Proteomic Analysis of the Soybean Symbiosome Identifies New Symbiotic Proteins (original) (raw)
Abstract
Legumes form a symbiosis with rhizobia in which the plant provides an energy source to the rhizobia bacteria that it uses to fix atmospheric nitrogen. This nitrogen is provided to the legume plant, allowing it to grow without the addition of nitrogen fertilizer. As part of the symbiosis, the bacteria in the infected cells of a new root organ, the nodule, are surrounded by a plant-derived membrane, the symbiosome membrane, which becomes the interface between the symbionts. Fractions containing the symbiosome membrane (SM) and material from the lumen of the symbiosome (peribacteroid space or PBS) were isolated from soybean root nodules and analyzed using nongel proteomic techniques. Bicarbonate stripping and chloroform-methanol extraction of isolated SM were used to reduce complexity of the samples and enrich for hydrophobic integral membrane proteins. One hundred and ninety-seven proteins were identified as components of the SM, with an additional fifteen proteins identified from peripheral membrane and PBS protein fractions. Proteins involved in a range of cellular processes such as metabolism, protein folding and degradation, membrane trafficking, and solute transport were identified. These included a number of proteins previously localized to the SM, such as aquaglyceroporin nodulin 26, sulfate transporters, remorin, and Rab7 homologs. Among the proteome were a number of putative transporters for compounds such as sulfate, calcium, hydrogen ions, peptide/dicarboxylate, and nitrate, as well as transporters for which the substrate is not easy to predict. Analysis of the promoter activity for six genes encoding putative SM proteins showed nodule specific expression, with five showing expression only in infected cells. Localization of two proteins was confirmed using GFP-fusion experiments. The data have been deposited to the ProteomeXchange with identifier PXD001132. This proteome will provide a rich resource for the study of the legume-rhizobium symbiosis.
Biological nitrogen fixation occurs through the activity of the enzyme nitrogenase, which is found only in certain prokaryotes, including those of the family Rhizobiaceae (termed rhizobia). The enzyme converts atmospheric N2 to ammonia, a biologically available form of nitrogen, but requires large amounts of ATP to fuel the conversion (1). Legumes, such as soybeans (Glycine max), are able to form an association with these nitrogen-fixing rhizobia. In this symbiotic relationship, N2 is fixed by the rhizobia and made available to the plant in exchange for organic acids and other nutrients. This mutually beneficial association occurs within specialized root organs termed nodules. Within the nodule infected cells, N2-fixing bacteroids (the symbiotic form of rhizobia) are enclosed in a plant-derived membrane to form organelle-like structures termed symbiosomes (2).
The symbiosome membrane (SM)1 originates from invaginated plasma membrane as the bacteria enter infected cells, but quickly becomes specialized as the symbiosis matures (3). Within symbiosomes of nodules, the rhizobia continue to multiply before differentiating into bacteroids in which symbiosis-related genes are induced (4). Symbiosomes thus result from the coordinated division of bacteria and growth of the surrounding SM, fed by the systems for endomembrane synthesis (5).
The SM surrounds one or more differentiated bacteroids, effectively excluding them from the plant cytosol. The region between the SM and bacteroids is termed the peribacteroid space (PBS). The SM is a physical barrier between the plant and the bacteroid and represents a regulation point for the movement of solutes between the symbionts, via an array of transporters and channels (4, 6).
It is estimated that in a mature infected cell, the SM surface area is many times that of the plasma membrane, allowing it to encapsulate the multiplying bacteroids (7). The expanding SM requires the synthesis of lipids and proteins in the infected cell (7). The composition of the SM is thought to vary during nodule development and senescence, to facilitate the dynamic transport requirements of the symbionts (3). Targeting to the symbiosome has been linked to an N-terminal signal sequence for several proteins (8–10), but no conserved N-terminal signal has been identified for SM proteins.
The principal nutrient transfer across the SM is the exchange of a plant carbon energy source, for nitrogen fixed by the bacteroid. This carbon source is derived from sucrose produced via photosynthesis, which is converted in the nodules to dicarboxylic acids (6). Dicarboxylates, probably malate, are then transported across the SM to the bacteroids (11). Although malate transport across the SM has been characterized biochemically (12, 13), a transport protein has not yet been identified on the SM of any of the legumes studied.
The main product of nitrogen fixation in bacteroids is ammonia, the majority of which is thought to be protonated to ammonium in the acidic PBS (14). There are two routes proposed for transport of fixed-N across the SM; as NH3 through the aquaglyceroporin NOD26 (15, 16) and as NH4+ through a monovalent cation channel (17). Although NOD26 is well described in soybean (16, 18–21), the protein catalyzing monovalent cation transport has not been identified.
Several additional transport processes on the SM have been identified, including proteins for transport of iron, zinc, calcium, and sulfate (22–27). In addition, the movement of hydrogen ions has been reported through the activity of an H+-ATPase (28–30).
The SM is expected to contain many more proteins that facilitate the interaction between the plant host and bacteroids. Identification and characterization of SM proteins has been limited to date and a comprehensive description of the protein content of this membrane is lacking. Previous attempts to characterize the proteome of legume SMs have yielded modest results, with the main barriers to overcome being the lack of completed reference genomes with which to compare sequencing results, and the intrinsic hydrophobic nature of SM proteins hindering their identification. Two proteomic studies have been performed on the G. max : Bradyrhizobium japonicum SM. Both studies occurred prior to the release of the soybean genome and thus were limited in their success at identifying SM proteins (31, 32). Proteomic studies of the SM in other legume-rhizobia symbioses (Lotus japonicus, Pisum sativum, and Medicago truncatula) have succeeded in identifying only a small number of SM proteins as they were done at a time when there was limited genomic information available for these legumes (33–36). In addition, all studies except Wienkoop and Saalbach (35) have relied on 2D-PAGE methodologies, which are known to hinder the subsequent detection by mass spectrometry of hydrophobic membrane proteins. Here, we report a more comprehensive sampling of SM proteins and also proteins from the PBS of soybean. Together, these proteomic analyses provide a valuable resource for future studies on the structure and function of the symbiosome in all legume-rhizobium symbioses.
EXPERIMENTAL PROCEDURES
Plant Growth and Protein Isolation
Soybeans (G. max cv. Stephens) were grown under natural light extended to 16 h day length with incandescent lighting in a temperature controlled glasshouse (26 °C day/20 °C night). Plants were grown in washed river sand and seed-inoculated with B. japonicum in peat (Nodulaid Group H, Becker Underwood, NSW, Australia), and again at 5 days postsowing. Nodules were harvested from roots at 32 days postinoculation. Nitrogen-fixing ability of the mature nodules was confirmed using an acetylene reduction assay as described in (37). SM was isolated from mature nitrogen-fixing soybean nodules using previously established procedures that yield membrane that is generally free of contamination from other organelles (31, 38). The SM protein fraction was further purified by either bicarbonate stripping (39) or chloroform-methanol extraction (40). Isolated SM protein pellets were suspended in 100 mm Na2CO3, then pelleted by ultracentrifugation to isolate stripped proteins. Following bicarbonate stripping, SM proteins were phenol extracted as described in Day et al. (38). For chloroform-methanol extraction, isolated SM proteins were suspended in 50 mm MOPS/NaOH, pH 7.5, with protease inhibitors (cOmplete Protease Inhibitor Mixture Tablets, Roche, Basel, Switzerland) and mixed with a 5:4 chloroform : methanol solution as described (40). After 30 min incubation on ice, soluble and insoluble proteins were recovered by diethyl ether precipitation and ultracentrifugation (86,000 rpm for 1 h). Isolated SM protein fractions were resuspended in 8 m urea/1% SDS buffer and stored at −20 °C prior to proteomic analysis.
The peribacteroid space fraction was isolated during the SM isolation protocol following disruption of isolated intact symbiosomes (38). PBS proteins were concentrated using Nanosep® centrifugal devices (PALL Life Sciences, Long Island, NY), collected, and stored at −20 °C.
For three biological replicates, sodium bicarbonate stripping removed peripheral proteins from the SM. To reduce the complexity of the SM preparations by further fractionation and to enhance the collection of more hydrophobic proteins, chloroform-methanol extraction was performed on a subsequent set of four biological replicates. These four biological replicates were also used to generate PBS samples. Proteins identified from sodium bicarbonate stripped and C:M extracted fractions are together referred to as the SM proteome. Proteins removed from the SM with bicarbonate stripping were analyzed as the SM peripheral proteome. PBS and SM peripheral proteins were concentrated using Nanosep® centrifugal devices (PALL Life Sciences) prior to proteomic analysis.
Western Blot Analysis
Ten micrograms of total nodule protein, nodule microsomal, SM (bicarbonate stripped and chloroform-methanol fractions), SM peripheral and PBS samples were separated by SDS-PAGE using Bio-Rad Mini-PROTEAN gel equipment. Separated proteins were transferred to PVDF membrane (Bio-Rad, Hercules, CA) for Western blotting, or stained with Coomassie brilliant blue G to visualize protein. Blots were stained with Ponceau then destained, blocked and probed with primary antibodies at appropriate dilutions (nodulin 26 1:1000, HDEL 1:200, and porin 1:1000). Nodulin 26 antibody was provided by Dan Roberts, Knoxville, TN (41), HDEL antibody was sourced from Santa Cruz Biotechnology, Inc (Dallas, TX) and porin antibody was obtained from Dr. Tom Elthon, Lincoln, NE via Harvey Millar, Perth, WA (42). Blots were rinsed twice with TBST (Tris-Buffered Saline with 0.3% Tween 20) and incubated with secondary antibody conjugated to horseradish peroxidase (1/10,000 dilution, Promega, Madison, WI) followed by four washes in TBST. Immunoreactive proteins were visualized by chemiluminescence using Immun-StarTM WesternCTM Chemiluminescence Kit (Bio-Rad, Hercules, CA) as per the manufacturer's instructions and documented with the GelDoc Imager (UVP, Upland, CA).
Sample Preparation and LCMS/MS
Protein concentration of samples was determined using LavaPep Protein Quantification Kit (Gel Company, San Francisco, CA). Each biological sample was prepared and analyzed in triplicate by LCMS/MS. Ten micrograms of protein for each technical replicate was reduced with TCEP (tris(2-chloroethyl) phosphate), alkylated with MMST (methyl methanethiosulfonate) and digested with porcine trypsin (Promega) overnight at 37 °C. Digested peptides were prepared for LCMS/MS by removing excess salts with a HLB SPE column (Waters) and excess detergent with a SCX Stage Tip (Thermo Fischer Scientific, Waltham, CA), according to the manufacturer's guidelines. Insoluble components were removed by centrifugation at 10,000 rpm for 10 min. Peptides were then resuspended in 0.1% formic acid.
Samples were separated by liquid chromatography (LC) and analyzed on an Analyst QSTAR ESI-QUAD-TOF mass spectrometer (Thermo Fischer Scientific, Waltham, CA). The LC component consisted of a 150 mm separation column (Zorbax Column 300SB C18), driven by Agilent Technologies (Santa Clara, CA) 1100 series nano/capillary liquid chromatography system. Peptides were separated over two hours (5% Acetonitrile, 40% Acetonitrile) and eluted directly into the mass spectrometer. The mass spectrometer was run in positive ion mode and MS scans ran over a range of m/z 400–1500 and at four spectra s−1. Precursor ions were selected for auto MS/MS at an absolute threshold of 500 and a relative threshold of 0.01, with a maximum of three precursors per cycle. Precursor charge-state selection and preference was set to 2+ and then 3+ and precursors selected by charge then abundance. Resulting MS spectra were opened with Analyst QS 2.0 software, and exported to MASCOT (Matrix Science, Boston, MA).
Data Analysis
The soybean proteome derived from version 1.1 of the soybean genome (available at www.phytozome.net, 73,320 entries) was searched for peptide matches using MASCOT. Up to one missed tryptic cleavage was tolerated, variable modifications were Oxidation (M) and Carbamidomethyl (C); peptide and MS/MS tolerance was set as 0.2 Da and peptide charge was set at 2+ and 3+ monoisotopic.
