Regular Article Macroautophagy is Defective in Mucolipin 1-Deficient Mouse Neurons (original) (raw)
. Author manuscript; available in PMC: 2015 Apr 10.
Published in final edited form as: Neurobiol Dis. 2010 Jun 28;40(2):370–377. doi: 10.1016/j.nbd.2010.06.010
Abstract
Mucolipidosis Type IV is a neurodegenerative lysosomal disease clinically characterized by psychomotor retardation, visual impairment, and achlorhydria. In this study we report the development of a neuronal cell model generated from cerebrum of _Mcoln1_-/- embryos. Prior functional characterization of MLIV cells has been limited to fibroblast cultures gleaned from patients. The current availability of the mucolipin-1 knockout mouse model _Mcoln1_-/- allows the study of mucolipin1-defective neurons, which is important since the disease is characterized by severe neurological impairment. Electron microscopy studies reveal significant membranous intracytoplasmic storage bodies, which correlate with the storage morphology observed in cerebral cortex of _Mcoln1_-/- P7 pups and E17 embryos. The _Mcoln1_-/- neuronal cultures show an increase in size of LysoTracker and Lamp1 positive-vesicles. Using this neuronal model system, we show that macroautophagy is defective in mucolipin-1 deficient neurons and that LC3-II levels are significantly elevated. Treatment with rapamycin plus protease inhibitors did not increase levels of LC3-II in _Mcoln1_-/- neuronal cultures, indicating that the lack of mucolipin-1 affects LC3-II clearance. P62/SQSTM1 and ubiquitin levels were also increased in _Mcoln1_-/- neuronal cultures, suggesting an accumulation of protein aggregates and a defect in macroautophagy which could help explain the neurodegeneration observed in MLIV. This study describes, for the first time, a defect in macroautophagy in mucolipin-1 deficient neurons, which corroborates recent findings in MLIV fibroblasts and provides new insight into the neuronal pathogenesis of this disease.
Keywords: Mucolipidosis Type IV, lysosomal disease, mucolipin 1, macroautophagy, neuronal storage
Introduction
Mucolipidosis Type IV (MLIV; OMIM 252650) is a progressive neurological lysosomal disease that usually presents during the first year of life with mental retardation, corneal opacities, elevated blood gastrin levels with achlorhydria, and delayed motor milestones (Berman ER, 1974; Frei KP, 1998). MLIV is an autosomal recessive disease with the majority of the cases (80%) reported in individuals of Ashkenazi Jewish (AJ) descent (Bargal, 2000). The carrier frequency in AJ is estimated to be 1:100 and the major AJ mutation, present on 72% of the AJ MLIV alleles, is an A–G transition at the 3′ acceptor site of intron 3 (Bargal, 2000; Bassi, 2000; Sun, 2000). The minor AJ mutation, found on 23% of the AJ MLIV alleles, is a 6434 bp genomic deletion that spans exons 1-6 and the first 12 bp of exon 7 (Goldin, 2004; Wang, 2002).
The _MCOLN1_-encoded protein (mucolipin-1, TRPML1) is a transmembrane protein with topological homology to other members of the TRP superfamily of ion channels. Several studies have begun to shed light on the function of mucolipin-1 as a calcium channel, however, the pathogenic mechanism by which loss of mucolipin-1 leads to cellular storage and neuronal cell dysfunction and death in MLIV is still poorly understood (Kiselyov, 2005; LaPlante, 2002; LaPlante, 2006; Manzoni, 2004; Miedel, 2006; Pryor, 2006; Thompson, 2007; Venkatachalam, 2006; Vergarajauregui, 2006).
Recently, an increased association between lysosomal diseases and macroautophagy dysfunction has been described. Macroautophagy is a constitutive, well orchestrated process responsible for bulk degradation of long-lived proteins and cellular organelles. Macroautophagy is essential for the survival of neural cells and its impairment is implicated in the pathogenesis of neurodegenerative disorders (Komatsu, 2006; Nedelsky, 2008; Ventruti, 2007). In some lysosomal diseases, the observed autophagic stress is also correlated with neuronal death and could be linked to the neurodegeneration, although the full extent of neuronal death in most lysosomal diseases has not yet been determined (Cao, 2006; Koike, 2005; Pacheco, 2007; Raben, 2009a; Raben, 2009b; Settembre, 2008a; Settembre, 2008b).
We previously generated the first murine model for MLIV by targeted knock-out of Mcoln1, and showed that the mouse model accurately replicates MLIV disease in humans. The _Mcoln1_-/- mice present with numerous dense inclusion bodies in all cell types in brain but particularly in neurons, severe retinal degeneration, elevated plasma gastrin, and vacuolization in parietal cells (Venugopal, 2007). More recently, a more detailed neuropathological characterization of the Mcoln1-/- mouse showed evidence of ganglioside accumulation throughout the CNS, presence of P62/SQSTM1 inclusions, reduced myelination in axonal tracts of the cerebrum and cerebellum and presence of axonal spheroids in white-matter tracts and Purkinje cell axons of the cerebellum (Micsenyi, 2009). We have recently shown that human MLIV fibroblasts have a defect in chaperone mediated autophagy (CMA) (Venugopal, 2009). Given the crosstalk between different autophagy pathways (Kaushik, 2008), herein we have investigated macroautophagy in a new neuronal culture model of MLIV. We show for the first time that macroautophagy is defective in _Mcoln1_-/- neurons, suggesting that defective autophagy contributes to the pathogenesis of neuronal loss in MLIV, the most devastating component of the human disease.
Materials and Methods
Establishment of mouse neuronal cultures
Embryonic day 17 (E17) embryos resulting from heterozygous mating were dissected from the mother via cesarean section. Mouse neuronal primary cultures were established as previously described (Lesuisse, 2002; Reis, 2006). Briefly, both hemispheres of the cerebrum were dissected and treated with 0.5% trypsin, 0.1% trypsin inhibitor, and 228 U/ml DNAse I for 1 min each at 37°C. The tissue was dissociated in neurobasal medium containing 2% B27 supplement and 1% penicillin/streptomycin by repeated passage through a P1000 and a P200 pipette. Cells were plated at a density of 2.106 cells/ml (∼2000 cells/mm2) and were cultured at 37°C with 5% CO2 and maintained in the same medium as above. Genotyping was done by PCR using genomic DNA from embryo tails as previously described (Venugopal, 2007).
