Two ancient human genomes reveal Polynesian ancestry among the indigenous Botocudos of Brazil (original) (raw)
. Author manuscript; available in PMC: 2015 Nov 3.
Published in final edited form as: Curr Biol. 2014 Oct 23;24(21):R1035–R1037. doi: 10.1016/j.cub.2014.09.078
Understanding the peopling of the Americas remains an important and challenging question. Here, we present 14C dates, and morphological, isotopic and genomic sequence data from two human skulls from the state of Minas Gerais, Brazil, part of one of the indigenous groups known as ‘Botocudos’. We find that their genomic ancestry is Polynesian, with no detectable Native American component. Radiocarbon analysis of the skulls shows that the individuals had died prior to the beginning of the 19th century. Our findings could either represent genomic evidence of Polynesians reaching South America during their Pacific expansion, or European-mediated transport.
A recent study of skeletal remains from Brazil belonging to the indigenous Botocudo peoples found that two male individuals presented a combination of mitochondrial DNA (mtDNA) variants common in present day Oceanian populations [1]. Although it was argued that these genetic traits were likely to be derived from runaway slaves brought to Brazil by Europeans, no direct-dating or genomic analyses were used to support this conclusion. The Botocudos, named after the wooden disks (‘botoques’) in their lower lips and ear lobes, were an indigenous group with presumably Native American origins occupying the coast and the interior of Eastern-Central Brazil until the late 18th century, when most were exterminated by European colonists after decades of violence. The population size and origin of the Botocudos remain unclear, but they are likely to have comprised several tribes who spoke a common Macro-Jê language at the time of European contact [2]. We conducted a variety of genomic, morphological and isotopic analyses of skeletal remains attributed to the Botocudos of Brazil.
We confined our analyses to four Botocudo individuals (Bot13, Bot15, Bot17 and Bot65; Supplemental information) after careful review of all available records at the Museu Nacional in Rio de Janeiro, Brazil, including an extensive archival study that pointed to the four crania being bona fide Botocudo, a designation which is also corroborated by the labels on the skulls (Figure 1A).
Figure 1. Radiocarbon and genetic evidence for Polynesian ancestry among Botocudos.
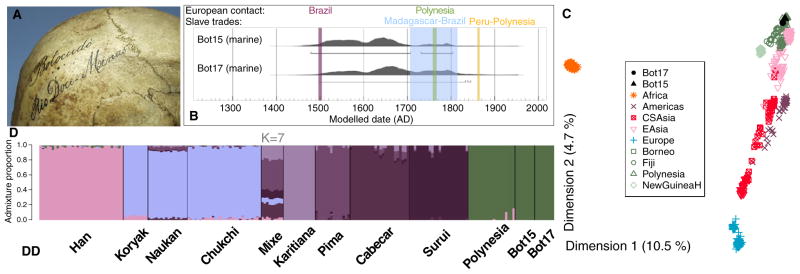
(A) Botocudo 17 skull. Lateral photograph of Bot17 showing a close-up of the labeling. (B) Calibrated 14C dates for Bot15 and Bot17. We assumed 60 ± 16% (Bot15) and 30 ± 16% (Bot17) marine carbon dietary uptake, respectively, and a reservoir age that is equivalent to the average world ocean reservoir to compute the calibrated 14C dates and associated 95% highest posterior density regions (horizontal lines). The colored bars indicate four pertinent historical events: the European discovery of Brazil (purple), the established European contact with Polynesian islands (green), the Madagascar-Brazil slave trade (blue), and the Peru-Polynesia slave trade (orange). The uncalibrated dates for the samples are Bot15 OxA-27184 408 ± 24BP and AAR-17522 417 ± 25BP and Bot17 AAR-17657 487 ± 25BP. (C) MDS plots. Multidimensional scaling (MDS) plots (dimension 1 and 2) based on an identity-by-state distance matrix between individuals from worldwide populations (SNP chip data) and the whole genome Bot15 and Bot17 data. Populations are colored by broader geographic areas as follows: Africa: Bambaran, Dogon, YRI; Europe: CEU, Iraqi, Slovenian; EAsia: CHB, JPT, Thai, Mongolian; CSAsia: Kyrgyzstan, Nepalese, Pakistanis; Americas: Bolivian, Totonac; NewGuineaH: New Guinea Highlands. The reference panel includes around 820,000 SNPs and the overlap between Bot15 or Bot17 and the reference panel is around 640,000 SNPs. Note that MDS plots obtained after removing transitions (reducing the effect of ancient DNA damage) are almost identical. Appropriate references and results for other datasets are shown in Supplemental information. (D) Clustering analysis. NGSadmix analysis for K = 7 based on SNP chip data and the Botocudo whole genome sequence data. The SNP chip data includes Asian (Han), Siberian (Koryak, Naukan, Chukchi) and Native American (Mixe, Karitiana, Pima, Cabecar, Surui) samples. The Bot15 and Bot17 genomes are represented with wider bars on the right-hand side. Appropriate references, results for other datasets and other values of K can be found in Supplemental Data.
