Reconstruction and Validation of Saccharomyces cerevisiae iND750, a Fully Compartmentalized Genome-Scale Metabolic Model (original) (raw)
Abstract
A fully compartmentalized genome-scale metabolic model of Saccharomyces cerevisiae that accounts for 750 genes and their associated transcripts, proteins, and reactions has been reconstructed and validated. All of the 1149 reactions included in this in silico model are both elementally and charge balanced and have been assigned to one of eight cellular locations (extracellular space, cytosol, mitochondrion, peroxisome, nucleus, endoplasmic reticulum, Golgi apparatus, or vacuole). When in silico predictions of 4154 growth phenotypes were compared to two published large-scale gene deletion studies, an 83% agreement was found between iND750's predictions and the experimental studies. Analysis of the failure modes showed that false predictions were primarily caused by iND750's limited inclusion of cellular processes outside of metabolism. This study systematically identified inconsistencies in our knowledge of yeast metabolism that require specific further experimental investigation.
Metabolic networks are commonly reconstructed either by hand or through the use of automated reconstruction tools based on comparative genomic analysis (Covert et al. 2001). A manual assembly is preferred because it allows for the inclusion of disparate data sources, such as genome annotations, gene expression experiments, enzymatic assays, and physiological data, rather than exclusively genomic data (Covert et al. 2001). Genome-scale metabolic networks have been manually reconstructed for several microorganisms, including Escherichia coli (Pramanik and Keasling 1997; Edwards and Palsson 2000; Reed et al. 2003), Helicobacter pylori (Schilling et al. 2002), Haemophilus influenzae (Edwards and Palsson 1999), and Saccharomyces cerevisiae (Förster et al. 2003a). These models have been shown to be useful for predicting optimal growth phenotypes in various media conditions, even for cases in which the organisms have been genetically modified (Kauffman et al. 2003; Price et al. 2003).
The metabolic network reconstructed by Förster and Famili (Förster et al. 2003a) was the first genome-scale model of yeast. It included 708 open reading frames (ORFs), corresponding to ∼12% of the ORFs identified in the S. cerevisiae genome according to the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org). The Förster–Famili model was also the first genome-scale model to capture one of the most important properties of a eukaryotic cell: compartmentalization. The 842 reactions5 included in the model were localized to the cytosol, mitochondria, or extracellular space. Predictions from this model have been verified by comparison with various sets of physiological data (Famili et al. 2003), gene essentiality data (Förster et al. 2003b), and growth perturbation experiments (P.C. Fu, N.C. Duarte, I. Famili, and B.Ø. Palsson, in prep.). Based on these studies, the following modifications and extensions have been made to improve the Förster–Famili model: (1) the localization of gene products were reevaluated to allow for five additional cellular compartments (peroxisome, nucleus, Golgi apparatus, vacuole, endoplasmic reticulum); (2) the functional assignments of the gene products were revised so that they are consistent with newly published results and are described in terms of elementally and charge balanced reactions, imposing cell-wide proton balance; and (3) associations between the genes, their products, and the metabolic reactions they catalyze were introduced in order to incorporate expression and genomic data.
The culmination of these changes is iND750; a fully compartmentalized S. cerevisiae metabolic model that summarizes our current understanding of the relationship between the ORFs, transcripts, and proteins that define the metabolic capabilities of yeast. The name “iND750” was chosen based on a new convention for naming computational models described in Reed et al. (2003). Briefly, the letter “i” designates an in silico model, “ND” are the initials of the scientist who reconstructed the network, and “750” is the total number of genes accounted for in the model. Similarly, the model of Förster and Famili is referred to as iFF708.
Genome-scale metabolic models such as the one described in this work can be used to calculate experimentally verifiable phenotypic predictions. One of the key scientific uses of these models is to enable systematic improvement of our current knowledge of metabolic networks by comparing model predictions to experimental data. This process corresponds to the notion of iterative model development (Palsson 2000), in which the comparisons between in silico predictions and in vivo data are used to identify potential improvements to the model, which then in turn can be used to design new experiments. In particular, quantitative data on growth rates of individual gene deletions strains under a number of experimental conditions can be directly compared with in silico gene deletions in order to probe specific subcomponents of the metabolic network in detail. These comparisons enable identification of potential problem areas in the network, allow verification of hypothesized metabolic reactions, and suggest specific experiments that can be used to verify components of the network, such as the enzymatic function of particular genes.
For yeast, the availability of whole-genome scale collections of gene deletion strains (both haploid and diploid) has resulted in a proliferation of high-throughput phenotyping studies (Winzeler et al. 1999; Birrell et al. 2001; Fleming et al. 2002; Giaever et al. 2002; Jorgensen et al. 2002; Steinmetz et al. 2002). Of particular interest to metabolism are those studies that specifically test growth on different media (Giaever et al. 2002; Steinmetz et al. 2002) because the relevant conditions can be recreated in silico and the model's predictions are at least qualitatively comparable to the experimental results. In an earlier study (Förster et al. 2003b), the predictions of iFF708 (Förster et al. 2003b) were compared with experimental data on gene essentiality (Winzeler et al. 1999), that is, whether a deletion strain grows when grown in rich media under aerobic conditions. The significantly improved genome-scale model of S. cerevisiae described in this article and the availability of gene deletion data for multiple relevant media conditions, that is, multiple carbon sources in addition to rich media (Giaever et al. 2002; Steinmetz et al. 2002), allows us to evaluate the predictive capability of iND750 as well as systemically suggest a number of potential improvements and extensions to our current model. In particular, the comprehensive gene-protein-reaction associations introduced in iND750 allow metabolic reactions to be removed from the model in an in silico deletion study according to the logical relationships among genes, transcripts, proteins, and reactions, a feature that was not available for simulations with the previous model (Förster et al. 2003a). The results of the present study illustrate the power of model-driven data analysis in connecting specific genotypic changes to phenotypic predictions in order to systematically improve our understanding of a biological system.
RESULTS
Reconstruction of iND750
An earlier metabolic model of S. cerevisiae (referred to here as iFF708; Förster et al. 2003a) served as a starting point for the reconstruction of iND750, a fully compartmentalized yeast model that requires a cell-wide proton balance and includes associations among its genes, proteins, and reactions. This section summarizes our changes to iFF708 as well as key properties of iND750.
Before genes and reactions from iFF708 were included in iND750, it was verified that they were consistent with recently published reports. The extent of the changes that were made to iFF708 to form iND750 is reflected in Table 1, which compares the number of genes, reactions, and metabolites in the two models. Nearly all of the genes in iFF708 are accounted for in iND750. The additional genes primarily encode tRNA synthetases (26 genes) and ATPases found in the vacuole and Golgi apparatus (13). Both models also share a large number of metabolites, although the compartmental location of the metabolites has not been considered in this comparison. Most of the metabolites added to iND750 are found in reactions that have been expanded, that is, reactions that were lumped in iFF708 and are now included as individual steps or with distinct metabolites in the new model. For example, the replacement of a generic ceramide metabolite with two specific moieties led to the introduction of ∼20 additional metabolites in subsequent reactions. The most notable difference between the models is in their reaction sets. Of iND750's 1149 reactions (counting isozymes as separate reactions, iND750 includes a total of 1489 reactions), only 56% are the same as those in iFF708, even after accounting for changes required for elemental and charge balancing of the reactions. Most of these changes are the result of iND750's five additional compartments; many of the reactions that were previously listed as cytosolic were reassigned to a new compartment, and >80 reactions were added to represent the metabolite exchange for these five compartments. Also, as mentioned earlier, many types of metabolic reactions were expanded, especially in fatty acid degradation, in which four individual steps in iND750 replaced the one lumped reaction for each fatty acid included in iFF708. Other changes that are not noted in Table 1 include the following: the introduction of a systemic definition of the associations among genes, proteins, and reactions; the removal of redundant compound abbreviations and duplicated reactions; and updates to gene names and Enzyme Commission (EC) numbers.
Table 1.