Immunofluorescence and confocal microscopy
Staining with 50 nM LysoTracker (Invitrogen) was done in live cells for 15 min at 37°C. Lamp1 staining was done with anti-lamp1 antibody clone 1DB4 (BD Biosciences) after fixation of neurons with 4% paraformaldehyde PFA (pH 7). Lamp1 antibody was diluted in 1% bovine serum albumin (BSA), 2% normal goat serum (NGS) and 0.05 % saponin to a final concentration of 0.25 ug/ml. Secondary antibody used was a rat IgG conjugated with Alexafluor 488 (Molecular Probes) diluted in 1% BSA, 2% NGS and 0.05 % saponin to a final concentration of 2 ug/ml. Coverslips were mounted with FluoromountG (Southern Biotech). Specimens were analyzed using a Leica SP5 confocal microscope. Quantification and measurement of LysoTracker-labeled vesicles was done using the MetaXpress cellular imaging analysis software (MDS Analytical Technologies), based on a single optical confocal plane per neuron. The chosen threshold of 2.1μm, to define large vesicles, was determined based on visual comparison of wild type and knockout images, followed by measurement of vesicle width.
Electron microscopy on embryonic neuronal primary cultures
Analysis was performed at the Electron Microscopy facility at the Massachusetts General Hospital Pathology Service. Briefly, after removal of the culture medium, cells were fixed in Karnovsky's KII Solution containing 2.5% glutaraldehyde, 2.0% PFA, 0.025% calcium chloride in a 0.1M sodium cacodylate buffer, pH 7.4 for 1 hour. Cells were then resuspended in warm 2% agar. After solidification, the agar block was processed routinely for electron microscopy in a Leica Lynx™ automatic tissue processor. Specimens were post-fixed in osmium tetroxide, stained en bloc with uranyl acetate, dehydrated in graded ethanol solutions, infiltrated with propylene oxide/Epon mixtures, embedded in pure Epon, and polymerized over night at 60°C. For electron microscopy of the cerebral cortex tissues obtained from dissected pups or embryos, the same methodology was used with the exception of the 2% agar block step which was omitted and fixation was overnight. One micron sections were cut, stained with toluidine blue, and examined by light microscopy. Thin sections were cut with an LKB 8801 ultramicrotome and diamond knife, stained with lead citrate, and examined in a Phillips 301 transmission electron microscope (Phillips Analytical). Images were captured with an AMT™ (Advanced Microscopy Techniques) digital CCD camera.
Western blot analysis
Cells were washed with PBS, harvested, lysed for 30 minutes at 4°C in lysis buffer containing 50mM NaPO4, pH 7.4, 0.5% Triton X-100, 10% glycerol, type I protease inhibitor cocktail set (Calbiochem), 1mM phenymethylsulfonyl fluoride and spun at 11,000 g for 10 minutes. Lysates were resolved using 16% or 8% Trisglycine gels (Invitrogen) prior to transfer to a polyvinylidene fluoride membrane (Bio-Rad). Detergent (1% NP-40) soluble and insoluble p62 fractions were isolated and analyzed as previously described (Vergarajauregui, 2008). Primary antibodies used were: rabbit polyclonal anti-autophagy APG86 (anti-LC3 AP 1802a) (1.25μg/mL) (Abgent), mouse monoclonal anti-neuronal nuclei (NeuN) (1μg/mL) (Millipore), rabbit polyclonal anti-p62/SQSTM1 (1:1000) (Biomol Int, LP), rabbit polyclonal anti-p70S6K (9ng/mL), rabbit polyclonal anti-phospho-p70S6K (Thr389) (109ng/mL), rabbit monoclonal anti-mTOR (30ng/mL), rabbit polyclonal anti-phospho-mTOR (Ser2448) (136ng/mL), rabbit polyclonal anti-ubiquitin antibody (68ng/mL), rabbit polyclonal anti-beclin-1 (58ng/mL) (Cell Signaling Technology). Anti-mouse or rabbit IgG secondary antibodies HRP-conjugated and peroxidase substrate were used following manufacturer's instructions (GE Healthcare). Bands were quantified using the Scanner/Software Quantity One 4.5.0 (Bio-Rad).
Statistical analysis
Results are expressed as the mean ± standard deviation throughout the text and figures and are representative of at least 3 independent experiments. Unpaired t test was performed using GraphPad Prism (GraphPad Software).
Results
Characterization of _Mcoln1_-/- mouse neuronal cultures
To further characterize the neuronal cell phenotype present in the MLIV mouse model, we established neuronal cultures from E17 cerebrum of _Mcoln1_-/- embryos and wild type littermates. It has been previously demonstrated that neuronal cultures established according to the protocol utilized in this study show expression of neuronal nuclei (NeuN), a neuronal marker, at day 10 of culture and do not show glial contamination until 20 days in culture. Therefore, to ensure neuronal purity in our experiments, cells were plated at a density of 1000 cells/mm2 and in all experiments cells were cultured for a maximum of 12 days (Brewer, 1993; Lesuisse, 2002). NeuN expression was analyzed by Western blot and similar levels were found in Mcoln1+/+ and _Mcoln1_-/- neuronal cultures (data not shown).
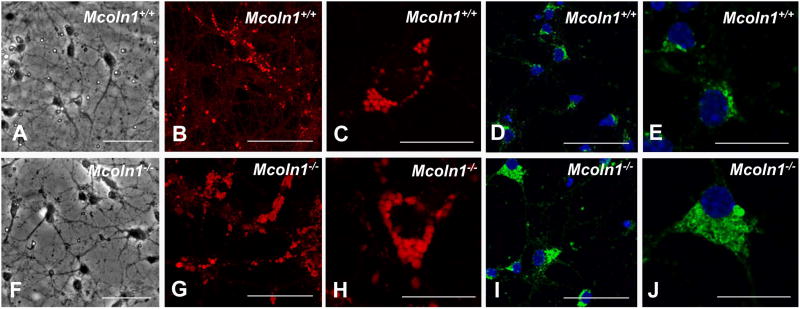
After 10 days in culture, phase contrast microscopy showed that neurons from Mcoln1+/+ and _Mcoln1_-/- neuronal cultures extended prominent neurites and showed no apparent difference in gross morphology (Fig. 1 A and F). Using LysoTracker to stain acidified vesicles of the lysosomal system, an increase in the size of vesicles was evident in _Mcoln1_-/- cultures (average vesicle count/cell was 42.2 ± 16.5 in Mcoln1+/+vs. 51.3 ± 26.3 in _Mcoln1_-/-;n=15; p=0.266; average count of large vesicles per cell (>2.1μm) was 0.46 ± 0.52 in Mcoln1+/+vs. 3.75 ± 2.5 in _Mcoln1_-/-; n=15; p<0.0001) (Fig. 1 B and C for Mcoln1+/+ vs. G and H for _Mcoln1_-/-). Lamp1 staining confirmed the findings observed with Lysotracker (Fig. 1 D and E for Mcoln1+/+vs. I and J for _Mcoln1_-/-). Neuronal cultures were also analyzed for the presence of autofluorescence, which has been previously observed in MLIV human skin fibroblasts (Goldin, 1995). As expected, the _Mcoln1_-/- neurons presented autofluorescence when compared to neurons established from Mcoln1+/+ littermate embryos (Fig. 1S - Supplemental data).