Three technical replicates were prepared for each sample, with multiple biological replicates analyzed for each sample type (sodium bicarbonate stripped SM: three biological replicates, C:M extracted SM: four biological replicates, SM Peripheral: two biological replicates and PBS: three biological replicates). Results were matched to the predicted soybean proteome (www.phytozome.net; 43) using MASCOT and visualized using Scaffold4 Proteome software (Proteome Software, Portland, OR). Significance thresholds were defined in Scaffold4 at 95% minimum peptide identification probability and 95% minimum protein identification probability. These probabilities are generated using the Peptide Prophet and Protein Prophet algorithms (44, 45), which convert the statistical significance output of MASCOT into a discriminate score.
To be considered a significant match, proteins had a minimum of two distinct peptides observed in one or more biological replicates. Where multiple proteins were identified with the same peptides, one unique peptide was required for a protein match to be considered significant (along with one or more shared peptides). Percent coverage was calculated based on coverage of the complete protein sequence by matched peptide queries. The false discovery rate (FDR) was calculated by Scaffold4, based on the method of Kall et al. (46) using a reversed decoy database. The protein FDR was 0.1% and peptide FDR was 0.46% (from merged results).
Bioinformatics
Information on protein function was compiled from the G. max genome annotation (www.phytozome.net) and from top matches in NCBI (www.ncbi.nlm.nih.gov). Proteins were grouped according to functional classification by MapMan (47). Previous SM and PBS proteome data sets (31, 33, 34, 36) were blasted against the soybean proteome (www.phytozome.net) to identify soybean homologs that were identified in this proteomic analysis.
The membrane topology of proteins was predicted by three bioinformatic suites: SOSUI (48), TMHMM (49), and TopPred2 (50). Subcellular localization was predicted using TargetP (51), PREDOTAR (52), Plant mpLOC (53), and MulitLOC (54). Where possible, a consensus location was determined, otherwise proteins were marked as unknown location. The presence of signal peptides in the proteins was assessed using SignalP (55) and GPI-anchors using GPI-SOM (56).
Cloning, Constructs, and Transformation
Soybean Glyma11g34600.1 (NPF5.25) and Glyma18g03790.1 (NPF5.29) open reading frames and the 2-kb 5′ regulatory sequences of Glyma11g34600.1 (NPF5.25), Glyma11g34613.1 (NPF5.24), Glyma09g31910.1, Glyma01g31910.2, Glyma07g39320.1, and Glyma09g21070.2 were amplified via PCR from 30 day old nodule cDNA and genomic DNA, respectively, using Phusion high-fidelity polymerase (New England Biolabs, Ipswich, MA). Primers are listed in Table I. PCR products were cloned into pENTR or pDONR entry vectors using either TOPO cloning (Invitrogen) or Gateway Recombination (Invitrogen). The Gateway cloning system (Invitrogen) was used to create genetic constructs for promoter-GUS and GFP fusion. Entry clones were recombined into the following destination vectors using LR Clonase (Invitrogen): pKGW-GGRR for promoter-GUS fusion and pK7WGFLhc3-R, creating N-terminal GFP-X fusions driven by the nodule specific leghemoglobin promoter (these vectors are modified from pKGWFS7 and pK7WGF2, respectively, obtained from Plant Systems Biology, Ghent University, Belgium; http://gateway.psb.ugent.be). _Agrobacterium rhizogenes_–based root transformation of G. max was performed according to Mohammadi-Dehcheshmeh et al. (57).
Table I. Primer sequences.
Sequences of primers used to amplify the promoter or coding regions of Glyma11g34600.1, Glyma11g34610.1, Glyma09g31910.1, Glyma01g31910.2, Glyma09g21070.2 and Glyma07g39320.1. Underlined sequences are gateway recombination sites. CDS is coding sequence.
| Candidate (amplified) | Forward primer | Reverse primer |
|---|---|---|
| Glyma11g34600.1 (CDS) | CACCATGGAGCAAGAAATGGAGAAGA | TCATTTTGCCACTGTCTCCA |
| Glyma18g03790.1 (CDS) | CACCATGAAGCAGGAAATGGAGAAG | TCATGCCACTGTATCCACC |
| Glyma11g34600.1 (promoter) | CACCGCTTAAAGTTTATGATCCTGTCATAAGTA | TTGACAATGCCAAAAGGG |
| Glyma11g34610.1 (promoter) | CACCCTCATAAGTATCAATTAATAAATTGGTTCC | TTCTTACTCCATTTGACAATGCC |
| Glyma09g31910.1 (promoter) | GGGGACAAGTTTGTACAAAAAAGCAGGCTGATGCATGAGGCATGGTAAC | GGGGACCACTTTGTACAAGAAAGCTGGGTGGTATCCATTTTGAAGCTCTAATC |
| Glyma01g31910.2 (promoter) | GGGGACAAGTTTGTACAAAAAAGCAGGCTGACGATCCGCTCCTAGTCAG | GGGGACCACTTTGTACAAGAAAGCTGGGTCCATGACACTCTTTGGATTC |
| Glyma09g21070.2 (promoter) | GGGGACAAGTTTGTACAAAAAAGCAGGCTAACCAACCATCCCAAAAAC | GGGGACCACTTTGTACAAGAAAGCTGGGTGTTAATATTCTCTCTCTCTTTCTCTCTC |
| Glyma07g39320.2 (promoter) | GGGGACAAGTTTGTACAAAAAAGCAGGCTGGGGTTATGACTTTGTCAGG | GGGGACCACTTTGTACAAGAAAGCTGGGTAAAGTGTAGCCATTTTTCC |
GUS Staining, Sectioning, and Microscopy
Transgenic nodules were collected, washed twice in 0.1 m sodium phosphate buffer (pH 7.2) and incubated in GUS buffer under vacuum at room temperature for 30 min to allow the buffer to replace oxygen in the tissue, and then at 37 °C for 1 h. Hand sections were mounted on microscope slides and analyzed using a Leica M205FA stereo microscope.
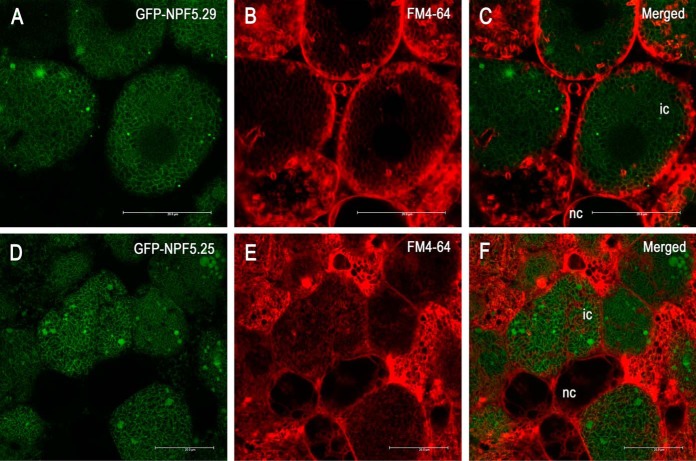
Confocal imaging of GFP-fused proteins was done on transgenic hand-sectioned nodules using a Leica SP5 II confocal microscope. Sections were counterstained with FM4–64 (30 mg/ml).
RESULTS AND DISCUSSION
Purity of Symbiosome Membrane Preparations
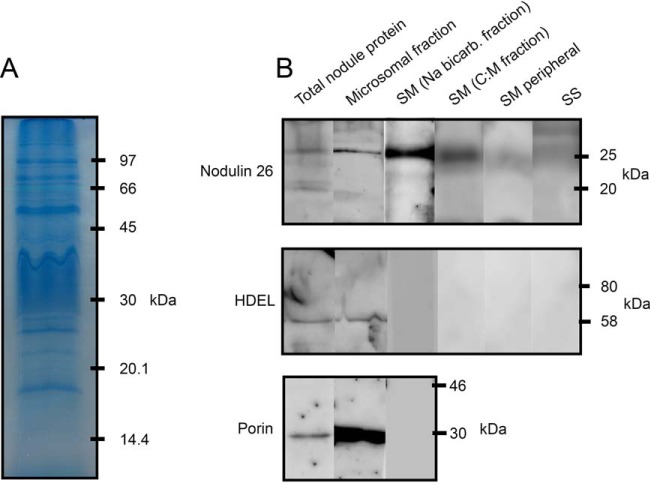
To evaluate the enrichment of SM during the isolation and fractionation procedure and to assess the purity of samples, nodule total protein, nodule microsomal fraction, sodium bicarbonate stripped SM, C:M extracted SM, SM peripheral, and PBS protein fractions were Western blotted and probed with marker antibodies for proteins with different subcellular locations (Fig. 1B). Fig. 1A demonstrates the SM proteins resolved by one-dimensional SDS-PAGE.
Fig. 1.
One dimensional SDS-PAGE of SM proteins and Western blot analysis of nodulin 26, HDEL and porin in nodule fractions. A, Ten micrograms of sodium bicarbonate stripped symbiosome membrane (SM) protein resolved on a 12% SDS-polyacrylamide gel and stained with Coomassie brilliant blue. B, Ten micrograms of protein from total nodule, microsomal, sodium bicarbonate stripped SM, C:M extracted SM, SM peripheral, and peribacteroid space (PBS) fractions were resolved on 12% SDS-polyacrylamide gels then transferred to PVDF membranes. Blots were blocked and probed with antibodies for either nodulin-26, HDEL or porin proteins.
Nodulin 26 was used as a marker for the SM as it is a well characterized SM protein (18, 19, 21, 58), whereas mitochondrial porin and HDEL are markers for mitochondria and endoplasmic reticulum (ER), respectively (59).
Nodulin 26 signal was observed in the microsomal fraction and both SM fractions (sodium bicarbonate stripped and C:M extracted) with highest intensity in the bicarbonate stripped sample. It was not observed in the PBS or SM peripheral fractions but a weak signal was detected in the total nodule preparation. A small number of nodulin 26 peptides were detected in peripheral samples by LCMS/MS, suggesting the proteomic analysis is more sensitive than Western blot analysis. Our immunoblot results showed similar enrichment of SM from the initial nodule extract as that seen by Catalano et al. (36) in preparations of SM from M. truncatula.
The mitochondrial porin (29 kDa) and HDEL (65 kDa) antibodies identified protein bands in the microsomal fraction and total nodule samples. No signal for antibody binding was observed in any of the SM or SM-related fractions. Together our results indicate enrichment from a total nodule homogenate to isolated SM that is relatively free from mitochondrial and ER contaminants, as determined previously via enzyme assays (31) for SM isolated using the same method.
We reviewed the data in the literature for localization of the proteins identified in our SM samples and also used bioinformatic programs to predict their subcellular localization (Table II). In general, the proteins identified were not given a localization in the prediction program, although a number were suggested to be directed to the ER or secretory pathway. Many of these proteins were predicted to contain signal peptides. Because there may be trafficking of proteins from the ER to the SM (see below) the ER/secretory pathway predictions may still allow targeting of proteins to the SM. Although the bioinformatic predictions of subcellular localization must be treated with caution as the existence of symbiotic membranes is not built into these programs it is of interest that few were suggested to be targeted to organelles. There is little information about how proteins are targeted to symbiosomes. Infected nodule cells, which contain symbiosomes, are a specialized cell type, thus, proteins with roles on the SM may have evolved from proteins with other cellular roles. It is possible that proteins normally localized in one organelle in other tissues may have been recruited to a new symbiotic role in infected cells of nodules or have dual roles in these cells. An example of this is the P-type ATPase (see below).
