Not WT Cas9 Activity.
The potential for enhanced spatiotemporal control of genome engineering events with split-Cas9 prompted us to investigate ways in which sgRNA-mediated dimerization of the α-helical and nuclease lobes could be perturbed. In particular, we reasoned that certain 3′-truncated or modified sgRNAs, which have weak affinity for the nuclease lobe (Fig. 2_E_) but still support robust DNA cleavage activity of WT Cas9 (4), would selectively inactivate split-Cas9 activity through their inability to effectively recruit and dimerize both lobes into a functional enzyme complex. Thus, the activity of split-Cas9 in cells could be made dependent upon inducible protein–protein dimerization domains (Fig. S6).
When we tested sgRNA variants that lacked one or both hairpins at the 3′ end for their ability to support in vitro cleavage, split-Cas9 activity was either severely compromised or completely abolished whereas WT Cas9 activity was slightly reduced relative to a full-length sgRNA (Fig. 3_C_, Fig. S7, and Table S1). A recent report found that sgRNAs in which only the first hairpin is deleted function robustly in cells (11), and we found that similar designs supported DNA cleavage activity of WT Cas9 but not split-Cas9 in vitro (Fig. S7). Thus, rationally designed variants of the sgRNA scaffold can be used to prevent RNA-mediated heterodimerization of the two lobes without compromising the intrinsic RNA-guided DNA-cleaving capabilities of Cas9.
Discussion
Here, we have successfully designed a split version of Cas9 that maintains the cleavage activity of the native enzyme. We demonstrated that the sgRNA is necessary and sufficient to dimerize the nuclease and α-helical lobes into an active complex and furthermore showed that multiple distinct sgRNA motifs interact with the lobes independently. Split-Cas9 is active for genome editing in cells, albeit at a reduced level relative to WT Cas9, and can be rendered nonfunctional through the removal of one or more of the hairpins at the 3′ end of sgRNA. Although optimization will help split-Cas9 to function effectively in cells, we have shown the potential to enable a variety of interesting and useful applications.
Split-protein systems have often been designed such that the functional unit is restored through the interactions of an exogenous pair of proteins (14). For example, DNA-binding and effector domains of modified TALEs have been fused to CRY2 and CIB1 to enable light-inducible control of gene expression (15). Alternatively, genes may be split such that a single functional polypeptide is the final product of mRNA transsplicing or intein-based protein splicing (16). Our strategy with split-Cas9 differs in that it faithfully recapitulates the functionality of full-length Cas9 using the very same RNA ligand that WT Cas9 requires. In this sense, sgRNA-mediated dimerization and activation of split-Cas9 may be viewed analogously to the sgRNA-mediated rearrangement and structural activation of WT Cas9.
Optimizing expression levels of the split-Cas9 components could increase their effectiveness. In particular, whereas the sgRNA should be kept limiting to avoid titrating the lobes away from the ternary complex, overall expression must be high enough to overcome the reduced affinity of both protein components for the sgRNA scaffold. Split-Cas9 could be regulated by the combinatorial use of promoters, restricting activity to highly specific subsets of tissues or creating a “coincidence detector” with two inducible promoters. Split-Cas9 could also be developed for use with adeno-associated viral vectors, where the smaller coding regions of each lobe would enable the use of effector or reporter domains that are currently prohibited by limited packaging capacity. We also suggest that split-Cas9 can be converted into a regulatable system using exogenous dimerization domains (Fig. S6). Fusing both lobes to domains that selectively dimerize upon chemical or optical induction, such as the abscisic acid-inducible PYL-ABI dimer (17) or the blue light-inducible CRY2-CIB1 dimer (18), would allow for enhanced spatiotemporal control of genome-engineering events (6). Dimerization domains may also increase the efficiency of complex formation by making lobe assembly independent of the sgRNA. We propose that the combined use of inducible dimerization domains with compromised sgRNA variants that enable DNA targeting, but not split-Cas9 assembly, would eliminate leaky activity in the absence of inducer while still allowing for robust activation, creating an extremely sensitive inducible system.
Finally, our study provides important insights into the structure–function relationship of the native Cas9 enzyme. The ability of sgRNA to act as a molecular scaffold in assembling two separate polypeptides highlights the crucial role that the sgRNA plays in orchestrating conformational rearrangements of WT Cas9. The separation of recognition and catalytic functions into two separate lobes may have evolved to eliminate nonspecific nuclease activity and control licensing of Cas9 for DNA interrogation. Our results also invite comparisons to the mechanisms of RNP assembly and DNA targeting in other CRISPR-Cas systems, particularly the type I-E Cascade interference complex. Although Cascade is composed of 11 distinct subunits, none of which possesses nuclease activity, the guide RNA (crRNA) plays a similarly critical role in scaffolding the assembly of distinct domains into a structure that is primed to engage DNA targets (19–21). Similar principles are likely to govern the assembly and activity of other CRISPR RNA-guided DNA-targeting complexes (2). Thus, distinct CRISPR-Cas systems may have evolved similar organizational strategies in parallel that use the guide RNA for structural assembly and conformational activation.