Figure 1.
Mcoln1+/+ and _Mcoln1_-/- embryonic neuronal cultures. (A-J) Mcoln1+/+ and _Mcoln1_-/- E17 embryonic-derived neuronal cultures after 10 days in vitro, (A and F) Phase contrast microscopy shows neuronal cultures from Mcoln1+/+ and _Mcoln1_-/-, respectively, LysoTracker-stained (red) cells showing presence of larger vesicles in _Mcoln1_-/- (G and H in _Mcoln1_-/-vs. B and C in Mcoln1+/+), Lamp1-immunolabelled cells (green) showing presence of larger vesicles in _Mcoln1_-/- (I and J in _Mcoln1_-/-vs. D and E in Mcoln1+/+). Bar indicates 50 microns (A, B, D, F, G and I) and 20 microns (C, E, H and J).
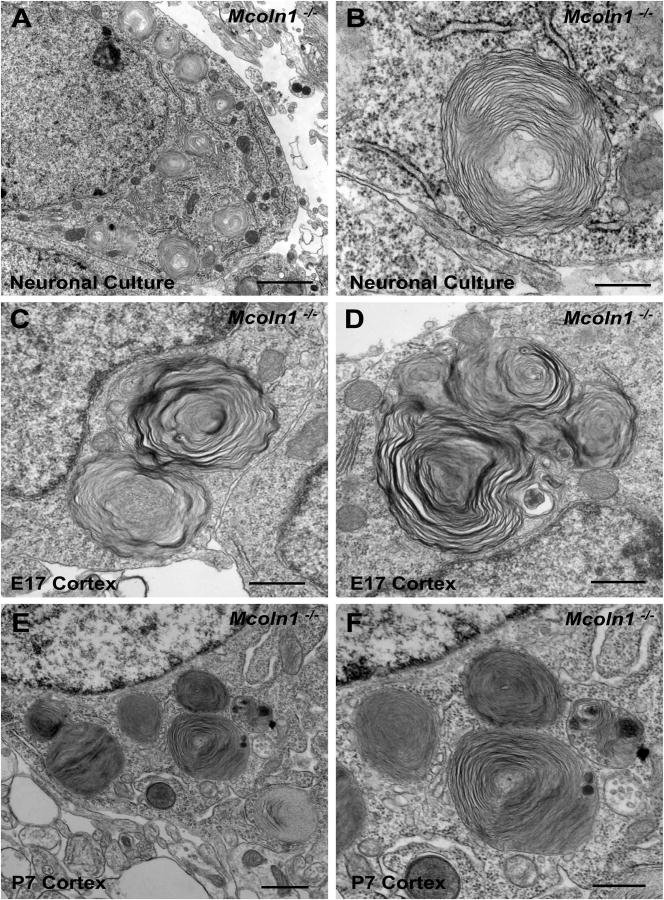
Based on the cellular pathology observed with LysoTracker staining, we further analyzed the MLIV neuronal pathology in neuronal cultures by electron microscopy, which revealed the presence of membranous cytoplasmic bodies (Fig. 2 A and B). In order to correlate the storage seen in culture to in vivo neuronal storage, we also performed electron microscopy directly in cerebral cortex collected from embryonic day 17 (E17) embryos and postnatal day 7 (P7) pups (corresponding in time to 10 day-cultured E17-embryonic neurons). As shown on figures 2 C, D (E17 embryos) and 2 E, F (P7 pups) there was an impressive presence of membranous cytoplasmic bodies, morphologically identical to the ones observed in neuronal embryonic cultures (Fig. 2 A and B). Neuronal cultures established from Mcoln1+/+ littermates as well as cortex dissected from postnatal day 7 Mcoln1+/+ pups were analyzed by electron microscopy and, as expected, did not reveal presence of any cytoplasmic storage bodies (Fig. 2S-supplemental data). Importantly, the EM data obtained from E17 and P7 cerebral cortex support the validity of using our neuronal primary cultures established from _Mcoln1_-/- embryos as tools for the study of cellular phenotypes and potential treatments in MLIV disease.
Figure 2.
_Mcoln1_-/- storage bodies in post natal day 7 _Mcoln1_-/- pups cortex _versus Mcoln1_-/- neuronal cultures. (A and B) (A) Electron microscopy of _Mcoln1_-/- embryonic neuronal cultures after 10 days in culture (bar indicates 2 microns) (B) Magnification view of storage body (bar indicates 500 nm). (C and D) (C) Electron microscopy of embryonic cortex from E17 embryos (bar indicate 2 microns) (D) Magnification view of storage body (bar indicates 500 nm). (E and F) (E) Electron microscopy of post natal day 7 pup _Mcoln1_-/- brain cortex (bar indicates 2 microns) (F) view of storage body (bar indicates 500 nm).
LC3-II levels are increased in Mcoln1-/- mouse neuronal cultures
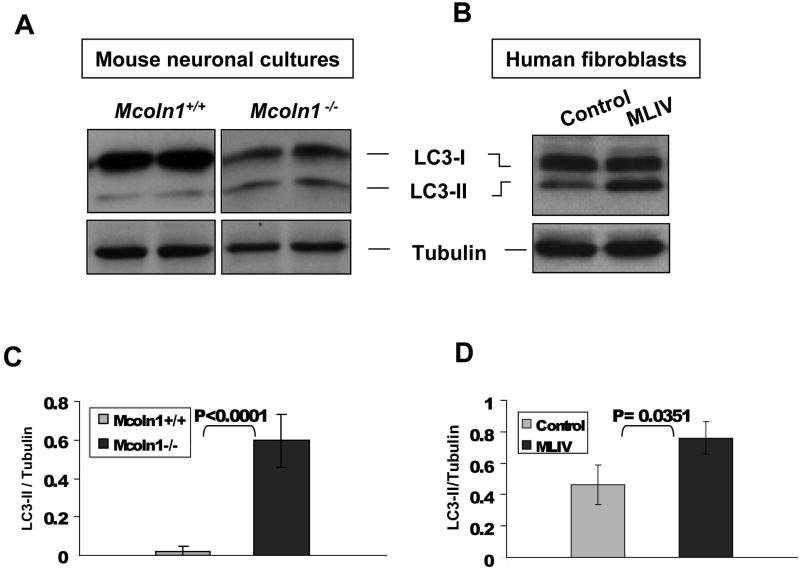
Given that many lysosomal diseases have been associated with altered macroautophagy, coupled with the fact that we have shown defective chaperone mediated autophagy in human fibroblasts derived from MLIV patients, the next step was to investigate the macroautophagy status in _Mcoln1_-/- neuronal cells (Cao, 2006; Koike, 2005; Nixon, 2008; Pacheco, 2007; Settembre, 2008a; Settembre, 2008b; Vergarajauregui, 2008). The microtubule-associated protein (LC3) is lipidated upon activation of autophagy. The lipidated form of the protein (LC3-II) associates with autophagosomal membranes and the pool associated with the internal face of the autophagosomal double membrane is rapidly degraded after autophagosome-lysosome fusion. LC3-II can be discerned from the non-lipidated form, LC3-I, by mobility shift on SDS PAGE (Kabeya, 2000; Tanida, 2008). Therefore, detection of LC3-II by immunoblotting has become a routine method for monitoring macroautophagy. Analysis of steady state LC3-II levels in neuronal cultures from _Mcoln1_-/- embryos showed increased levels of LC3-II relative to levels seen in the Mcoln1+/+, suggesting that macroautophagy is altered in _Mcoln1_-/- neuronal cells (Fig. 3 A and C). Human fibroblasts isolated from control and MLIV patients were also analyzed and LC3-II levels were increased in MLIV, correlating our findings obtained from the murine neuronal cultures to human MLIV cells (Fig. 3 B and D). It is important to note, however, that given the pathology of the disease, demonstration of cellular defects in neurons is crucial to understanding the profound neuronal phenotype in MLIV.