Comparison of iFF708 and iND750
| iFF708 | iND750 | % Conserved | |
|---|---|---|---|
| Genes | 708 | 750 | 94 |
| Metabolitesa | 584 | 646 | 90 |
| Unique reactionsb | 842 | 1149 | 56c |
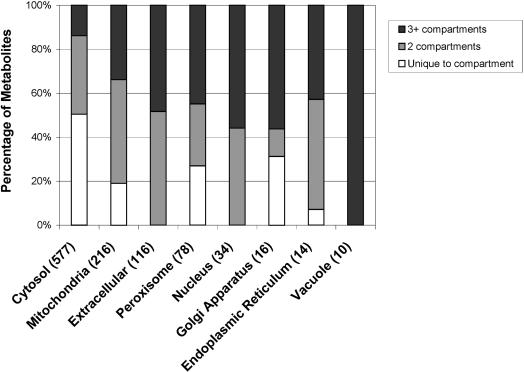
iND750 accounts for eight cellular localizations, three of which were included in iFF708 (extracellular space, cytosol, and mitochondria) and five additional compartments (peroxisome, nucleus, Golgi apparatus, endoplasmic reticulum, and vacuole). To evaluate the connectivity of these compartments, iND750's 646 distinct metabolites were analyzed according to their compartmental location (Fig. 1). Most notably, ∼90% of the metabolites appear in cytosolic reactions. Half of these metabolites are unique to the cytosol; this large percentage is not surprising because reactions were assigned to the cytosol by default (unless there was evidence to the contrary.) The cytosol contains nearly all of the metabolites shared between two compartments because the metabolites must pass through it to be exchanged between the compartments. The seven other compartments vary significantly in their number of metabolites and connectivity. For example, >75 metabolites can be found in the mitochondria, extracellular space, and peroxisome. All of the metabolites in the extracellular space are shared with other compartments, whereas the mitochondria and peroxisome have a defined set of unique metabolites that do not appear in other compartments. The nucleus, Golgi apparatus, endoplasmic reticulum, and vacuole have <35 metabolites, almost all of which can be found in multiple compartments.
Figure 1.
Distribution of S. cerevisiae iND750's 646 unique metabolites in its eight compartments. The number of metabolites found in each compartment is shaded based on its connectivity. Metabolites that are unique to a particular compartment are shown in white; metabolites found in two compartments are shaded in grey; and metabolites found in three or more compartments are shaded in black.
Developing a fully compartmentalized S. cerevisiae network required the addition of many intercompartmental transport reactions. Table 2 shows the 297 transport reactions included in iND750. The majority of these reactions represent transport across the plasma and mitochondrial membranes (for a note on the representation of these membranes, see Methods). The primary transport mechanisms across the plasma membrane and the intracellular membranes are noticeably different. Nearly two-thirds of the metabolites exchanged between the cytosol and the extracellular space occur by symport, typically a primary metabolite and proton transported in the same direction, whereas most of the metabolites exchanged between the intracellular compartments are transported by diffusion. The membranes also vary in their number of gene-associated reactions. The plasma membrane has the largest proportion of gene-associated reactions (∼50%), whereas the nuclear, endoplasmic reticular, Golgi apparatus, and vacuolar membranes do not have any. As a result, many of the transport reactions across the intracellular membranes had to be inferred based on reactions known to take place in these compartments.
Table 2.
Comparison of Transport Reactions Included in iND750
| Transport mechanism (# gene-associated) | |||||
|---|---|---|---|---|---|
| No. of reactions | Diffusion | Symport | Antiport | Other | |
| Extracellular Transport | 113 | 36 (9) | 74 (46) | 3 (1) | 0 |
| Mitochondrial Transport | 101 | 65 (0) | 21 (2) | 14 (13) | 1 (1) |
| Peroxisomal Transport | 39 | 19 (2) | 6 (0) | 5 (0) | 9 (9) |
| Nuclear Transport | 23 | 18 (0) | 5 (0) | 0 | 0 |
| Endoplasmic Reticular Transport | 10 | 9 (0) | 1 (0) | 0 | 0 |
| Vacuolar Transport | 7 | 5 (0) | 2 (0) | 0 | 0 |
| Golgi Apparatus Transport | 4 | 3 (0) | 0 | 1 (0) | 0 |
All of the compartments in iND750 were assumed to have a pH of 7.2. Consequently, the charge and formulae of all metabolites were determined by their ionization form at this pH. By including water molecules and protons in iND750's reactions, >99% could be written so that they were both elementally and charge balanced. The few imbalanced reactions are typically those catalyzed by enzymes with a mechanism that is not fully understood, such as biotin synthase (E.C. 2.8.1.6). Structuring the reactions in this manner forces the proton production and consumption to be balanced within each compartment and thus in the entire cell. This global proton balancing has implications for cellular growth, as has been demonstrated for E. coli grown on various carbon sources (Reed et al. 2003).
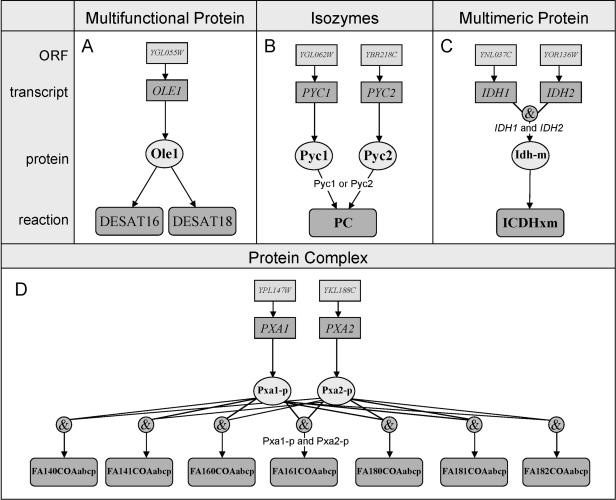
Unlike iFF708, which does not systematically represent the relationship between its genes and reactions, iND750's gene-protein-reaction associations can be viewed as graphical representations of the logical relationships between its ORFs, transcripts, proteins, and reactions. For example, proteins classified as multifunctional can catalyze more than one reaction (Fig. 2A). Distinct proteins that can individually catalyze a reaction are defined as isozymes (Fig. 2B). Multimeric proteins are defined as those formed by more than one transcript (Fig. 2C). Finally, a protein complex is a set of proteins that are required to catalyze a reaction (Fig. 2D). A catalog of all of iND750's gene-protein-reaction associations can be found in the Supplemental material. A similar genome-scale set of gene-protein-reaction associations has been established for Escherichia coli (Reed et al. 2003).
Figure 2.
Examples of gene-protein-reaction associations that represent the detailed logical relationships between open reading frames (ORFs), transcripts, proteins, and reactions in the model. (A) A multifunctional protein, such as Ole1p, can catalyze more than one reaction. (B) Pyc1p and Pyc2p are examples of isozymes, or proteins that can catalyze the same reaction independently. (C) Idhp is an example of a multimeric protein; it is formed by the association of two transcripts. (D) Proteins Pxa1p and Pxa2p form a protein complex. Both proteins are required to catalyze the reactions. All the gene-protein-reaction associations in Saccharomyces cerevisiae iND750 are found in the Supplemental materials as well as at http://systemsbiology.ucsd.edu.
Gene Deletion Study
The genome-scale compartmentalized metabolic model of S. cerevisiae described above was validated and interrogated in detail by comparing model predictions for deletion strain phenotypes with published results from two large-scale growth phenotyping studies (Giaever et al. 2002; Steinmetz et al. 2002) for seven different media conditions. The media conditions included in this study were aerobic growth on glucose minimal media (MMD) and on rich media with six different carbon sources: glucose (YPD), galactose (YPGal), glucose-ethanol-glycerol mixed media (YPDGE), glycerol (YPG), ethanol (YPE), and lactate (YPL). Four of the carbon sources allow fermentative growth (MMD, YPD, YPGal, and YPDGE) and three allow only nonfermentative growth (YPG, YPE, and YPL). In addition to the seven media conditions described above, one of the experimental studies (Giaever et al. 2002) also separately reported genes for which deletions strains could not be constructed (essential genes) and genes with deletion that leads to a slow growing strain on rich media (slow growth genes). The data sets from the two different experimental studies are partially overlapping, because the genes with deleterious phenotypes under the YPD condition in Steinmetz et al. (2002) should agree with the essential/slow growth genes in Giaever et al. (2002). However, because the experimental designs and data analysis methods used in the two studies were different, the two gene lists do not necessarily always agree.
In silico gene deletions were performed by using established procedures using flux balance analysis (FBA; Varma and Palsson 1994; Bonarius et al. 1997; Price et al. 2003) as described in the Methods section. The media conditions for the simulations were set to match the experimental conditions as closely as possible (see Methods). The experimental data was obtained from the two different data sources (Giaever et al. 2002; Steinmetz et al. 2002) and preprocessed as described in the Methods section. Lists of essential and slow growth genes (as described above) as well as phenotyping data for the MMD and YPGal media were obtained from Giaever et al. (2002), and the phenotyping data for the remaining conditions were obtained from Steinmetz et al. (2002). To make the in vivo data and in silico predictions comparable, both were converted from continuous-value relative fitness scores to a discrete viable/retarded growth assessment for each gene deletion strain and condition as described in the Methods. This data transformation was done as it was not expected that the in silico predicted growth rates would necessarily quantitatively match in vivo fitness scores obtained from the experimental data. In addition, the two experimental studies used different approaches to measure the fitness of each deletion strain so that these two different fitness scores had to be made comparable.
