Phosphorylated ubiquitin chain is the genuine Parkin receptor (original) (raw)
PINK1-phosphorylated ubiquitin chain is the genuine Parkin receptor that recruits Parkin to depolarized mitochondria.
Abstract
PINK1 selectively recruits Parkin to depolarized mitochondria for quarantine and removal of damaged mitochondria via ubiquitylation. Dysfunction of this process predisposes development of familial recessive Parkinson’s disease. Although various models for the recruitment process have been proposed, none of them adequately explain the accumulated data, and thus the molecular basis for PINK1 recruitment of Parkin remains to be fully elucidated. In this study, we show that a linear ubiquitin chain of phosphomimetic tetra-ubiquitin(S65D) recruits Parkin to energized mitochondria in the absence of PINK1, whereas a wild-type tetra-ubiquitin chain does not. Under more physiologically relevant conditions, a lysosomal phosphorylated polyubiquitin chain recruited phosphomimetic Parkin to the lysosome. A cellular ubiquitin replacement system confirmed that ubiquitin phosphorylation is indeed essential for Parkin translocation. Furthermore, physical interactions between phosphomimetic Parkin and phosphorylated polyubiquitin chain were detected by immunoprecipitation from cells and in vitro reconstitution using recombinant proteins. We thus propose that the phosphorylated ubiquitin chain functions as the genuine Parkin receptor for recruitment to depolarized mitochondria.
Introduction
Genetic studies on the hereditary form of Parkinson’s disease have identified genes relevant to disease pathogenesis. PTEN-induced putative kinase 1 (PINK1; also known as PARK6) and PARKIN (also known as PARK2) have been identified as the causal genes responsible for hereditary recessive early onset Parkinsonism (Kitada et al., 1998; Valente et al., 2004). To date, there is significant evidence supporting a functional link between PINK1, Parkin, and mitochondrial quality control. PINK1 is a serine/threonine kinase that specifically accumulates on and is activated by mitochondria with a decreased membrane potential (membrane potential = ΔΨm; Matsuda et al., 2010; Narendra et al., 2010b; Okatsu et al., 2012b, 2013). PINK1 is an upstream factor of Parkin (Clark et al., 2006; Park et al., 2006; Yang et al., 2006) that activates the latent ubiquitin ligase (E3) activity of Parkin (Matsuda et al., 2010) and recruits it to depolarized mitochondria (Geisler et al., 2010; Narendra et al., 2010b; Vives-Bauza et al., 2010; Ziviani et al., 2010). Parkin then catalyzes ubiquitin transfer from ubiquitin-charged E2 enzymes (UBE2A, UBE2N, UBE2L3, or UBE2D2/3) to various substrates on depolarized mitochondria (Tanaka et al., 2010; Chan et al., 2011; Haddad et al., 2013; Sarraf et al., 2013; Fiesel et al., 2014; Geisler et al., 2014). As a consequence, inferior mitochondria with low ΔΨm are quarantined and degraded via the proteasome and autophagy (Narendra et al., 2008; Matsuda et al., 2010; Okatsu et al., 2010; Chan et al., 2011; Yoshii et al., 2011). Mechanistic insights into PINK1-mediated Parkin activation have recently been revealed. Parkin is an intramolecular autoinhibitory E3 (Chaugule et al., 2011; Chew et al., 2011) that usually has its catalytic Cys431 core occluded by a RING0 domain (Riley et al., 2013; Trempe et al., 2013; Wauer and Komander, 2013). PINK1 phosphorylation of Ser65 in the both ubiquitin-like domain of Parkin and ubiquitin triggers removal of Parkin autoinhibition by phosphorylated ubiquitin, which then results in the conversion of phosphorylated Parkin to the fully active form (Kondapalli et al., 2012; Shiba-Fukushima et al., 2012; Iguchi et al., 2013; Kane et al., 2014; Kazlauskaite et al., 2014; Koyano et al., 2014).
Although significant progress has been made in elucidating the Parkin activation mechanism, the molecular basis for Parkin recruitment to depolarized mitochondria by PINK1 remains obscure. PINK1-phosphorylated mitofusin was recently reported as a Parkin receptor on damaged mitochondria (Chen and Dorn, 2013); however, the recruitment of Parkin to depolarized mitochondria and acceleration of mitochondrial degradation equivalent to wild-type (WT) cells in mitofusin1/2 double knockout (KO) MEFs seem to contradict this mitofusin receptor model (Narendra et al., 2008; Chan et al., 2011). Moreover, other data on Parkin translocation are difficult to interpret using this hypothesis. The catalytically inactive Parkin C431S mutant results in a dead-end intermediate via ubiquitin-oxyester conjugation on Ser431 (Iguchi et al., 2013; Lazarou et al., 2013). Parkin(C431S) is thus folded correctly but dysfunctional in E3, and it fails to translocate to depolarized mitochondria, which suggests that the ubiquitin ligase activity of Parkin is required for mitochondrial translocation (Lazarou et al., 2013; Zheng and Hunter, 2013). Under these conditions, we have no consensus on whether phosphorylated mitofusin is the genuine Parkin receptor on depolarized mitochondria. Thus the largest unresolved issue in this field at present is to elucidate the mechanism by which Parkin is recruited to damaged mitochondria. Here we report that a PINK1 phosphorylated ubiquitin chain is the genuine Parkin receptor. This proposal enables us to reasonably explain many aspects of Parkin recruitment.
Results
K63- and K48-linked polyubiquitin chains are phosphorylated by PINK1
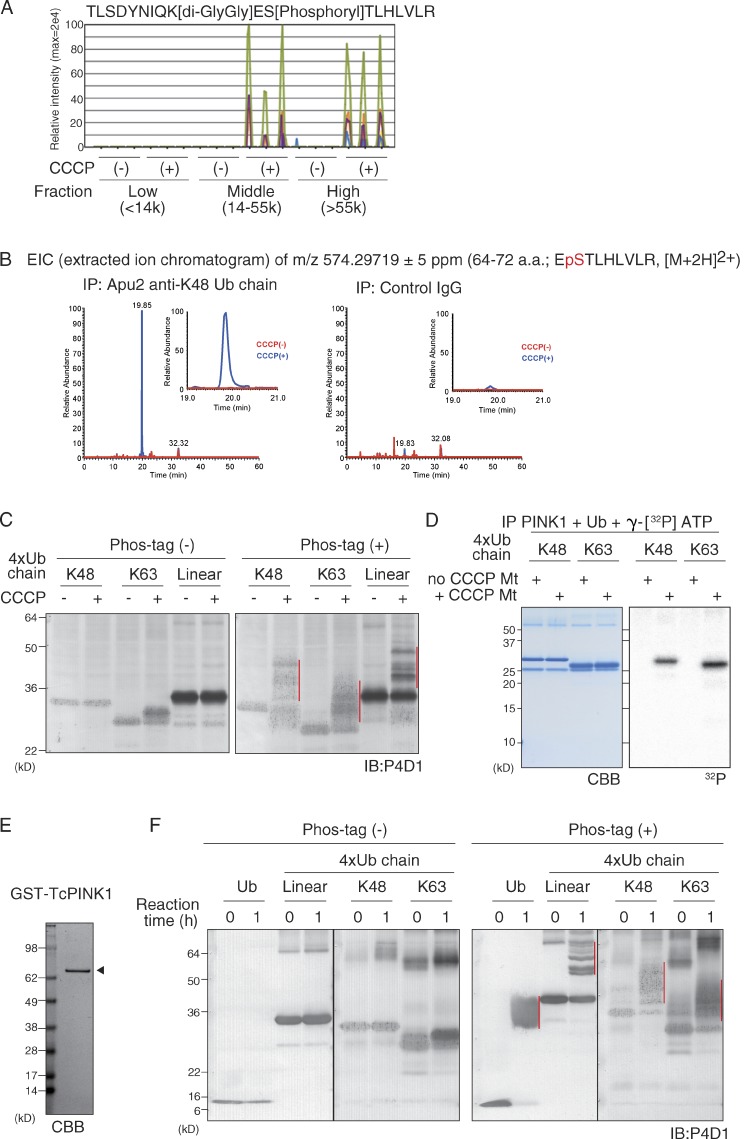
In our previous paper, we showed that phosphorylated ubiquitin lacking the C-terminal diglycine motif, which is crucial for conjugation to the substrate and polyubiquitin chain formation, remains capable of activating Parkin E3 activity (Koyano et al., 2014). This result indicates that neither polyubiquitin chain formation nor substrate conjugation of phosphorylated ubiquitin is required for Parkin activation. Nevertheless, when the absolute level of phosphorylated ubiquitin in cell lysates was determined by mass spectrometry (MS) analysis, a significant amount of phosphorylated ubiquitin was detected in the middle (14,000–55,000) and the high (>55,000) molecular weight fractions (Koyano et al., 2014). Because ubiquitin is a small protein (∼9 kD), it is reasonable to assume that the aforementioned signal was derived from substrate-conjugated phosphorylated ubiquitin and/or ubiquitin chain containing phosphorylated ubiquitin. We thus examined whether the phosphorylated ubiquitin chain exists in cells after mitochondrial uncoupler (carbonyl cyanide m-chlorophenylhydrazine [CCCP]) treatment. The major polyubiquitin chain is constituted via ubiquitin–ubiquitin conjugation on Lys48 (K48) or Lys63 (K63). Because the position of ubiquitin phosphorylation (S65) is very close to K63, we can directly verify and analyze incorporation of a phosphate in the K63-linked polyubiquitin chain by MS analysis. When we searched the MS data for a peptide signal corresponding to both S65 phosphorylation and a K63-GlyGly branch, which is a vestige of K63-linked polyubiquitylation, the signal was detected in the high and the middle molecular weight fractions of lysates prepared from CCCP-treated cells in three independent experiments (Fig. 1 A). This signal was absent in control cells not treated with CCCP and the low (<14,000) molecular weight fraction of CCCP-treated cells (Fig. 1 A). In contrast, the MS signal derived from unmodified ubiquitin, S65-phosphoryated ubiquitin without the K63-GlyGly branch, or a K63-linked chain-forming nonphosphorylated ubiquitin was observed in all fractions, CCCP-treated fractions, and the high and middle molecular weight fractions, respectively (Fig. S1, A–C). We thus confidently concluded that the K63-linked polyubiquitin chain is phosphorylated only in CCCP-treated cells.
Figure 1.
Detection of a PINK1 phosphorylated ubiquitin chain in cells after a decrease in ΔΨm. (A) Mass-spectrometric (MS) analysis identified peptides with a phosphorylated S65 and a K63-GlyGly branch in the middle (14,000–55,000) and high (>55,000), but not low (<14,000), molecular weight fractions of cell lysates after CCCP treatment. The data shown are from a single MS analysis of three independently prepared samples. (B) The extracted m/z 574.29719 ion chromatogram corresponds to the doubly charged ubiquitin phosphopeptide EpSTLHLVLR, which was identified in immunoprecipitates using an Apu2 anti-K48–linked polyubiquitin chain antibody but not control IgG. This experiment was completed once (n = 1). (C) Retarded-mobility bands corresponding to K48-linked, K63-linked, and linear tetra-ubiquitin chains (red vertical lines) were observed in Phos-tag PAGE only after incubation with mitochondria isolated from CCCP-treated cells. P4D1, anti-ubiquitin antibody. IB, immunoblotting. (D) Incorporation of 32P in the recombinant K48-linked and K63-linked tetra-ubiquitin chains was specifically detected when incubated with immunoprecipitated PINK1 (IP-PINK1) from digitonin-solubilized depolarized mitochondria (+ CCCP Mt). (E and F) _Tc_PINK1 (indicated by the arrowhead in E) purified from E. coli was incubated with mono-ubiquitin or linear, K48-linked, and K63-linked tetra-ubiquitin chains. Retarded-mobility bands for all of the ubiquitin chains (F; red vertical lines) were observed in Phos-tag PAGE. Note that mobility does not reflect the molecular weight of proteins in Phos-tag PAGE (Kinoshita et al., 2012), and thus molecular weight markers are not shown. Black lines indicate that intervening lanes have been spliced out.
It is difficult to demonstrate phosphorylation in K48-linked polyubiquitin chains by MS analysis because a long peptide harboring both the S65 phosphorylation and the K48-GlyGly branch are not detected. As an alternative approach, we immunoprecipitated K48-linked polyubiquitin chains using a linkage-specific ubiquitin antibody, Apu2 (Newton et al., 2008), and examined the immunoprecipitated product by MS analysis for the S65-phosphorylated peptide. The MS signal derived from a peptide with the K48-GlyGly branch was detected in the high-molecular-weight fractions of Apu2 immunoprecipitates but not in control IgG immunoprecipitates, which indicates successful immunoprecipitation of K48-linked polyubiquitin chains (Fig. S1, D and E). The S65-phosphorylated peptide was detected only in Apu2 immunoprecipitates from CCCP-treated cells (Fig. 1 B), which suggests that K48-linked polyubiquitin chain is also phosphorylated after CCCP treatment in cells.