Figure 3.
LC3-II levels in embryonic neuronal cultures (A and C) and human MLIV-affected fibroblasts (B and D). Cell lysates were analyzed by Western blot with anti-LC3A Abgent antibody. LC3-II levels were determined using the average of band densitometry value, divided by β-tubulin (n=4 for primary neuronal cultures and MLIV fibroblasts, n=2 for control fibroblast).
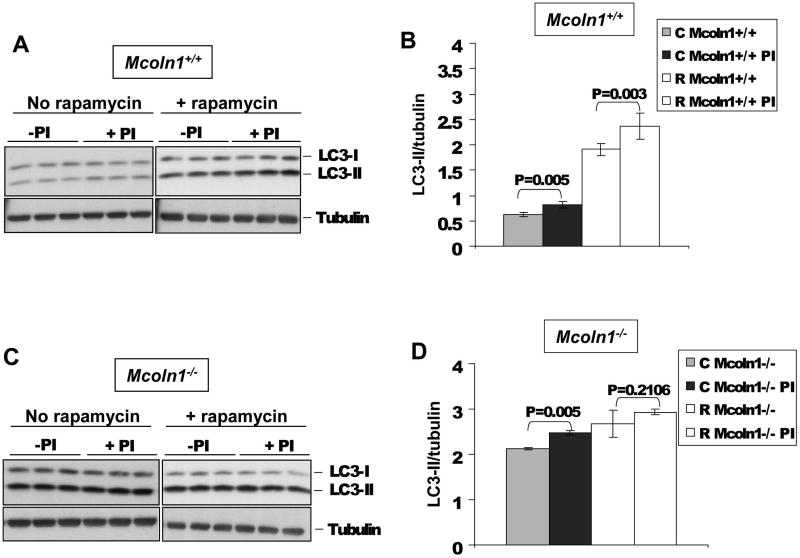
To investigate the status of LC3-II clearance in _Mcoln1_-/- neuronal cells we analyzed lysosomal LC3-II turnover after treatment with protease inhibitors (Hamacher-Brady, 2006). Since intra-autophagosomal components, such as LC3-II, are degraded by lysosomal hydrolases, treatment of cells with protease inhibitors will promote an accumulation of LC3-II. On the other hand, if there is already an autophagosomal maturation or lysosomal defect present, protease inhibitor treatment may not show a striking protective effect on LC3-II levels. Neuronal cultures isolated from Mcoln1+/+ and _Mcoln1_-/- embryos were cultured for 10 days and then treated with protease inhibitors (10 μg/ml E64d and 10 μg/ml Pepstatin), plus or minus 250 nM rapamycin for 8 hours. Rapamycin is an mTOR-dependent inhibitor of p70S6K that was used alone or in combination with the protease inhibitors to challenge the cells by stimulating autophagy above its basal activity level. _Mcoln1_-/- cells responded in a similar way as the Mcoln1+/+ cells after PI treatment when not challenged by rapamycin (Fig. 4). However, in the presence of rapamycin, LC3-II levels in _Mcoln1_-/- cells were not remarkably elevated upon protease inhibitor treatment, compared to no protease inhibitor treatment (Fig. 4 C and D). Notably, similar results were obtained when neuronal cultures were treated for 16 hours with rapamycin and protease inhibitors (data not shown). These results suggest that TRPML1 deficiency affects LC3-II lysosomal turnover in neuronal cultures.
Figure 4.
LC3-II turnover in Mcoln1+/+ (A and B) and _Mcoln1_-/- (C and D) neuronal cultures before and after protease inhibitors (Pepstatin and E64) and rapamycin treatments. On graphs B and D, LC3-II levels were determined using the average of band densitometry value, divided by β-tubulin (n=3).
Macroautophagy regulation in the _Mcoln1_-/- mouse neuronal cultures
A previous study done in MLIV human fibroblasts showed macroautophagy activation as a result of increased de novo autophagosome formation combined with delayed fusion of autophagosomes with late endosomes/lysosomes (Vergarajauregui, 2008). Therefore we sought to assay the signaling pathways regulating macroautophagy in our novel neuronal culture model of MLIV.
Beclin-1 levels increase basal autophagy by stimulating autophagosome formation. We observed a trend of increased Beclin-1 levels in _Mcoln1_-/- neurons, but this was not statistically significant (Fig. 3S, A and B – Supplemental data). Mammalian target of rapamycin (mTOR) acts to negatively regulate autophagy, therefore a decrease in mTOR function increases basal levels of autophagy. We have analyzed levels of phosphorylated mTOR as well as p70S6 Kinase (p70S6K) phosphorylation at Thr389, an immediate downstream target of mTOR, in both Mcoln1+/+ and _Mcoln1_-/- neuronal cells and no significant differences were observed (Fig. 3S, C-F – Supplemental data). Together, these data do not support a significant stimulation of macroautophagy caused by TRPML1 deficiency.