To facilitate analysis of the results, each in silico phenotype prediction was classified into one of four categories following the convention used in Förster et al. (2003b): True positive (TP; experimentally and in silico viable), true negative (TN; experimentally and in silico growth retarded), false positive (FP; experimentally growth retarded, in silico viable), and false negative (FN, experimentally viable, in silico growth retarded). Deletion phenotype predictions for at least one condition were done for 682 of the total of 750 genes in the model, and the predictions were classified as described above. No experimental deletion data were available for the remaining genes.
The number of model predictions in each of the four categories described above for each growth condition as well as the overall totals for all conditions taken together are shown in Table 3. A total of 4154 comparisons between in silico and in vivo deletions were analyzed in this study, representing, to date, the largest evaluation of the predictive power of genome-scale metabolic models. The overall correct prediction rate was 82.6%, which is similar to that obtained in more limited studies with other organisms as well as yeast (Edwards and Palsson 1999, 2000; Schilling et al. 2002; Förster et al. 2003b). The TP rate (TP predictions/total number of in vivo normal growth phenotypes) was 96.6%, indicating that the model correctly captures the built-in redundancy in metabolism in that most gene deletions have no phenotypic effect under most conditions. However, the FP rate (FP predictions/total number of in vivo deleterious phenotypes) was 77.0%, showing that less than one-fourth of slow growth phenotypes were predicted correctly.
Table 3.
Overall Results for the Comparison Between In Silico and In Vivo Gene Deletions
| Essential (118) | Slow (83) | MMD (564) | YPGal (564) | YPD (565) | YPDGE (565) | YPG (565) | YPE (565) | YPL (565) | All (4154) | |
|---|---|---|---|---|---|---|---|---|---|---|
| TP | 0 | 0 | 439 | 476 | 474 | 465 | 466 | 461 | 466 | 3247 |
| FP | 81 | 67 | 74 | 69 | 73 | 64 | 62 | 60 | 61 | 611 |
| TN | 37 | 16 | 35 | 7 | 3 | 17 | 23 | 23 | 22 | 183 |
| FN | 0 | 0 | 16 | 12 | 15 | 19 | 14 | 21 | 16 | 113 |
| Total | 118 | 83 | 564 | 564 | 565 | 565 | 565 | 565 | 565 | 4154 |
| Unique false | 81 | 3 | 17 | 4 | 15 | 7 | 2 | 6 | 4 | 139a |
| Correct rate | 31.4 | 19.3 | 84.0 | 85.6 | 84.4 | 85.3 | 86.5 | 85.7 | 86.4 | 82.6 |
| TP rate | — | — | 96.5 | 97.5 | 96.9 | 96.1 | 97.1 | 95.6 | 96.7 | 96.6 |
| FP rate | 68.6 | 80.7 | 73.4 | 85.5 | 96.1 | 79.0 | 72.9 | 72.3 | 73.5 | 77.0 |
The correct prediction rates for each media condition (after correcting the MMD and YPGal data to include the slow growth predictions) were very similar, ranging from 84.0% for MMD to 86.6% for YPG. Surprisingly, the FP rate on glucose (YPD) was higher than the rate on other substrates even though the mechanisms of glucose utilization are much better established than those of, for example, glycerol or lactate. Most of the FP predictions were for genes defined as essential or slow growth on rich media, for which the FP rates were 68.6% and 80.7%, respectively. Largely the same set of genes were responsible for the false predictions in all media conditions, indicating that most of the model inaccuracies were not condition dependent. However, there were a number of false predictions unique to each condition, ranging from two (YPG) to 17 (MMD). These unique false predictions are particularly useful for model improvement as they may potentially suggest specific improvements to the model as described in the Discussion.
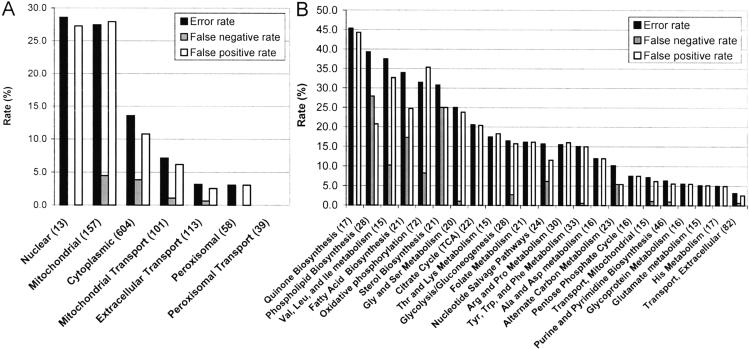
To further investigate the sources of the false predictions, we analyzed their distribution with respect to cellular compartments and metabolic subsystems. The overall false prediction rates as well as FN and FP rates for genes in particular cellular compartments are shown in Figure 3A. Genes localized to the nucleus and mitochondria had the highest false prediction rate (28.6% and 27.4%), whereas this rate was much lower (13.5%) for cytosolic genes. Figure 3B shows the false prediction rates for genes associated with major metabolic subsystems included in the model. The highest false prediction rates were obtained for genes involved in quinone biosynthesis (45.3%), followed by phospholipid biosynthesis (39.3%) and branched chain amino acid biosynthesis (37.5%). Subsystems with high false prediction rates also included oxidative phosphorylation (31.4%), mirroring the tendency for mitochondrial gene deletions to be falsely predicted as seen in Figure 3A. The lowest false prediction rates were obtained for genes involved in extracellular transport (3.2%), histidine metabolism (5.0%), and glutamate metabolism (5.1%). Overall, the distribution of the false predictions can be seen to be quite uneven, with a few metabolic subsystems accounting for the majority of the problems.
Figure 3.
False prediction percentages for genes in particular cellular compartments (A) and particular metabolic subsystems (B). The overall error rate is the percentage of false predictions out of all of the predictions. The false-negative (FN) rate is the percentage of FN predictions out of all predictions in which the experimental data show normal growth. The false-positive (FP) rate is the percentage of FP predictions out of all predictions in which the experimental data show retarded growth. Genes that participate in transport functions between compartments are classified according to Table 2. Compartments with at least 10 genes and metabolic subsystems with at least 15 genes are included.
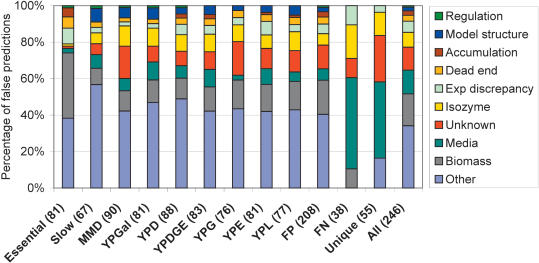
The reasons for the false prediction for each of the 246 genes with false predictions under one or more conditions were individually evaluated by both studying relevant literature on previously determined mutant phenotypes for the gene and by interrogating its role in the metabolic model. The results of this evaluation for each of the media conditions separately as well as for all false positives and false negatives, for false predictions under a unique condition, and for all false predictions together are shown in Figure 4. The primary sources of false predictions were organized into 10 different categories (detailed in the caption for Fig. 4). Overall, more than half of the false predictions can be accounted for by the involvement of the genes in other cellular processes in addition to metabolism (33.7%) and problems in the biomass composition assumed in the in silico deletion study (17.5%). Interestingly, the reasons for FP and FN predictions were quite different, with majority of the FPs arising for the abovementioned reasons, whereas the majority of FNs could be traced to uncertainty in the in silico media composition (50.0%) and issues related to the gene-protein-reaction relationships in the model (18.4%). The different media conditions had similar distribution of the sources of false predictions, but the majority of the false predictions that arose because of missing in silico biomass components were related to essential genes. The sources of false predictions for genes with a unique false prediction under one experimental condition were also quite different from the overall pattern, with a particularly high fraction of false predictions with no clear reason for the false assessment (25.5%).
Figure 4.
Breakdown of the false predictions by the source of false prediction. The reasons for false predictions are as follows: transcriptional regulation (regulation), model structure, accumulation of toxic intermediate in vivo (accumulation), dead ends in the model (dead end), discrepancy in the experimental data (exp discrepancy), gene–protein reaction associations (isozyme), unknown, in silico media composition (media), in silico biomass composition (biomass), and other cellular processes not included in the model (other). Results are shown for each experimental condition, including essential genes (essential) and slow growth genes (slow) on rich media. In addition, the distributions of the sources of false predictions are shown for false-positive (FP), false-negative (FN), and unique false predictions (unique) separately.