There are two possible mechanisms that generate phosphorylated K63-linked and K48-linked ubiquitin chains in cells: free ubiquitin is phosphorylated by PINK1 and then incorporated into the ubiquitin chain by E3s, or the ubiquitin chain itself is directly phosphorylated by PINK1. We thus examined whether PINK1 phosphorylates the polyubiquitin chain. To detect potentially phosphorylated ubiquitin chains, phosphate-affinity (Phos-tag) PAGE in which the phosphorylated form of proteins can be easily distinguished from the nonphosphorylated form as a slower migrating band was used (Kinoshita et al., 2006). When recombinant K48-linked, linear, and K63-linked tetra-ubiquitin chains were incubated with isolated mitochondria from CCCP-treated or untreated cells, retarded-mobility bands of all ubiquitin chains were observed only with the CCCP-treated mitochondria (Fig. 1 C). To demonstrate PINK1-catalyzed phosphorylation of the ubiquitin chain more convincingly, PINK1 was immunoprecipitated from digitonin-solubilized mitochondria (referred to as IP-PINK1 hereafter), and recombinant K48-linked and K63-linked tetra-ubiquitin chains were incubated with IP-PINK1 and γ-[32P]ATP. Incorporation of 32P in the recombinant ubiquitin chains was specifically detected by IP-PINK1 from depolarized mitochondria (Fig. 1 D). Moreover, when recombinant Tribolium castaneum PINK1 (TcPINK1; Woodroof et al., 2011) was purified from Escherichia coli (Fig. 1 E) and incubated with K48-linked, linear, and K63-linked tetra-ubiquitin chains, retarded-mobility bands of all tetra-ubiquitin chains was observed in Phos-tag PAGE (Fig. 1 F). We thus concluded that ubiquitin chains are directly phosphorylated by PINK1, although it remains unclear whether PINK1-phosphorylated free ubiquitin could be incorporated into the ubiquitin chain by E3s.
Catalytically active Parkin restores the mitochondrial localization of a catalytically inactive Parkin mutant in trans
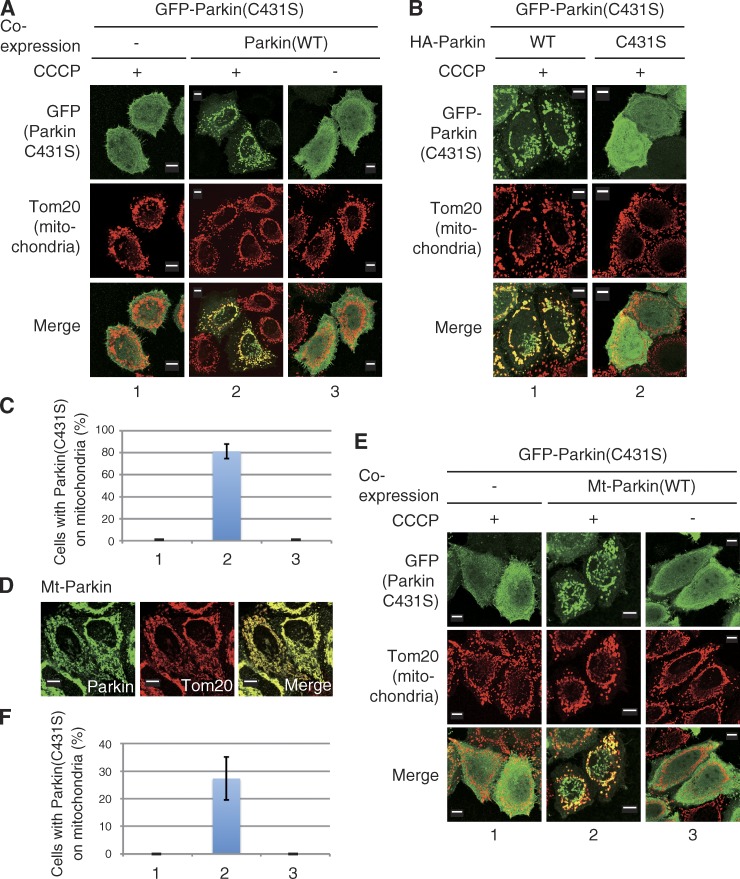
Parkin catalyzes ubiquitylation via a unique “RING/HECT hybrid” mechanism. To function as an E3, Parkin generates an intermediate form via a ubiquitin-thioester conjugation at Cys431 (C431), the catalytic core of Parkin (Wenzel et al., 2011; Iguchi et al., 2013; Lazarou et al., 2013; Riley et al., 2013; Zheng and Hunter, 2013). Two research groups reported previously that Parkin harboring a C431 mutation (such as C431S) failed to translocate to depolarized mitochondria, whereas coexpression of WT Parkin or a Parkin R275W mutant (a unique pathogenic mutant that exhibits normal localization on depolarized mitochondria but has an ∼50% decrease in substrate ubiquitylation and p62 recruitment; Okatsu et al., 2010; Lazarou et al., 2013) could rescue the translocation defect of the Parkin(C431S) mutant in trans (Lazarou et al., 2013; Zheng and Hunter, 2013). Lazarou et al. (2013) hypothesized that self-association activity of Parkin allowed the C431S mutant to bind the R275W Parkin mutant and translocate with it onto depolarized mitochondria (Lazarou et al., 2013). We confirmed that a GFP-Parkin(C431S) mutant did not translocate to depolarized mitochondria after 1 h of treatment with CCCP, whereas coexpression of WT Parkin clearly complemented mislocalization of C431S in the presence of CCCP (Fig. 2, A and C). Coexpression of the mutant Parkin(C431S) did not complement the GFP-Parkin(C431S) localization defect; this excludes the possibility that complementation is a mass action phenomenon or somehow requires Parkin lacking a GFP tag (Fig. 2 B). To examine the self-association–mediated mitochondrial localization of Parkin(C431S), we next constructed a mitochondria-targeting Parkin (Mt-Parkin) that artificially localizes on the outer mitochondrial membrane (OMM). MitoNEET is an integral OMM protein with an N-terminal anchor sequence that tethers the protein to the OMM such that it is oriented toward the cytoplasm (Wiley et al., 2007). We thus fused the membrane-anchoring domain of MitoNEET to the N terminus of Parkin to generate Mt-Parkin. Although Mt-Parkin clearly localizes on mitochondria (Fig. 2 D), Parkin(C431S) was not transported to mitochondria by Mt-Parkin alone, and CCCP treatment was still required for mitochondrial localization of Parkin(C431S) (Fig. 2, E and F). We propose two mechanisms to explain these data. The first posits that intermolecular self-association of Parkin is mediated by PINK1, thus CCCP treatment is a prerequisite for mitochondrial localization of Parkin(C431S), as suggested by Lazarou et al. (2013). An alternative mechanism is that WT Parkin-catalyzed ubiquitylation of mitochondria is involved in Parkin(C431S) recruitment to the mitochondria. This second mechanism has also been suggested by Zheng and Hunter (2013). Because the latter model can logically explain why Parkin localization on depolarized mitochondria is impaired by E3 activity–deficient mutations or a decrease in ubiquitin-conjugating enzyme (E2s) activity (Haddad et al., 2013; Hasson et al., 2013; Fiesel et al., 2014), we further investigated the latter possibility.
Figure 2.
WT Parkin complements the mislocalization of a catalytically inactive Parkin mutant in trans. (A) HeLa cells expressing GFP-Parkin(C431S) (catalytically inactive mutant) with or without WT HA-Parkin were treated with CCCP (10 µM, 1.5 h), and immunostained with anti-GFP and anti-Tom20 antibodies. (B) Coexpression of HA-Parkin(C431S) did not complement the localization defect of GFP-Parkin(C431S). (C) The number of cells with GFP-Parkin(C431S) localized to the mitochondria was counted in 100 cells. Numbers on the horizontal axis correspond to those of A. Bars represent the mean ± SD values of three experiments (error bars). (D) Mitochondrial localization of Mt-Parkin (Parkin fused with a MitoNEET mitochondrial outer membrane–targeting signal). (E) Cells expressing GFP-Parkin(C431S) (catalytically inactive mutant) with or without Mt-Parkin were treated with CCCP (10 µM, 1 h), and immunostained with anti-GFP and anti-Tom20 antibodies. (F) The number of cells with GFP-Parkin(C431S) localized to the mitochondria was counted as in C. Numbers on the horizontal axis correspond to those of E. Bars, 10 µm.
A mitochondrial localized ubiquitin chain recruits Parkin to energized mitochondria
Previous assays using linkage-specific anti-polyubiquitin chain antibodies (Okatsu et al., 2010), a linkage-specific ubiquitin chain binding probe (van Wijk et al., 2012; Chen et al., 2013), and MS analyses (Chan et al., 2011) suggested that Parkin catalyzes the formation of both K48- and K63-linked poly-ubiquitin chains to substrates on depolarized mitochondria, but preferentially catalyzes formation of the K63-linked polyubiquitin chain. Exogenous expression of ubiquitin variants also suggested that the K27- and K63-linked ubiquitin chains are formed on depolarized mitochondria by Parkin (Geisler et al., 2010). We revealed that PINK1 phosphorylates ubiquitin chains (Fig. 1), and Parkin-catalyzed ubiquitylation on mitochondria is implicated in the recruitment of Parkin(C431S), an E3-negative mutant (Fig. 2). We thus hypothesized that the phosphorylated ubiquitin chain is the Parkin receptor on depolarized mitochondria.
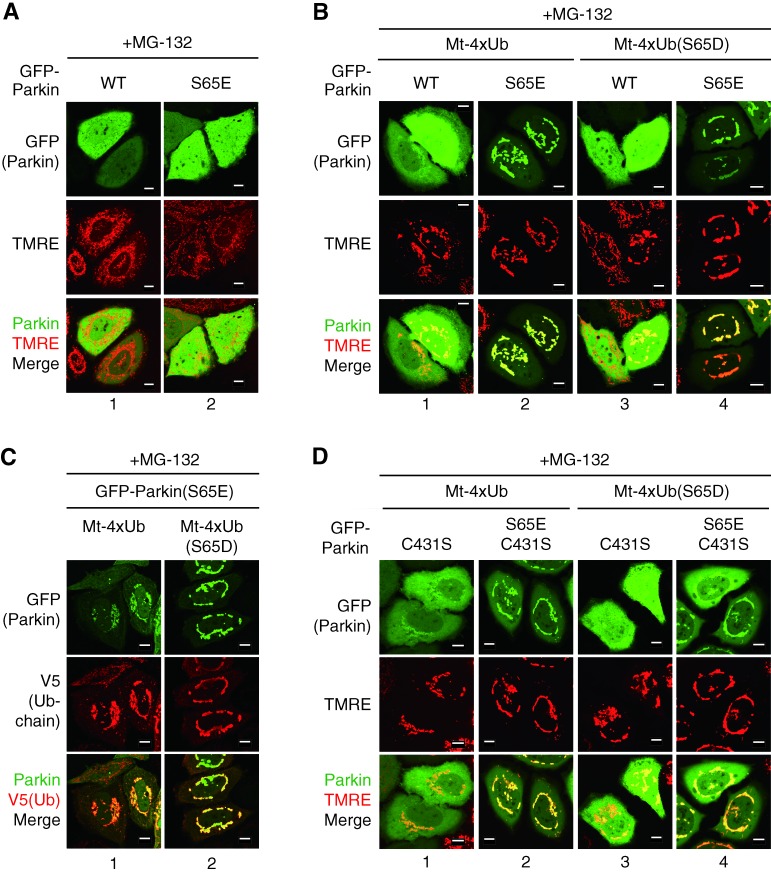
The tertiary structure of the K63-linked ubiquitin chain resembles a linear ubiquitin chain (Komander et al., 2009), thus we examined whether linear ubiquitin chains on normal mitochondria, with or without phosphomimetic mutations, recruit Parkin. To prevent cleavage by ubiquitin processing enzymes, a G76V mutation was introduced into ubiquitin. We tandemly arranged the mitochondria-targeting domain of Tom20, a V5-tag, and four copies of ubiquitin(G76V) to make a mitochondria-localized linear ubiquitin chain (referred hereafter as Mt-4×Ub). Mt-4×Ub, however, was poorly detected in cells using either immunoblotting or immunocytochemistry. Because the K63-linked ubiquitin chain, which structurally resembles the tandem ubiquitin chain used in this study, potentially serves as a targeting signal for the 26S proteasome (Saeki et al., 2009), we tried proteasomal inhibition to stabilize Mt-4×Ub. Treatment with the proteasome inhibitor MG132 restored and stabilized the expression of both 4×Ub (Fig. S2 A, lanes 1 and 2) and Mt-4×Ub (lanes 5 and 6) as well as other cellular ubiquitin conjugates (Fig. S2 B). The distribution of Mt-4×Ub to the mitochondria was confirmed in both immunocytochemistry (Fig. S2 C, 1 and 2) and fractionation experiments (Fig. S2 D, lanes 1 and 2). Similarly, we constructed a mitochondria-targeting tetra phosphomimetic ubiquitin chain (referred hereafter as Mt-4×Ub(S65D)) that also localizes on mitochondria after MG132 treatment (Fig. S2, C and D). We then determined if the mitochondria-localized ubiquitin chains could recruit Parkin to normal (energized) mitochondria. Because Ser65 of Parkin is phosphorylated by PINK1 after a reduction in ΔΨm (Kondapalli et al., 2012; Shiba-Fukushima et al., 2012; Iguchi et al., 2013), we constructed both WT and a phosphomimetic mutant (S65E) of Parkin. MG132 treatment alone did not alter the dispersed cytosolic localization of GFP-Parkin(WT) or GFP-Parkin(S65E) (Fig. 3 A), and the subcellular localization of GFP-Parkin(WT) did not change when Mt-4×Ub or Mt-4×Ub(S65D) were coexpressed in cells (Fig. 3 B, 1 and 3), even though the ubiquitin chain was expressed (Fig. S3 A). Expression of Mt-4×Ub in cells caused a tendency to perinuclear accumulation of mitochondria (Fig. 3 B), as reported previously (Okatsu et al., 2010). In stark contrast, phosphomimetic GFP-Parkin(S65E) localized on energized (tetramethylrhodamine ethyl ester [TMRE]-stainable] mitochondria when coexpressed with Mt-4×Ub and Mt-4×Ub(S65D) after MG132 treatment (Fig. 3 B, 2 and 4). Colocalization of phosphomimetic Parkin and the mitochondrial ubiquitin chain was confirmed by immunocytochemistry (Fig. 3 C). We next examined whether a mitochondria-targeting linear ubiquitin chain was capable of recruiting catalytically inactive Parkin(C431S) to mitochondria. The cytosolic localization of GFP-Parkin(C431S) did not change in the presence of Mt-4×Ub or Mt-4×Ub(S65D) in cells (Fig. 3 D, 1 and 3) and did not overlap with the ubiquitin chain (Fig. S3 A). We previously reported that Ser65 in the Parkin(C431S) mutant is phosphorylated after CCCP treatment (Iguchi et al., 2013). We thus constructed a catalytically inactive phosphomimetic Parkin mutant, GFP-Parkin(S65E/C431S), that localized on mitochondria when coexpressed with Mt-4×Ub and Mt-4×Ub(S65D) (Fig. 3 D, 2 and 4). Mitochondria in all of the expression studies were stained with TMRE (a ΔΨm-dependent dye), which suggests that ΔΨm is conserved. Colocalization of Parkin(S65E/C431S) and the mitochondrial ubiquitin chain was also confirmed (Fig. S3 B, 2 and 4). All data shown in Fig. 3 suggest that the mitochondrial polyubiquitin chain recruited phosphorylated Parkin from the cytosol to the mitochondria.