Sequestosome 1 (P62/SQSTM1) and ubiquitin levels are increased in Mcoln1-/- mouse neuronal cultures
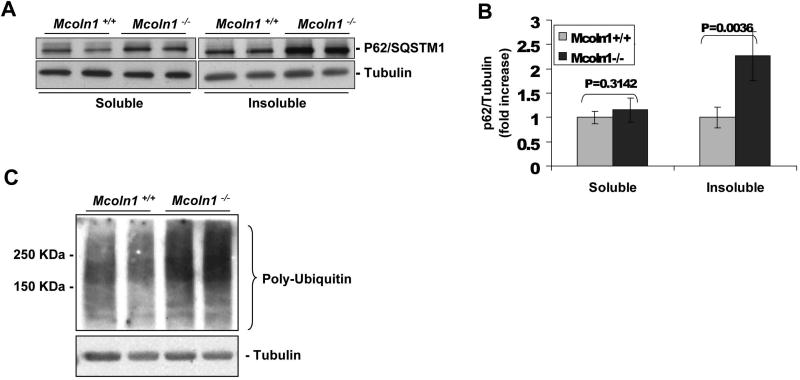
Our LC3 data suggest that TRPML1 deficiency leads to a defect in macroautophagy. P62/SQSTM1 has been demonstrated to be upregulated as a result of autophagy inhibition (Klionsky, 2008; Korolchuk, 2009). P62/SQSTM1 binds to ubiquitin and LC3-II, and is required for both the formation and the degradation of polyubiquitinated bodies by autophagy (Mizushima, 2007; Pankiv, 2007; Rubinsztein, 2009). Therefore, we sought to assay P62/SQSTM1 levels in the _Mcoln1_-/- neuronal culture. P62/SQSTM1 is degraded by autophagy and quantification of detergent-insoluble protein fractions is used as a way to measure levels of protein aggregation in cells (Tanemura, 2002; Vergarajauregui, 2008). Our results from immunoblots revealed a more than 2-fold increase in P62/SQSTM1 levels in the NP-40-insoluble cell fraction in _Mcoln1_-/- compared to Mcoln1+/+ neurons (Fig. 5 A and B). These findings are in agreement with a previous study using MLIV-affected human fibroblasts (Micsenyi, 2009; Vergarajauregui, 2008). No significant changes were observed in P62/SQSTM1 levels in the NP-40 soluble fraction in _Mcoln1_-/- compared to Mcoln1+/+ (Fig. 5 A and B). An increase in protein aggregation suggests that poly-ubiquitinated proteins may be accumulating in _Mcoln1_-/- neurons therefore recruiting more P62/SQSTM1 to poly-ubiquitin positive aggregates. In fact, we observed a large increase in poly-ubiquitin chain levels in _Mcoln1_-/- compared to Mcoln1+/+ neurons (Fig. 5 C). The accumulation of P62/SQSTM1 and poly-ubiquitin indicates a defect in macroautophagy, consistent with the deficiency in LC3-II clearance observed in _Mcoln1_-/- neuronal cells. Aggregation of ubiquitinated proteins is observed in many neurodegenerative disorders and ultimately could contribute to the neurodegeneration seen in MLIV disease.
Figure 5.
P62/SQSTM1 in detergent-insoluble cell fraction (A and B) and Ubiquitin (C) levels are upregulated in _Mcoln1_-/- mouse neuronal cultures. P62/SQSTM1 levels in detergent-soluble and insoluble fractions were determined using the average of each band densitometry value, divided by β-tubulin (n=4).
Taken together, our data shows that _Mcoln1_-/- embryonic neuronal cultures present with significant membranous intracytoplasmic storage bodies, which correlate with the storage morphology observed in cerebral cortex of _Mcoln1_-/- P7 pups and E17 embryos. Macroautophagy is abnormal in _Mcoln1_-/- neuronal cultures given the observed increase in steady-state LC3-II levels and alteration in LC3-II clearance. In addition, we observed an increase in P62/SQSTM1 levels in cell fractions enriched for protein aggregates and elevated levels of polyubiquitin were detected in _Mcoln1_-/- neuronal cultures.
Discussion
Autophagy is essential for cell renewal, especially in non-dividing cells such as neurons, where the turnover of intracellular proteins may be critical for cell survival. Several reports have described defects in autophagy in living neurons through the utilization of neuronal culture systems in neurodegenerative disorders, e.g. Parkinson's, Alzheimer's and Huntington's diseases, though neuronal autophagy remains poorly understood (Larsen, 2002; Rami, 2009). No report to date has used neuronal cultures for the study of macroautophagy in MLIV. Here we describe for the first time the establishment and characterization of _Mcoln1_-/- neuronal cultures and show that cellular storage is readily evident by 10 days in vitro in neurons derived from cerebrum of 17 day _Mcoln1_-/- embryos. Importantly, electron microscopy of cortex tissue from _Mcoln1_-/- embryonic day 17 and post natal day 7 pups shows a similar pattern of membranous cytoplasmic bodies, strongly validating the usefulness of _Mcoln1_-/- embryonic neuronal cultures for the current study as well as for the testing of potential treatments for MLIV.
The storage bodies observed at very early age, either in embryonic or postnatal day seven mice, were somewhat different from what we had previously observed and reported in aged _Mcoln1_-/- mice brain. Ten month-old _Mcoln1_-/- mice present with a mixture of compact lamellae and a granular matrix, with the latter occasionally presenting as a crystalline array (Venugopal, 2007). These storage bodies were formed as a large commingled mass concentrated within localized regions of the neuronal perikaryon (Venugopal, 2007). In the present study, the vast majority of the early storage observed consisted of larger membranous bodies dispersed throughout the cytoplasm, with less compacted lamellae. This difference in ultrastructural morphology of early stage versus aged storage material is suggestive that MLIV storage changes in appearance over time. Changes in storage morphology over time are also observed in other lysosomal diseases and are likely due to the progress of the disease and influenced by neuronal age (Crawley, 2007; Koike, 2000). At embryonic stage and postnatal day seven, neurons are still developing and have a different rate of metabolism when compared to aged neurons, which also contributes to differences in storage body morphology.
Recently, many lysosomal diseases have been associated with altered macroautophagy, e.g. Batten Disease, Niemann Pick Type C, MLIV, Multiple Sulfatase Deficiency, Mucopolysaccharidosis type IIIA and Pompe disease (Cao, 2006; Koike, 2005; Pacheco, 2007; Raben, 2009a; Raben, 2009b; Settembre, 2008a; Settembre, 2008b; Vergarajauregui, 2008). A generalized model to explain the occurrence of cell death in lysosomal disease has been suggested, based on the observation that MLIV cells, as well as those from Mucolipidosis types II and III patients, show significant mitochondrial fragmentation and decreased efficiency of mitochondrial Ca2+ buffering (Jennings, 2006; Kiselyov, 2007). Since lysosomes are major players in the autophagic recycling of mitochondria, the initial cellular observations led the authors to hypothesize that impaired lysosomal function affects mitochondrial recycling, which in turn causes accumulation of fragmented mitochondria and an inability to effectively buffer cytoplasmic Ca2+. This reduced buffering capacity, in turn, may make cells more sensitive to pro-apoptotic signals. More recently, a proposed model for MLIV states that the defect in macroautophagy accounts for the neurodegeneration, via an increase in protein aggregation and organelle damage, with consequent autophagic stress and neuronal death (Vergarajauregui, 2008). It is likely that a defect in autophagy plays an important role in the neurodegenerative process that functions in many lysosomal diseases.