DISCUSSION
The new genome-scale metabolic model iND750 is an expansion of the initial genome-scale metabolic reconstruction of S. cerevisiae (Förster et al. 2003a). Unlike its predecessor, iND750 is fully compartmentalized, accounting for eight localizations (extracellular space, cytosol, mitochondria, peroxisome, nucleus, Golgi apparatus, endoplasmic reticulum, and vacuole). The expanded network also includes gene-protein-reaction associations that represent the logical relationships among iND750's 750 genes and their corresponding transcripts, proteins, and functional activities. Finally, the 1149 unique reactions that describe iND750's metabolic capabilities have been formulated so that they are both elementally and charge balanced to allow for the enforcement of a cell-wide proton balance. It was verified separately that iND750 is capable of predicting whole-cell functions such as P/O ratios and byproduct secretion rates under a variety of conditions with similar or improved accuracy compared with iFF708 (Famili et al. 2003; data not shown).
After successfully reconstructing iND750, a large-scale phenotyping experiment was performed in silico to comprehensively evaluate its performance. The network's predictions of growth phenotypes of knockout strains for seven different media conditions were found to agree with 3430 of the 4154 phenotypes reported in two large-scale in vivo deletion studies. The overall prediction performance in this study (82.6% correct phenotypes) was lower than that obtained in the previous study (Förster et al. 2003b) with iFF708 (85%), in which data on gene essentiality only was used. When predictions by the two models were compared in detail, it was found that iND750 made correct predictions on gene essentiality in six cases that were incorrectly predicted by iFF708. On the other hand, iND750 made incorrect predictions for two genes with essentiality that was correctly predicted by iFF708. The main reasons for the overall lower prediction accuracy of iND750 were the larger number of media conditions considered in this study and the inclusion of a number of new genes in iND750 that have a role in other cellular processes in addition to metabolism.
The results of the in silico deletion study showed surprisingly large variability in the false prediction rates between genes in different compartments. The nuclear and mitochondrial compartments had the highest overall error rate, and most of these errors were FP predictions. Because the mitochondria were shown to have a distinct set of metabolites (Fig. 1), it seems surprising that iND750 may not have fully captured its unique role in cellular growth. Further analysis of the failure modes in terms of pathways revealed that these false predictions might be due to the fact that the model does not accurately represent mitochondrial maintenance. Also, in this model, we have assumed that the outer mitochondrial membrane is like a sieve, allowing free diffusion of metabolites; however, there is evidence to suggest that the permeability of outer mitochondrial membrane may be regulated (Mannella 1992). This variation in permeability may have important implications for controlling energy metabolism (Vander Heiden et al. 2000). Peroxisomal reactions were one of the most significant additions to iFF708 as the peroxisome has its own defined set of metabolites and plays a crucial role in the degradation of fatty acids. The high correct prediction rate obtained for peroxisomal genes (96.9%) indicates that the model fairly accurately accounts for the metabolic function of this important cellular compartment.
Because iND750 represents the current understanding of metabolism in yeast as completely as possible within a stoichiometric model, analysis of its failure modes is important because they can be used to highlight inconsistencies in the body of information used in the reconstruction. The 10 categories of sources of false predictions described in the caption for Figure 4 are discussed in detail below, with appropriate examples given for each category. The classifications of each false prediction into these categories as well as detailed explanations for the false prediction are included in the Supplemental material to this article. In many cases, the false predictions led to direct suggestions of how to potentially improve the model or of specific experiments that could be performed to further improve our understanding of yeast metabolism. In the following, we will focus on examples that were not described in the earlier study (Förster et al. 2003b) or on examples in which the interpretation of the reason for the false prediction has changed due to a more complete model or data available under multiple experimental conditions.
Model Structure
Analysis of the sources of false predictions revealed five genes for which the false predictions are probably due to missing or extraneous functionalities in the model. POS5 (coding for a mitochondrial NADH/NADPH kinase) deletion resulted in a FP prediction, because the model can produce NADPH in mitochondria using other mechanisms, whereas it has been recently shown experimentally that Pos5p is the major source of mitochondrial NADPH (Outten and Culotta 2003). The FP predictions for ERG2, ERG3, and ERG6 are due to a bypass in the model in ergosterol metabolism that allows direct synthesis of ergosterol from zymosterol. Although this bypass has been suggested to exist in yeast (Parks 1978), based on the current study it appears that this alternate route in yeast does not bypass Erg2p, Erg3p, and Erg6p. The mitochondrial pyrophosphatase PPA2 deletion is a FP, because the model can use a cytosolic pyrophosphatase instead and transport phosphate and pyrophosphate between the two compartments. If this transport capacity were limited, as it is likely to be in vivo, the PPA2 deletion would result in a lower growth rate due to limitation in mitochondrial metabolism. All these false predictions directly suggest potential changes to the actual structure of the model and also possibly the need to reevaluate our understanding of the specific parts of yeast metabolism as in the case of the ergosterol biosynthetic pathway.
Gene-Protein-Reaction Associations
The false predictions relating to gene-protein-reaction associations are primarily due to either potentially missing isozymes (FNs) or the existence of a dominant isozyme with activity that can not be fully compensated for by the other isozymes (FPs). The Gal2p galactose transporter is an example of the latter class, as it is known that other hexose transporters can also transport galactose (Wieczorke et al. 1999), but based on the comparison between simulation results and experimental data, it appears that these transporters are insufficient to maintain maximal galactose uptake. A typical example of the latter class is the FN prediction for Bat2p transaminase, which was found to be due to the lack of valine transamination functionality of the Bat1p isozyme in the model. This function has not been experimentally proven (Kispal et al. 1996), but based on the results presented here, it appears likely that BAT1 gene product can catalyze valine transamination in addition to other transamination reactions. The false predictions that were due to gene-protein-reaction associations suggest modifications to the model that relate to how the gene-to-enzymatic function mapping occurs in vivo.
Regulatory Mechanisms
The lack of incorporation of regulatory mechanisms in the model could only clearly explain false model predictions for two of the genes—CDC19 (pyruvate kinase) and ADH1 (alcohol dehydrogenase). Both of these genes have isozymes that are capable of catalyzing the same reaction but are known to be down-regulated under the particular condition in which the FP prediction was done. The lack of regulatory restraints in the current model could also partially explain the observed general pattern of higher false prediction rates for conditions with glucose as the main carbon source, as one would expect that the model would otherwise be more accurate for glucose than for less well characterized carbon sources. Because of the extensive metabolic reprogramming in glucose-grown cells using different glucose repression mechanisms, regulation plays a more significant role on glucose-containing media than on other media conditions. In future generations of constraint-based metabolic models, transcriptional regulation will be at least qualitatively incorporated in the models (Covert and Palsson 2002) so that regulatory effects will be more accurately accounted for.
Dead Ends in the Model
For eight genes with FP predictions, the reaction catalyzed by the gene product leads to a dead end in the model, whereas in vivo the product of the reaction clearly is necessary for cellular function. This result indicates that either the model is missing some metabolic functions or there are gaps in the literature in understanding specific metabolic subsystems. Many of the dead ends are in phospholipids metabolism in which the corresponding genes participate in the biosynthesis of complex phospholipids that are not used within the model, but that are probably converted to essential membrane phospholipids. Not all the dead ends in the model result in false predictions so that the eight-gene subset provides direct suggestions for further experimental work necessary for understanding the role of the currently unused metabolites in yeast cellular function.
Accumulation of Toxic Intermediates
In a few cases, the primary reason for a FP prediction by the model appears to be the accumulation of a toxic intermediate in vivo when a particular enzyme further down the pathway is removed. For example, although folate biosynthesis is not required in rich media, genes involved in the biosynthetic pathway (FOL1/FOL2/FOL3/DFR1) are essential, which is most likely due to toxicity of dihydropteorate (DHP), a precursor in the pathway (Bayly and Macreadie 2002). Similarly, in vivo _MET22_-null mutant accumulates phosphoadenylyl sulfate (PAPS), which is cytotoxic (Thomas et al. 1990). The in silico model does not account for nonspecific chemical toxicity effects because these are usually not directly related to the metabolic function, but it is also possible that the model allows balancing of a toxic intermediate even if this would not happen in vivo and hence fails to predict the deleterious phenotype.