Figure 3.
Ubiquitin chains recruit phosphomimetic Parkin to energized mitochondria in cells. (A) HeLa cells expressing GFP chimeras of WT Parkin or a phosphomimetic (S65E) mutant were treated with MG-132 (10 µM, 3 h), stained with the ΔΨm-dependent dye TMRE, and observed with a fluorescence microscope. (B) HeLa cells coexpressing GFP chimeras of WT Parkin or a phosphomimetic (S65E) mutant with a WT or phosphomimetic (S65D) Mt-4×Ub (tandem tetra-ubiquitin fused with a targeting signal for mitochondrial outer membrane) were observed as in A. (C) Cells expressing GFP-Parkin(S65E) with WT or phosphomimetic (S65D) Mt-4×Ub were subjected to immunocytochemistry using an anti-V5 antibody that detects Mt-4×Ub. (D) HeLa cells coexpressing the indicated GFP-Parkin mutants and Mt-4×Ub were treated with MG-132 (10 µM, 3 h), stained with TMRE, and observed with a fluorescence microscope. Bars, 10 µm.
Only the phosphomimetic ubiquitin chain can recruit Parkin to the mitochondria
Fig. 3 suggests that phosphorylated Parkin interacts with the polyubiquitin chain regardless of the phosphorylation status of ubiquitin. However, that interpretation may be tenuous, as ubiquitin chain formation is integral to many cellular processes such as proteasomal degradation, DNA repair response, membrane trafficking, and the nuclear factor κB (NF-κB) pathway. According to the model, if Parkin is phosphorylated by PINK1, phosphorylated Parkin might be recruited to the other cellular pathways involved in ubiquitin chain formation.
We speculated that PINK1 had nothing to do with the Parkin recruitment experiments (Fig. 3) because PINK1 is degraded on energized mitochondria in a ΔΨm-dependent manner (Matsuda et al., 2010; Narendra et al., 2010b), and maintenance of ΔΨm was confirmed by the potentiometric dye TMRE (Fig. 3, B and D). However, cells were treated with MG132 to stabilize the mitochondrial polyubiquitin chains (Fig. S2). MG132 inhibits the final step in PINK1 degradation by the N-end rule pathway and causes accumulation of the cleaved form of PINK1 (Narendra et al., 2010b; Yamano and Youle, 2013). To rule out the possible effect of PINK1 completely, we generated a PINK1 KO HeLa cell line by clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9-mediated genome editing (Cong et al., 2013). HeLa cells transfected with plasmids for PINK1 KO by the CRISPR/Cas9 system were subjected to single clone selection, of which we obtained four clonal cells, identified as #1–4 (Fig. S4 A). Among the putative _PINK1_-KO HeLa cells, two clones (clone #1 and #4) showed complete inhibition of GFP-Parkin translocation and autoubiquitylation (Fig. S4, A and B, lanes 2 and 5). Endogenous PINK1 was undetectable in the PINK1 KO cell lines (Fig. S4 B), and genomic deletion of PINK1 was confirmed by direct sequencing (not depicted). Furthermore, complementation of exogenous PINK1 in clone #4 restored the translocation and autoubiquitylation of GFP-Parkin to WT levels (Fig. S4, C–E). We thus used this clone in all subsequent experiments.
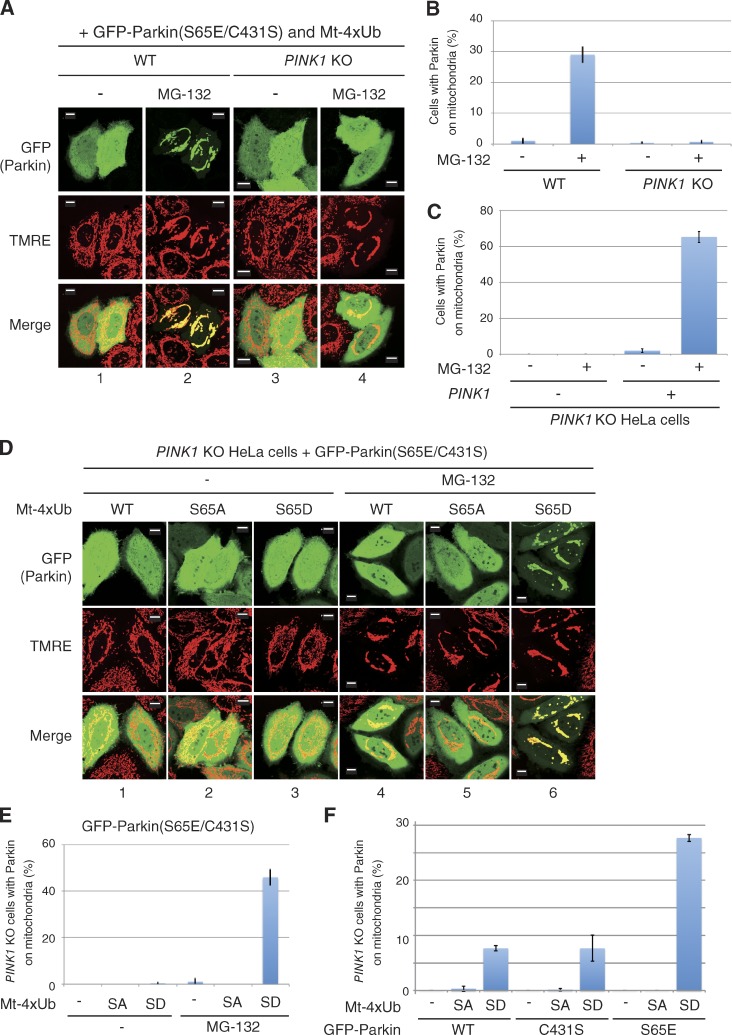
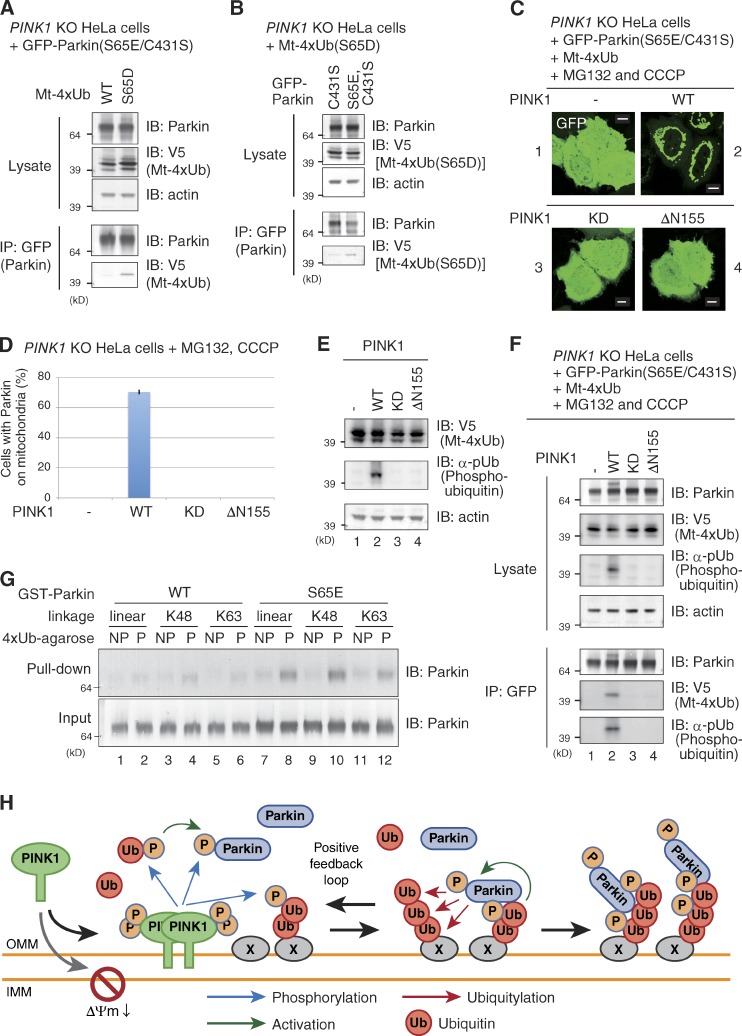
We reevaluated the mitochondrial localization of Parkin in PINK1 KO cells coexpressing Mt-4Ub. The C431S mutation was introduced to prevent ubiquitin chain remodeling by Parkin, and the S65E mutation was introduced to overcome the prerequisite for Parkin phosphorylation. Although GFP-Parkin(S65E/C431S) was recruited to mitochondria by Mt-4×Ub after MG132 treatment in WT HeLa cells (Fig. 4 A, panels 1 and 2), no mitochondrial localization of GFP-Parkin(S65E/C431S) was observed in PINK1 KO HeLa cells expressing Mt-4×Ub (Fig. 4 A, 3 and 4; and Fig. 4 B). Reintroduction of WT PINK1 complemented the mitochondrial localization of GFP-Parkin (S65E/C431S) in the presence of Mt-4×Ub (Fig. 4 C). These results strongly suggest that mitochondrial localization of phosphomimetic Parkin by the mitochondria-localized WT ubiquitin chain is attributable to phosphorylation by undegraded PINK1. Interestingly, even in the PINK1 KO cells, the mitochondria-localized phosphomimetic ubiquitin chain (Mt-4×Ub(S65D)) could recruit Parkin(S65E/C431S) to energized mitochondria (Fig. 4 D, 6; and Fig. 4 E), whereas a linear ubiquitin chain composed of either WT or phosphorylation-deficient mutant (S65A) ubiquitin failed to recruit Parkin(S65E/C431S) to the mitochondria (Fig. 4 D, 4 and 5; and Fig. 4 E). We also examined the subcellular localization of WT, phosphomimetic (S65E), and catalytically inactive (C431S) Parkin under the same conditions. Parkin(S65E), but not WT Parkin or Parkin(C431S), was also recruited to mitochondria by Mt-4×Ub(S65D) (Fig. 4 F). The results shown in Fig. 4 indicate that the phosphomimetic polyubiquitin chain itself is sufficient for mitochondrial localization of phosphomimetic Parkin even in the absence of PINK1, which suggests that the phosphorylated ubiquitin chain is the mitochondrial receptor for phosphorylated Parkin.
Figure 4.
Phosphomimetic ubiquitin chains specifically recruit phosphomimetic Parkin to mitochondria even in the absence of PINK1. (A) WT and PINK1 KO HeLa cells expressing GFP-Parkin(S65E/C431S) and Mt-4×Ub(WT) were treated with MG-132 (10 µM, 3 h), stained with TMRE, and observed with a fluorescence microscope. Accumulation of linear ubiquitin causes mitochondrial aggregation as reported previously (Okatsu et al., 2010). Bars, 10 µm. (B) The number of cells with GFP-Parkin(S65E/C431S) localized to the mitochondria. Bars represent the mean ± SD (error bars) values of 100 cells in three independent experiments. (C) PINK1 is essential for Mt-4×Ub(WT) recruitment of phosphomimetic Parkin. Mt-4×Ub and MG-132-dependent mitochondrial localization of GFP-Parkin(S65E/C431S) in PINK1 KO HeLa cells with or without PINK1 reintroduction is shown. Cell counting was performed as in B. Error bars represent mean ± SD. (D) PINK1 KO HeLa cells coexpressing GFP-Parkin(S65E/C431S) with WT, phosphorylation-deficient (S65A), or phosphomimetic (S65D) Mt-4×Ub with or without MG-132. Bars, 10 µm. (E) Statistical analysis of D. SA, S65A; SD, S65D. Error bars represent mean ± SD. (F) PINK1 KO HeLa cells expressing the indicated GFP-Parkin mutants and Mt-4×Ub after MG-132 treatment were observed with a fluorescence microscope, and the number of cells with Parkin-positive mitochondria was determined as in B. Error bars represent mean ± SD. SA, S65A; SD, S65D.