Materials and Methods
Cloning and Protein Purification.
The expression vector for purification of the nuclease lobe was generated by Around the Horn (ATH) PCR using a preexisting pET-based expression vector for S. pyogenes Cas9. The final construct encodes an N-terminal decahistidine-maltose binding protein (His10-MBP) tag, a tobacco etch virus (TEV) protease cleavage site, residues 1–57, a glycine-serine-serine linker, and residues 729–1,368. The vector for the catalytically inactive dNuclease lobe was generated by ATH PCR of a similar dCas9 (D10A/H840A) vector. The vector for expression of the α-helical lobe was generated by PCR amplification of S. pyogenes Cas9 residues 56–714 and assembly of the resulting fragment into a His10-MBP expression vector via ligation-independent cloning.
Each protein was overexpressed in E. coli BL21 Rosetta 2(DE3) (EMD Biosciences) by growing in 2xYT medium at 37 °C to an optical density of 0.5, inducing with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and growing an additional 16 h at 18 °C. Cells were lysed by sonication in a buffer containing 50 mM Tris, pH 7.5, 500 mM NaCl, 1 mM Tris(2-carboxyethyl)phosphine (TCEP), 5% (vol/vol) glycerol, and a protease inhibitor mixture (Roche). The clarified lysate was bound in batch to nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen). The resin was washed extensively with 20 mM Tris, pH 7.5, 500 mM NaCl, 1 mM TCEP, 10 mM imidazole, and 5% (vol/vol) glycerol; and the bound protein was eluted in 20 mM Tris, pH 7.5, 500 mM NaCl, 1 mM TCEP, 300 mM imidazole, and 5% (vol/vol) glycerol. The His10-MBP affinity tag was removed with His6-tagged TEV protease during overnight dialysis against 20 mM Tris, pH 7.5, 500 mM NaCl, 1 mM TCEP, and 5% (vol/vol) glycerol. The protein was then flowed over Ni-NTA agarose to remove TEV protease and the cleaved affinity tag.
The α-helical lobe was dialyzed for 2 h against 20 mM Tris, pH 7.5, 125 mM KCl, 1 mM TCEP, and 5% (vol/vol) glycerol, and purified on a 5-mL HiTrap SP Sepharose column (GE Healthcare), with elution over a linear gradient from 125 mM to 1 M KCl. The α-helical lobe was further purified by size-exclusion chromatography on a Superdex 200 16/60 column (GE Healthcare) in 20 mM Tris, pH 7.5, 200 mM KCl, 1 mM TCEP, and 5% (vol/vol) glycerol. The nuclease lobe was purified via size-exclusion chromatography immediately after the ortho-Ni-NTA step. The dNuclease lobe was purified as described for the nuclease lobe, except the size-exclusion chromatography was performed with 20 mM Tris, pH 7.5, 500 mM KCl, 1 mM TCEP, and 5% (vol/vol) glycerol. Full-length Cas9 was purified as previously described (8).
In Vitro Transcription of sgRNA.
Linearized plasmid DNA was used as a template for in vitro transcription of full-length, Δhairpins1-2 and Δhairpin2 λ1 sgRNA. The appropriate region of the sgRNA, along with a T7 RNA polymerase promoter sequence, was PCR-amplified and restriction-cloned into EcoRI/BamHI sites of a pUC19 vector, and the resulting vector was digested with BamHI to enable run-off transcription. The Δhairpin1 and Δspacer–nexus λ1 sgRNA, as well as λ1 crRNA and tracrRNA, were transcribed from a single-stranded DNA template with an annealed T7 promoter oligonucleotide. The DNA template for EMX1 sgRNA was produced by overlapping PCR as previously described (13).