Media Composition
Uncertainties in the in silico media compositions used to mimic the experimental conditions were the primary source of 32 false predictions, most of which were FNs. There are two separate sources of errors that can be identified: (1) wrong media composition, and (2) incorrect numerical values of maximum uptake rates of key nutrients. The former category includes examples such as TPS1/2 (trehalose 6-phosphate synthase/phosphatase), which both are FP predictions on rich media due to the fact that the in silico YP medium contains trehalose, and hence, these genes that are essential for trehalose biosynthesis are not needed. However, it has been shown that trehalose is indeed a major component of the yeast extract medium (Zhang et al. 2003) so that the FP prediction is probably due to the inability of the yeast to use the trehalose in the media in the experimental deletion studies. The latter category of errors is typically manifested as a function of either the major carbon source or oxygen uptake rate or both. For many genes involved in mitochondrial respiration, either lowering or raising the oxygen uptake rate would result in better predictive power. However, the maximum oxygen uptake rates in a batch culture are hard to estimate as they depend both on the degree of aeration provided and on the growth rate–dependent limitations due to the Crabtree effect (for details, see Methods). Most of the false predictions unique to a specific experimental condition could be traced back to uncertainties in the in silico media composition or maximal uptake rates, indicating that a more careful evaluation of these failure modes would require performing the in vivo deletion studies in well-defined media conditions that can be reproduced more accurately in silico.
Biomass Composition
As noted already in our earlier deletion study using iFF708 (Förster et al. 2003b), the biomass composition used in the model is a major source of false predictions as it determines which metabolites are considered to be essential for cellular function and in what relative quantities these metabolites have to be produced. The biomass composition is derived primarily from experimental data on the composition of yeast cells growing in the exponential phase, and it only includes the major biomass components, as measuring trace components is difficult (Förster et al. 2003a). Typical examples of FP predictions by the model are all genes involved in heme and quinone biosynthesis as these cofactors are obviously essential for cellular function. However, although the model uses these and other cofactors, they are recycled in the reactions, and unless there is a drain of cofactors to the biomass, they do not need to be synthesized de novo. An example of FN predictions that relate to the in silico biomass composition are certain genes in membrane lipid and steroid biosynthesis. Although some of the lipids are essential, they can often be used interchangeably by the cell so that any particular type of lipid or sterol may not be essential as long as sufficient overall amount of, for example, phospholipids is produced. Because the model biomass requires fixed amounts of certain types of phospholipids and steroids, this leads to FN predictions. The false predictions due to the biomass composition could be easily corrected by including trace amounts of essential cofactors in the biomass and allowing more flexible usage of phospholipids and steroids, but it would be difficult to estimate exactly the relative amounts of the metabolites required without further experimentation.
Other Cellular Processes
The single most common source of false predictions in this study was the involvement of metabolic genes in other cellular processes that are not accounted for in the current model. As mentioned earlier, the model does not currently include mRNA and protein synthesis, and thus, all pathways resulting in the biosynthesis of various RNA species such as transfer RNAs are dead ends, although these functions are clearly essential for cellular function. Because methods for incorporating protein synthesis into the constraint-based modeling framework have been developed (Allen and Palsson 2003; Allen et al. 2003), in future versions of the model, these currently missing functionalities can be accounted for. Another type of FP prediction that arises from the involvement of metabolic genes in other cellular processes is the role of these genes in overall cellular maintenance. For example, FP predictions were made for all vacuolar ATPase components, as their disruption in vivo results in major problems in pH balancing and the current model does not yet implement full pH balancing between compartments. Similarly, although the model does correctly predict the phenotypes for deletions of ATP synthase subunits on nonfermentable carbon sources, on fermentable carbon sources the model does not require the mitochondrial ATP synthase, although in vivo this functionality is required for general mitochondrial maintenance. As the constraint-based framework is extended to include other types of cellular processes in addition to metabolism and regulation, it can be expected that many of the false predictions will be corrected and that the comparison between in silico and in vivo gene deletions will provide valuable assistance for the expanded model building.
Discrepancies in Experimental Data
There were 16 genes with false predictions for which apparent discrepancies in experimental data were found. These included cases such as PRO3, which is listed as an essential gene in one study (Giaever et al. 2002) but appears to be nonessential in the other study (Steinmetz et al. 2002). In addition to discrepancies between the two genome-wide deletion studies, there were also genes with a phenotype in the large-scale studies that disagreed with the reported phenotype in the literature (e.g., _THR1_-null mutant should only be a threonine auxotroph and should grow on rich media). False predictions for cases in which apparent discrepancies in experimental data were found were not further analyzed as it was not clear which data set would be the most trustworthy.
Unknown Sources of False Predictions
There were 31 genes with predicted false phenotypes that could not be explained by any of the reasons listed above, even after careful evaluation of both the model and experimental data. Many of the genes in this list are related to a few separate metabolic subsystems with false phenotypic predictions under specific media conditions, indicating that there may be important unidentified biochemical mechanisms present in these systems. An especially interesting example is the high number of false predictions related to methionine and homocysteine biosynthesis, which have been extensively studied both in yeast and in higher eukaryotes because of the role of homocysteine in cardiovascular and neurodegenerative diseases (Lievers et al. 2003; Mattson and Haberman 2003). The key gene in this system is MET6, which codes for the methionine synthetase responsible for converting homocysteine into methionine. This deletion has no phenotype on rich media in vivo, but the model predicted the deletion to be lethal due to inability to balance homocysteine in absence of the methionine synthetase reaction. However, the model currently accounts for all of the biochemical transformations with homocysteine either as a reactant or product that are known to be present in yeast, indicating that there may still be some unknown mechanism by which homocysteine balancing is accomplished in vivo. The genes with false predictions with no clearly identifiable reason for the false result provide clues to areas in which further experimental work is clearly needed in order to improve our understanding of eukaryotic metabolism.
Taken together, the detailed analysis of model failures presented above resulted in the 27 direct suggestions for improving the current model listed in Table 4 either by changing its reaction structure or the gene-protein-reaction associations. Some of these suggestions are straightforward, such as making a component of a complex nonessential for the enzymatic function, whereas others, such as restricting phosphate transport across the mitochondrial membrane, would be somewhat more challenging to implement. For all of the 27 cases, the model represents the current knowledge on metabolic biochemistry, genetics, and physiology as given the currently available information, and the changes relate primarily to how the available information is interpreted. These suggestions demonstrate how model-driven evaluation of experimental data (in this case gene deletion phenotypes) can be used to systematically fine tune the model and hence improve our understanding of a particular biological system.
Table 4.