We further examined if Parkin recruitment by Mt-4×Ub(S65D) triggers downstream events such as the recruitment of p62 and LC3 for mitochondrial perinuclear aggregation and autophagic degradation (Narendra et al., 2008, 2010a; Geisler et al., 2010; Okatsu et al., 2010; Itakura et al., 2012). We found that exogenous GFP-LC3 and endogenous p62 accumulate not only on Parkin-recruiting mitochondria in PINK1 KO cells expressing Mt-4×Ub(S65D) but also on mitochondria in Mt-4×Ub(WT)–expressing cells that cannot recruit Parkin. Finally, we realized that even in cells lacking Parkin expression, GFP-LC3 and p62 were recruited to energized mitochondria decorated with Mt-4×Ub(S65D) or Mt-4×Ub(WT) (Fig. S5, A and B). These results indicate that the linear ubiquitin chain on mitochondria, which is equivalent to the most downstream product of the PINK1–Parkin pathway, is sufficient to recruit the autophagic machinery (LC3 and p62) irrespective of PINK1 and Parkin.
Parkin-dependent accumulation of phosphorylated ubiquitin on depolarized mitochondria
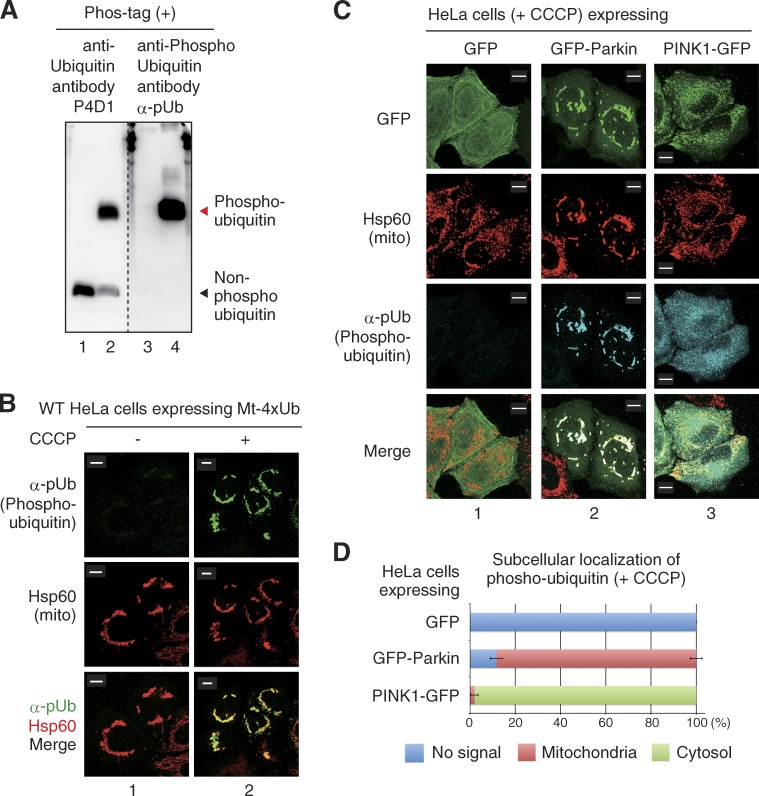
Although tandem chains of phosphomimetic ubiquitin (ubiquitin(S65D) mutant) can recruit Parkin to energized mitochondria independently of PINK1 (Fig. 4), we wanted to investigate Parkin recruitment activity under more physiological conditions that used phosphorylated ubiquitin rather than phosphomimetic ubiquitin. To monitor the subcellular localization of phosphorylated ubiquitin, we prepared a specific affinity-purified rabbit polyclonal antibody for phosphorylated ubiquitin (α-pUb). Immunoblotting experiments with nonphosphorylated and phosphorylated ubiquitin revealed that the α-pUb antibody specifically recognized phosphorylated ubiquitin (Fig. 5 A). We next confirmed the specificity of α-pUb in immunocytochemical experiments. Expression of Mt-4×Ub in WT HeLa cells causes perinuclear accumulation of mitochondria (Fig. 3; Okatsu et al., 2010); however, α-pUb failed to stain these aggregated mitochondria (Fig. 5 B, 1). In contrast, α-pUb detected clustered mitochondria when cells expressing Mt-4×Ub were treated with CCCP (Fig. 5 B, 2), revealing that the α-pUb detected only phosphorylated ubiquitin even in cytochemistry. When HeLa cells expressing sole GFP or exogenous Parkin-GFP were treated with CCCP and stained with α-pUb, phosphorylated ubiquitin accumulated on depolarized mitochondria in a Parkin-dependent manner (Fig. 5 C, 1 and 2). In contrast, in exogenous PINK1-expressing HeLa cells, the phosphorylated ubiquitin signal increased but was not restricted on mitochondria. Instead, the signal was dispersed throughout the cytosol (Fig. 5 C, 3), revealing that localization of phosphorylated ubiquitin on depolarized mitochondria is facilitated by Parkin (Fig. 5 D). These results are consistent with the recent report by Ordureau et al. (2014) finding that Parkin-dependent ubiquitylation is required for accumulation of phosphorylated ubiquitin on mitochondria.
Figure 5.
Parkin-dependent accumulation of phosphorylated ubiquitin on depolarized mitochondria. (A) Preparation of the anti-phosphorylated ubiquitin antibody, α-pUb. Non-phosphorylated ubiquitin control (lanes 1 and 3) and ubiquitin phosphorylated by recombinant _Tc_PINK1 (lanes 2 and 4) were subjected to Phos-tag PAGE. The membrane was separated in two at the broken line and immunoblotted with an anti-ubiquitin antibody (P4D1) or the anti–phospho-ubiquitin antibody, α-pUb. α-pUb specifically reacts with phosphorylated ubiquitin. Note that mobility does not reflect the molecular weight of proteins in Phos-tag PAGE (Kinoshita et al., 2012) and thus molecular weight markers are not shown. (B) Specificity of α-pUb in cytochemical experiments. HeLa cells expressing Mt-4×Ub ± CCCP were stained with α-pUb and Hsp60 (mitochondrial marker). Mt-4×Ub containing aggregated mitochondria were detected by α-pUb only after CCCP treatment. (C) HeLa cells expressing sole GFP (1), GFP-Parkin (2), or PINK1-GFP (3) were treated with CCCP and were stained with α-pUb. Phosphorylated ubiquitin is detectable after overproduction of Parkin or PINK1; however, restricted accumulation of phosphorylated ubiquitin on mitochondria is dependent on Parkin. (D) The number of cells with no signal (blue), a mitochondria-localized signal (red), or a cytosolic signal (green) for phosphorylated ubiquitin was determined in 100 cells. Bars represent the mean ± SD (error bars) values of three experiments. Bars, 10 µm.
Ectopic phosphorylated ubiquitin can recruit Parkin to alternative compartments in cells
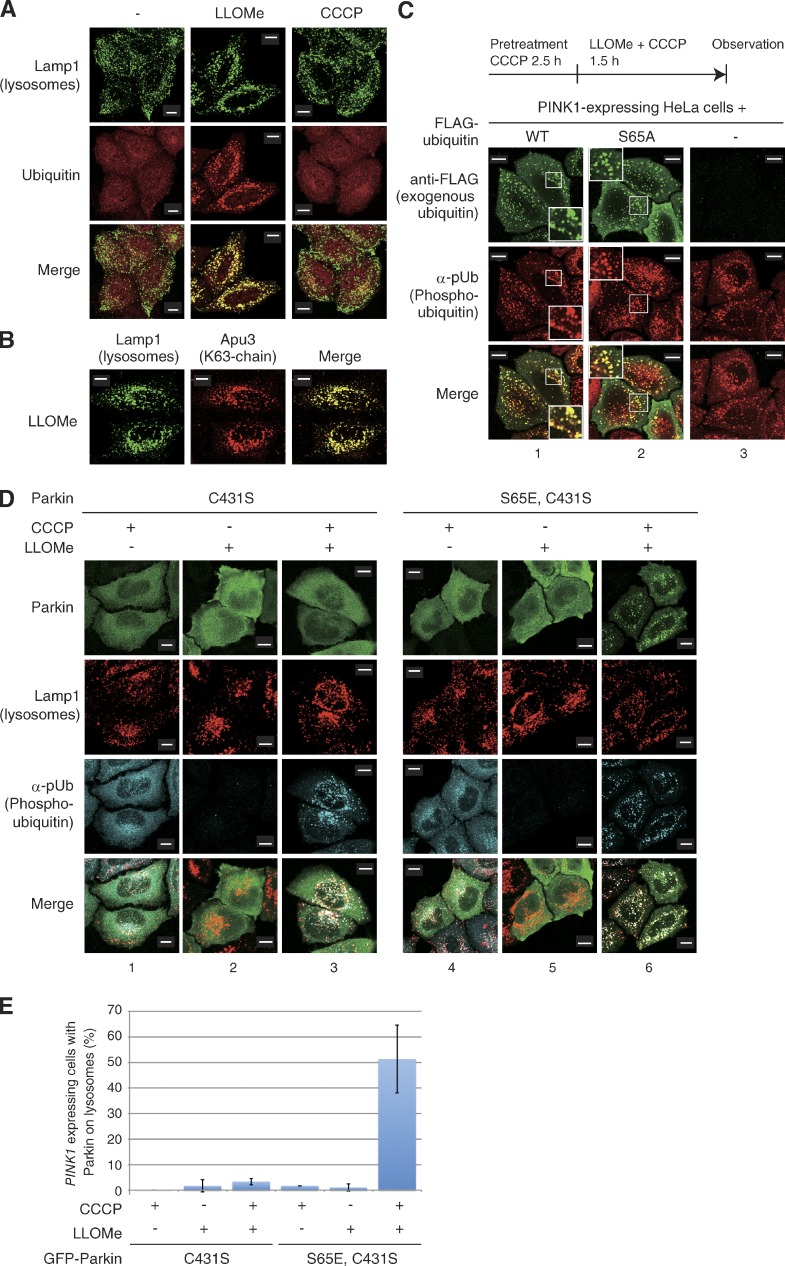
We then examined whether the K48- or K63-linked chain of phosphorylated ubiquitin rather than a linear phosphomimetic polyubiquitin chain recruits Parkin. To achieve this, we established an inducible cellular system in which phosphorylated ubiquitin is targeted to the lysosome. When cells are treated with the lysosomotropic reagent l-leucyl-l-leucine methyl ester (LLOMe), the LLOMe is converted by a lysosomal thiol protease dipeptidyl peptidase I into a membranolytic form (Leu-Leu)n-OMe (n > 3) that causes lysosomal rupture. Damaged lysosomes are promptly and heavily ubiquitylated (Maejima et al., 2013). We confirmed that ubiquitin is targeted and accumulated on the lysosomal membrane after 1 h of LLOMe treatment (Fig. 6 A). Immunocytochemistry using a linkage-specific anti-ubiquitin chain antibody (Apu3) suggests that a K63-linked polyubiquitin chain is formed on the ruptured lysosome (Fig. 6 B), as is the case with depolarized mitochondria in Parkin-expressing cells (Okatsu et al., 2010). We thus expected that the accumulation of phosphorylated ubiquitin in the cytosol would lead to passive incorporation into the ubiquitin chain at the lysosomal membrane after LLOMe treatment. Indeed, when PINK1-expressing HeLa cells were pretreated with CCCP (promotes cytosolic accumulation of phosphorylated ubiquitin as Fig. 5 C) and subsequently treated with both CCCP and LLOMe, phosphorylated ubiquitin colocalized on lysosomes with exogenous Flag-ubiquitin (Fig. 6 C, 1; and Fig. 6 D, 3). Even though the unphosphorylatable Flag-ubiquitin(S65A) mutant was used, Flag-positive foci were also detected using an anti-phosphorylated ubiquitin antibody (Fig. 6 C, 2). This result suggests that phosphorylated ubiquitin along with unmodified ubiquitin molecules was targeted to the ruptured lysosome after LLOMe treatment for passive incorporation into the ubiquitin chain. We then examined whether Parkin is recruited to lysosomes after LLOMe-mediated accumulation of phosphorylated ubiquitin. This experiment, however, presents a challenge, in that CCCP treatment results in the targeting of Parkin to mitochondria accompanied with accumulation of phosphorylated ubiquitin on mitochondria (Fig. 5 C, 2). Indeed, when cells expressing WT or phosphomimetic (S65E) Parkin were pretreated with CCCP, phosphorylated ubiquitin as well as WT and phosphomimetic (S65E) Parkin localized on depolarized mitochondria irrespective of LLOMe treatment (Fig. S5 C). However, as shown in Fig. 2 A, the C431S E3–deficient mutation of Parkin uncouples mitochondrial localization of Parkin from CCCP treatment (Fig. 2 A, 1). Parkin(C431S) however still targeted the site where the phosphorylated-ubiquitin chain accumulates, such as depolarized mitochondria under WT Parkin-coexpressing conditions (Fig. 2 A, 2). In cells expressing Parkin(C431S), phosphorylated ubiquitin accumulated only after CCCP treatment but was dispersed throughout cells (Fig. 6 D, 1 and 2). When Parkin(C431S)-expressing cells were treated first with CCCP and then with LLOMe, phosphorylated ubiquitin localized to the lysosome, but with minimal colocalization of Parkin(C431S) (Fig. 6 D, 3). Because phosphorylation of Parkin Ser65 accelerates interactions with phosphorylated ubiquitin (Fig. 4; Koyano et al., 2014), we used a Parkin(S65E,C431S) double mutant to examine the subcellular localization. As is the case with GFP-Parkin(C431S) (Fig. 2), GFP-Parkin(S65E/C431S) alone did not localize to mitochondria after CCCP treatment (Fig. S5 D, 1), whereas coexpression of WT Parkin restored its mislocalization (Fig. S5 D, 2), which suggests that the WT Parkin–catalyzed ubiquitin chain and CCCP recruits Parkin(S65E/C431S) to depolarized mitochondria. Importantly, Parkin(S65E/C431S) was dispersed throughout the cytoplasm after treatment with either CCCP or LLOMe alone (Fig. 6 D, 4 and 5), whereas Parkin(S65E/C431S) was recruited to phosphorylated ubiquitin-localized lysosomes after treatment with both CCCP and LLOMe (Fig. 6 D, 6; and Fig. 6 E). We thus concluded that the phosphorylated polyubiquitin chain in cells, which is more physiologically relevant than the phosphomimetic linear polyubiquitin chain, is sufficient for Parkin recruitment.