Table II. Proteins identified by LCMS/MS from various fractions of the soybean symbiosome. A minimum of two peptides were identified in one or more biological samples for all proteins indicated. Proteins are grouped according to functional classification by MapMan. Information about protein function is compiled from the genome annotation (www.phytozome.net) and from the top matches in NCBI (www.ncbi.nlm.nih.gov). Protein names are as annotated in Phytozome (www.phytozome.net) and data are compiled from the August 2012 release of the G. max genome (except peribacteroid space (PBS) which is from the previous genome release). An asterisk (*) indicates that the protein, or a close homologue, has been identified in a previous symbiosome membrane (SM) or PBS proteomic study. The presence of a predicted GPI-anchor sequence (56) within the protein is indicated by G. The presence of a predicted signal peptide (55) on the protein is indicated by SP. Sample/s in which the protein was present are denoted by the total number of spectral counts for each sample in columns corresponding to (SM) (bicarbonate stripped and chloroform-methanol extractions pooled), SM peripheral (SMP) and PBS. Percentage coverage (%C) is the maximum percentage of a protein to which peptides have been mapped in a biological sample and #P indicates the number of unique peptides assigned to the protein match. Membrane topology was predicted by three bioinformatic suites: (i) SOSUI (http://harrier.nagahama-i-bio.ac.jp/sosui/), (ii) TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) and (iii) TopPred2 (http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html), with the number of predicted transmembrane domains indicated. Predicted localisation (L) is a composite result of several sub-cellular localization prediction algorithms (TARGETP, PREDOTAR, Plant mpLOC and MulitLOC) and proteins were classified as either Chloroplastic (C), Endoplasmic Reticulium/Secretory Pathway (ER/SP), Mitochondrial (M), Peroxisomal (P), or Unknown (U). Gene expression, expressed as normalized reads/Kb/Million, in nodule tissue (N) is indicated for each gene and the expression profile across other soybean tissues (E) is denoted significant (S; summed transcript number > 40 across non-nodule tissues), low (L; summed transcript number < 40 across non-nodule tissues), or ns where expression is nodule-specific (66).
| Potential function | Protein name | SM | SMP | PBS | %C | #P | i | ii | iii | L | N | E |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C and CHO Metabolism | ||||||||||||
| Serine hydroxymethyl-transferase 3 | Glyma13g29410.1 | 46 | 63 | 21 | 17 | 6 | 0 | 0 | 1 | C | 460 | S |
| Sucrose synthase 4 | Glyma17g05067.1* | 18 | 2 | 5 | 4 | 0 | 0 | 1 | U | |||
| Triosephosphate isomerase | Glyma03g34300.1 | 7 | 15 | 3 | 0 | 0 | 0 | C | 129 | S | ||
| Transketolase | Glyma03g03200.1 | 6 | 4 | 2 | 0 | 0 | 1 | C | 35 | S | ||
| Trehalase 1 | Glyma05g36580.1SP | 10 | 7 | 3 | 0 | 1 | 1 | ER/SP | 43 | S | ||
| Glycolysis | ||||||||||||
| Glyceraldehyde-3-phosphate dehydrogenase | Glyma18g01330.1 | 6 | 8 | 2 | 0 | 0 | 0 | U | 291 | S | ||
| Glyceraldehyde-3-phosphate dehydrogenase of plastid 2 | Glyma16g09020.1 | 12 | 7 | 2 | 0 | 0 | 0 | C | 70 | S | ||
| Fermentation | ||||||||||||
| Aldehyde dehydrogenase (NAD+) | Glyma14g24140.1 | 62 | 8 | 4 | 1 | 0 | 1 | C | 11 | S | ||
| Mitochondrial | ||||||||||||
| ADP/ATP carrier 2 | Glyma15g42900.1* | 40 | 17 | 8 | 0 | 3 | 3 | U | 171 | S | ||
| F0F1-type ATP synthase, beta subunit | Glyma10g41330.1* | 49 | 22 | 9 | 4 | 0 | 0 | 0 | M | 93 | S | |
| Lactate/malate dehydrogenase family protein | Glyma12g19520.1* | 27 | 35 | 5 | 0 | 0 | 3 | M | 171 | S | ||
| Lactate/malate dehydrogenase family protein | Glyma06g34190.1* | 18 | 26 | 1 | 0 | 0 | 3 | M | 197 | S | ||
| Malate dehydrogenase | Glyma05g01010.1* | 2 | 6 | 2 | 0 | 0 | 3 | C | 24 | S | ||
| Mitochondrial carrier protein | Glyma08g16420.1* | 37 | 17 | 1 | 0 | 3 | 4 | U | 63 | S | ||
| Mitochondrial substrate carrier family protein | Glyma05g29050.1 | 4 | 9 | 2 | 0 | 0 | 3 | U | 51 | S | ||
| Mitochondrial substrate carrier family protein | Glyma06g07310.1 | 17 | 6 | 2 | 0 | 0 | 4 | U | 126 | L | ||
| Mitochondrial substrate carrier family protein | Glyma08g24070.1 | 6 | 6 | 2 | 0 | 3 | 3 | U | 44 | L | ||
| Ubiquinol–cytochrome c reductase. | Glyma05g07020.1 | 5 | 16 | 3 | 1 | 0 | 2 | M | 8 | S | ||
| Cell wall | ||||||||||||
| Cellulose synthase | Glyma08g44310.1 | 13 | 7 | 4 | 7 | 8 | 7 | U | 82 | S | ||
| Cellulose synthase-like B3 | Glyma12g31800.1 | 5 | 4 | 3 | 2 | 5 | 6 | U | 58 | L | ||
| FASCICLIN-like arabinogalactan protein 17 precursor | Glyma12g31691.1G, SP | 24 | 9 | 2 | 0 | 1 | 1 | U | ||||
| FASCICLIN-like arabinogalactan-protein 10 | Glyma09g40420.1 G, SP | 23 | 9 | 3 | 3 | 0 | 3 | ER/SP | 82 | S | ||
| Glycosyl hydrolase family 3 protein | Glyma15g13620.1 G, SP | 19 | 21 | 11 | 5 | 2 | 1 | 2 | ER/SP | 14 | L | |
| Pectin lyase-like superfamily protein | Glyma07g07290.1SP | 28 | 18 | 5 | 1 | 0 | 1 | ER/SP | 90 | ns | ||
| Xyloglucan:-xyloglucosyl transferase | Glyma13g01140.1SP | 95 | 15 | 3 | 1 | 1 | 1 | ER/SP | 3 | L | ||
| Lipid metabolism | ||||||||||||
| Favodoxin-like quinone reductase 1 | Glyma07g30750.1 | 26 | 19 | 3 | 0 | 0 | 1 | U | 14 | S | ||
| Neutral/alkaline non-lysosomal ceramidase | Glyma18g07680.2G, SP | 51 | 9 | 5 | 3 | 1 | 0 | 3 | U | 10 | L | |
| Neutral/alkaline non-lysosomal ceramidase | Glyma02g47420.1 G, SP | 21 | 3 | 2 | 3 | 0 | 5 | U | 6 | S | ||
| Phospholipase D1 | Glyma01g36680.1 | 18 | 8 | 5 | 0 | 0 | 0 | U | 11 | S | ||
| Quinone reductase family protein | Glyma01g38400.1 | 9 | 10 | 2 | 0 | 0 | 1 | U | 16 | S | ||
| UDP-Glycosyltransfer-ase superfamily protein | Glyma03g19751.1 | 5 | 5 | 3 | 0 | 0 | 0 | U | ||||
| Nitrogen metabolism | ||||||||||||
| Glutamate dehydrogenase 3 | Glyma02g07940.1 | 9 | 6 | 2 | 0 | 0 | 1 | U | 0 | L | ||
| Glutamine synthetase; nodulin 61 | Glyma10g06810.1 SP | 35 | 8 | 6 | 13 | 4 | 1 | 0 | 2 | ER/SP | 417 | L |
| NADH-dependent glutamate synthase 1 | Glyma06g13280.2 | 3 | 1 | 3 | 0 | 0 | 5 | U | 54 | S | ||
| Amino acid metabolism | ||||||||||||
| d-3-phosphoglycerate dehydrogenase | Glyma10g40750.1 | 55 | 9 | 4 | 0 | 0 | 0 | C | 184 | S | ||
| d-3-phosphoglycerate dehydrogenase | Glyma13g44970.1 | 15 | 5 | 2 | 0 | 0 | 0 | C | 634 | L | ||
| Glycine dehydrogenase (decarboxylating) | Glyma14g10820.1 | 7 | 3 | 1 | 0 | 0 | 3 | M | 10 | S | ||
| Glycine dehydrogenase (decarboxylating) | Glyma17g34690.1 | 10 | 3 | 3 | 0 | 0 | 3 | M | 103 | S | ||
| Hormone metabolism | ||||||||||||
| Auxin Conjugate Hydrolase M20/M25/M40 family protein | Glyma08g21040.1 | 81 | 64 | 35 | 25 | 6 | 0 | 1 | 3 | ER/SP | 345 | ns |
| Carotenoid cleavage dioxygenase 1 | Glyma13g27220.1 | 21 | 9 | 4 | 1 | 1 | 1 | U | 11 | S | ||
| Lipoxygease l-4 | Glyma13g42330.1 | 38 | 6 | 5 | 1 | 0 | 1 | U | 164 | S | ||
| Stress | ||||||||||||
| Adenine nucleotide alpha hydrolases-like superfamily protein | Glyma02g17470.3 | 24 | 22 | 2 | 0 | 0 | 1 | U | 1278 | S | ||
| Chaperone protein htpG family protein | Glyma14g40320.1 SP | 6 | 2 | 2 | 1 | 0 | 1 | ER/SP | 16 | S | ||
| Chitinase A | Glyma20g30450.1G, SP | 47 | 4 | 21 | 20 | 3 | 0 | 1 | 2 | ER/SP | 72 | L |
| Domain of unknown function (DUF221) | Glyma02g43910.1 | 11 | 6 | 4 | 10 | 9 | 10 | U | 6 | S | ||
| Domain of unknown function (DUF221) | Glyma07g39320.1 | 15 | 4 | 3 | 9 | 10 | 10 | ER/SP | 13 | S | ||
| Heat shock protein 70 (Hsp 70) family protein | Glyma05g36600.1* | 19 | 4 | 3 | 0 | 1 | 1 | U | 4 | S | ||
| Heat shock protein 70 (Hsp 70) family protein | Glyma18g52650.2* | 12 | 5 | 2 | 0 | 0 | 2 | U | 14 | L | ||
| Respiratory burst NADPH oxidase | Glyma10g29280.1 | 78 | 11 | 9 | 4 | 4 | 6 | U | 4 | L | ||
| Respiratory burst NADPH oxidase | Glyma20g38000.2 | 56 | 8 | 2 | 4 | 4 | 6 | U | 5 | L | ||
| Redox | ||||||||||||
| Catalase 2 | Glyma04g01920.1 | 7 | 5 | 2 | 0 | 0 | 0 | U | 89 | S | ||
| Cytochrome B5 | Glyma04g41010.1 | 33 | 39 | 1 | 1 | 1 | 1 | M | 122 | S | ||
| Cytochrome B5 isoform E | Glyma06g13840.5 | 29 | 28 | 3 | 1 | 1 | 1 | M | 117 | S | ||
| Leghemoglobin | Glyma10g34280.1* | 25 | 29 | 40 | 2 | 0 | 0 | 0 | U | 12309 | L | |
| Leghemoglobin | Glyma20g33290.1* | 51 | 10 | 25 | 40 | 2 | 0 | 0 | 0 | U | 7046 | ns |