We performed an initial DNA screening by shotgun sequencing the four individuals, finding that only Bot15 and Bot17 yielded a endogenous human DNA content higher than 1% (5.6% and 12.5%, respectively), thus allowing for whole genome sequencing (Supplemental information). In total, we obtained genetic data from the following experiments: mtDNA capture for Bot15; single nucleotide polymorphism (SNP) capture for 6,000 ancestry informative markers (AIMs) for Bot15; and whole genome shotgun sequencing for Bot15 and Bot17 (to an average genomic depth of 1.2X and 1.5X, respectively). We found features in the data characteristic of ancient DNA (Supplemental information). All the genetic data point towards two individuals with Polynesian ancestry and no detectable Native American ancestry. For example, when conducting a classical multidimensional scaling (MDS) analysis on identity-by-state (IBS) distance-of-genotype data of worldwide populations, including the Botocudo whole genome data, we found that the ancient genomes fall within the Polynesian populations (Figure 1C). Moreover, clustering analyses suggest that Bot15 and Bot17 have no detectable Native American ancestry and share the same components as the Polynesian population. When assuming seven ancestral populations (K = 7), the Polynesians form their own cluster and more than 99.9% of the genomes of Bot15 and Bot17 were assigned to this Polynesian cluster (Figure 1D).
Accelerator mass spectrometry (AMS) radiocarbon dating of five teeth from the four Botocudo individuals was carried out in two independent laboratories. When calibrating the 14C dates with the Southern Hemisphere curve [3], the 95% highest posterior density regions (HPD) were 1452–1510 AD and 1579–1620 AD for Bot15 (bimodal distribution), and 1419–1477AD for Bot17. While there is a general lack of baseline isotope data for the relevant areas and from archaeological remains, the combined evidence from the δ13C, δ15N and δ34S values when compared to Brazilian fauna and flora, as well as archaeological and modern human bone samples, suggests that a marine reservoir effect cannot be fully excluded (Supplemental information). The 95% HPDs of the marine corrected dates are 1479–1708 AD and 1730–1804 AD for Bot15, and 1496–1842 AD for Bot17 (Figure 1B). More work on foodwebs and carbon cycling in the Minas Gerais area is needed to determine if a marine reservoir correction for the 14C dates is indeed required; hence, the marine-corrected age estimates presented must be considered highly conservative. Strontium analysis and craniometry results are detailed in the Supplemental information.
Several scenarios have been previously proposed that are of potential relevance for explaining our result that two Botocudo individuals have Polynesian ancestry for both the mitochondrial and nuclear genomes [1]: the Polynesia–Peru slave trade [4], the Madagascar–Brazil slave trade, voyaging on European ships either as crew, passengers, or stowaways, and Polynesian voyaging. Regardless, the scenario must necessarily invoke a certain number of Polynesian migrants — presumably more than two, as we detected Polynesian ancestry in two out of 35 Botocudo individuals in the Museu Nacional collection. A detailed investigation of these possibilities can be found in the Supplemental information. The 1862–1864 AD Peru–Polynesia slave trade can be excluded, given that the 14C calibrated dates for the skulls predate the beginning of this trade. The Madagascar–Brazil slave trade is of relevance, as Madagascar is known to have been peopled by Southeast Asians [5]. However, we can further exclude this hypothesis as recent genomic data have demonstrated that the Malagasy ancestors admixed with African populations prior to the slave trade [5], and no such ancestry is detected in our Botocudo sample (Supplemental information). Furthermore, Madagascar was peopled by Southeast Asian and not Polynesian populations.
While Fernão de Magalhães (Magellan) first spotted some seemingly uninhabited Polynesian islands in 1521 AD, the lack of precise navigational techniques [6], as well as war between European nations, meant that many of those islands were not visited by Europeans again for at least another 200 years. Therefore, trade involving Euroamerican ships in the Pacific only began after 1760 AD [7]. By 1760 AD, Bot15 and Bot17 were already deceased with a probability of 0.92 and 0.81, respectively (Supplemental information), making this scenario unlikely. Although improbable also because it would involve both individuals making it to the interior of Brazil, we cannot exclude this scenario.