Many neurodegenerative diseases are associated with an increase in P62/SQSTM1 positive structures (Bjorkoy, 2005; King, 2009; Kuusisto, 2003; Kuusisto, 2001; Wooten, 2006). Moreover, it has been shown that P62/SQSTM1 levels can be used as markers of autophagic activity (Klionsky, 2008). Recent studies have examined P62/SQSTM1 levels in MLIV patient fibroblasts and have suggested a link between the increase in P62/SQSTM1 positive structures and the neurodegeneration seen in MLIV disease, however this study has been done in fibroblasts only (Vergarajauregui, 2008). Our data shows a more than 2-fold increase in P62/SQSTM1 levels in the detergent-insoluble cell fraction, combined with a significant increase in poly-ubiquitin levels. Together, these data suggest an accumulation of protein aggregates and a defect in macroautophagy which could help explain the neurodegeneration observed in MLIV (Micsenyi, 2009). The pathological impact of P62/SQSTM1 accumulation in neuronal death has not yet been clearly elucidated. However, a recent study identified P62/SQSTM1 as a key player in mediating extrinsic apoptosis signaling, a pathway initiated by specific proapoptotic ligands outside the cell (Jin, 2009). P62/SQSTM1 is thought to act as a recruiting scaffold for polyubiquitinated caspase 8 and translocation of the protein to ubiquitin rich foci. Consequently, P62/SQSTM1 increase may act to complete the cycle and could explain the link between defective macroautophagy and neuronal cell death by activating the caspase pathway involved in extrinsic apoptosis signaling. This result is in agreement with the idea that neurodegeneration observed in MLIV is due to increased cellular sensitivity to proapoptotic signals (Jennings, 2006; Kiselyov, 2007).
Our data indicates that there is a defect in LC3-II clearance in Mcoln1-/- neuronal cultures and this defect may play an important role in the macroautophagy impairment observed in these cells. To date, two reports of macroautophagy studies in other MLIV model systems have yielded unequivocal results. It has been shown that markers for autophagosomes (GFP-ATG8) and lysosomes (LysoTracker) were colocalized in the MLIV Drosophila model in the same way as the wild type controls, suggesting that the fusion of autophagosome with lysosomes is not defective in this model of MLIV (Venkatachalam, 2008). However, our results are in agreement with recent studies done in human MLIV fibroblasts, indicating that autophagosome degradation is indeed delayed in MLIV cells (Vergarajauregui, 2008). The increase in P62/SQSTM1 and poly-ubiquitin levels in combination with our results showing a defect in LC3-II clearance corroborates the idea that LC3-II accumulation in MLIV is possibly due to a defect in the macroautophagy pathway.
It is possible that the defect in macroautophagy observed in MLIV-diseased neurons is an indirect effect of impaired lysosomal function. Loss of TRPML1 in the lysosomes may prevent or slow degradation. This may cause the organelle to become overloaded, thereby delaying the delivery of vesicles to lysosomes for degradation. However, our group has recently shown that chaperone mediated autophagy (CMA) is defective in human MLIV fibroblasts and that this defect is not just a consequence of the lysosomal alteration, but rather is due to the functional association of the mutated TRPML1 with members of the CMA machinery (Orenstein, 2010; Venugopal, 2009). A potential crosstalk between autophagic pathways has been demonstrated, suggesting that the defect in CMA observed in MLIV could stimulate macroautophagy, as a compensatory mechanism (Kaushik, 2008; Massey, 2006). However, considering our findings that LC3-II clearance is delayed in MLIV-diseased neurons, macroautophagy alone is not sufficient to reduce lysosomal storage and compensate for the defect in CMA observed in MLIV. The molecular mechanisms that regulate crosstalk among different autophagic pathways are still under investigation and the potential autophagic crosstalk present in MLIV is yet to be determined.
In summary, our studies show that neuronal cultures isolated from the Mcoln1-/- embryos are rich in intracytoplasmic membranous bodies, which correlates with the storage bodies observed in cortex from day 17 embryos and day 7 pups. Mcoln1-/- neuronal cultures show an alteration in macroautophagy, seen by a significant increase in LC3-II levels. In addition, LC3-II clearance was affected in this MLIV neuronal model. Interestingly, we observe accumulation of P62/SQSTM1 in the detergent-insoluble fraction and poly-ubiquitin, which indicates the presence of protein aggregates and may contribute to the neurodegeneration seen in MLIV.
The data presented here describes for the first time macroautophagy impairment in Mucolipin-1 deficient neurons and the characterization of this novel neuronal cellular model provides us with the means to test future therapies aimed at preventing or reversing the abnormal lysosomal storage and associated cellular pathology. Macroautophagy is a vital cellular pathway that has been shown to be defective in other lysosomal storage disorders and suggested to be a common pathogenic mechanism. However, each of these diseases is clinically distinguishable and caused by genetic mutations in different genes. Therefore, despite lysosomal dysfunction and the observed defects in macroautophagy, the developmental pathogenesis of each disease must be unique. Unraveling the differences between these disorders, and not only the similarities, is vital to eventually understanding and treating these devastating diseases.
Supplementary Material
01
Figure 1S. Autofluorescence in _Mcoln1_-/- neuronal cultures. (A) Autofluorescence detected with 488 nm excitation in (A) Mcoln1+/+vs. (B) _Mcoln1_-/-. Arrows point to autofluorescent bodies.
02
Figure 2S. Mcoln1+/+ neuronal cultures electron microscopy. (A) Electron microscopy of post natal day 7 pup Mcoln1+/+ brain cortex (bar indicates 2 microns) (B) Electron microscopy of Mcoln1+/+ embryonic neuronal cultures after 10 days in culture (bar indicates 500 nm).
03
Figure 3S. Macroautophagy regulation in _Mcoln1_-/- mouse neuronal cultures. Neuronal cultures from Mcoln1+/+ and _Mcoln1_-/- were analyzed by western blot for expression of (A and B) Beclin-1, (C and D) phospho-mTOR and mTOR, (E and F) phospho-p70S6K (Thr 389) and p70S6K. Beclin-1 levels were determined using the average of each band densitometry value, divided by β-tubulin (n=6). P70S6K and mTOR analyses were done using the average of each band densitometry value of phospho-p70S6K and phospho-mTOR, divided by total levels of P70S6K and mTOR, respectively (n=4).