| Leghemoglobin | Glyma10g34260.1* | 51 | 10 | 34 | 40 | 4 | 0 | 0 | 0 | U | 6913 | ns |
| Leghemoglobin | Glyma10g34290.1* | 74 | 59 | 43 | 50 | 5 | 0 | 0 | 0 | U | 55714 | L |
| Protein disulfide isomerase-like 1–2 | Glyma04g42690.1* SP | 18 | 10 | 4 | 1 | 1 | 1 | ER/SP | 48 | S | ||
| Protein disulfide isomerase A1 | Glyma06g12090.1* SP | 17 | 8 | 2 | 1 | 0 | 1 | ER/SP | 13 | S | ||
| Nucleotide metabolism | ||||||||||||
| Adenylosuccin-ate lyase | Glyma02g42130.1 | 25 | 5 | 4 | 1 | 0 | 0 | C | 31 | L | ||
| Adenylosuccin-ate lyase | Glyma14g06780.1 | 24 | 8 | 5 | 4 | 1 | 0 | 0 | C | 62 | S | |
| Glutamine phosphoribosyl-pyrophosphate amidotransferase | Glyma04g00930.1 | 19 | 4 | 2 | 0 | 0 | 0 | C | 932 | L | ||
| Phosphoribosyl-amidoimidazole-succinocarbox-amide synthase | Glyma14g35690.1 | 4 | 8 | 2 | 0 | 0 | 0 | U | 183 | L | ||
| Phosphoribosyl-amine–glycine ligase | Glyma10g29780.1 | 3 | 7 | 2 | 0 | 0 | 1 | U | 163 | S | ||
| Phosphoribosyl-aminoimidazole carboxylase | Glyma20g36934.1 | 18 | 7 | 3 | 0 | 0 | 1 | C | ||||
| Phosphoribosyl-aminoimidazole carboxylase | Glyma10g30611.1 | 20 | 16 | 7 | 1 | 0 | 0 | 2 | C | |||
| Phosphoribosy-lformylglycin-amidine synthase | Glyma18g04070.1 | 14 | 4 | 1 | 0 | 0 | 2 | C | 76 | S | ||
| Phosphoribosyl-formylglycin-amidine synthase | Glyma11g34241.1 | 34 | 22 | 5 | 6 | 0 | 0 | 1 | C | |||
| Uricase (nodulin 35) | Glyma10g23790.1 | 167 | 107 | 32 | 66 | 15 | 0 | 0 | 0 | P | 2632 | S |
| RNA regulation | ||||||||||||
| Remorin family protein | Glyma08g01590.1 | 53 | 20 | 4 | 0 | 0 | 0 | U | 323 | ns | ||
| Remorin family protein | Glyma05g37990.2 | 32 | 15 | 1 | 0 | 0 | 0 | U | 243 | ns | ||
| DNA synthesis | ||||||||||||
| Histone superfamily protein | Glyma02g38921.1 | 6 | 15 | 2 | 0 | 0 | 0 | U | ||||
| Protein synthesis | ||||||||||||
| GTP binding elongation factor Tu family protein | Glyma05g11630.2 | 30 | 7 | 2 | 0 | 0 | 0 | U | 113 | S | ||
| Large subunit ribosomal protein L12e | Glyma10g06040.1 | 24 | 22 | 3 | 0 | 0 | 0 | U | 30 | S | ||
| Ribosomal protein L5 B | Glyma07g06580.3 | 4 | 11 | 2 | 0 | 0 | 1 | U | 25 | S | ||
| Protein targeting | ||||||||||||
| Insulinase (Peptidase family M16) protein | Glyma05g36040.1 | 4 | 4 | 2 | 0 | 0 | 2 | M | 5 | S | ||
| Insulinase (Peptidase family M16) protein | Glyma07g01720.1 | 4 | 4 | 2 | 0 | 0 | 2 | M | 19 | S | ||
| Insulinase (Peptidase family M16) protein | Glyma08g46020.1 | 3 | 4 | 2 | 0 | 0 | 1 | M | 98 | S | ||
| Protein postranslational modification | ||||||||||||
| Protein kinase superfamily protein | Glyma05g02080.1 | 6 | 5 | 1 | 0 | 0 | 1 | U | 16 | S | ||
| Protein kinase superfamily protein | Glyma10g44212.1 | 23 | 27 | 6 | 0 | 0 | 0 | U | ||||
| Protein phosphatase 2C | Glyma05g24410.1 | 64 | 18 | 1 | 0 | 0 | 0 | U | 10 | S | ||
| Protein phosphatase 2C | Glyma08g19090.1 | 87 | 30 | 7 | 0 | 0 | 0 | U | 11 | S | ||
| Protein phosphatase 2C family protein | Glyma06g10820.1 | 9 | 11 | 2 | 0 | 0 | 0 | U | 6 | L | ||
| Serine/threonine protein kinase | Glyma19g27110.1 | 29 | 8 | 2 | 0 | 0 | 0 | U | 23 | L | ||
| Protein degradation | ||||||||||||
| Aspartyl protease | Glyma15g41420.1 SP | 140 | 49 | 66 | 31 | 7 | 0 | 0 | 4 | ER/SP | 278 | ns |
| Eukaryotic aspartyl protease family protein | Glyma08g17680.1 SP | 13 | 27 | 20 | 4 | 0 | 0 | 2 | ER/SP | 108 | ns | |
| Matrix metalloproteinase | Glyma01g04370.1G, SP | 19 | 9 | 3 | 2 | 1 | 1 | ER/SP | 5 | ns | ||
| Membrane-anchored ubiquitin-fold protein 2 | Glyma08g26430.1 | 5 | 18 | 2 | 0 | 0 | 0 | U | 3 | S | ||
| Prolyl oligopeptidase family protein | Glyma20g23350.1 | 3 | 3 | 1 | 1 | 1 | 1 | U | 17 | S | ||
| Saposin-like aspartyl protease family protein | Glyma10g28370.1 SP | 10 | 5 | 2 | 1 | 1 | 1 | ER/SP | 21 | S | ||
| Serine carboxypeptid-ase-like 50 | Glyma07g34300.1 SP | 5 | 7 | 2 | 0 | 1 | 3 | ER/SP | 21 | L | ||
| Subtilase family protein | Glyma17g14270.1 SP | 35 | 15 | 5 | 0 | 0 | 5 | ER/SP | 95 | ns | ||
| Subtilisin/kexin-related serine protease | Glyma05g03760.1 | 49 | 102 | 98 | 30 | 13 | 1 | 1 | 4 | U | 310 | ns |
| Subtilisin/kexin-related serine protease | Glyma14g06970.1 SP | 6 | 43 | 21 | 7 | 1 | 0 | 5 | ER/SP | 315 | L | |
| Subtilisin/kexin-related serine protease | Glyma19g44060.1 | 73 | 14 | 8 | 1 | 1 | 4 | ER/SP | 429 | L | ||
| Ubiquitin family protein | Glyma01g03570.1 | 51 | 30 | 4 | 0 | 0 | 0 | U | 71 | S | ||
| Ubiquitin family protein | Glyma05g38330.2 | 20 | 26 | 3 | 0 | 0 | 0 | U | 14 | S | ||
| Protein folding | ||||||||||||
| Chaperonin precursor | Glyma08g18760.1 | 43 | 11 | 10 | 4 | 0 | 0 | 0 | C | 57 | S | |
| Chaperonin-60alpha | Glyma11g20180.1 | 35 | 12 | 6 | 0 | 0 | 0 | U | 27 | S | ||
| Heat shock protein 60 | Glyma10g25630.1 | 41 | 7 | 4 | 0 | 0 | 1 | M | 6 | S | ||
| Protein glycosylation | ||||||||||||
| Oligosaccharyl-transferase | Glyma11g12800.1 | 6 | 6 | 2 | 1 | 1 | 2 | U | 11 | S | ||
| Signalling | ||||||||||||
| ATP-binding protein | Glyma14g39550.1 SP | 60 | 11 | 2 | 2 | 2 | 2 | ER/SP | 44 | S | ||
| Autoinhibited Ca2+ -ATPase, isoform 8 | Glyma09g06890.2 | 11 | 4 | 3 | 10 | 9 | 7 | U | 3 | S | ||
| BCL-2-associated athanogene 7 | Glyma09g36560.1 | 18 | 12 | 4 | 0 | 0 | 0 | U | 91 | S | ||
| Calcium-dependent lipid-binding (CaLB domain) family protein | Glyma03g02370.1 | 11 | 4 | 2 | 3 | 3 | 4 | ER/SP | 9 | S | ||
| Calcium-dependent lipid-binding (CaLB domain) family protein | Glyma10g35410.1 | 5 | 6 | 3 | 1 | 1 | 2 | ER/SP | 82 | L | ||
| Calcium-dependent lipid-binding (CaLB domain) family protein | Glyma11g11470.1 | 6 | 7 | 3 | 1 | 1 | 1 | ER/SP | 14 | S | ||
| Calnexin | Glyma04g38000.1 SP | 17 | 11 | 1 | 3 | 3 | 3 | ER/SP | 28 | S | ||
| Calnexin | Glyma06g17060.1 SP | 20 | 11 | 4 | 3 | 3 | 3 | ER/SP | 32 | S | ||
| Calreticulin 1b | Glyma10g28890.2 SP | 20 | 2 | 4 | 0 | 0 | 1 | ER/SP | 107 | S | ||
| Endomembrane-type CA-ATPase 4 | Glyma03g33240.1 | 62 | 8 | 2 | 8 | 8 | 9 | U | 29 | S | ||
| Endomembrane-type CA-ATPase 4 | Glyma19g35960.1 | 57 | 10 | 8 | 8 | 8 | 10 | U | 37 | S | ||
| GTPase Rab homolog G3D (Rab7) | Glyma07g00660.1* | 11 | 10 | 2 | 0 | 0 | 0 | U | 5 | S | ||
| Glyma08g21940 | 6 | S | ||||||||||
| Glyma11g12630 | 9 | S | ||||||||||
| Glyma12g04830* | 30 | S | ||||||||||
| GTPase Rab2, small G protein superfamily | Glyma09g01950.1 | 18 | 17 | 3 | 0 | 0 | 0 | U | 5 | S | ||
| Glyma15g12880.1 | 5 | S | ||||||||||
| Glyma02g10450.1 | 9 | S | ||||||||||
| Leucine-rich repeat ATP-binding protein kinase | Glyma02g41160.2 SP | 35 | 11 | 5 | 2 | 2 | 2 | ER/SP | 20 | S | ||
| Ras-related small GTP-binding family protein (Rab8, RABE1) | Glyma10g43590 | 19 | 10 | 2 | 0 | 0 | 0 | U | 13 | S | ||
| Glyma11g15120 | 7 | S | ||||||||||
| Glyma12g07070 | 15 | S | ||||||||||
| Glyma18g52450 | 7 | S | ||||||||||
| Glyma20g23210 | 10 | S | ||||||||||
| Cell vesicle transport | ||||||||||||
| SNARE protein Syntaxin 131 | Glyma13g38370.1 | 16 | 13 | 3 | 1 | 1 | 1 | U | 6 | L | ||
| Transport ATPase | ||||||||||||
| P-type H+ATPase | Glyma19g02270.2* | 196 | 17 | 14 | 10 | 8 | 9 | U | 13 | L | ||
| P-type H+ATPase | Glyma04g34370.1* | 120 | 9 | 2 | 10 | 8 | 12 | U | 11 | S | ||
| P-type H+ATPase | Glyma06g20200.1* | 59 | 9 | 2 | 10 | 8 | 12 | U | 3 | S | ||
| Peptides and oligopeptide transport | ||||||||||||
| GmNPF8.6 Oligopeptide transport related | Glyma02g38970.1* | 19 | 9 | 4 | 11 | 10 | 11 | U | 31 | ns | ||
| GmNPF5.25 Oligopeptide transport related | Glyma11g34600.1* | 21 | 6 | 5 | 12 | 10 | 11 | U | 61 | ns | ||
| GmNPF1.2 Oligopeptide transport related | Glyma08g04160.4 | 51 | 9 | 5 | 12 | 10 | 11 | U | 155 | ns | ||
| GmNPF5.24 Oligopeptide transport related | Glyma11g34613.1* | 27 | 5 | 1 | 12 | 10 | 12 | U | ||||
| GmNPF5.29 Oligopeptide transport related | Glyma18g03790.1* | 17 | 3 | 1 | 11 | 10 | 11 | U | 25 | L | ||
| GmYSL7A Yellow Stripe Like 7 | Glyma11g31870.1 | 4 | 4 | 2 | 12 | 14 | 13 | U | 160 | ns | ||
| ABC Transporters | ||||||||||||
| GmABCA2 ABC transporter subfamily A | Glyma04g34140.2 | 16 | 3 | 2 | 7 | 6 | 6 | U | 52 | L | ||
| GmABCA7 ABC transporter subfamily A | Glyma04g34130.1 | 37 | 7 | 6 | 6 | 7 | 6 | U | 205 | L | ||
| GmABCB20 ABC transporter subfamily B | Glyma02g10530.1 | 11 | 2 | 2 | 11 | 13 | 10 | U | 2 | L | ||