Transcription reactions (1 mL) were conducted in buffer containing 50 mM Tris, pH 8.1, 25 mM MgCl2, 0.01% Triton X-100, 2 mM spermidine, and 10 mM DTT, along with 5 mM each of ATP, GTP, CTP, and UTP, 100 μg/mL T7 polymerase, and ∼1 μM DNA template. Reactions were incubated at 37 °C overnight and subsequently treated with 5 units of DNase (Promega) for 1 h. Reactions were then quenched with 800 μL of 95% (vol/vol) formamide, 0.05% bromophenol blue, and 20 mM EDTA, and loaded onto a 7 M urea 10% (wt/vol) polyacrylamide gel. The appropriate band was excised, and the RNA was eluted from the gel overnight in diethylpyrocarbonate (DEPC)-treated water. The sgRNA was ethanol-precipitated and resuspended in DEPC-treated water. Concentrations were determined by A260nm using a NanoDrop (Thermo Scientific). For filter-binding assays, the sgRNA was dephosphorylated with calf intestinal phosphatase (New England Biolabs) before radiolabeling with T4 polynucleotide kinase (New England Biolabs) and γ-[32P]-ATP (Perkin-Elmer). The radiolabeled sgRNA was gel-purified as described above.
Split-Cas9 Complex Reconstitution.
Split-Cas9 complexes were reconstituted before cleavage and binding assays at 37 °C for 10 min in a buffer containing 20 mM Tris, pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, and 5% (vol/vol) glycerol. For binding assays, reactions containing equimolar amounts of the dNuclease lobe, the α-helical lobe, and in vitro-transcribed sgRNA. For cleavage assays, reactions contained equimolar amounts of the nuclease lobe and sgRNA (or crRNA:tracrRNA), with a twofold molar excess of the α-helical lobe.
DNA Cleavage Assays.
All cleavage assays were performed in 1× cleavage buffer, which contained 20 mM Tris, pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, and 5% (vol/vol) glycerol. Preformed complexes were diluted in cleavage buffer, and reactions were initiated with the addition of radiolabeled dsDNA substrates. Final reaction concentrations were 100 nM protein:RNA complex and ∼1 nM radiolabeled DNA target. The concentration of Cas9 was chosen to be sufficiently above the _K_d for the sgRNA such that complex assembly is unlikely to be rate-limiting, except in the case of split-Cas9 and the Δhairpins1-2 sgRNA (_K_d >100 nM). Reactions proceeded at room temperature, and aliquots were removed at selected time points and quenched with an equal volume of buffer containing 50 mM EDTA, 0.02% bromophenol blue, and 90% (vol/vol) formamide. Reaction products were resolved by 7 M urea-PAGE, gels were dried, and DNA was visualized by phosphorimaging and quantified using ImageQuant software (GE Healthcare). The percentage of DNA cleaved was determined by dividing the amount of cleaved DNA by the sum of uncleaved and cleaved DNA. Kinetic analysis was performed using Prism (GraphPad Software). Reported observed rate constants (_k_obs) are the average of three independent experiments ± SEM. Graphed values are the averaged time points of three independent experiments, with error bars representing the SD.
Electrophoretic Mobility-Shift Assays.
All binding assays were performed in 1× binding buffer, which contains 20 mM Tris, pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% (vol/vol) glycerol, 50 μg/mL heparin, 0.01% Tween 20, and 100 μg/mL BSA. Preformed complexes were diluted into 1× binding buffer, after which radiolabeled dsDNA substrates were added to a final concentration of <0.2 nM. Reactions were incubated at room temperature for 60 min and then resolved at 4 °C on a native 8% polyacrylamide gel containing 0.5× Tris/borate/EDTA (TBE) and 5 mM MgCl2. Gels were dried, and DNA was visualized by phosphorimaging and quantified using ImageQuant software (GE Healthcare). The fraction of DNA bound (amount of bound DNA divided by the sum of free and bound DNA) was plotted versus concentration of protein and fit to a binding isotherm using Prism (GraphPad Software).
Filter-Binding Assays.
All filter-binding assays were performed in 1× RNA binding buffer, which contains 20 mM Tris, pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% (vol/vol) glycerol, 0.01% Igepal CA-630, 10 μg/mL yeast tRNA, and 10 μg/mL BSA. WT Cas9, α-helical lobe, nuclease lobe, or an equimolar mix of both lobes (split-Cas9) were incubated with <0.02 nM radiolabeled sgRNA for 60 min at room temperature. Tufryn (Pall Corporation), Protran (Whatman), and Hybond-N+ (GE Healthcare) membranes were soaked in buffer containing 20 mM Tris, pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, and 5% (vol/vol) glycerol and arranged on a dot blot apparatus. Binding reactions were separated through the membranes by the application of vacuum, and, after drying, the membranes were visualized by phosphorimaging and quantified using ImageQuant Software (GE Healthcare). The fraction of sgRNA bound was plotted versus the concentration of protein and fit to a binding isotherm using Prism (GraphPad Software). Reported _K_d values are the average of three independent experiments, and errors represent the SEM.