Figure 6.
Ectopic lysosomal expression of phosphorylated ubiquitin chains recruit phosphomimetic Parkin. (A and B) Lysosomes were ubiquitylated via a K63-linked ubiquitin chain after LLOMe treatment. WT HeLa cells without Parkin expression were treated with LLOMe or CCCP, and were subjected to immunocytochemistry using anti-Lamp1 (lysosomal marker), anti-ubiquitin, and anti-K63–linked ubiquitin chain (Apu3) antibodies. (C) HeLa cells stably expressing PINK1 were transfected with WT Flag-ubiquitin or the phosphorylation-deficient Flag-ubiquitin(S65A) mutant. Cells were treated with CCCP and LLOMe before immunocytochemical staining using anti-Flag and α-pUb antibodies. The phosphorylated ubiquitin signal merged well with that of unphosphorylatable Flag-ubiquitin(S65A), suggesting that phosphorylated ubiquitin is passively incorporated into the ubiquitin chain on ruptured lysosomes. However, cells stably expressing PINK1-3×Flag, 2H8 antibody did not detect PINK1-3×Flag (3) because 2H8 reacts with only N-terminally fused mono Flag tag. Higher magnification views (2×) of the boxed areas are shown in the insets. (D) HeLa cells stably expressing PINK1 were transfected with the indicated Parkin mutants, treated with CCCP, LLOMe, or both, and then immunostained with the indicated antibodies. (E) The number of cells with Parkin-positive lysosomes under the indicated experimental conditions. Bars represent the mean ± SD (error bars) values of 100 cells in three independent experiments. Bars, 10 µm.
Ubiquitin Ser65 phosphorylation is essential for Parkin recruitment onto depolarized mitochondria
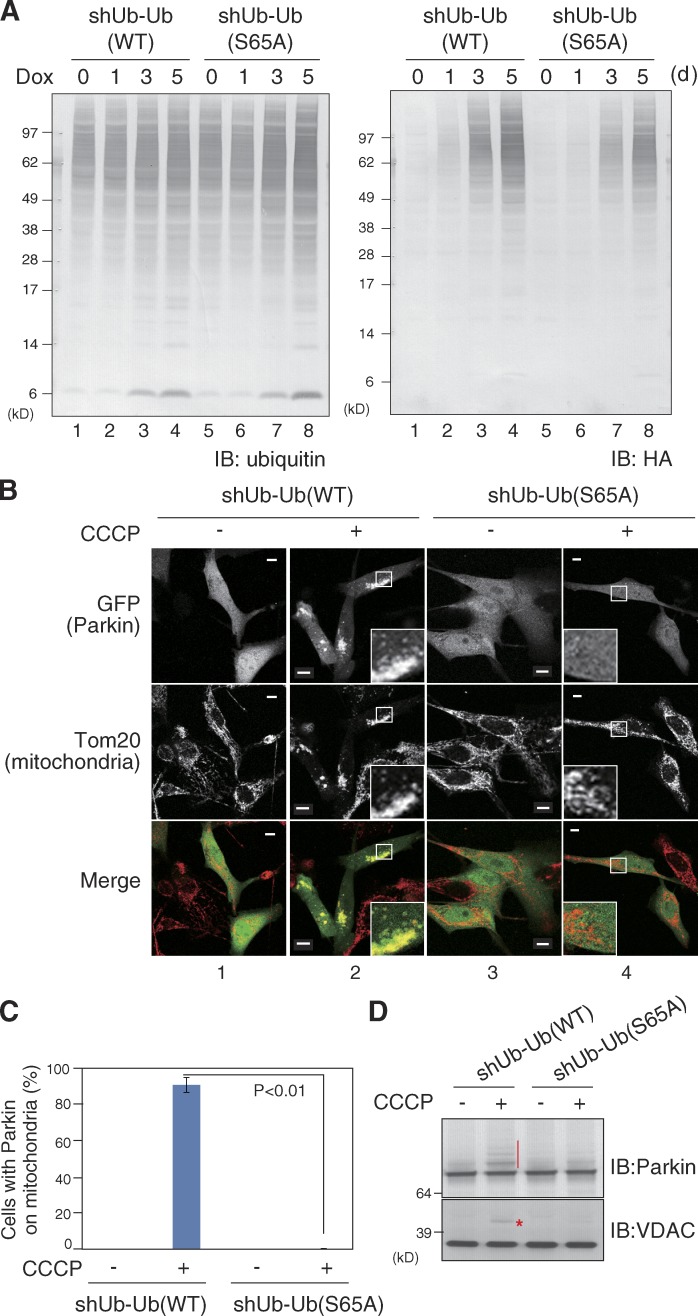
To further explore the importance of ubiquitin Ser65 phosphorylation in Parkin recruitment, we tried to replace genomic ubiquitin with a phosphorylation-deficient S65A mutant in mammalian cells, and examined mitochondrial translocation of Parkin. Ubiquitin is encoded by four genes (RPS27A, UBA52, UBB, and UBC) in the human genome, and is synthesized as a fusion protein with essential ribosomal subunits (S27a and L40) or tandem-repeated polyubiquitin chains. We used U2OS-shUb cells in which all of the genomic ubiquitin genes are knocked down by specific shRNAs, whereas two ubiquitin genes (RPS27A and UBA52 encoding single ubiquitin fused with essential ribosomal subunit S27a or L40) are complemented as an shRNA-resistant form (Xu et al., 2009). The ubiquitin S65A mutation was introduced into these retransformed RPS27A and UBA52 genes, and the subcellular localization of GFP-Parkin was examined after CCCP treatment. Complementary shRNA-resistant ubiquitin possesses an HA tag that allowed us to confirm that the WT and mutant ubiquitins are expressed at similar levels in U2OS-shUb cells harboring WT ubiquitin (referred as shUb-Ub(WT)) or only the ubiquitin(S65A) mutant (referred to as shUb-Ub(S65A); Fig. 7 A). GFP-Parkin in shUb-Ub(WT) cells localized to the cytosol under steady-state conditions and translocated normally to depolarized mitochondria after CCCP treatment (Fig. 7 B, 1 and 2). In contrast, GFP-Parkin translocation was almost completely inhibited in shUb-Ub(S65A) cells (Fig. 7 B, 3 and 4; and Fig. 7 C). The clear inhibition on E3 activity of Parkin monitored by autoubiquitylation of GFP-Parkin and VDAC ubiquitylation was also observed in shUb-Ub(S65A) cells (Fig. 7 D). The results derived from this ubiquitin replacement cellular system confirmed that ubiquitin phosphorylation at Ser65 is indeed essential for Parkin translocation.
Figure 7.
Parkin translocation to depolarized mitochondria was abrogated in U2OS-shUb cells in which the endogenous ubiquitin was replaced with S65A phosphorylation-deficient mutants. (A) All genomic ubiquitin genes in U2OS-shUb cells were knocked down and complemented with either HA-tagged WT ubiquitin (shUb-Ub(WT)) or a phosphorylation-deficient ubiquitin(S65A) mutant (shUb-Ub(S65A)) as the tetracycline (Dox)-inducible system. Both complemented ubiquitin proteins are fused with L40 and S27a (essential ribosomal proteins). To quantify the total and complementary ubiquitin levels, shUb-Ub(WT) and shUb-Ub(S65A) cells were subjected to immunoblotting using anti-ubiquitin or anti-HA antibodies. (B) Subcellular localization of GFP-Parkin in shUb-Ub(WT) or shUb-Ub(S65A) cells. Translocation of GFP-Parkin to depolarized mitochondria was observed in shUb-Ub(WT) cells, but was completely depressed in shUb-Ub(S65A) cells. Bars, 10 µm. Insets show 4× magnified views of the boxed regions. (C) The rate of Parkin mitochondrial localization in 100 cells was determined. Bars represent the mean ± SD (error bars) of three independent experiments. (D) Autoubiquitylation of GFP-Parkin and VDAC ubiquitylation in shUb-Ub(WT) or shUb-Ub(S65A) cells. Both ubiquitylation types observed after CCCP treatment in shUb-Ub(WT) cells (red vertical bar and asterisk) were substantially suppressed in shUb-Ub(S65A) cells.
Phosphomimetic ubiquitin chain physically interacts with Parkin in vitro
Finally, we investigated whether the phosphorylated ubiquitin chain physically interacts with Parkin. First, we performed coimmunoprecipitation experiments under the same conditions as in Fig. 4. HeLa cells lacking PINK1 were cotransfected with GFP-Parkin(S65E/C431S) and Mt-4×Ub or Mt-4×Ub(S65D), and Parkin was immunoprecipitated using anti-GFP antibody-coupled agarose beads. Consistent with the immunocytochemical data (Fig. 4), specific coimmunoprecipitation of Mt-4×Ub(S65D) with GFP-Parkin(S65E/C431S), but not Mt-4×Ub, was observed (Fig. 8 A). We confirmed the Parkin S65E dependency of the immunoprecipitation experiments (Fig. 8 B). These results indicate an interaction between the phosphomimetic ubiquitin chain and phosphomimetic Parkin. Next, we examined the interaction between the phosphorylated (not phosphomimetic) ubiquitin chain and Parkin. When PINK1 KO HeLa cells expressing GFP-Parkin(S65E/C431S) were transfected with Mt-4×Ub (WT) and treated with MG132 and CCCP, Parkin did not localize on depolarized mitochondria (Fig. 8 C, 1) because the nonphosphorylated ubiquitin chain is insufficient for recruitment of Parkin to mitochondria. However, further introduction of WT PINK1 lead to GFP-Parkin(S65E/C431S) localization on mitochondria (Fig. 8 C, 2; and Fig. 8 D), but neither a kinase-dead (KD) nor a dysfunctional N-terminal deletion (ΔN155) mutant of PINK1 restored mitochondrial localization (Fig. 8, C and D). Immunoblotting using the anti-phosphorylated ubiquitin antibody α-pUb (Fig. 5) confirmed phosphorylation of the Mt-4×Ub chain when GFP-Parkin(S65E/C431S) was recruited to the mitochondria (Fig. 8 E, lane 2). We also examined the interaction between Parkin and the phosphorylated ubiquitin chain. Under these experimental conditions, unphosphorylated Mt-4×Ub did not immunoprecipitate with GFP-Parkin(S65E/C431S) (Fig. 8 F, lane 1), whereas phosphorylated Mt-4×Ub did coimmunoprecipitate (Fig. 8 F, lane 2), which indicates a physical interaction. To demonstrate this interaction between Parkin and the phosphorylated ubiquitin chain more directly, we performed an in vitro pull-down assay using recombinant proteins. We expressed and purified WT GST-Parkin and phosphomimetic GST-Parkin(S65E) from E. coli as previously reported (Trempe et al., 2013). Recombinant poly-ubiquitin chains consisting of a linear, K48, or K63 linkage were conjugated to agarose and were phosphorylated beforehand using recombinant TcPINK1 (Fig. 1 E). WT GST-Parkin or GST-Parkin(S65E) were co-incubated with the phosphorylated ubiquitin chains for 1 h at 4°C, and then the ubiquitin chain–conjugated agarose was collected by centrifugation. WT GST-Parkin was rarely pulled down by the ubiquitin chain–conjugated agarose (Fig. 8 G, lanes 1–6). In contrast, GST-Parkin(S65E) was specifically pulled down by phosphorylated linear, K48-linked, and K63-linked ubiquitin chains (Fig. 8 G, lanes 8, 10, and 12), but not their nonphosphorylated forms (lanes 7, 9, and 11). Because phosphorylation clearly accelerates the ability of recombinant ubiquitin chains to bind recombinant Parkin (Fig. 8 G), we concluded that the phosphorylated polyubiquitin chain directly interacts with Parkin.
Figure 8.
Physical interaction between phosphorylated polyubiquitin chains and Parkin. (A and B) Immunoprecipitation shows that phosphomimetic Parkin and phosphomimetic linear ubiquitin chains interact in cells. GFP-Parkin(S65E/C431S) immunoprecipitated from cell lysates was immunoblotted using an anti-V5 antibody that detects Mt-4×Ub. The interaction depends on phosphomimetic mutation of both the ubiquitin chain (A) and Parkin (B). (C and D) A mitochondria-localized linear ubiquitin chain (Mt-4×Ub) recruits Parkin in a CCCP- and PINK1-dependent manner. PINK1 KO HeLa cells were transfected and treated as described in the text, and the GFP-Parkin(S65E/C431S)-derived signal was observed with a fluorescence microscope (C). Bars, 10 µm. The rate of Parkin mitochondrial localization was determined (D). KD, kinase dead; ΔN155, lacking the first 155 N-terminal amino acids. Bars represent the mean ± SD (error bars) values of 100 cells in three independent experiments. (E) Phosphorylation of the linear ubiquitin chain was confirmed by immunoblotting using α-pUb. (F) Interaction between phosphomimetic Parkin and a phosphorylated polyubiquitin chain. GFP-Parkin(S65E/C431S) was immunoprecipitated from PINK1 KO HeLa cells transfected and treated as described in the text, and immunoblotted with an anti-V5 antibody for Mt-4×Ub and α-pUb. (G) Direct interaction between recombinant phosphomimetic Parkin and a phosphorylated ubiquitin chain. Polyubiquitin chain–conjugated agarose beads were phosphorylated by _Tc_PINK1. Recombinant GST-Parkin(WT) or the phosphomimetic GST-Parkin(S65E) mutant were incubated with the agarose beads and examined for GST-Parkin capture by the phosphorylated ubiquitin-conjugated agarose. (H) Model for phosphorylated polyubiquitin chain recruitment of Parkin to damaged mitochondria. See the text for details.