| GmABCG39 ABC transporter subfamily G (PDR) | Glyma10g34700.2 | 16 | 4 | 4 | 10 | 13 | 13 | U | 6 | L | ||
| GmABCG11 ABC transporter subfamily G (white-brown complex) | Glyma08g07580.1 | 20 | 4 | 2 | 4 | 6 | 6 | U | 37 | L | ||
| Sugar Transport | ||||||||||||
| Voltage-dependent anion channel | Glyma08g40800.1* | 15 | 21 | 1 | 0 | 0 | 1 | U | 14 | S | ||
| Voltage-dependent anion channel | Glyma09g37570.1* | 4 | 12 | 1 | 0 | 0 | 2 | U | 30 | S | ||
| Voltage-dependent anion channel | Glyma18g16260.1* | 22 | 21 | 5 | 0 | 0 | 0 | U | 25 | S | ||
| Voltage-dependent anion channel | Glyma18g49070.1* | 7 | 21 | 4 | 0 | 0 | 0 | U | 47 | S | ||
| Amino acid transport | ||||||||||||
| GmAPC1 Amino acid permease | Glyma09g21070.2 | 22 | 10 | 4 | 14 | 14 | 13 | U | 70 | L | ||
| Sulfate transport | ||||||||||||
| Sulfate/bicarbon-ate/oxalate exchanger SAT-1 (SST1 Lotus homologue) | Glyma07g09710.1 | 55 | 3 | 1 | 10 | 12 | 10 | U | 209 | S | ||
| Sulfate/bicarbon-ate/oxalate exchanger SAT-1 (SST1 Lotus homologue) | Glyma09g32110.3 | 105 | 13 | 6 | 10 | 10 | 10 | U | 355 | ns | ||
| Phosphate transport | ||||||||||||
| Phosphate transporter 1–1, MFS | Glyma10g33030.1 | 9 | 7 | 2 | 9 | 11 | 12 | U | 38 | S | ||
| Phosphate transporter 1–4 | Glyma10g04230.1 | 10 | 5 | 2 | 11 | 11 | 12 | U | 13 | L | ||
| NIP Transport | ||||||||||||
| Nod26 aquaporin | Glyma08g12650.1 | 929 | 35 | 20 | 5 | 6 | 6 | 6 | U | 3167 | S | |
| Miscellaneous transport | ||||||||||||
| Nucleotide transporter 1 | Glyma15g01420.1 | 50 | 10 | 5 | 10 | 9 | 8 | C | 29 | L | ||
| Secretory carrier membrane protein | Glyma06g45160.1 | 90 | 10 | 4 | 4 | 4 | 4 | U | 190 | S | ||
| Secretory carrier membrane protein 3 | Glyma12g11820.1 | 78 | 10 | 2 | 4 | 4 | 4 | U | 63 | S | ||
| Other | ||||||||||||
| C2 domain protein binding | Glyma09g05100.1 | 2 | 7 | 2 | 0 | 1 | 2 | U | 67 | S | ||
| Cupredoxin superfamily protein | Glyma17g14730.1G, SP | 12 | 10 | 4 | 2 | 1 | 2 | ER/SP | 7 | S | ||
| Cytochrome P450 CYP2 subfamily | Glyma07g20430.1* | 29 | 14 | 6 | 1 | 1 | 1 | ER/SP | 64 | ns | ||
| Domain of unknown function (DUF221) | Glyma13g29270.1 | 6 | 3 | 2 | 10 | 9 | 9 | U | 2 | S | ||
| Domain of unknown function (DUF588) | Glyma01g31910.2 | 42 | 11 | 2 | 4 | 3 | 4 | U | 247 | ns | ||
| Domain of unknown function (DUF588) | Glyma03g05230.1 | 11 | 23 | 2 | 0 | 3 | 3 | U | 247 | L | ||
| Domain of unknown function (DUF3411) | Glyma11g09920.1 | 23 | 7 | 2 | 0 | 0 | 3 | C | 62 | S | ||
| Protein of unknown function (DUF3411) | Glyma18g44970.1 | 8 | 5 | 2 | 0 | 2 | 4 | C | 48 | S | ||
| Early nodulin-like protein 10 | Glyma02g36580.1G, SP | 44 | 9 | 2 | 2 | 0 | 2 | ER/SP | 454 | ns | ||
| EKN rich protein | Glyma05g25020.1 | 244 | 66 | 7 | 0 | 0 | 0 | U | 372 | L | ||
| EKN rich protein | Glyma08g08140.1 | 317 | 77 | 6 | 0 | 0 | 0 | U | 798 | L | ||
| Endomembrane protein 70 | Glyma08g20640.1 | 4 | 3 | 2 | 10 | 10 | 10 | ER/SP | 12 | S | ||
| FAD/NAD(P)-binding oxidoreductase family protein | Glyma11g18320.1 | 13 | 6 | 2 | 0 | 0 | 0 | U | 12 | S | ||
| Fatty acid amide hydrolase | Glyma08g00535.1 | 37 | 11 | 5 | 0 | 1 | 4 | U | ||||
| Ferretin 1 | Glyma18g43650.2 | 13 | 10 | 1 | 0 | 0 | 0 | C | 44 | S | ||
| Glucosidase 1 | Glyma05g27890.1 | 189 | 114 | 142 | 28 | 21 | 0 | 1 | 4 | U | 149 | L |
| Glycosyl hydrolase family 17 protein | Glyma14g08200.2G, SP | 2 | 6 | 2 | 1 | 1 | 2 | U | 69 | S | ||
| Glycosyl hydrolase family 17 protein | Glyma14g16830.1G | 36 | 11 | 4 | 1 | 1 | 1 | ER/SP | 14 | S | ||
| Glycosyl hydrolase family 17 protein | Glyma16g04680.1G, SP | 12 | 6 | 3 | 2 | 2 | 2 | ER/SP | 4 | S | ||
| Glycosyl hydrolase family 31 protein | Glyma15g14150.1 | 7 | 5 | 3 | 1 | 0 | 3 | U | 21 | S | ||
| Lipase/lipooxygenase, PLAT/LH2 family protein | Glyma11g38220.1 SP | 26 | 10 | 2 | 1 | 0 | 0 | ER/SP | 75 | S | ||
| LITAF-domain-containing protein | Glyma08g47500.1 | 16 | 23 | 2 | 1 | 1 | 1 | U | 276 | S | ||
| Membrane-associated progesterone binding protein 3 | Glyma09g25940.1 | 2 | 14 | 2 | 2 | 1 | 1 | U | 67 | S | ||
| Mo25 family protein | Glyma17g01180.2 | 3 | 5 | 2 | 0 | 0 | 0 | U | 330 | S | ||
| Nodulin | Glyma20g02921.1 SP | 21 | 9 | 2 | 1 | 1 | 1 | ER/SP | ||||
| Nucleotide-diphospho-sugar transferase | Glyma16g06180.1 | 5 | 5 | 2 | 1 | 1 | 1 | U | 129 | ns | ||
| Oligosaccharyl-transferase subunit Ribophorin II | Glyma03g32140.3 SP | 9 | 7 | 3 | 4 | 4 | 5 | ER/SP | 3 | S | ||
| Outer envelope pore protein 24 | Glyma09g18920.1 | 8 | 14 | 3 | 0 | 0 | 0 | U | 18 | S | ||
| Patched family protein | Glyma04g01901.1 SP | 6 | 2 | 2 | 13 | 12 | 12 | ER/SP | ||||
| Peroxidase | Glyma16g27890.1 | 34 | 24 | 22 | 6 | 2 | 1 | 1 | ER/SP | 191 | ns | |
| Peroxidase superfamily protein | Glyma14g38210.1 SP | 33 | 19 | 5 | 0 | 0 | 1 | ER/SP | 14 | L | ||
| PLAC8 family protein | Glyma05g37590.1 | 28 | 13 | 3 | 0 | 1 | 3 | U | 482 | S | ||
| PLAC8 family protein | Glyma08g01990.1 | 9 | 7 | 2 | 0 | 1 | 4 | U | 31 | S | ||
| PLAC8 family protein | Glyma08g04830.1 | 167 | 5 | 20 | 3 | 0 | 0 | 1 | U | 1146 | L | |
| PLAC8 family protein | Glyma09g31910.1 | 310 | 21 | 5 | 2 | 1 | 1 | U | 1015 | ns | ||
| Protein of unknown function (DUF2359) | Glyma05g27090.1 | 2 | 4 | 2 | 0 | 0 | 1 | U | 10 | S | ||
| Protein of unknown function (DUFB2219) | Glyma13g25150.1 | 35 | 10 | 4 | 0 | 0 | 1 | C | 3 | L | ||
| Proteinase inhibitor, propeptide | Glyma02g18320.1 SP | 7 | 12 | 26 | 3 | 1 | 1 | 1 | ER/SP | 133 | S | |
| Purple acid phosphatases superfamily protein | Glyma18g17541.1 SP | 25 | 9 | 4 | 0 | 0 | 0 | ER/SP | ||||
| SPFH/Band 7/PHB protein family | Glyma05g01360.1* | 728 | 9 | 49 | 14 | 0 | 0 | 0 | U | 484 | S | |
| SPFH/Band 7/PHB protein family | Glyma13g05120.3* | 417 | 38 | 1 | 0 | 0 | 1 | U | 18 | S | ||
| SPFH/Band 7/PHB protein family | Glyma19g02370.1* | 444 | 38 | 7 | 0 | 0 | 0 | U | 37 | S | ||
| SPFH/Band 7/PHB protein family Nod53b flotillin | Glyma06g06930.1* | 475 | 23 | 43 | 18 | 0 | 0 | 0 | U | 342 | L |
The SM proteome does contain several membrane proteins that have been localized to the SM by methods other than proteomic analysis. These include nodulin 26 (18, 19), remorin (60) and H+-ATPases (28, 29, discussed further below, 61).
Possible Contaminants of the SM Preparation
As the SM is partially derived from the ER and Golgi (62), it might be expected that some proteins would be present in all membranes. For example, the soybean calnexin protein (Glyma06g17060.1) was identified in this proteomic analysis in the SM along with the related ER protein calreticulin (Glyma10g28890.2, Table II). ER lumen proteins calreticulin, BiP, and protein disulfide isomerases (PDI), identified here, have all been previously identified on the SM of other legumes (34, 36). The infected cell is tightly packed with symbiosomes and has high requirements for protein synthesis; consequently, abundant ER proteins may adhere to the SM during isolation, although it is equally possible that these proteins are associated with the SM because of fusion of vesicles derived from the ER (see below). It will be important to further validate the localization of proposed SM proteins by other means in the future.
Surprisingly, a number of soluble proteins were present in the SM fractions and we assume that these are contaminants that are in high abundance in nodules and that adhere to the SM during isolation of symbiosomes. These include a number of purine biosynthesis enzymes, uricase, and malate dehydrogenase. Purine biosynthesis is important in soybean nodules for assimilation of fixed nitrogen as ureides and the enzymes involved are localized to both plastids and mitochondria in infected cells of cowpea nodules (63). Uricase is also important for nitrogen assimilation and is localized in peroxisomes of uninfected nodule cells (64). The genes encoding these enzymes are expressed at high levels in nodule tissue (65, 66) and the activity of many of the enzymes is high, because of the high requirements for nitrogen assimilation in nodules (67). It is therefore likely that the peptides identified for these enzymes represent a relatively low level of contamination from plastids or mitochondria (63) or peroxisomes from uninfected cells (64) that, at least for mitochondria, is not detectable through the immunological analysis described above.