Negative-Stain Electron Microscopy and Image Processing.
We prepared and imaged negatively stained samples of the α-helical lobe, nuclease lobe, and split-Cas9 (α-helical lobe and nuclease lobe) with and without sgRNA as described previously (4). Data were acquired using a Tecnai F20 Twin transmission electron microscope operated at 120 keV at a nominal magnification of 80,000× (1.45 Å at the specimen level) using low-dose exposures (∼20 _e_−Å−2) with a randomly set defocus ranging from −0.7 to −1.6 μm. A total of 150–200 images of each Cas9 sample was automatically recorded on a Gatan 4k × 4k CCD camera using the MSI-Raster application within LEGINON (22). Low-resolution negative-stain class averages of Lid particles from the yeast 26S proteasome (23) were used as references for template-based particle picking. The Lid complex was used as a template to avoid selection bias because it bears minimal to no structural resemblance to Cas9. Cas9 complexes were extracted using a 224 × 224-pixel box size. These particles were subjected to 2D reference-free alignment and classification using multivariate statistical analysis and multireference alignment in IMAGIC (24).
Cas9 and Split-Cas9 RNP Assembly and Nucleofection.
The split-Cas9 RNP was prepared immediately before the experiment by incubating both lobes with sgRNA at molar ratios of 1.2:1:1.2 (α-helical lobe:nuclease lobe:sgRNA) for 10 min at 37 °C in 20 mM Hepes, pH 7.5, 150 mM KCl, 1 mM MgCl2, 10% (vol/vol) glycerol, and 1 mM TCEP. The nucleofections were carried out as previously described for Cas9, using 10, 30, and 100 pmol of RNP complex for ∼2 × 105 cells (13). Where indicated, cells were synchronized with 200 ng/mL nocodazole for 17 h before nucleofection. Neither WT Cas9 nor the split-Cas9 lobes had nuclear localization signals, which may have led to reduced editing levels, particularly for the unsynchronized cells.
Analysis of In-Cell Genome-Editing Efficiency.
Determination of the percentage of indels induced at the target region was performed as previously described (13). In brief, 640-nt regions of the EMX1 locus containing the target sites were PCR-amplified, and the resulting products were denatured, reannealed, and digested with T7 endonuclease I (New England Biolabs), which cleaves mismatched heteroduplex DNA (25). The products were resolved on a 2% (wt/vol) agarose gel containing SYBR Gold (Life Technologies), and band intensities were determined using Image Lab (Bio-Rad Laboratories). Editing efficiencies were determined using the formula {1 – [1 – (b + c/a + b +c)]1/2} × 100, where a is the band intensity of DNA substrate and b and c are the cleavage products (26).
Supplementary Material
Supplementary File
Acknowledgments
We thank B. Castellano and M. Kaplan for assisting with cloning and protein purification; E. Nogales for use of EM resources and expert assistance in EM analysis; and members of the J.A.D. laboratory for helpful discussions. S.H.S. is a National Science Foundation Graduate Research Fellow and a National Defense Science and Engineering Graduate Research Fellow. A.V.W. is a National Science Foundation Graduate Research Fellow. B.T.S. is a Roche Postdoctoral Fellow, RPF 311. J.A.D. is a Howard Hughes Medical Institute investigator.
Footnotes
Conflict of interest statement: A.V.W., S.H.S. and J.A.D. have filed a related patent.
References
- 1.Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54(2):234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12(7):479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39(21):9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10(10):957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doudna JA, Charpentier E. Genome editing: The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 8.Jinek M, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343(6176):1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishimasu H, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513(7519):569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briner AE, et al. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 2014;56(2):333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;4:3. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shekhawat SS, Ghosh I. Split-protein systems: Beyond binary protein-protein interactions. Curr Opin Chem Biol. 2011;15(6):789–797. doi: 10.1016/j.cbpa.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500(7463):472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lienert F, et al. Two- and three-input TALE-based AND logic computation in embryonic stem cells. Nucleic Acids Res. 2013;41(21):9967–9975. doi: 10.1093/nar/gkt758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang F-S, Ho WQ, Crabtree GR. Engineering the ABA plant stress pathway for regulation of induced proximity. Sci Signal. 2011;4(164):rs2. doi: 10.1126/scisignal.2001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy MJ, et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7(12):973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiedenheft B, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477(7365):486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson RN, et al. Structural biology: Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345(6203):1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulepati S, Héroux A, Bailey S. Structural biology: Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345(6203):1479–1484. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151(1):41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482(7384):186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116(1):17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 25.Kim HJ, Lee HJ, Kim H, Cho SW, Kim J-S. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19(7):1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File