Discussion
Parkin catalyzes ubiquitylation of depolarized mitochondria to facilitate removal of the damaged mitochondria from the cells. Under steady-state conditions, Parkin is maintained in an inert state by two independent mechanisms. First, Parkin is broadly distributed in the cytosol under normal conditions, and is spatially separated from mitochondrial substrates. Second, the E3 activity of Parkin is kept latent, thus Parkin is enzymatically impeded from functioning in mitochondrial ubiquitylation. However, when ΔΨm decreases, Parkin is promptly recruited to depolarized mitochondria and its E3 activity is simultaneously reestablished, which allows ubiquitylation of mitochondrial substrates on OMM. Although PINK1 exerts essential roles in both steps and its kinase activity is indispensable, the molecular basis for how PINK1 recruits Parkin selectively to depolarized mitochondria has not been fully elucidated. Various models for the PINK1-mediated Parkin recruitment process have been proposed, but, until now, none have adequately explained the accumulated data. PINK1 phosphorylation of Parkin at Ser65 (Kondapalli et al., 2012; Shiba-Fukushima et al., 2012; Iguchi et al., 2013) is thought to convert Parkin to a membrane-bound form. This model is sufficient to explain why phosphorylation-deficient mutants of Parkin (such as S65A) significantly, but not completely, inhibit Parkin recruitment (Shiba-Fukushima et al., 2012; Zhang et al., 2014), and why the kinase activity of PINK1 is required for Parkin mitochondrial localization (Geisler et al., 2010; Matsuda et al., 2010; Narendra et al., 2010b). This model, however, inadequately explains selective recruitment of Parkin to depolarized mitochondria when both depolarized and polarized mitochondria are present (Narendra et al., 2008; Matsuda et al., 2010; Yang and Yang, 2011). As an alternative model, phosphorylation of additional PINK1 mitochondrial substrates increases the affinity of mitochondria for Parkin, such that Parkin is recruited to the same mitochondria. This model is compatible with the requirement for PINK1 kinase activity in Parkin recruitment, and with the finding that Parkin is selectively directed to a subset of mitochondria when energized and depolarized mitochondria coexist. Indeed, it has been reported that PINK1-phosphorylated mitofusin is a Parkin receptor on damaged mitochondria (Chen and Dorn, 2013). It is difficult however to reconcile the mitofusin model with contradictory data reporting Parkin recruitment to depolarized mitochondria in mitofusin1/2 double KO MEFs (Narendra et al., 2008; Chan et al., 2011). Moreover, even if mitofusin or some other mitochondria-localized PINK1 substrates function as a Parkin receptor, it is difficult to explain how PINK1 targeting to peroxisomes or lysosomes, which lack such a genuine PINK1 substrate, still recruit Parkin to the respective organelles (Lazarou et al., 2012). The last and the simplest hypothesis is that PINK1 physically interacts with Parkin and thus is the Parkin receptor. This model easily explains why PINK1 is essential for mitochondrial Parkin localization and is sufficient for Parkin targeting to organelles (e.g., peroxisome) other than mitochondria (Lazarou et al., 2012). However, although PINK1 forms a stable high-molecular-weight complex composed of a PINK1 dimer and TOM machineries in response to a decrease in ΔΨm (Lazarou et al., 2012; Okatsu et al., 2013), association of Parkin with this complex was not detected, and the size of the PINK1 complex does not changed in the absence of Parkin (Lazarou et al., 2012). This result suggests that PINK1 and Parkin do not stably associate in cells. An even more fundamental deficiency in all three models is their inability to explain why the E3 activity of Parkin itself is important for depolarized mitochondrial localization (Lazarou et al., 2013; Zheng and Hunter, 2013; Fig. 2).
In this paper, we present important findings that address the aforementioned deficiencies. First, phosphorylated polyubiquitin chains exist in cells when ΔΨm is decreased (Fig. 1). Second, a linear ubiquitin chain of phosphomimetic tetra-ubiquitin(S65D) recruits Parkin to energized mitochondria in the absence of PINK1, whereas a linear ubiquitin chain of WT tetra-ubiquitin does not (Fig. 4). Third, under more physiological conditions, a lysosomal polyubiquitin chain containing phosphorylated ubiquitin can recruit phosphomimetic Parkin to the lysosome (Fig. 5). In addition, physical interactions between phosphomimetic Parkin and phosphorylated ubiquitin were detected by immunoprecipitation both from cells and after in vitro reconstitution using recombinant proteins (Fig. 8). We thus propose a completely novel model for Parkin recruitment in which the phosphorylated ubiquitin chain functions as the genuine Parkin receptor.
Several research groups have reported that substrate specificity of Parkin is rather weak, and that Parkin ubiquitylates various OMM proteins (Chan et al., 2011; Narendra et al., 2012; Sarraf et al., 2013), including VDAC, mitofusin1/2, RHOT1/2(Miro), hexokinase I, and TOMM20 (Gegg et al., 2010; Geisler et al., 2010; Poole et al., 2010; Tanaka et al., 2010; Ziviani et al., 2010; Rakovic et al., 2011; Wang et al., 2011; Liu et al., 2012; Okatsu et al., 2012a; Bingol et al., 2014). As reported independently by Ordureau et al. (2014), we can easily imagine that Parkin catalyzes the formation of ubiquitin chains on various OMM proteins, and that these ubiquitin chains function as substrates for PINK1 phosphorylation, which then recruit and activate more Parkin molecules. This is a powerful feed-forward mechanism that can explain why the E3 activity of Parkin is required for efficient recruitment to depolarized mitochondria (Fig. 2), namely that the E3 activity is not required for initiation of the signal but rather for amplification. This model is also compatible with the confounding results that show PINK1 targeting to a heterogeneous organelle (e.g., peroxisomes or lysosomes, where genuine PINK1 substrates are not expected to exist) can recruit Parkin to the respective organelles, even though PINK1 is unlikely to be the Parkin receptor (Lazarou et al., 2012). Ubiquitylation is commonly used as a signal for various organelle-specific signaling pathways, and thus there are many E3s and ubiquitylated cargo proteins on every organelle. For example, on peroxisomes two E3s (Pex2 and Pex10p–Pex12p complex) catalyze ubiquitylation for protein import into the peroxisomal matrix and Pex5 retrotranslocation (Platta et al., 2009; Okumoto et al., 2014). A diverse number of membrane proteins are ubiquitylated in the endosome–lysosome pathway for down-regulation (Clague et al., 2012). The presence of these ubiquitylated proteins on various organelles might be phosphorylated by ectopically targeted PINK1, which then transmits the signal to Parkin. Similarly, for Parkin recruitment on depolarized mitochondria, the initiating signal for Parkin recruitment seems to be the already-existing mitochondrial ubiquitylated proteins mediated by other E3s. However, once the already-existing ubiquitylated protein is phosphorylated by PINK1 on depolarized mitochondria, it acts as a beacon for Parkin activation and recruitment, which then triggers local phosphorylated-ubiquitin signal amplification via a positive feedback loop (summarized in Fig. 8 H). However, in our model many details remain to be refined; i.e., we cannot exclude the possibility that Parkin preferentially incorporates phosphorylated ubiquitin over unmodified ubiquitin to increase the efficiency of this amplification process. Further analysis is needed to clarify this issue. While our manuscript was under review, Shiba-Fukushima et al. (2014) independently published a paper with a similar conclusion.
In the present study, using a specific antibody for phosphorylated ubiquitin, and unique experimental procedures that promote lysosomal targeting of phosphorylated ubiquitin, we demonstrate that an ectopically localized phosphorylated polyubiquitin chain can recruit Parkin to the corresponding organelle. This is, to our knowledge, the first direct evidence that a phosphorylated polyubiquitin chain is the Parkin receptor in cells. We and others have reported that phosphorylated ubiquitin functions as a Parkin activator by de-repressing autoinhibition of Parkin E3 activity (Kane et al., 2014; Kazlauskaite et al., 2014; Koyano et al., 2014), but unexpectedly, phosphorylated ubiquitin also functions in the Parkin recruitment process. We believe that ubiquitin phosphorylation enables us to understand comprehensively how PINK1 regulates Parkin.
Materials and methods
Plasmids and antibodies
Plasmids used in this study are summarized in Table S1. The following primary antibodies were used. Mouse primary antibodies: anti-Ubiquitin (clone P4D1, 1:500; Santa Cruz Biotechnology Inc.), anti-V5 (product number [PN] R960-25, 1:1,000; Life Technologies), anti-actin (clone AC-40, 1:500; Sigma-Aldrich), anti-Parkin (clone PRK8, 1:2,000; Sigma-Aldrich), anti-DYKDDDDK (clone 2H8, 1:500; TransGenic Inc.), anti-GFP (clone 3E6, 1:1,000; Life Technologies), anti-Lamp1 (clone H4A3, 1:200; Santa Cruz Biotechnology, Inc.), anti-HA (clone TANA2, 1:1,000; MBL), and anti-VDAC (PN Ab-2, 1:1,000; EMD Millipore). Rabbit primary antibodies: anti-Ubiquitin (PN Z-0458, 1:500; Dako), anti-Tom20 (PN FL-145, 1:250 for immunoblotting or 1:2,000 for immunostaining; Santa Cruz Biotechnology, Inc.), anti-PINK1 (PN BC100-494, 1:1,000; Novus Biologicals), and anti-K63–linked ubiquitin chain (clone Apu3, 1:250; EMD Millipore). Goat primary antibodies: anti-LDH (PN ab2101, Abcam, 1:500) and anti-Hsp60 (PN sc1052, Santa Cruz Biotechnology, Inc., 1:250). To generate the anti-phosphorylated ubiquitin antibody, rabbit was immunized by antigen peptide C (for carrier protein conjugation)-NIQKE(pS)TLH, and the serum was subjected to affinity purification. The following secondary antibodies were used: various mouse, rabbit, or goat anti-IgG antibodies conjugated with alkaline phosphatase (1:5,000; Santa Cruz Biotechnology, Inc.) or with horseradish peroxidase (1:5,000; Jackson ImmunoResearch Laboratories, Inc.) for immunoblotting, and various mouse or rabbit anti-IgG antibodies conjugated with Alexa Fluor 488, 568, and 647 (1:2,000; Life Technologies) for immunostaining. Anti-GFP mouse mAb-agarose (D153-8, MBL) and anti-K48–linked ubiquitin chain rabbit antibody (clone Apu2; EMD Millipore) were used for immunoprecipitation. Recombinant linear, K48-linked, and K63-linked tetra-ubiquitin chains and ubiquitin-conjugated agarose beads were purchased from Boston Biochem.
Protein expression and purification from E. coli
Plasmids expressing GST-Parkin and GST-TcPINK1 were provided by J.F. Trempe (McGill University, Montreal, Canada). E. coli BL21-CodonPlus(DE3)-RIL (Agilent Technologies) were transformed with plasmids harboring the T7 RNA polymerase promoter for GST-TcPINK1, GST-Parkin(WT), or GST-Parkin(S65E) expression, and cultured at 37°C until an optical density at 600 nm of ∼0.5–0.8 was obtained. The cells were then cooled to 16°C and treated with 100 µM (for GST-TcPINK1) or 25 µM (for GST-Parkin) IPTG for 18 h at 16°C. For expression of GST-Parkin, 25 µM ZnCl2 was added to the culture medium. Cells were collected by centrifugation, resuspended in TBS (50 mM Tris-HCl, pH 7.5, 120 mM NaCl, 1 mM DTT, and protease inhibitor cocktail) containing 0.5% Tween-20, and sonicated on ice. After centrifugation to pellet cellular debris, the lysate was bound to glutathione–Sepharose 4B (GE Healthcare) and recombinant GST-fused proteins were eluted with TBS containing 20 mM glutathione.
Cells and transfection
HeLa and U2OS cells were cultured at 37°C with 5% CO2 in DMEM (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Equitech-BIO. Inc.), 1× penicillin-streptomycin-glutamine (Life Technologies), 1× nonessential amino acids (Life Technologies), and 1× sodium pyruvate (Life Technologies). For U2OS cells, tetracycline-free fetal bovine serum was used (see the “Ubiquitin replacement in cells” section below for detailed information). Cells were transfected with Fugene 6 (Roche and Promega).
Immunofluorescence and TMRE staining
For immunofluorescence experiments, cells were fixed with 4% paraformaldehyde, permeabilized with 50 µg/ml digitonin, and then stained with the primary and secondary antibodies described earlier in the Materials and methods. To monitor the mitochondrial membrane potential, cells were treated with 50 nM TMRE (Sigma-Aldrich) for 30 min, washed three times, and subjected to live cell imaging to detect TMRE fluorescence at room temperature. Cells were imaged using a confocal laser-scanning microscope (LSM710 and LSM780; Carl Zeiss) with Plan-Apochromat 20×/0.8 NA M27, 40×/0.95 NA Korr M27, and 63×/1.40 NA oil differential interference contrast M27 objective lenses. The ZEN imaging software (Carl Zeiss) was used for image acquisition. Image contrast and brightness were adjusted in Photoshop Elements version 10 (Adobe).