Leghemoglobins (Lb) are the most abundant plant proteins in nodules and are essential for successful BNF (70). Four Lb proteins were detected in our fractions (Glyma10g34280, Glyma20g33290, Glyma10g34260, and Glyma10g34290). Lbs are known to be a cytosolic protein in infected cells and are encoded by the highest expressed genes in nodules (65, 66), so their presence in this and other SM proteomes (33, 34) is likely to be caused by contamination, a function of their high abundance in infected cells.
We detected peptides for a malate dehydrogenase (Glyma12g19520) in PBS samples (27 spectra in three biological replicates). A nodule-enhanced form of malate dehydrogenase was identified in alfalfa, but its subcellular localization was not identified (68). The protein we found in the PBS is predicted using bioinformatic programs to be mitochondrial, but previous proteomic analyses have identified homologs in symbiosomes (33, 34). We also found a small number of malate dehydrogenase peptides in the SM fractions (results not shown) but these were in low abundance and may reflect the transit of the enzyme to the PBS. Whether the nodule-enhanced form of malate dehydrogenase is mitochondrial or symbiosome localized requires further investigation.
Several rhizobial proteins also contaminated the SM fractions (Table III). Interestingly, several outer membrane rhizobial proteins were identified, suggesting perhaps that some bacteroid outer membranes rupture during SM preparation and contaminate the final SM sample. Proteins such as NifHD and FixA are abundant soluble proteins in bacteroids and clearly not localized on the SM. Again, it seems likely that some abundant soluble proteins have become associated with the SM during isolation. These may arise from symbiosomes and bacteroids damaged in the initial homogenization.
Table III. List of proteins identified in Bradyrhizobium japonicum by LCMS/MS from the soybean SM. A minimum of two unique peptides were identified in one or more biological samples for all proteins indicated. Symbiosome membrane (SM) (bicarbonate stripped and chloroform-methanol extractions pooled). Percentage coverage (%C) is the maximum percentage of a protein to which peptides have been mapped in a biological sample. Membrane topology has been predicted by three bioinformatic suites: (i) SOSUI, (ii) TMHMM and (iii) TopPred2, with number of predicted Trans-Membrane Domains indicated.
| Accession number | Description | SM | %C | i | ii | iii |
|---|---|---|---|---|---|---|
| gi|27383032 | ABC transporter substrate-binding protein | 2 | 16 | 0 | 0 | 1 |
| gi|384213970 | Acetyl-CoA acetyltransferase | 7 | 29 | 0 | 0 | 1 |
| gi|27376888 | AhpC gene product | 5 | 19 | 0 | 0 | 0 |
| gi|27378020 | Amino acid binding protein | 6 | 61 | 0 | 1 | 2 |
| gi|1209038 | FixA | 3 | 11 | 0 | 0 | 0 |
| gi|27377170 | GroEL gene product | 85 | 12 | 0 | 0 | 0 |
| gi|384221462 | GroES gene product | 69 | 12 | 0 | 0 | 0 |
| gi|27376941 | Hypothetical protein | 23 | 51 | 0 | 0 | 0 |
| gi|27380266 | Hypothetical protein | 19 | 9 | 0 | 0 | 0 |
| gi|27381635 | Hypothetical protein | 10 | 8 | 0 | 0 | 3 |
| gi|12620578 | ID352 | 9 | 10 | 0 | 1 | 1 |
| gi|12620644 | ID525 | 10 | 30 | 0 | 0 | 4 |
| gi|12620707 | ID693 | 30 | 5 | 1 | 0 | 2 |
| gi|27376854 | NifD gene product | 23 | 12 | 0 | 0 | 0 |
| gi|27376880 | NifH gene product | 68 | 9 | 0 | 0 | 0 |
| gi|152324 | Nitrogen fixation protein | 16 | 3 | 0 | 0 | 0 |
| gi|27376422 | Outer membrane protein | 9 | 12 | 0 | 1 | 2 |
| gi|27376315 | Outer-membrane immunogenic protein | 7 | 7 | 1 | 2 | 1 |
| gi|27379812 | Outer-membrane immunogenic protein | 12 | 9 | 2 | 0 | 0 |
| gi|27379978 | Outer-membrane immunogenic protein | 41 | 11 | 0 | 1 | 0 |
| gi|27382806 | Outer-membrane immunogenic protein | 59 | 7 | 0 | 1 | 0 |
| gi|27382260 | Peptidoglycan-associated lipoprotein | 17 | 1 | 0 | 0 | 1 |
| gi|27382263 | TolB gene product | 5 | 7 | 0 | 0 | 1 |
Some of the integral membrane proteins identified, including a nucleotide transporter I (Glyma15g01420.1), are predicted to be localized to plastids, but they could also represent valid SM proteins if this function is also required in the symbiosome and dual localization or modification of an originally plastid function occurred. Mitochondrial substrate carrier family members were identified on the SM but this family is not exclusively localized on mitochondrial membranes (69) and the bioinformatic programs used were unable to predict a subcellular location for the proteins (Table II). Independent experimental validation of the SM localization will be required to resolve these questions.
Proposed Functions for Identified Proteins
In total, 197 proteins were identified in the SM proteome, with a further six proteins found only in the peripheral membrane protein fraction, eight proteins identified only in PBS samples, and one protein identified in both PBS and SM peripheral protein samples (Table II). In Table II proteins identified in the SM proteome were grouped according to their proposed functions within the cell (MapMan predictions) with the data for percentage coverage of the protein by identified peptides, the number of unique peptides identified and the sample in which they were identified. Localization, signal peptide and GPI-anchor predictions compiled with expression data from one of the two soybean transcriptomes (64) are also included (Table II). Selected proteins are discussed below.
Protein Folding and Degradation
Several proteins involved in protein assembly and degradation processes were identified on the SM in this study, including two members of the protein disulfide isomerise (PDI) family (Glyma04g42690 and Glyma06g12090). This family of proteins have ubiquitous expression across soybean tissues, are localized to the ER lumen in other tissues and are involved in the proper folding and quality control of storage proteins (71). Import of proteins into the symbiosome would likely require the same processes and, as the structure and composition of the SM is most closely related to the ER (3), these PDI proteins may have been co-opted for this role during the symbiosis. PDIs have previously been identified in all proteomic analyses of the SM (31, 33, 34, 36).
Members of the protein degradation class feature strongly in the PBS but were also found in the SM proteome. Four members of the subtilase family were identified, three (Glyma17g14270, Glyma05g03760, Glyma14g06970) most clearly localized (based on number of peptides identified) in the PBS and one (Glyma19g44060) associated only with the SM. Many of these proteins are predicted to have GPI anchors (Table II) as expected for extracellular proteases. Because the SM has the same orientation as the plasma membrane, the inside of the symbiosome can be regarded as equivalent to the apoplast (3). The genes encoding all these proteins show high nodule-specific expression according to the soybean transcriptome (66). Subtilases are serine peptidases whose members may be involved in nonselective degradation of proteins or as proprotein convertases (72). They are involved in a range of processes including peptide hormone processing, plant interactions with microorganisms, seed germination and distribution of stomata (72). A number of subtilase genes are induced when L. japonicus is infected by mycorrhiza and rhizobia (73) and silencing of some of these genes reduced mycorrhizal colonization (74). The proteins encoded by Glyma19g44060.1 and Glyma05g03760.1 are closest to Arabidopsis homologs thought to have nonselective activity (AtSBT1.7 [Ara12, AtSLP1]; AtSBT1.6 both MEROPs database [http://merops.sanger.ac.uk/(75)] S08A), whereas for Glyma14g06970 the closest Arabidopsis homolog (AtSBT1.2 [SDD1], MEROPs database, S08A) is thought to affect stomata distribution and density by processing an unknown peptide to generate a signal molecule (76).
Two aspartate proteinases (Glyma15g41420, Glyma08g17680) were also detected in the PBS. The Phaseolus vulgaris ortholog of Glyma15g41420, Nodulin 41, was recently localized in uninfected cells (77) and the possibility that it is a contaminant in the PBS in this study cannot be ruled out. However, the closest Arabidopsis homolog, constitutive disease resistance 1, has an apoplastic localization (78), which is consistent with a PBS localization in nodules. Because the SM has the same orientation as the plasma membrane, the inside of the symbiosome can be regarded as equivalent to the apoplast (3). Constitutive disease resistance 1 is thought to be involved in generating a peptide signal to induce defense responses (78). The identification of both subtilases and aspartate proteases in the PBS suggests an important role for these enzymes, perhaps in generating peptide signals. There is evidence for activity of nodule-specific cysteine-rich peptides in terminal differentiation of bacteroids in legumes such as M. truncatula (79) and although terminal differentiation does not occur in soybean (80), peptide signals may be involved in other processes including communication between the symbionts.
Membrane Trafficking
Members of three subfamilies of the small GTPase Rab family were present in the SM proteome but because of their conserved amino acid sequences, the peptides identified could not be ascribed to a protein encoded by one particular soybean gene. The RabG (Rab7) peptides are present in proteins encoded by four different soybean genes (Table II). All these genes are expressed in nodules but Glyma12g04830 has the most nodule-enhanced expression (64). RabB (Rab2) peptides are present in proteins encoded by Glyma09g01950 and/or Glyma15g12880. RabE (Rab8) peptides are present in proteins encoded by six soybean genes (Table II). Of these, Glyma12g07070 and Glyma15g12880 have the highest expression in nodules, although both are also expressed in most other soybean tissues. Rabs are involved in vesicular transport within cells. Rab1 and Rab7 have previously been implicated in SM biogenesis in soybean (81) and Rab7 proteins were identified on the M. truncatula and L. japonicus SM (34). Rab7 is a marker for the late endosome/prevacuolar compartment (PVC) and tonoplast and is essential for PVC-to-vacuole trafficking and vacuole biogenesis (82). Although M. truncatula symbiosomes gain Rab7 (but not the early endosome marker Rab5) they do not develop into a lytic compartment because they do not acquire vacuolar SNAREs (soluble N-ethylmaleimide sensitive factor attachment protein receptor) until nodules start to senesce (83). Instead, a plasma membrane SNARE SYP132 is present on the SM from early in development. It suggests the involvement of an exocytosis-derived process in SM formation, which was proved by functional analysis of two VAMP72 homologs in M. truncatula nodules (84). Therefore, a unique identity of the SM could allow the membrane to intercept specific secretory traffic to the plasma membrane and specific endocytic/biosynthetic traffic toward the vacuole (83). The presence of Rab8 and Rab2 small GTPases, that are thought to be involved in trafficking of vesicles from the Golgi and the ER, respectively, to the plasma membrane (85–87), further supports the idea of the SM as a chimeric membrane.
SNARE proteins such as syntaxins are also involved in vesicle fusion and we have identified a protein related to syntaxin 131 in the soybean SM (SYP131; Glyma13g38370, 13% peptide coverage). SYP131 is part of the clade that includes the Medicago SM syntaxin SYP132 (88, see below). These syntaxins are considered plasma membrane SNAREs in nonsymbiotic tissues (89) but MtSYP132 is localized to regions of the plasma membrane close to the infection thread and infection droplet membranes as well as on the SM (88, 90). Whether GmSYP131 is localized on membranes other than the SM is not known, but the gene encoding it is expressed in other plant tissues and its expression is not enhanced significantly in nodules (66), suggesting that it may have a role on the plasma membrane in nonsymbiotic cells.
In the arbuscular mycorhhizal symbiosis, secretory vesicles normally targeted to the plasma membrane can be redirected to the periarbuscular membrane (derived from and contiguous with the plasma membrane) at a specific time in the symbiosis, to form the specialized symbiotic membrane (91). This is analogous to the SM and the presence of the Rab small GTPases and syntaxins suggests that perhaps a similar reorientation of the secretory system is used to create the specialized membrane that is the SM. This might also explain how proteins with roles on both the plasma membrane and SM are targeted to the SM when required although a particular targeting sequence is not obvious.