Polynesian ancestors originated from East Asia and on their migration eastwards interacted with and admixed with local New Guineans before colonizing the Pacific. In recent years, evidence has continued to accumulate in favor of a Polynesian and South American contact [8], although the issue has remained mired in controversy (e.g., [9]). It has been established that the Polynesian Pacific expansion from Southeast Asia covered distances of thousands of kilometers, reaching New Zealand, Hawaii and Easter Island — an area approximately the size of North America — between ca. 1200 and 1300 AD [10]. It is hard to explain how the first possible genomic evidence of such Polynesian contact with South America would be found in Brazil rather than on the west coast of South America, yet we cannot exclude this scenario either.
Whether brought by Europeans or the result of the Polynesian expansion, the fact remains that some Brazilian Botocudos carried distinctive Polynesian genetic signatures. We hope that further sampling will provide a more definitive answer to this intriguing finding.
Supplementary Material
S1
Acknowledgments
We would like to thank the laboratory technicians at the Danish National High-throughput DNA Sequencing Centre for technical assistance; Martin Kircher, Thorfinn Sand Korneliussen, Johannes Krause, David Reich, Erik Thorsby for helpful discussion; Toomas Kivisild, Jinchuan Xing, Andreas Wollstein, David Reich for early access and/or assistance with their data. GeoGenetics members were supported by the Lundbeck Foundation, the Danish National Research Foundation (DNRF94) and the European Union (FP7/2007-2013/317184 and 319209). A.S.M. was supported by a fellowship from the Swiss National Science Foundation (PBSKP3_143529); M.D. by the US National Science Foundation (grant DBI-1103639); P.L.J. by the National Institutes of Health (grant K99 GM104158); V.F.G. by a Strategic Training for Advanced Genetic Epidemiology (STAGE) fellowship, University of Toronto.
Footnotes
References
- 1.Gonçalves VF, Stenderup J, Rodrigues-Carvalho C, Silva HP, Gonçalves-Dornelas H, Líryo A, Kivisild T, Malaspinas AS, Campos PF, Rasmussen M, et al. Identification of Polynesian mtDNA haplogroups in remains of Botocudo Amerindians from Brazil. Proc Natl Acad Sci USA. 2013;110:6465–6469. doi: 10.1073/pnas.1217905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wied-Neuwied MAP. Beitraege zur Naturgeschichte von Brasilien 1832
- 3.McCormac FG, Hogg AG, Blackwell PG, Buck CE, Higham TFG, Reimer PJ. SHCal04 Southern Hemisphere calibration, 0-11.0 cal kyr BP. Radiocarbon. 2004;46:1087–1092. [Google Scholar]
- 4.Maude HE. Slavers in Paradise: The Peruvian Slave Trade in Polynesia, 1862–1864 Redwood City. California: Stanford University Press; 1981. [Google Scholar]
- 5.Pierron Denis, Razafindrazaka Harilanto, Pagani Luca, Ricaut Francois-Xavier, Antao Tiago, Capredon Melanie, Sambo Clement, Radimilahy Chantal, Rakotoarisoa Jean-Aime, Blench Roger M, et al. Genome-wide evidence of Austronesian-Bantu admixture and cultural reversion in a hunter-gatherer group of Madagascar. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1321860111. 201321860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maude HE. Spanish discoveries In the Pacific. J Polynesian Soc. 1959;68:285–326. [Google Scholar]
- 7.Thomas N. Islanders: The Pacific in the Age of Empire. Yale University Press; 2010. [Google Scholar]
- 8.Jones TL, Storey SS, Matisoo-Smith EA, Ramírez-Aliaga JM. Polynesians in America. Pre-Columbian Contact with the New World. Lanham, Maryland: Altamira Press; 2011. [Google Scholar]
- 9.Gongora J, Rawlence NJ, Mobegi VA, Jianlin H, Alcalde JA, Matus JT, Hanotte O, Moran C, Austin JJ, Ulm S, et al. Indo-European and Asian origins for Chilean and Pacific chickens revealed by mtDNA. Proc Natl Acad Sci USA. 2008;105:10308–10313. doi: 10.1073/pnas.0801991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilmshurst JM, Hunt TL, Lipo CP, Anderson AJ. High-precision radiocarbon dating shows recent and rapid initial human colonization of East Polynesia. Proc Natl Acad Sci USA. 2011;108:1815–1820. doi: 10.1073/pnas.1015876108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1