Suggested Changes to Model Structure Based on the Gene Deletion Study
| ORF | Gene | Reason for false predictiona | Suggested change and comments |
|---|---|---|---|
| YPL188W | POSS | Mod | Change the model so that only Pos5p can provide NADPH in mitochondria |
| YMR267W | PPA2 | Mod | Force the model to use Ppa2p instead of the cytoplasmic isoforms by restricting phosphate transport out of the mitochondria. |
| YMR202W | ERG2 | Mod | Modify the interconversion between zymosterol and ergosterol biosynthesis to require ERG2. |
| YLR056W | ERG3 | Mod | See ERG2. |
| YML008C | ERG6 | Mod | See ERG2. |
| YDR178W | SDH4 | Iso | Make Sdh4p a nonessential part of the succinate dehydrogenase complex. |
| YML123C | PHO84 | Iso | There are multiple alternative isozymes for the phosphate transporters, but Pho84p should be the dominant one. |
| YBR069C | TAT1 | Iso | There are multiple alternative isozymes for amino acid transporters in the model, but they need to be made less efficient than Tat1p. |
| YLR081W | GAL2 | Iso | Model includes other isozymes (HXT genes) that are not nearly as efficient for gal transport, so disabling their gal transport ability should result in a correct prediction. |
| YMR105C | PGM2 | Iso | Pgm2p is major isoform of phosphoglucomutase; do not allow the minor isoform (Pgm1p) to fully compensate for loss of Pgm2p. |
| YHR137W | ARO9 | Iso | Aro8p should be able to compensate for ARO9 deletion on minimal media; modify the gene-protein-reaction association to reflect this. |
| YGL125W | MET13 | Iso | Met13p is the dominant isozyme; do not allow isozyme (Met12p) to compensate fully for the loss of Met13p. |
| YHR046C | INM1 | Iso | Add the gene product of YDR287W as an isozyme for Inm1p. |
| YHR001WA | QCR10 | Iso | This subunit should be made a nonessential part of the cytochrome bc1 complex since it only plays a structural role. |
| YFR033C | QCR6 | Iso | Deletion of QCR6 does not have significant effect on the formation or stability of cytochrome bc complex so that it should not play an essential role in complex formation. |
| YKL067W | YNK1 | Iso | Null mutant retains 10% of nucleoside diphosphate kinase activity. Sources of remaining enzyme activity are unknown. Reaction without gene associations should be added to the model to represent these unidentified enzymes. |
| YLR304C | ACO1 | Iso | The isozyme coded by YJL200C should not be able to fully compensate for ACO1 deletion. |
| YNL052W | COX5A | Iso | Cox5Ap is the dominant isoform; Cox5Bp should not be able to fully compensate. |
| YKL148C | SDH1 | Iso | Sdh1p should not be considered to be an essential part of the succinate dehydrogenase complex. |
| YGL008C | PMA1 | Iso | This is the major isoform of the cytosolic ATPase, but in the model a minor isoform (which contains Pma2p instead of Pma1p) can compensate for the function. Do not allow the minor isoform to fully compensate for the loss of the major isoform. |
| YLR342W | FKS1 | Iso | There are three alternate isozymes in the model, but Fks1p should be made the dominant isozyme. |
| YHR183W | GND1 | Iso | This is the major isozyme (80% of activity); other isozymes should be made less efficient. |
| YLR044C | PDC1 | Iso | There are three alternate isozymes in the model, but PDC1 deletion alone is sufficient to reduce pyruvate decarboxylase activity significantly enough to result in a slow growth phenotype. Should have Pdc1p as the major isozyme. |
| YJR148W | BAT2 | Iso | BAT2 single deletion should not be lethal as there is a mitochondrial isozyme (Bat1p); double deletion should be lethal. Bat1p currently does not catalyze valine transamination so this functionality should be added. |
| YCL009C | ILV6 | Iso | Ilv6p is the regulatory subunit of phenylalanine transaminase. This subunit should be made nonessential for the enzymatic function. |
| YAL038W | CDC19 | Reg | Pyk2p isozyme should only be expressed under conditions of very low glycolytic flux. |
| YOL086C | ADH1 | Reg | This isozyme (out of five) should be the only one active under severely glucose repressed conditions. |
Conclusions
We have shown that multicompartmental in silico metabolic models of eukaryotic cells with elementally and charge balanced reactions can be successfully built. In addition, these models can be used to compute growth phenotypes of organisms with altered genotypes in various media conditions. The growth phenotypes computed with the compartmentalized eukaryotic model were found to be consistent with 83% of the in vivo results. Detailed case-by-case analysis of the false predictions led to the identification of gaps or inconsistencies in our knowledge base that require either changes in the model structure or further experimental investigation. This high correct prediction rate demonstrates the growing predictive power of constraint-based metabolic models even under variable environmental conditions and the overall importance of network topology in determining phenotypic consequences of genotypic changes.
METHODS
Model Reconstruction
The S. cerevisiae genome-scale metabolic network reconstructed by Förster and Famili (iFF708; Förster et al. 2003a) was used as a basis for the development of iND750. Starting with the list of ORFs included in iFF708, the corresponding gene names, Enzyme Commission (EC) numbers, and reactions were all reevaluated to check their consistency with recently published reports. Special attention was given to compartmentalization, elemental and charge balancing of reactions, and the relationships among genes, proteins, and reactions, which are discussed below. The updated metabolic network was then constructed by using the SimPheny software package (Genomatica).
Compartmentalization
Because reactions in iFF708 were restricted to only the cytosol, mitochondria, and extracellular space, the localization of each gene product was revised to take into consideration the five additional compartments included in iND750 (peroxisome, endoplasmic reticulum, Golgi apparatus, nucleus, and vacuole). Information on the localization of the gene products was primarily taken from the SGD (Weng et al. 2003) and Comprehensive Yeast Genome Database (Mewes et al. 2002). If there was little or no evidence that a gene product was found in a particular compartment, then it was assumed to be located in the cytosol. An additional assumption was also needed for membrane proteins because oftentimes there was no evidence regarding the location of their catalytic domains. Unless there was evidence to the contrary, it was assumed that reactions catalyzed by membrane proteins occurred in the cytosol. Finally, all of the compartments were modeled as if there were only one boundary between the cytosol and its lumen. For example, because the intercompartmental space of the mitochondria is considered to be equivalent to the cytosol in its metabolite and ion concentrations (Voet et al. 1999), proteins that are localized to these regions are considered cytosolic. Similarly, the cell wall and periplasmic space are both treated as part of extracellular compartment.
Intercompartmental Transport
Additional transport reactions were needed to describe the exchange of compounds between the eight cellular compartments of iND750. The transport processes across the plasma membrane have been well studied; many genes have been identified that encode transport proteins (for a comprehensive list, see Walker 1998; Dickinson and Schweizer 1999). These genes and their documented transport mechanisms have been included in iND750. In addition, many metabolites are known to diffuse across the yeast cell wall (Walker 1998; Dickinson and Schweizer 1999). For those compartments in which there was little information about transport processes, most of the exchange reactions had to be inferred. A primary assumption was that a particular compound was transported across various membranes by a similar process. For example, because tyrosine is known to cross the plasma membrane via proton symport, it was also assumed to be transported across the peroxisomal membrane by the same mechanism. Transport reactions were also inferred based on the known characteristics of some membranes, such as the nuclear membrane, which contains pores that allow substrates <9 nm or 60 kD to pass freely (Allen et al. 2000). Consequently, most of the compounds transported into and out of the nucleus are exchanged by simple diffusion.
Elemental and Charge Balancing
The reactions in iND740 are elementally and charge balanced. The formula and charge of the metabolites were determined based on their ionization state at a pH of 7.2. For simplicity, all of the compartments were assumed to have the same pH. By introducing ionized compounds, water molecules and protons that participate in the reactions are explicitly accounted for so that the reactions had no net charge change and obeyed elemental balances. Water molecules were allowed to freely diffuse into all of the compartments. However, the protons could only enter and leave the various compartments by participating in active transport reactions. Thus, the production and consumption of protons had to be balanced within each compartment.
Gene-Protein-Reaction Associations
Unlike the iFF708, which only considered one-to-one associations between genes and reactions, the logical relationships among genes, proteins, and reactions are all modeled in iND750. To do this, the entry of each gene was examined to see if there was any evidence that its gene product was multifunctional, an isozyme, a protein subunit, or a participant in a protein complex. Multifunctional proteins were defined as those that can catalyze more than one reaction (Fig. 2A). Distinct proteins that could catalyze the same reaction were labeled as isozymes (Fig. 2B). Proteins were classified as multimeric if more than one transcript was required to catalyze an enzymatic function (Fig. 2C). Key words used to identify multimeric proteins were “chains” or “subunits” of proteins. Proteins could also form complexes; this is defined as a functional entity in which proteins from different transcripts must act together to catalyze a reaction (Fig. 2D). There were also more complex cases in which a protein belonging to a complex was made up of subunits, such as in the fatty acid synthase complex.
The reaction and metabolite lists, metabolic network maps, and gene-protein-reaction associations for S. cerevisiae iND750 can be found in the Supplemental material as well as at http://systemsbiology.ucsd.edu.
Gene Deletion Study
In Silico Gene Deletions
The constraint-based framework for computing metabolic phenotypes based on a description of the reaction stoichiometry of the metabolic network of an organism has been described elsewhere (Covert et al. 2001; Edwards et al. 2002), but the basic computational approach used in this work is described briefly below. The allowed solution space for steady-state metabolic fluxes is determined by the null space of the stoichiometric matrix, reaction directionality constraints (reversibility), and the maximal reaction rates for each reaction (if known). These constraints describe a convex solution space, which contains all the allowed flux distributions for the metabolic network. A particular flux distribution under a particular environmental condition is found by using flux-balance analysis (FBA), which hypothesizes that the organism will optimize its metabolic fluxes to maximize (or minimize) some objective function. In this study the objective function corresponds to the measured biomass composition. The optimal solution is found by using standard linear programming techniques.
To simulate the effect of a single gene deletion in the genome-scale metabolic model, the fluxes through the reactions indicated by the gene-protein-reaction associations as being dependent on the particular gene product were constrained to be zero, and FBA was performed to find the predicted growth rate of the in silico deletion strain. If the deletion is deleterious in silico, it results in an optimal solution with lower growth flux than that obtained for the wild-type in silico model. A major challenge associated in mimicking as closely as possible the in vivo experimental conditions used in the high-throughput deletion studies is formulation of the in silico media used in the study. For defined media this is relatively straightforward as the media composition is known and the maximal nutrient uptake rates for each individual nutrient can be found from relevant literature. However, all but one of the experiments that generated the data used in this study were performed by using complex media (yeast extract-peptone or YP) of which the composition is not known. This necessitates making assumptions of both the media composition and the individual uptake rates for all the nutrients.