Transport
Nodulin 26
Peptides corresponding to nodulin 26 (Glyma08g12650) were detected in all SM samples analyzed in this study, with up to 20% coverage of the protein (Table II). Spectra corresponding to this protein were the most abundant in the proteomic analysis, as expected for a dominant SM protein. Nodulin 26 was detected in the previous proteomic analysis of the soybean SM (31), but was not in SM proteomes from L. japonicus, M. truncatula, or pea (P. sativum). Nodulin 26 is exclusively localized to the SM and because of its prevalence is widely used as a marker for the membrane. Nodulin 26 was first identified as an integral membrane transporter of soybean SM (19) and is a member of the major intrinsic protein/aquaporin (MIP/AQP) channel family. It is estimated to constitute 10% of the protein content of the SM (21, 58). Nodulin 26 acts as a multifunctional aquaglyceroporin, with Xenopus oocyte studies showing it can facilitate the movement of glycerol and formamide (18, 21). Other studies have shown that it can also facilitate ammonia transport across the SM (16) and can act as a docking station for cytosolic glutamine synthetase (20). Glutamine synthetase (Glyma10g06810) was detected in both the SM and SM peripheral proteomes, and interestingly in the PBS proteome. Its detection in the PBS is unexpected as the C terminus of nodulin 26, to which glutamine synthetase binds, is cytosolic (92). The detection of both glutamine synthetase and nodulin 26 across all our samples, however, provides further support for their suggested roles in ammonia release from the symbiosome (16, 20).
Sulfate Transporters
Two putative sulfate transporter proteins were identified in the SM proteome (Glyma09g32110 and Glyma07g09710) with 8 and 6% coverage, respectively, from identified peptides (Table II). These proteins, classified as sulfate/bicarbonate/oxalate exchangers, are homologous to the L. japonicus SST1 protein. Sulfur is a component of the metallo-clusters of nitrogenase, essential for the reduction of nitrogen, and must be actively transported across membranes (23). LjSST1 was identified from a _fix_− mutant in L. japonicus and complemented a yeast strain deficient in sulfate transport (23). LjSST1 is also one of the few transporters that has been previously identified on the SM through proteomic analysis (34). Krusell et al. (23) reported that LjSST1 expression is essential for symbiotic nitrogen fixation; knockout mutants grow normally in nonsymbiotic conditions but are unable to produce functioning nodules when inoculated with Mesorhizobium loti.
Transcriptome data shows expression of Glyma09g32110 and Glyma07g09710 in soybean is specific to nodule tissue, where they are highly expressed (65, 66). Detection of peptides corresponding to the soybean homologs here provides evidence for a role in the symbiosis in soybean as well as L. japonicus. Studies using 35SO4− and isolated soybean symbiosomes failed to detect sulfate uptake (Day, unpublished data) and in this context, it should be noted that some members of the SST family, though not phylogenetically close to these soybean candidates, can transport other metabolites in addition to sulfate, including molybdate (93). Molybdenum is an essential component of the nitrogenase enzyme and an SM molybdate transporter is yet to be identified.
Energization of the SM
Three related P-type H+-ATPases were identified in the soybean SM proteome (Glyma04g34370, Glyma06g20200, and Glyma19g02270) with 16%, 16%, and 15% peptide coverage, respectively. A number of other H+-ATPases share peptides with these proteins so in fact there may be many different proteins that play this role on the SM. The soybean transcriptome suggests that at least 13 H+-ATPases genes are expressed in nodules. Of those with unique peptides Glyma19g02270 and Glyma04g34370 show highest nodule expression (66). However, expression levels in other tissues are similar to that of nodules suggesting that the same proteins have this activity in symbiotic and nonsymbiotic tissues. This agrees with data of Blumwald et al. (29) that suggests that the H+-ATPase on symbiosomes and the plasma membrane of uninfected soybean root cells were not immunologically distinct, although they saw some differences in activity. Because presumably the activity of H+-ATPase on the SM reflects the activity of a number of different proteins, the differences in activity might reflect the different combination of H+-ATPase proteins on the SM and root plasma membrane. A P-type H+-ATPase was detected on the SM of soybean using specific antibody labeling (29) and found in the SM proteomes in L. japonicus and M. truncatula (34, 36). P-type H+-ATPases are considered to have an important role in the development of the symbiotic association both to acidify the symbiosome space to promote protonation of NH3, as well as to energize the SM by establishing an electrochemical gradient across the membrane that is necessary for the secondary transport of other solutes (reviewed in 14). Interestingly, the related V-type ATPases are also in the SM proteome of pea and L. japonicus (33, 34), but could not be detected by immunolocalization on the soybean SM (29). The absence of V-type ATPases in this study, together with Fedorova et al. 's (29) results, suggest that soybeans may differ from other legumes in their SM ATPase requirements.
Calcium Transport
Three Ca2+-ATPases were identified in the SM proteome: Glyma09g06890, Glyma03g33240, and Glyma19g35960. It has been suggested that symbiosomes may behave as calcium stores in infected cells (61). Calcium uptake is an active (ATP-driven) process and an ATP-driven Ca2+-pump has been biochemically characterized on the SM of broad bean (61). As for the P-type H+-ATPases, the Ca2+-ATPases identified here are expressed broadly across soybean tissues (65, 66), suggesting recruitment to a new role and location as part of the symbiosis.
Nitrogen/Carbon Transport
Five orthologs of the Arabidopsis NTR/PTR Family Transporters (NPF; 94) were identified in the SM proteome in this study: Glyma02g38970 (GmNPF8.6), Glyma08g04160 (GmNPF1.2), Glyma11g34600 (GmNPF5.25), Glyma11g34613 (GmNPF5.24), and Glyma18g03790 (GmNPF5.29). The NPF proteins identified here have eleven (GmNPF8.6 and GmNPF5.29) or twelve (GmNPF1.2, GmNPF5.24, and GmNPF5.25) transmembrane domains (SOSUI algorithm prediction). A role for these transporters on the symbiosome membrane is further supported by RNA-seq transcriptome data, which reports gene expression in nodule tissue samples only (66) or at very low levels in other tissues compared with nodule (65). Proteins homologous to GmNPF5.24, GmNPF5.25 and GmNPF5.29 have also been identified in the L. japonicus SM proteome (34). These transporters fall into the same subfamily as a di- and tri-peptide transporter from the NPF family, AtNPF5.2 (AtPTR3; 94, 95), whereas GmNPF8.6 is in the same subfamily as dipeptide transporters AtNPF8.1 (PTR1), AtNPF8.2 (PTR5), and AtNPF8.3 (PTR2; 94, 96, 97).
Members of the NPF transport a range of nitrogen-based compounds (98). AtNPF6.3 (AtNTR1.1, CHL1), one of 53 proteins in the NPF of Arabidopsis, can transport nitrate (99) and auxin (100) as can the M. truncatula homolog MtNRT1.3 (101, 102). In this context, indole acetic acid uptake by isolated soybean symbiosomes as reported (103) may be relevant. NPF proteins with dual transport functions are implicated in nutrient sensing within the plant, in addition to high- and low-affinity nitrate uptake (100). Other members of the NPF in Arabidopsis transport glucosinolate defense compounds in seeds (104). In the nonlegume Alnus glutinosa, AgDCAT1 was localized to the symbiotic interface and shown to transport dicarboxylates when expressed in E. coli (105), though its closest homologs are characterized as nitrate transporters (e.g. AtNPF6.3). This suggests homology alone cannot be used to predict solute specificity in this family. Because the main transfer of carbon from plant host to bacteroid in the symbiosis is through the dicarboxylate malate (106), members of any transporter family capable of malate transport on the SM are of particular interest.
Transport of nitrogen containing compounds is of interest in legumes, especially as nodule development is suppressed in the presence of nitrate (107). In all plants, nitrogen plays an important regulatory role, particularly in lateral root formation and nodulation. An NPF family member in M. truncatula (MtNPF1.7 previously called LATD/NIP), classified in the same subfamily as GmNPF1.2, is essential for the development and maintenance of lateral roots and release of rhizobia into the symbiosome (108–110). Heterologous expression experiments have suggested that MtNIP/LATD encodes a nitrate transporter, but its function in nodules could not be directly replaced by its Arabidopsis homolog NTR1.1 (111).
There is also recent evidence to suggest that bacteroids in the pea : Rhizobium leguminasarum symbiosis may be auxotrophs for branched-chain amino acids, relying on the plant host to provide these solutes (112). Transported peptides may serve as a source of these amino acids, rescuing the bacteroids from their branched-chain amino acid deficiency. Also identified on the SM was Glyma09g21070, a member of the cationic amino acid transporter (CATs) subfamily of the amino acid-polyamine-choline family of amino acid transporters. Expression of Glyma09g21070 appears nodule specific.
ATP-binding Cassette Family Transporters
Five proteins with homology to the ATP-binding cassette (ABC) superfamily were identified in the SM proteome: Glyma04g34140 (GmABCA2), Glyma04g34130 (GmABCA7), Glyma02g10530 (GmABCB20), Glyma08g07580 (GmABCG11), and Glyma10g34700 (GmABCG39). GmABCA2, GmABCA7, and GmABCG11 have expression that is high and relatively specific to nodule tissue, whereas GmABCB20 and GmABCG39 have a more diverse expression pattern across soybean tissues (65, 66). ABC transporters can act as importers or exporters and are driven by ATP hydrolysis. There are 133 members of this family in Arabidopsis, distributed over eight subclasses, but only 22 members have been characterized functionally (reviewed in 113). Plant ABC transporters have been localized to a range of subcellular membranes such as those of the vacuoles, chloroplasts, mitochondria, ER, and peroxisomes, as well as to the plasma membrane. They fulfil a range of functions within the plant and roles have been established in the transport of hormones, lipids, metals, secondary metabolites, and xenobiotics (reviewed in 114). The first member of the ABCA subfamily characterized, AtABCA9, has recently been demonstrated to mediate the transport of fatty acids for lipid synthesis in the endoplasmic reticulum (115). A number of members of the ABCB subfamily are auxin efflux carriers (116), whereas AtABCB14 is a malate importer (117; as opposed to the protein expected to export malate out of the cytosol and into the symbiosome). Members of the ABCG subfamily in Arabidopsis have a number of different roles including transport of strigolactones in development of the plant-mycorrhizal symbiosis (118), transport of lipids and waxes involved in production of the cuticle and in vascular development (119–121), and cadmium and lead export to aid in cell detoxification (122, 123). GmABCG11 is a half-sized transporter that would function as a dimer. Its closest Arabidopsis homolog is WBC11 (AtABCG11), which forms both hetero- and homodimers in its role in transport of cuticular lipids and sterols (119–121). GmABCG39 is a full-size ABCG transporter with 82% similarity to AtABCG39 and AtABC34. AtABC39 is localized on the plasma membrane and mediates resistance to paraquat although there is no direct evidence that it transports this compound (124).
Lipid Raft Proteins
Several band 7/flotillin-like type proteins were identified in this study (Glyma05g01360, Glyma06g06930, and Glyma19g02370). There is 62% coverage of peptides for Glyma06g06930 and 65% for Glyma05g01360 (Table II). The genes encoding both are expressed at high levels in nodule tissue with limited expression in the other tissues (Table II, 63, 64). The proteins share a common motif, the SPFH (stomatin, prohibitin, flotillin, and HflK/C) domain. Flotillin-like proteins have previously been identified on the SM in pea as well as soybean (31, 33) and play an important role in the infection process in legume-rhizobia symbioses (125). Glyma06g06930, the soybean homolog of M. truncatula FLOT4, contains a conserved flotillin domain, a subgroup of the band-7 like proteins. Flotillin domain proteins are lipid raft-associated. Lipid raft-microdomains on plant membranes are dynamic, sterol and lipid rich protein assemblies that serve as centers for membrane trafficking and signaling events as they interact with a range of different proteins (126, 127). FLOT4 is up-regulated in a strongly nod-factor dependent manner during early symbiotic events and has been localized to the infection thread membrane and the plasma membrane in root nodules (125). FLOT4 silenced plants form fewer nodules that do not fix nitrogen efficiently (125). Although a role for flotillin has been established in the infection thread process, this study suggests it has a continuing presence on the symbiosome membrane in soybean.