Mitochondria enriched fractionation
Cells were suspended in fractionation buffer (250 mM NaCl, 20 mM Hepes-NaOH, pH 8, and protease and phosphatase inhibitor cocktail [Roche]) and disrupted by 20 passages through a 25-gauge needle with a 1-ml syringe. Debris was removed by centrifugation at 1,000 g for 7 min, and the supernatant was centrifuged (10,000 g, 4°C, 10 min) to precipitate the mitochondria-rich fraction.
Phos-tag SDS-PAGE and immunoblotting
To detect phosphorylated proteins, SDS-PAGE with 50 µM Phos-tag acrylamide (Wako Pure Chemical Industries) and 100 µM MnCl2 was used. Mn2+ was removed from the electrophoresed gels by gentle shaking in transfer buffer with 0.01% SDS and 1 mM EDTA for 10 min. The gels were then washed for another 10 min in transfer buffer with 0.01% SDS but lacking EDTA according to the manufacturer’s protocol. Proteins were transferred to PVDF membranes and detected with the indicated antibodies.
Generation of PINK1 KO HeLa cell line
PINK1 KO HeLa cells were established using the CRISPR/Cas9 system (GeneArt CRISPR Nuclease System; Life Technologies). The vector in this kit encodes a Cas9 nuclease expression cassette and a guide RNA cloning cassette. The PINK1 guide RNA (5′-ACAAAGTGGCGGCCGATTGT-3′) was selected using a web site for CRISPR design (crispr.mit.edu). HeLa cells were transfected with the GeneArt CRISPR nuclease vector and a pSilencer5.1-H1 Retro empty vector (for selecting puromycin resistant cells) for 24 h. The cells were treated with 5 µg/ml puromycin for 48 h, then maintained in fresh puromycin-free medium. Monoclonal cell lines were generated through limiting dilutions. The putative PINK1 KO HeLa cell lines were validated by genomic DNA sequencing and assaying for Parkin recruitment activity. To exclude the influence of off-target effects, the Parkin recruitment defect in the KO cell line was confirmed by complementation with exogenous PINK1.
In vitro labeling of ubiquitin chains by γ-[32P]ATP
For immunoprecipitation experiments, mitochondria from HeLa cells stably expressing PINK1-3×Flag were collected, resuspended in cell-free assay buffer (20 mM Hepes-KOH, pH 7.5, 220 mM sorbitol, 10 mM KAc, and 70 mM sucrose), solubilized with 10 mg/ml digitonin (Wako) for 15 min at 4°C, and reacted with anti-FLAG M2 agarose (Sigma-Aldrich) for 1 h at 4°C. The resulting immunoprecipitates were washed repeatedly with the same buffer and collected by centrifugation. The immunoprecipitated PINK1 was then incubated with recombinant K48-linked or K63-linked tetra-ubiquitin chains (2 µg; Boston Biochem) and 100 µM γ-[32P]ATP (5 µCi) in 30 µl of kinase buffer (20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, and 1 mM DTT) for 30 min at 30°C. The reaction was stopped by adding Laemmli’s sample buffer and boiling. One-third of the sample was subjected to 17% SDS-PAGE and CBB staining. Phosphorylated proteins were visualized by autoradiography.
In vitro kinase assay and ubiquitin–Parkin interaction analysis
Recombinant ubiquitin and polyubiquitin chains (60 µM) were incubated with GST-TcPINK1 (2 µM) in kinase reaction buffer 1 (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgSO4, 2 mM ATP, 2 µM DTT, and 1% glycerol) for 30 min at 30°C. The reaction was stopped by boiling for 5 min at 98°C. The reacted ubiquitin and polyubiquitin chains were subjected to Phos-tag PAGE. To examine the physical interaction between GST-Parkin and phosphorylated ubiquitin, polyubiquitin chain–conjugated agarose beads (15 µM) were incubated with _Tc_PINK1 (0.5 µM) in kinase reaction buffer 2 (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgSO4, 2 mM ATP, and 1% glycerol) for 1 h at 30°C to promote phosphorylation and then boiled for 10 min at 90°C to deactivate _Tc_PINK1. The resulting phospho-ubiquitin chain–conjugated agarose beads (0.36 µM) were then incubated with recombinant GST-Parkin(WT) or phosphomimetic GST-Parkin(S65E) in reaction buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1% glycerol, 1 mM DTT, and 0.2% Triton X-100) for 1 h at 4°C. The phosphorylated ubiquitin-conjugated agarose beads were collected by centrifugation, washed three times, and boiled at 98°C for 5 min with SDS buffer. Captured GST-Parkin was detected by immunoblotting.
Liquid chromatography–tandem MS (LC-MS/MS) analysis of ubiquitin
Mass spectrometric analysis was performed as previously reported (Koyano et al., 2014), with some modification. To identify endogenous S65 phosphorylated ubiquitin with a K63-GlyGly branch, whole cell lysates of intact PINK1-expressing HeLa cells ± CCCP treatment were subjected to SDS-PAGE and stained with CBB, then in-gel trypsin digestion was performed. Gels were extensively washed with MilliQ water (EMD Millipore), the low (<14,000)-, middle (14,000–55,000)-, or high (>55,000)-molecular-weight fractions of gels were excised, cut into 1–2 mm pieces, and destained with 1 ml of 50 mM ammonium bicarbonate (AMBC) buffer containing 50% acetonitrile (ACN) with agitation for 1 h. A final 100% ACN wash was performed to ensure complete gel dehydration. In-gel digestion was prepared by diluting modified sequencing grade trypsin (Promega) and lysyl endopeptidase (Wako Pure Chemical Industries) with 50 mM AMBC buffer, pH 8.0, containing 5% ACN. The digestion solution was added to the gel pieces and incubated overnight at 37°C. Digests were quenched and extracted by addition of 50 µl of 50% ACN containing 0.1% trifluoroacetic acid (TFA) solution for 1 h by shaking. The digested peptides were recovered into fresh Protein LoBind tubes and an additional extraction was performed with 70% ACN containing 0.1% TFA solution for 30 min. The extracted peptides were concentrated to 20 µl using a SpeedVac system (Thermo Fisher Scientific). The concentrated peptides were prepared in 0.1% TFA. The resultant peptides were analyzed on a nanoflow UHPLC instrument (Easy nLC 1000, Q-Exactive MS and nano electrospray ion source; Thermo Fisher Scientific) with the raw data processed using Xcalibur (Thermo Fisher Scientific). MS spectra were analyzed using Protein Discoverer software version 1.3 (Thermo Fisher Scientific). The fragmentation spectra were searched against the UniProt database with the MASCOT search engine. Data were prepared with PinPoint software version 1.3 (Thermo Fisher Scientific).
LC-MS/MS analysis of immunoprecipitated K48-linked polyubiquitin chains
Lysates of HeLa cells stably expressing PINK1-3×Flag were incubated with Apu2 (a rabbit mAb for the K48-linked polyubiquitin chain) or anti-SAMM50 (a rabbit mAb serving as a control IgG) coupled to 10 µl of protein G–Sepharose 4FF for 2 h at 4°C. The beads were washed three times with TNE-N+ buffer and boiled in SDS sample buffer. Immunoprecipitated proteins were resolved by SDS-PAGE and stained with CBB. The high-molecular-weight fractions (>60,000) in the gel were excised, and in-gel digestion using trypsin and lysyl endopeptidase (Wako Pure Chemical Industries) was performed as described previously (Koyano et al., 2014), with some modification. In detail, gels were washed with MilliQ water. The bands were then cut into 1-mm2 pieces and destained with 25 mM AMBC buffer containing 50% ACN with agitation for 10 min. A final 100% ACN wash was performed to ensure complete gel dehydration. In-gel digestion solution was prepared by diluting modified sequencing-grade trypsin and lysyl endopeptidase with 50 mM AMBC buffer containing 5% ACN. The solution was added to the gel pieces and incubated overnight at 37°C. Digests were quenched and extracted by addition of 50% ACN and 1% formic acid mixture for 30 min by shaking. The digested peptides were recovered into fresh Protein LoBind tubes (Eppendorf). The extracted peptides were concentrated and prepared in 2% ACN and 0.1% TFA mixture. The resultant peptides were analyzed on a UHPLC system coupled to a Q Exactive mass spectrometer (Thermo Fisher Scientific). To generate extracted ion chromatograms, the raw data were processed using Xcalibur software (Thermo Fisher Scientific) and directly analyzed against the SwissProt database using Proteome Discoverer (Thermo Fisher Scientific) with the Mascot search engine (Matrix Science).
Ubiquitin replacement in cells
To replace genomic ubiquitin with a phosphorylation-deficient S65A mutant in mammalian cells, we modified the cellular system established by Z.J. Chen (Xu et al., 2009). Ubiquitin knockdown U2OS cells and the WT ubiquitin rescue plasmid were provided by Z.J. Chen (University of Texas Southwestern Medical Center, Dallas, TX). In brief, U2OS cells were stably integrated with a tetracycline-inducible shRNA vector for all four ubiquitin genes RPS27A, UBA52, UBB, and UBC (referred to as U2OS-shUb cells hereafter). Target sequences for shRNA are ACACCATTGAGAATGTCAA (UBA52 and UBC) and AGGCCAAGATCCAGGATAA (RPS27A and UBB). Because the Ub shRNA vector harbors a puromycin resistance gene, the U2OS-shUb cells were selected accordingly. The cells were then transfected with a rescue plasmid harboring a neomycin resistance gene and an shRNA-resistant ribosomal subunit-fusion ubiquitin (WT or S65A) that is induced by tetracycline. The cDNAs harboring silent mutations were subcloned into a modified pcDNA3 vector in which two TetO2 sites were inserted between the CMV promoter and the start codon. The rescue plasmids contained two expression cassettes in which RNAi-resistant WT or S65A ubiquitin was expressed under the control of the tetracycline-inducible promoter. The second ubiquitin fusion gene is driven by the IRES (internal ribosomal entry site) sequence and contains an N-terminal HA epitope. The RNAi-resistant mutations in the transgenes are ATACAATCGAAAACGTCAA (Uba52) and AAGCTAAAATTCAGGACAA (RPS27A). After selection with G418 (400 µg/ml; Sigma-Aldrich), U2OS-shUb cells with the shRNA-resistant ubiquitin (WT or S65A) were treated with doxycycline (1 µg/ml), and used in experiments.
Immunoprecipitation
HeLa cells expressing GFP-Parkin proteins were lysed with TNE-N+ buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and protease and phosphatase inhibitor cocktail [Roche]). The lysates were centrifuged at 16,500 g for 10 min, and the GFP-fused protein in the supernatant was purified by immunoprecipitation with anti-GFP mAb-agarose (MBL). Immunoprecipitation of the K48-linkged polyubiquitin chain was performed as well using an Apu2 antibody (EMD Millipore).
Online supplemental material
Fig. S1 shows MS-based analysis of K48- and K63-linked polyubiquitin chains, supporting our conclusion that they are phosphorylated by PINK1 in cells. Fig. S2 indicates that tandem tetra-ubiquitin chain targeting to mitochondria is stabilized by treatment with the proteasome inhibitor MG-132. Fig. S3 shows the colocalization between phosphomimetic Parkin and phosphomimetic tetra-ubiquitin chain targeting to mitochondria. Fig. S4 shows the evidence for generation of a PINK1 KO HeLa cell line. Fig. S5 shows immunocytochemical data indicating recruitment of LC3 and p62 by a mitochondrial ubiquitin chain, and suggesting recruitment of Parkin by mitochondrial phosphorylated ubiquitin chain. Table S1 lists the plasmids used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201410050/DC1.
Supplementary Material
Supplemental Material
Acknowledgments
We are grateful to J.F. Trempe for plasmids, N. Tani (Kumamoto University) for help with the LC-MS/MS analysis, Z.J. Chen for the ubiquitin knockdown system, and T. Ueno (Juntendo University) for valuable discussion during the lysosomal experiments.
This work was supported by Japan Science and Technology Agency PRESTO, Japan Society for the Promotion of Science (JSPS) KAKENHI grant no. 23687018, Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI grant nos. 24111557 and 25112522, and the Tomizawa Jun-ichi and Keiko Fund for Young Scientist (to N. Matsuda); by MEXT KAKENHI grant no. 24112008 (to Y. Saeki); by JSPS KAKENHI grant no. 23-6061 (to K. Okatsu); by KAKENHI grant no. 21000012 (to K. Tanaka); by the Japan Foundation for Applied Enzymology (to H. Kosako): and by the Takeda Science Foundation (to H. Kosako, K. Tanaka, and N. Matsuda).
The authors declare no competing financial interests.
Author contributions: K. Okatsu, F. Koyano, and M. Kimura carried out immunocytochemistry, biochemical analysis, and immunoblotting experiments. H. Kosako performed 32P labeling experiments. F. Koyano, H. Kosako, and Y. Saeki performed mass spectrometric analysis. K. Tanaka and N. Matsuda designed the research and analyzed the data. N. Matsuda wrote the manuscript with help and supervision from K. Tanaka. All authors contributed to data analysis and preparation of the manuscript.