Acknowledgments
This work was supported by the National Institutes of Health [NS39995 to S.A.S., HD045561 to S.U.W.]; the Mucolipidosis Type 4 Foundation [C.C.M.]; and by the Executive Committee on Research, Massachusetts General Hospital [C.C.M.]. We thank Mr. Martin K. Selig at the Massachusetts General Hospital, Department of Pathology, for the outstanding electron microscopy work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bargal R, Avidan N, Ben-Asher E, Olender Z, Zeigler M, Frumkin A, et al. Identification of the gene causing mucolipidosis type IV. Nat Genet. 2000;26:118–23. doi: 10.1038/79095. [DOI] [PubMed] [Google Scholar]
- Bassi MT, Manzoni M, Monti E, Pizzo MT, Ballabio A, Borsani G. Cloning of the gene encoding a novel integral membrane protein, mucolipidin-and identification of the two major founder mutations causing mucolipidosis type IV. Am J Hum Genet. 2000;67:1110–20. doi: 10.1016/s0002-9297(07)62941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman ER LN, Shapira E, Merin S, Levij IS. Congenital corneal clouding with abnormal systemic storage bodies: a new variant of mucolipidosis. J Pediatr. 1974;84:519–26. doi: 10.1016/s0022-3476(74)80671-2. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin -induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35:567–76. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Cao Y, Espinola JA, Fossale E, Massey AC, Cuervo AM, MacDonald ME, et al. Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J Biol Chem. 2006;281:20483–93. doi: 10.1074/jbc.M602180200. [DOI] [PubMed] [Google Scholar]
- Crawley AC, Walkley SU. Developmental analysis of CNS pathology in the lysosomal storage disease alpha-mannosidosis. J Neuropathol Exp Neurol. 2007;66:687–97. doi: 10.1097/nen.0b013e31812503b6. [DOI] [PubMed] [Google Scholar]
- Frei KP PN, Crutchfield KE, Altarescu G, Schiffmann R. Mucolipidosis type IV: characteristic MRI findings. Neurology. 1998;51:565–9. doi: 10.1212/wnl.51.2.565. [DOI] [PubMed] [Google Scholar]
- Goldin E, Blanchette-Mackie EJ, Dwyer NK, Pentchev PG, Brady RO. Cultured skin fibroblasts derived from patients with mucolipidosis 4 are auto-fluorescent. Pediatr Res. 1995;37:687–92. doi: 10.1203/00006450-199506000-00003. [DOI] [PubMed] [Google Scholar]
- Goldin E, Stahl S, Cooney AM, Kaneski CR, Gupta S, Brady RO, et al. Transfer of a mitochondrial DNA fragment to MCOLN1 causes an inherited case of mucolipidosis IV. Hum Mutat. 2004;24:460–5. doi: 10.1002/humu.20094. [DOI] [PubMed] [Google Scholar]
- Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–87. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- Jennings JJ, Jr, Zhu JH, Rbaibi Y, Luo X, Chu CT, Kiselyov K. Mitochondrial aberrations in mucolipidosis type IV. J Biol Chem. 2006;22:39041–50. doi: 10.1074/jbc.M607982200. [DOI] [PubMed] [Google Scholar]
- Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–35. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Massey AC, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell. 2008;19:2179–92. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A, Al-Sarraj S, Shaw C. Frontotemporal lobar degeneration with ubiquitinated tau-negative inclusions and additional alpha-synuclein pathology but also unusual cerebellar ubiquitinated p62-positive, TDP-43-negative inclusions. Neuropathology. 2009;29:466–71. doi: 10.1111/j.1440-1789.2008.00966.x. [DOI] [PubMed] [Google Scholar]
- Kiselyov K, Chen J, Rbaibi Y, Oberdick D, Tjon-Kon-Sang S, Shcheynikov N, et al. TRP-ML1 is a lysosomal monovalent cation channel that undergoes proteolytic cleavage. J Biol Chem. 2005;280:43218–23. doi: 10.1074/jbc.M508210200. [DOI] [PubMed] [Google Scholar]
- Kiselyov K, Soyombo A, Muallem S. TRPpathies. J Physiol. 2007;578:641–53. doi: 10.1113/jphysiol.2006.119024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M, Nakanishi H, Saftig P, Ezaki J, Isahara K, Ohsawa Y, et al. Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J Neurosci. 2000;20:6898–906. doi: 10.1523/JNEUROSCI.20-18-06898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, et al. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroidlipofuscinoses (Batten disease) Am J Pathol. 2005;167:1713–28. doi: 10.1016/S0002-9440(10)61253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC. A novel link between autophagy and the ubiquitin-proteasome system. Autophagy. 2009;5:862–3. doi: 10.4161/auto.8840. [DOI] [PubMed] [Google Scholar]
- Kuusisto E, Parkkinen L, Alafuzoff I. Morphogenesis of Lewy bodies: dissimilar incorporation of alpha-synuclein, ubiquitin, and p62. J Neuropathol Exp Neurol. 2003;62:1241–53. doi: 10.1093/jnen/62.12.1241. [DOI] [PubMed] [Google Scholar]
- Kuusisto E, Salminen A, Alafuzoff I. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport. 2001;12:2085–90. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- LaPlante JM, Falardeau J, Sun M, Kanazirska M, Brown EM, Slaugenhaupt SA, et al. Identification and characterization of the single channel function of human mucolipin-1 implicated in mucolipidosis type IV, a disorder affecting the lysosomal pathway. FEBS Lett. 2002;532:183–7. doi: 10.1016/s0014-5793(02)03670-0. [DOI] [PubMed] [Google Scholar]
- LaPlante JM, Sun M, Falardeau J, Dai D, Brown EM, Slaugenhaupt SA, et al. Lysosomal exocytosis is impaired in mucolipidosis type IV. Mol Genet Metab. 2006;89:339–48. doi: 10.1016/j.ymgme.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Larsen KE, Sulzer D. Autophagy in neurons: a review. Histol Histopathol. 2002;17:897–908. doi: 10.14670/HH-17.897. [DOI] [PubMed] [Google Scholar]
- Lesuisse C, Martin LJ. Long-term culture of mouse cortical neurons as a model for neuronal development, aging, and death. J Neurobiol. 2002;51:9–23. doi: 10.1002/neu.10037. [DOI] [PubMed] [Google Scholar]
- Manzoni M, Monti E, Bresciani R, Bozzato A, Barlati S, Bassi MT, et al. Overexpression of wild-type and mutant mucolipin proteins in mammalian cells: effects on the late endocytic compartment organization. FEBS Lett. 2004;567:219–24. doi: 10.1016/j.febslet.2004.04.080. [DOI] [PubMed] [Google Scholar]
- Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2006;103:5805–10. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micsenyi MC, Dobrenis K, Stephney G, Pickel J, Vanier MT, Slaugenhaupt SA, et al. Neuropathology of the Mcoln1(-/-) knockout mouse model of mucolipidosis type IV. J Neuropathol Exp Neurol. 2009;68:125–35. doi: 10.1097/NEN.0b013e3181942cf0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedel MT, Weixel KM, Bruns JR, Traub LM, Weisz OA. Posttranslational cleavage and adaptor protein complex-dependent trafficking of mucolipin-1. J Biol Chem. 2006;281:12751–9. doi: 10.1074/jbc.M511104200. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–5. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Nedelsky N, PK Todd, Taylor JP. Autophagy and the ubiquitin-proteasome system: Collaborators in neuroprotection. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbadis.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA, Yang DS, Lee JH. Neurodegenerative lysosomal disorders: a continuum from development to late age. Autophagy. 2008;4:590–9. doi: 10.4161/auto.6259. [DOI] [PubMed] [Google Scholar]
- Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: Molecular mechanisms and physiological relevance. Semin Cell Dev Biol. 2010 Feb;:20. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco CD, Kunkel R, Lieberman AP. Autophagy in Niemann-Pick C disease is dependent upon Beclin-1 and responsive to lipid trafficking defects. Hum Mol Genet. 2007;16:1495–503. doi: 10.1093/hmg/ddm100. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Pryor PR, Reimann F, Gribble FM, Luzio JP. Mucolipin-1 is a lysosomal membrane protein required for intracellular lactosylceramide traffic. Traffic. 2006;7:1388–98. doi: 10.1111/j.1600-0854.2006.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raben N, Baum R, Schreiner C, Takikita S, Mizushima N, Ralston E, et al. When more is less: excess and deficiency of autophagy coexist in skeletal muscle in Pompe disease. Autophagy. 2009a;5:111–3. doi: 10.4161/auto.5.1.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raben N, Shea L, Hill V, Plotz P. Monitoring autophagy in lysosomal storage disorders. Methods Enzymol. 2009b;453:417–49. doi: 10.1016/S0076-6879(08)04021-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rami A. Review: autophagy in neurodegeneration: firefighter and/or incendiarist? Neuropathol Appl Neurobiol. 2009;35:449–61. doi: 10.1111/j.1365-2990.2009.01034.x. [DOI] [PubMed] [Google Scholar]
- Reis SA, Oostra BA, Willemsen R. Isolation of mouse neuritic mRNAs. J Mol Histol. 2006;37:79–86. doi: 10.1007/s10735-006-9036-7. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, Kaushik S, et al. In search of an “autophagomometer”. Autophagy. 2009;5:585–9. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, et al. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008a;17:119–29. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Rubinsztein DC, Ballabio A. Lysosomal storage diseases as disorders of autophagy. Autophagy. 2008b;4:113–4. doi: 10.4161/auto.5227. [DOI] [PubMed] [Google Scholar]
- Sun M, Goldin E, Stahl S, Falardeau JL, Kennedy JC, Acierno JS, Jr, et al. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum Mol Genet. 2000;9:2471–8. doi: 10.1093/hmg/9.17.2471. [DOI] [PubMed] [Google Scholar]
- Tanemura K, Murayama M, Akagi T, Hashikawa T, Tominaga T, Ichikawa M, et al. Neurodegeneration with tau accumulation in a transgenic mouse expressing V337M human tau. J Neurosci. 2002;22:133–41. doi: 10.1523/JNEUROSCI.22-01-00133.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Ueno T, Kominami E. LC3 and Autophagy. Methods Mol Biol. 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- Thompson EG, Schaheen L, Dang H, Fares H. Lysosomal trafficking functions of mucolipin-1 in murine macrophages. BMC Cell Biol. 2007;8:54. doi: 10.1186/1471-2121-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam K, Hofmann T, Montell C. Lysosomal localization of TRPML3 depends on TRPML2 and the mucolipidosis-associated protein TRPML1. J Biol Chem. 2006;281:17517–27. doi: 10.1074/jbc.M600807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam K, Long AA, Elsaesser R, Nikolaeva D, Broadie K, Montell C. Motor deficit in a Drosophila model of mucolipidosis type IV due to defective clearance of apoptotic cells. Cell. 2008;135:838–51. doi: 10.1016/j.cell.2008.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventruti A, Cuervo AM. Autophagy and neurodegeneration. Curr Neurol Neurosci Rep. 2007;7:443–51. doi: 10.1007/s11910-007-0068-5. [DOI] [PubMed] [Google Scholar]
- Venugopal B, Browning MF, Curcio-Morelli C, Varro A, Michaud N, Nanthakumar N, et al. Neurologic, Gastric, and Opthalmologic Pathologies in a Murine Model of Mucolipidosis Type IV. Am J Hum Genet. 2007;81:1070–83. doi: 10.1086/521954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal B, Mesires NT, Kennedy JC, Curcio-Morelli C, Laplante JM, Dice JF, et al. Chaperone-mediated autophagy is defective in mucolipidosis type IV. J Cell Physiol. 2009;219:344–53. doi: 10.1002/jcp.21676. [DOI] [PubMed] [Google Scholar]
- Vergarajauregui S, Connelly PS, Daniels MP, Puertollano R. Autophagic dysfunction in mucolipidosis type IV patients. Hum Mol Genet. 2008;17:2723–37. doi: 10.1093/hmg/ddn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergarajauregui S, Puertollano R. Two di-leucine motifs regulate trafficking of mucolipin-1 to lysosomes. Traffic. 2006;7:337–53. doi: 10.1111/j.1600-0854.2006.00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZH, Zeng B, Pastores GM, Raksadawan N, Ong E, Kolodny EH. Rapid detection of the two common mutations in Ashkenazi Jewish patients with mucolipidosis type IV. Genet Test. 2002;5:87–92. doi: 10.1089/109065701753145529. [DOI] [PubMed] [Google Scholar]
- Wooten MW, Hu X, Babu JR, Seibenhener ML, Geetha T, Paine MG, et al. Signaling, Polyubiquitination, Trafficking, and Inclusions: Sequestosome 1/p62's Role in Neurodegenerative Disease. J Biomed Biotechnol. 2006;2006:62079. doi: 10.1155/JBB/2006/62079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
Figure 1S. Autofluorescence in _Mcoln1_-/- neuronal cultures. (A) Autofluorescence detected with 488 nm excitation in (A) Mcoln1+/+vs. (B) _Mcoln1_-/-. Arrows point to autofluorescent bodies.
02
Figure 2S. Mcoln1+/+ neuronal cultures electron microscopy. (A) Electron microscopy of post natal day 7 pup Mcoln1+/+ brain cortex (bar indicates 2 microns) (B) Electron microscopy of Mcoln1+/+ embryonic neuronal cultures after 10 days in culture (bar indicates 500 nm).
03
Figure 3S. Macroautophagy regulation in _Mcoln1_-/- mouse neuronal cultures. Neuronal cultures from Mcoln1+/+ and _Mcoln1_-/- were analyzed by western blot for expression of (A and B) Beclin-1, (C and D) phospho-mTOR and mTOR, (E and F) phospho-p70S6K (Thr 389) and p70S6K. Beclin-1 levels were determined using the average of each band densitometry value, divided by β-tubulin (n=6). P70S6K and mTOR analyses were done using the average of each band densitometry value of phospho-p70S6K and phospho-mTOR, divided by total levels of P70S6K and mTOR, respectively (n=4).