In this study the YP medium was assumed to contain in addition to the defined carbon source (glucose, galactose, ethanol, glycerol, lactate), ammonium sulfate as a nitrogen source, phosphate, necessary salts (K, Na, Ca), all 20 amino acids, and all four nucleotide bases. In addition, based on recent results on the composition of yeast extract (Zhang et al. 2003), we also assumed that the media contains significant amounts of trehalose that can be used by the cells. The maximal uptake rates for the carbon sources were obtained from the literature (Strathern et al. 1982; Sutherland et al. 1997; Casal et al. 1999; Diderich et al. 1999; Malluta et al. 2000), and the uptake rates for the other nutrients were set to be high enough not to be strongly growth limiting, but low enough so that the primary carbon source was still used. In addition to the primary carbon source uptake rate, the maximum oxygen uptake rate is also an important parameter in the simulations. Here, we adjusted the maximum oxygen uptake rate in such a way that for the wild-type strain under particular media conditions, the rate is close to that measured experimentally for the same growth rate (van Hoek et al. 1998). For example, for YPD and YPGal media, the oxygen uptake rate was set to a relatively low value to mimic the Crabtree effect that limits the overall oxidative capacity of yeast at high growth rates. The in silico set-up also accounts for the auxotrophic markers present in the in vivo strains by deleting these marker genes (HIS3, LEU2, URA3) and supplementing the medium with the correct nutritional supplements (histidine, leucine, uracil).
We convert the continuous in silico relative growth rate (growth rate of a particular deletion strain/mean of growth rates of all deletion strains under a particular condition) obtained for each strain into a discrete normal growth/deleterious prediction by considering strains with relative growth rates >1 SD below the mean relative growth rate of all strains to have deleterious phenotype and the remaining strains to have a normal growth phenotype. The large-scale deletion computations in this study were performed by using the MATLAB APIs to the LINDO (Lindo Systems, Inc.) linear programming package, and the detailed evaluation of the deletions was done within the SimPheny framework described above.
In Vivo Gene Deletion Data Preprocessing
The phenotyping data used in this study was obtained in two studies (Giaever et al. 2002; Steinmetz et al. 2002) by using a comprehensive collection of yeast deletion strains to perform competitive growth experiments under a number of different experimental conditions. The strains in these studies were pooled, and the relative abundance of strain-specific DNA tags after growth for a specific number of generations in a particular condition was measured by using hybridization to a custom high-density oligonucleotide array. As the metabolic model described in this article predicts growth rates for individual deletion strains given a defined media composition and maximal nutrient uptake rates, whereas the experimental data was from competitive growth experiments with undefined nutrient uptake rates and in most cases complex media, the experimental data and model predictions were not directly comparable. The two experimental studies also used different designs and data analysis procedures so that the data sets required different preprocessing steps described below.
Essential Genes on Rich Medium
A list of genes required for growth on rich glucose medium for which deletion strains could not be generated was downloaded from the Supplemental Web site to Giaever et al. (2002).
Slow Growth Genes on Rich Medium
A list of genes with significant slow growth phenotype in rich glucose medium together with a quantitative fitness defect score was downloaded from the Supplemental Web site to Giaever et al. (2002). The process used to compute the fitness defect scores is described on the Supplemental Web site to Giaever et al. (2002).
Glucose Minimal and YPGal Media
Fitness scores for these two media conditions were downloaded from Giaever et al. (2002). For both conditions the data contain four separate measurements for each strain, two after five generations of competitive growth and two after 15 generations of competitive growth. For each measurement the data indicate whether the deletion strain is sensitive to the condition (i.e., slower growth than the average deletion strain) or refractive to the condition (i.e., faster growth than the average strain). In addition, for each measurement there is a score indicating the likelihood of observing the experimental condition measurements given the background distribution in a control condition (YPD). Scores >100 in the 15 generations' measurements and scores >20 in the five generations' measurements were considered to be highly significant in Giaever et al. (2002). In Giaever et al. (2002) deletion strains considered to be sensitive to the condition, with scores exceeding the threshold in all four conditions considered to be slow growth. We used an alternative somewhat less stringent metric in order to identify deletions with borderline deleterious effect. This metric is the average of the normalized scores (for each measurement score is divided by the relevant significance threshold) over all experiments (only sensitive predictions are considered). Strains for which the value of this metric is >1.0 are considered to have a potentially deleterious phenotype and were not counted as false predictions if the in silico model predicted retarded growth. Because of the design of the study, deletion strains that were determined to be slow growth on rich medium did not have a deleterious phenotype on the other media, but for the purposes of comparing the experimental data to in silico predictions, the slow growth genes were considered to have a deleterious phenotype on MMD and YPGal media.
YPD, YPDGE, YPE, YPG, and YPL Media
Fitness scores for homozygous deletions strains under these media conditions were downloaded from the Supplemental Web site to Steinmetz et al. (2002). For each condition the mean of the measured fitness scores for each deletion strain was used as the experimental fitness measure. Fitness scores <1.0 correspond to strains growing slower than the average strain in the pool, and fitness scores >1.0 correspond to strains growing faster than the average strain. We designate strains with fitness scores >1 SD below the mean of gene fitness scores for each condition to be slow growth under that particular condition.
Acknowledgments
We acknowledge the National Science Foundation (MCB98-73384 and BES98-14092) and the Finnish Fulbright Foundation (ASLA-Fulbright Graduate Scholarship to M.J.H.) for financial support. We thank Iman Famili, Jochen Förster, and Markus W. Covert for stimulating discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2250904\. Article published online before print in June 2004.
Footnotes
Supplemental material is available online at www.genome.org. The reaction and metabolite lists, metabolic network maps, and gene-protein-reaction associations for Saccharomyces cerevisiae iND750 can be found at http://systemsbiology.ucsd.edu.
5
If different isozymes are counted as separate reactions, there is a total of 1175 reactions in the Förster-Famili model; the set of reactions discussed here is a count of the unique enzymatic and transport reactions determined from the published reaction list.
References
- Allen, T.D., Cronshaw, J.M., Bagley, S., Kiseleva, E., and Goldberg, M.W. 2000. The nuclear pore complex: Mediator of translocation between nucleus and cytoplasm. J. Cell. Sci. 113**:** 1651–1659. [DOI] [PubMed] [Google Scholar]
- Allen, T.E. and Palsson, B.Ø. 2003. Sequenced-based analysis of metabolic demands for protein synthesis in prokaryotes. J. Theor. Biol. 220**:** 1–18. [DOI] [PubMed] [Google Scholar]
- Allen, T.E., Herrgard, M.J., Liu, M., Qiu, Y., Glasner, J.D., Blattner, F.R., and Palsson, B.Ø. 2003. Genome-scale analysis of the uses of the Escherichia coli genome: Model-driven analysis of heterogeneous data sets. J. Bacteriol. 185**:** 6392–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly, A.M. and Macreadie, I.G. 2002. Cytotoxicity of dihydropteroate in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 213**:** 189–192. [DOI] [PubMed] [Google Scholar]
- Birrell, G.W., Giaever, G., Chu, A.M., Davis, R.W., and Brown, J.M. 2001. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc. Natl. Acad. Sci. 98**:** 12608–12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonarius, H.P.J., Schmid, G., and Tramper, J. 1997. Flux analysis of underdetermined metabolic networks: The quest for the missing constraints. Trends Biotechnol. 15**:** 308–314. [Google Scholar]
- Casal, M., Paiva, S., Andrade, R.P., Gancedo, C., and Leao, C. 1999. The lactate-proton symport of Saccharomyces cerevisiae is encoded by JEN1. J. Bacteriol. 181**:** 2620–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert, M.W. and Palsson, B.Ø. 2002. Transcriptional regulation in constraints-based metabolic models of Escherichia coli. J. Biol. Chem. 277**:** 28058–28064. [DOI] [PubMed] [Google Scholar]
- Covert, M.W., Schilling, C.H., Famili, I., Edwards, J.S., Goryanin, I.I., Selkov, E., and Palsson, B.Ø. 2001. Metabolic modeling of microbial strains in silico. Trends Biochem. Sci. 26**:** 179–186. [DOI] [PubMed] [Google Scholar]
- Dickinson, J.R. and Schweizer, M. 1999. The metabolism and molecular physiology of Saccharomyces cerevisiae. Taylor & Francis, Philadelphia.