Footnotes
Abbreviations used in this paper:
ACN
acetonitrile
CRISPR
clustered regularly interspaced short palindromic repeat
KO
knockout
LC-MS/MS
liquid chromatography–tandem mass spectrometry
LLOMe
l-leucyl-l-leucine methyl ester
MS
mass spectrometry
Mt-Parkin
mitochondria-targeting Parkin
OMM
outer mitochondrial membrane
TFA
trifluoroacetic acid
TMRE
tetramethylrhodamine ethyl ester
WT
wild type
References
- Bingol B., Tea J.S., Phu L., Reichelt M., Bakalarski C.E., Song Q., Foreman O., Kirkpatrick D.S., and Sheng M.. 2014. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 510:370–375. [DOI] [PubMed] [Google Scholar]
- Chan N.C., Salazar A.M., Pham A.H., Sweredoski M.J., Kolawa N.J., Graham R.L., Hess S., and Chan D.C.. 2011. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 20:1726–1737 10.1093/hmg/ddr048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaugule V.K., Burchell L., Barber K.R., Sidhu A., Leslie S.J., Shaw G.S., and Walden H.. 2011. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 30:2853–2867 10.1038/emboj.2011.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., and Dorn G.W. II. 2013. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 340:471–475 10.1126/science.1231031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhong Y., Wang Y., Xu S., Liu Z., Baskakov I.V., Monteiro M.J., Karbowski M., Shen Y., and Fang S.. 2013. Ubiquitination-induced fluorescence complementation (UiFC) for detection of K48 ubiquitin chains in vitro and in live cells. PLoS ONE. 8:e73482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew K.C., Matsuda N., Saisho K., Lim G.G., Chai C., Tan H.M., Tanaka K., and Lim K.L.. 2011. Parkin mediates apparent E2-independent monoubiquitination in vitro and contains an intrinsic activity that catalyzes polyubiquitination. PLoS ONE. 6:e19720 10.1371/journal.pone.0019720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague M.J., Liu H., and Urbé S.. 2012. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev. Cell. 23:457–467 10.1016/j.devcel.2012.08.011 [DOI] [PubMed] [Google Scholar]
- Clark I.E., Dodson M.W., Jiang C., Cao J.H., Huh J.R., Seol J.H., Yoo S.J., Hay B.A., and Guo M.. 2006. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 441:1162–1166 10.1038/nature04779 [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., and Zhang F.. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiesel F.C., Moussaud-Lamodière E.L., Ando M., and Springer W.. 2014. A specific subset of E2 ubiquitin-conjugating enzymes regulate Parkin activation and mitophagy differently. J. Cell Sci. 127:3488–3504 10.1242/jcs.147520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg M.E., Cooper J.M., Chau K.Y., Rojo M., Schapira A.H., and Taanman J.W.. 2010. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 19:4861–4870 10.1093/hmg/ddq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S., Holmström K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., and Springer W.. 2010. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12:119–131 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
- Geisler S., Vollmer S., Golombek S., and Kahle P.J.. 2014. The ubiquitin-conjugating enzymes UBE2N, UBE2L3 and UBE2D2/3 are essential for Parkin-dependent mitophagy. J. Cell Sci. 127:3280–3293 10.1242/jcs.146035 [DOI] [PubMed] [Google Scholar]
- Haddad D.M., Vilain S., Vos M., Esposito G., Matta S., Kalscheuer V.M., Craessaerts K., Leyssen M., Nascimento R.M., Vianna-Morgante A.M., et al. . 2013. Mutations in the intellectual disability gene Ube2a cause neuronal dysfunction and impair parkin-dependent mitophagy. Mol. Cell. 50:831–843 10.1016/j.molcel.2013.04.012 [DOI] [PubMed] [Google Scholar]
- Hasson S.A., Kane L.A., Yamano K., Huang C.H., Sliter D.A., Buehler E., Wang C., Heman-Ackah S.M., Hessa T., Guha R., et al. . 2013. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 504:291–295 10.1038/nature12748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi M., Kujuro Y., Okatsu K., Koyano F., Kosako H., Kimura M., Suzuki N., Uchiyama S., Tanaka K., and Matsuda N.. 2013. Parkin-catalyzed ubiquitin-ester transfer is triggered by PINK1-dependent phosphorylation. J. Biol. Chem. 288:22019–22032 10.1074/jbc.M113.467530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., Koyama-Honda I., and Mizushima N.. 2012. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J. Cell Sci. 125:1488–1499 10.1242/jcs.094110 [DOI] [PubMed] [Google Scholar]
- Kane L.A., Lazarou M., Fogel A.I., Li Y., Yamano K., Sarraf S.A., Banerjee S., and Youle R.J.. 2014. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205:143–153 10.1083/jcb.201402104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A., Kondapalli C., Gourlay R., Campbell D.G., Ritorto M.S., Hofmann K., Alessi D.R., Knebel A., Trost M., and Muqit M.M.. 2014. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460:127–139 10.1042/BJ20140334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., Takiyama K., and Koike T.. 2006. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics. 5:749–757 10.1074/mcp.T500024-MCP200 [DOI] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., and Koike T.. 2012. Phos-tag SDS-PAGE systems for phosphorylation profiling of proteins with a wide range of molecular masses under neutral pH conditions. Proteomics. 12:192–202 10.1002/pmic.201100524 [DOI] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., and Shimizu N.. 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 392:605–608 10.1038/33416 [DOI] [PubMed] [Google Scholar]
- Komander D., Reyes-Turcu F., Licchesi J.D., Odenwaelder P., Wilkinson K.D., and Barford D.. 2009. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 10:466–473 10.1038/embor.2009.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondapalli C., Kazlauskaite A., Zhang N., Woodroof H.I., Campbell D.G., Gourlay R., Burchell L., Walden H., Macartney T.J., Deak M., et al. . 2012. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2:120080 10.1098/rsob.120080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., et al. . 2014. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 510:162–166. [DOI] [PubMed] [Google Scholar]
- Lazarou M., Jin S.M., Kane L.A., and Youle R.J.. 2012. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell. 22:320–333 10.1016/j.devcel.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M., Narendra D.P., Jin S.M., Tekle E., Banerjee S., and Youle R.J.. 2013. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J. Cell Biol. 200:163–172 10.1083/jcb.201210111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Sawada T., Lee S., Yu W., Silverio G., Alapatt P., Millan I., Shen A., Saxton W., Kanao T., et al. . 2012. Parkinson’s disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet. 8:e1002537 10.1371/journal.pgen.1002537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima I., Takahashi A., Omori H., Kimura T., Takabatake Y., Saitoh T., Yamamoto A., Hamasaki M., Noda T., Isaka Y., and Yoshimori T.. 2013. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32:2336–2347 10.1038/emboj.2013.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. . 2010. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189:211–221 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.F., and Youle R.J.. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Kane L.A., Hauser D.N., Fearnley I.M., and Youle R.J.. 2010a. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 6:1090–1106 10.4161/auto.6.8.13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., and Youle R.J.. 2010b. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8:e1000298 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Walker J.E., and Youle R.. 2012. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb. Perspect. Biol. 4:a011338 10.1101/cshperspect.a011338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K., Matsumoto M.L., Wertz I.E., Kirkpatrick D.S., Lill J.R., Tan J., Dugger D., Gordon N., Sidhu S.S., Fellouse F.A., et al. . 2008. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 134:668–678 10.1016/j.cell.2008.07.039 [DOI] [PubMed] [Google Scholar]
- Okatsu K., Saisho K., Shimanuki M., Nakada K., Shitara H., Sou Y.S., Kimura M., Sato S., Hattori N., Komatsu M., et al. . 2010. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 15:887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K., Iemura S., Koyano F., Go E., Kimura M., Natsume T., Tanaka K., and Matsuda N.. 2012a. Mitochondrial hexokinase HKI is a novel substrate of the Parkin ubiquitin ligase. Biochem. Biophys. Res. Commun. 428:197–202 10.1016/j.bbrc.2012.10.041 [DOI] [PubMed] [Google Scholar]
- Okatsu K., Oka T., Iguchi M., Imamura K., Kosako H., Tani N., Kimura M., Go E., Koyano F., Funayama M., et al. . 2012b. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat. Commun. 3:1016 10.1038/ncomms2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K., Uno M., Koyano F., Go E., Kimura M., Oka T., Tanaka K., and Matsuda N.. 2013. A dimeric PINK1-containing complex on depolarized mitochondria stimulates Parkin recruitment. J. Biol. Chem. 288:36372–36384 10.1074/jbc.M113.509653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto K., Noda H., and Fujiki Y.. 2014. Distinct modes of ubiquitination of peroxisome-targeting signal type 1 (PTS1) receptor Pex5p regulate PTS1 protein import. J. Biol. Chem. 289:14089–14108 10.1074/jbc.M113.527937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A., Sarraf S.A., Duda D.M., Heo J.M., Jedrychowski M.P., Sviderskiy V.O., Olszewski J.L., Koerber J.T., Xie T., Beausoleil S.A., et al. . 2014. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell. 56:360–375 10.1016/j.molcel.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee S.B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J.M., and Chung J.. 2006. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 441:1157–1161 10.1038/nature04788 [DOI] [PubMed] [Google Scholar]
- Platta H.W., El Magraoui F., Bäumer B.E., Schlee D., Girzalsky W., and Erdmann R.. 2009. Pex2 and pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol. Cell. Biol. 29:5505–5516 10.1128/MCB.00388-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole A.C., Thomas R.E., Yu S., Vincow E.S., and Pallanck L.. 2010. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS ONE. 5:e10054 10.1371/journal.pone.0010054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovic A., Grünewald A., Kottwitz J., Brüggemann N., Pramstaller P.P., Lohmann K., and Klein C.. 2011. Mutations in PINK1 and Parkin impair ubiquitination of Mitofusins in human fibroblasts. PLoS ONE. 6:e16746 10.1371/journal.pone.0016746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley B.E., Lougheed J.C., Callaway K., Velasquez M., Brecht E., Nguyen L., Shaler T., Walker D., Yang Y., Regnstrom K., et al. . 2013. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat. Commun. 4:1982 10.1038/ncomms2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki Y., Kudo T., Sone T., Kikuchi Y., Yokosawa H., Toh-e A., and Tanaka K.. 2009. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 28:359–371 10.1038/emboj.2008.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf S.A., Raman M., Guarani-Pereira V., Sowa M.E., Huttlin E.L., Gygi S.P., and Harper J.W.. 2013. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 496:372–376 10.1038/nature12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba-Fukushima K., Imai Y., Yoshida S., Ishihama Y., Kanao T., Sato S., and Hattori N.. 2012. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2:1002 10.1038/srep01002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba-Fukushima K., Arano T., Matsumoto G., Inoshita T., Yoshida S., Ishihama Y., Ryu K.Y., Nukina N., Hattori N., and Imai Y.. 2014. Phosphorylation of mitochondrial polyubiquitin by PINK1 promotes Parkin mitochondrial tethering. PLoS Genet. 10:e1004861 10.1371/journal.pgen.1004861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.F., Karbowski M., and Youle R.J.. 2010. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191:1367–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe J.F., Sauvé V., Grenier K., Seirafi M., Tang M.Y., Ménade M., Al-Abdul-Wahid S., Krett J., Wong K., Kozlov G., et al. . 2013. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 340:1451–1455 10.1126/science.1237908 [DOI] [PubMed] [Google Scholar]
- Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. . 2004. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 304:1158–1160 10.1126/science.1096284 [DOI] [PubMed] [Google Scholar]
- van Wijk S.J., Fiskin E., Putyrski M., Pampaloni F., Hou J., Wild P., Kensche T., Grecco H.E., Bastiaens P., and Dikic I.. 2012. Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains in cells. Mol. Cell. 47:797–809 10.1016/j.molcel.2012.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. . 2010. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA. 107:378–383 10.1073/pnas.0911187107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Winter D., Ashrafi G., Schlehe J., Wong Y.L., Selkoe D., Rice S., Steen J., LaVoie M.J., and Schwarz T.L.. 2011. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 147:893–906 10.1016/j.cell.2011.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T., and Komander D.. 2013. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 32:2099–2112 10.1038/emboj.2013.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel D.M., Lissounov A., Brzovic P.S., and Klevit R.E.. 2011. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 474:105–108 10.1038/nature09966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley S.E., Murphy A.N., Ross S.A., van der Geer P., and Dixon J.E.. 2007. MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity. Proc. Natl. Acad. Sci. USA. 104:5318–5323 10.1073/pnas.0701078104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodroof H.I., Pogson J.H., Begley M., Cantley L.C., Deak M., Campbell D.G., van Aalten D.M., Whitworth A.J., Alessi D.R., and Muqit M.M.. 2011. Discovery of catalytically active orthologues of the Parkinson’s disease kinase PINK1: analysis of substrate specificity and impact of mutations. Open Biol. 1:110012 10.1098/rsob.110012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Skaug B., Zeng W., and Chen Z.J.. 2009. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFα and IL-1β. Mol. Cell. 36:302–314 10.1016/j.molcel.2009.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K., and Youle R.J.. 2013. PINK1 is degraded through the N-end rule pathway. Autophagy. 9:1758–1769 10.4161/auto.24633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.Y., and Yang W.Y.. 2011. Spatiotemporally controlled initiation of Parkin-mediated mitophagy within single cells. Autophagy. 7:1230–1238 10.4161/auto.7.10.16626 [DOI] [PubMed] [Google Scholar]
- Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J.W., Yang L., Beal M.F., Vogel H., and Lu B.. 2006. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. USA. 103:10793–10798 10.1073/pnas.0602493103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii S.R., Kishi C., Ishihara N., and Mizushima N.. 2011. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J. Biol. Chem. 286:19630–19640 10.1074/jbc.M110.209338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Lee S., Peng Y., Bunker E., Giaime E., Shen J., Zhou Z., and Liu X.. 2014. PINK1 triggers autocatalytic activation of Parkin to specify cell fate decisions. Curr. Biol. 24:1854–1865 10.1016/j.cub.2014.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., and Hunter T.. 2013. Parkin mitochondrial translocation is achieved through a novel catalytic activity coupled mechanism. Cell Res. 23:886–897 10.1038/cr.2013.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviani E., Tao R.N., and Whitworth A.J.. 2010. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl. Acad. Sci. USA. 107:5018–5023 10.1073/pnas.0913485107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material