- Diderich, J.A., Schepper, M., van Hoek, P., Luttik, M.A., van Dijken, J.P., Pronk, J.T., Klaassen, P., Boelens, H.F., de Mattos, M.J., van Dam, K., et al. 1999. Glucose uptake kinetics and transcription of HXT genes in chemostat cultures of Saccharomyces cerevisiae. J. Biol. Chem. 274**:** 15350–15359. [DOI] [PubMed] [Google Scholar]
- Edwards, J.S. and Palsson, B.Ø. 1999. Systems properties of the Haemophilus influenzae Rd metabolic genotype. J. Biol. Chem. 274**:** 17410–17416. [DOI] [PubMed] [Google Scholar]
- Edwards, J.S. and Palsson, B.Ø. 2000. The Escherichia coli MG1655 in silico metabolic genotype: Its definition, characteristics, and capabilities. Proc. Natl. Acad. Sci. 97**:** 5528–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, J.S., Covert, M., and Palsson, B. 2002. Metabolic modelling of microbes: The flux-balance approach. Environ. Microbiol. 4**:** 133–140. [DOI] [PubMed] [Google Scholar]
- Famili, I., Förster, J., Nielsen, J., and Palsson, B.Ø. 2003. Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc. Natl. Acad. Sci. 100**:** 13134–13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming, J.A., Lightcap, E.S., Sadis, S., Thoroddsen, V., Bulawa, C.E., and Blackman, R.K. 2002. Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341. Proc. Natl. Acad. Sci. 99**:** 1461–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster, J., Famili, I., Fu, P.C., Palsson, B.Ø., and Nielsen, J. 2003a. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 13**:** 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster, J., Famili, I., Fu, P.C., Palsson, B.Ø., and Nielsen, J. 2003b. Large-scale evaluation of in silico gene knockouts in Saccharomyces cerevisiae. Omics 7**:** 193–202. [DOI] [PubMed] [Google Scholar]
- Giaever, G., Chu, A.M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418**:** 387–391. [DOI] [PubMed] [Google Scholar]
- Jorgensen, P., Nishikawa, J.L., Breitkreutz, B.J., and Tyers, M. 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297**:** 395–400. [DOI] [PubMed] [Google Scholar]
- Kauffman, K.J., Prakash, P., and Edwards, J.S. 2003. Advances in flux balance analysis. Curr. Opin. Biotechnol. 14**:** 491–496. [DOI] [PubMed] [Google Scholar]
- Kispal, G., Steiner, H., Court, D.A., Rolinski, B., and Lill, R. 1996. Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein. J. Biol. Chem. 271**:** 24458–24464. [DOI] [PubMed] [Google Scholar]
- Lievers, K.J., Kluijtmans, L.A., and Blom, H.J. 2003. Genetics of hyperhomocysteinaemia in cardiovascular disease. Ann. Clin. Biochem. 40**:** 46–59. [DOI] [PubMed] [Google Scholar]
- Malluta, E.F., Decker, P., and Stambuk, B.U. 2000. The Kluyver effect for trehalose in Saccharomyces cerevisiae. J. Basic Microbiol. 40**:** 199–205. [DOI] [PubMed] [Google Scholar]
- Mannella, C.A. 1992. The “ins” and “outs” of mitochondrial membrane channels. Trends Biochem. Sci. 17**:** 315–320. [DOI] [PubMed] [Google Scholar]
- Mattson, M.P. and Haberman, F. 2003. Folate and homocysteine metabolism: Therapeutic targets in cardiovascular and neurodegenerative disorders. Curr. Med. Chem. 10**:** 1923–1929. [DOI] [PubMed] [Google Scholar]
- Mewes, H.W., Frishman, D., Guldener, U., Mannhaupt, G., Mayer, K., Mokrejs, M., Morgenstern, B., Munsterkotter, M., Rudd, S., and Weil, B. 2002. MIPS: A database for genomes and protein sequences. Nucleic Acids Res. 30**:** 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outten, C.E. and Culotta, V.C. 2003. A novel NADH kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae. EMBO J. 22**:** 2015–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson, B.Ø. 2000. The challenges of in silico biology. Nat. Biotechnol. 18**:** 1147–1150. [DOI] [PubMed] [Google Scholar]
- Parks, L.W. 1978. Metabolism of sterols in yeast. CRC Crit. Rev. Microbiol. 6**:** 301–341. [DOI] [PubMed] [Google Scholar]
- Pramanik, J. and Keasling, J.D. 1997. Stoichiometric model of Escherichia coli metabolism: Incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol. Bioeng. 56**:** 398–421. [DOI] [PubMed] [Google Scholar]
- Price, N.D., Papin, J.A., Schilling, C.H., and Palsson, B.Ø. 2003. Genome-scale microbial in silico models: The constraints-based approach. Trends Biotechnol. 21**:** 162–169. [DOI] [PubMed] [Google Scholar]
- Reed, J.L., Vo, T.D., Schilling, C.H., and Palsson, B.Ø. 2003. An expanded genome-scale model of Escherichia coli K-12 (_i_JR904 GSM/GPR). Genome Biol. 4**:** R54.51–R54.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling, C.H., Covert, M.W., Famili, I., Church, G.M., Edwards, J.S., and Palsson, B.Ø. 2002. Genome-scale metabolic model of Helicobacter pylori 26695. J. Bacteriol. 184**:** 4582–4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz, L.M., Scharfe, C., Deutschbauer, A.M., Mokranjac, D., Herman, Z.S., Jones, T., Chu, A.M., Giaever, G., Prokisch, H., Oefner, P.J., et al. 2002. Systematic screen for human disease genes in yeast. Nat. Genet. 22**:** 22. [DOI] [PubMed] [Google Scholar]
- Strathern, J.N., Jones, E.W., and Broach, J.R. 1982. The molecular biology of the yeast Saccharomyces: Metabolism and gene expression. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Sutherland, F.C., Lages, F., Lucas, C., Luyten, K., Albertyn, J., Hohmann, S., Prior, B.A., and Kilian, S.G. 1997. Characteristics of Fps1-dependent and -independent glycerol transport in Saccharomyces cerevisiae. J. Bacteriol. 179**:** 7790–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, D., Barbey, R., and Surdin-Kerjan, Y. 1990. Gene-enzyme relationship in the sulfate assimilation pathway of Saccharomyces cerevisiae: Study of the 3′-phosphoadenylylsulfate reductase structural gene. J. Biol. Chem. 265**:** 15518–15524. [PubMed] [Google Scholar]
- Vander Heiden, M.G., Chandel, N.S., Li, X.X., Schumacker, P.T., Colombini, M., and Thompson, C.B. 2000. Outer mitochondrial membrane permeability can regulate coupled respiration and cell survival. Proc. Natl. Acad. Sci. 97**:** 4666–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoek, P., Flikweert, M.T., van der Aart, Q.J., Steensma, H.Y., van Dijken, J.P., and Pronk, J.T. 1998. Effects of pyruvate decarboxylase overproduction on flux distribution at the pyruvate branch point in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 64**:** 2133–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma, A. and Palsson, B.Ø. 1994. Metabolic flux balancing: Basic concepts, scientific and practical use. Bio/Technology 12**:** 994–998. [Google Scholar]
- Voet, D., Voet, J.G., and Pratt, C.W. 1999. Fundamentals of biochemistry. Wiley, New York.
- Walker, G.M. 1998. Yeast physiology and biotechnology. J. Wiley & Sons, Chichester, NY.
- Weng, S., Dong, Q., Balakrishnan, R., Christie, K., Costanzo, M., Dolinski, K., Dwight, S.S., Engel, S., Fisk, D.G., Hong, E., et al. 2003. Saccharomyces Genome Database (SGD) provides biochemical and structural information for budding yeast proteins. Nucleic Acids Res. 31**:** 216–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorke, R., Krampe, S., Weierstall, T., Freidel, K., Hollenberg, C.P., and Boles, E. 1999. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464**:** 123–128. [DOI] [PubMed] [Google Scholar]
- Winzeler, E.A., Shoemaker, D.D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J.D., Bussey, H., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285**:** 901–906. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Reddy, J., Buckland, B., and Greasham, R. 2003. Toward consistent and productive complex media for industrial fermentations: Studies on yeast extract for a recombinant yeast fermentation process. Biotechnol. Bioeng. 82**:** 640–652. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://systemsbiology.ucsd.edu; Gene-protein-reaction associations for Saccharomyces cerevisiae iND750.
- http://www.yeastgenome.org; Saccharomyces Genome Database.