Two remorin proteins, Glyma08g01590 and Glyma05g37990, were identified in the SM proteome with 19 and 15% peptide coverage respectively (Table II). The genes encoding both these proteins show nodule-specific expression (Table II) Remorin proteins are plant specific and are localized to lipid rafts on membranes (128). Remorins have been implicated in regulatory functions in the symbiosis and their localization to lipid rafts on the SM confirmed (60). They were identified on the SM in the L. japonicus, and pea proteomes (33, 34). Identification here presents further evidence of a regulatory role for remorin proteins in the mature SM.
Other Proteins of Interest
Glyma11g31870 (GmYSL7), a member of the Yellow stripe-like (YSL) family that is part of the oligopeptide transporter family, was identified on the SM, with 4.3% peptide coverage of the protein. GmYSL7 was identified first in soybean nodules through a PCR based approach (22) and the soybean transcriptome suggests nodule-specific expression (63, 64). YSL proteins in dicots typically transport metals such as iron, copper, and manganese complexed with nicotianamine (NA) (reviewed in 129, 130). However, the closest Arabidopsis homolog, YSL7, has recently been shown to transport the Pseudomonas virulence factor, Syringolin A, which is a peptide derivative, with transport of Syringolin A inhibited by tri- to octapeptides (131). Syringolin A has similar chemical properties, size and net charge to metal-NA complexes (131) that are the usual substrate for YSL transporters, but whether AtYSL7 can also transport metal-NA was not established.
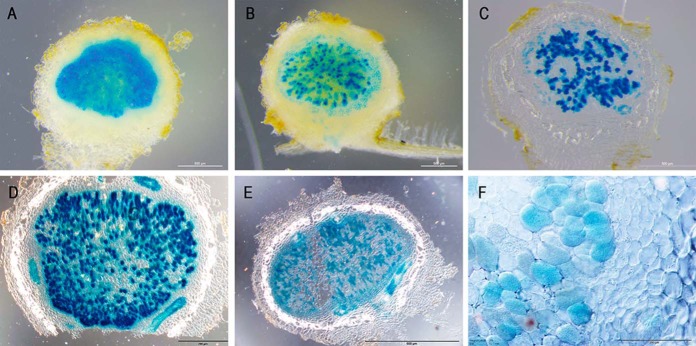
Four proteins with a PLAC8 superfamily motif (Glyma09g31910, Glyma08g04830, Glyma05g37590, and Glyma08g01990) are found on the SM. Expression of Glyma09g31910 and Glyma08g04830 is extremely high and virtually specific to nodule tissue, whereas Glyma05g37590 and Glyma08g01990 are expressed over a range of tissue types, with Glyma05g37590 enhanced five times in nodules compared with roots (65, 66). Glyma09g31910 and Glyma08g04830 are in a clade of the PLAC8 family, known as _p_lant _c_admium _r_esistance (PCR) proteins and _f_ruit _w_eight 2.2-_l_ike (FWL). Of particular interest, given the requirement for metal transport into the symbiosome (132, 133), is the reported role of two members of this clade from Arabidopsis, AtPCR1 and AtPCR2, that appear to be involved in the export of heavy metals from root cells (134, 135). This would translate to an import of metal into the symbiosome and the presence of homologous proteins on the SM suggests a possible role in maintaining adequate nutrition for the isolated bacteroids through import of a variety of metal cations. Zinc transport across the SM is also mediated through the ZIP1 transporter in soybean (136), so the PLAC8 transporters may present an additional transport mechanism to aid in maintaining zinc homeostasis. Ferrous iron transport into isolated symbiosomes was inhibited by cadmium and copper, perhaps indicating that a system for the transport of all three metals exists on the SM (27). There is also evidence that PCR proteins, such as BjPCR1, can mediate calcium ion transport (137, 138). Another role postulated for PLAC8 proteins is in regulating cell number and so fruit size (139, 140). Whether this role is governed by metal transport as observed for AtPCR1 and 2 or Ca2+ transport as recorded for BjPCR1 is not known (141). In soybean, Glyma09g31910, named FWL1, was recently investigated (142). Silencing of the gene resulted in decreased nodule numbers with structural aberrations and heterochromatin condensation in infected cells. Promoter-GUS analysis suggested expression was highest in the nodule epidermis and cortex. Our results suggest expression is almost exclusively in infected cell in nodules (Fig. 2, see below). Clearly there is more work needed to understand the role of this family in nodules.
Fig. 2.
Spatial activity of selected gene promoters in soybean nodules 30 days after inoculation with B. japonicum. Nodules expressing the 2-kb 5′ regulatory sequence of A, Glyma11g34600.1 (GmNPF5.25); B, Glyma11g34613.1 (GmNPF5.24); C, Glyma09g31910.1; D, Glyma01g31910.2; E, Glyma09g21070.2; and F, Glyma07g39320.1 fused to the GUS reporter gene were sectioned and incubated in GUS staining buffer. Cells expressing the GUS reporter gene appear blue following staining, highlighting the location of promoter activity. Scale bars represent 500 μm for A, B, C; 1 mm for D, E; and 200 μm for F.
Expression Analysis for Selected Genes
Analysis of the RNAseq data for soybean (64) shows 11% of proteins localized to the symbiosome membrane in this study are encoded by genes that are specifically expressed in nodules. A further 10% show expression 10-fold higher in nodules than any other tissues. Many of these specifically expressed genes fall into the transport and protein degradation categories, suggesting specific roles for these classes of proteins within the symbiosis. We investigated where the genes encoding six of the SM proteins were expressed using promoter GUS fusions. All genes showed infected cell expression in nodules as expected if the protein product is localized to the SM (Fig. 2). Glyma11g34613.1 (GmNPF5.24), Glyma09g31910.1, Glyma01g31910.2, and Glyma07g39320.1 had expression specifically in these cells, whereas Glyma09g21070.2, and probably Glyma11g34600.1 (GmNPF5.25), showed expression in both infected and uninfected (Fig. 2). This correlated well with the transcriptome data for soybean (64) that suggests nodule specific expression for all genes except Glyma07g39320.1. Because symbiosomes are only present in infected cells the specific infected cell expression for most of these genes supports the role of the protein product on the SM.
As many of the genes with specific expression have clear duplicated copies expressed in other tissues, it seems that there has been subfunctionalization and, at least, regulatory neofunctionalization for these genes because the two genome duplication events in soybean (143). Polyploidy in soybean has possibly allowed the specialization of particular genes to their role in the symbiosis, producing signals for infected cell specific expression as seen for five of the six genes investigated above. This may have led to neofunctionalization in a functional sense to make the symbiosis more efficient and to produce specific targeting signals that allow these SM proteins to reach their final location in the cell. The data for cell specific expression and subcellular localization will provide a basis for further study in this area.
Confirmation of Localization to the SM for GmNPF5.25 and 5.29
To confirm localization of putative SM proteins we analyzed their subcellular localization in infected cells of soybean nodules. GmNPF5.25 and GmNPF5.29 were fused to the N terminus of green fluorescent protein (GFP). We generated transgenic roots that expressed the GFP fusion constructs. Confocal microscopy showed that GFP-tagged proteins are located on symbiosomes (Fig. 3). The pattern of labeling closely resembles previous labeling of the SM in soybean nodules (29). Similar results were obtained for the products of Glyma11g31870.1 and Glyma08g04160.4 (results not shown). Nodules were costained with the lipophilic dye FM4–64. FM4–64 staining allows visualization of membranes of infected cells. Analysis of fluorescence intensity in the region of interest clearly showed colocalization of GFP-tagged proteins with the SM (supplemental Fig. S1).
Fig. 3.
Localization of GmNPF5.29 A, B, C, and GmNPF5.25 D, E, F, to the soybean SM. Confocal images of soybean nodules expressing GFP fused to the N-terminal of GmNPF5.29, A, and GmNPF5.25, D. The SM is counterstained with, FM4–64, a lipophilic membrane stain, B and E. Overlapping GFP and FM4–64 signals are presented in the merged images, C and F. Scale bars represent 20 μm (A–F).
For GmNPF5.29 the localization to the SM using GFP fusion was strong validation of our proteomic results because this was one of the lowest confidence proteins among those identified in the SM proteome. We had identified only two peptides for this protein in the proteome, one that was shared with other NPF family members.
Concluding Remarks
This is the most comprehensive proteomic study to date of the symbiosome membrane and the contents of the soluble space enclosed within that membrane. It confirms some previous studies and extends them substantially to identify new proteins that are likely to be involved in the transport of solutes across the symbiosome membrane and, through this transport, the regulation of communication between the symbiotic partners. We have shown that a subset of the genes encoding members of the SM proteome are expressed in infected cells of nodules, often specifically, and shown that some of these localize to the SM, using GFP-fusion analysis. Our results pave the way for functional analysis of these proteins and the further elucidation of mechanisms underpinning the function of the symbiotic organelle.
Supplementary Material
Supplemental Data
Acknowledgments
We thank Dr. Ben Crossett (University of Sydney, Australia) for technical advice on proteomic aspects of this work, and to Prof. Harvey Millar and Dr. Nicolas Taylor (University of Western Australia, Australia) for their technical assistance with our initial proteomic experiments. We acknowledge the facilities, and the scientific and technical assistance, of the Australian Microscopy and Microanalysis Research Facility at the Sydney Microscopy and Microanalysis facility, The University of Sydney. The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium (144) via the PRIDE partner repository with the data set identifier PXD001132 and DOI 10.6019/PXD001132.
Footnotes
Author contributions: VCC designed and conducted experiments, analyzed data and wrote the manuscript; PCL designed and conducted some of the experiments, analyzed data and edited the manuscript; AG conducted the GFP-fusion localization and some of the promoter-GUS experiments and edited the manuscript; CC and EMB conducted some of the promoter-GUS experiments and edited the manuscript; DAD planned the study, contributed resources, analyzed the data and edited the manuscript; PMCS planned the study, contributed resources and facilities, analyzed the data and wrote the manuscript.
* This research was funded by ARC Discovery Grants to DAD and PMCS (DP0772452) and PMCS (DP120102780). EMB was supported by a Grains Research and Development Corporation scholarship.
1 The abbreviations used are:
SM
symbiosome membrane
PBS
peribacteroid space
ABC
ATP-binding cassette
YSL
yellow stripe-like.
REFERENCES
- 1.Halbleib C. M., Ludden P. W. (2000) Regulation of Biological Nitrogen Fixation. J. Nutr.130, 1081–1084 [DOI] [PubMed] [Google Scholar]
- 2.Roth E., Jeon K., Stacey G. (1988) Homology in endosymbiotic systems: The term “symbiosome.” In: Palacios R., Verma D. P. S., eds. Molecular Genetics of Plant Microbe Interactions, pp. 220–225, American Phytopathology Society Press, St Paul, MN [Google Scholar]
- 3.Whitehead L. F., Day D. A. (1997) The peribacteroid membrane. Physiol. Plant. 100, 30–44 [Google Scholar]
- 4.Udvardi M., Poole P. S. (2013) Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64, 781–805 [DOI] [PubMed] [Google Scholar]
- 5.Robertson J. G., Lyttleton P. (1984) Division of peribacteroid membranes in root nodules of white clover. J. Cell Sci. 69, 147–157 [DOI] [PubMed] [Google Scholar]
- 6.Udvardi M. K., Day D. A. (1997) Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 493–523 [DOI] [PubMed] [Google Scholar]
- 7.Roth L. E., Stacey G. (1989) Cytoplasmic membrane systems involved in bacterium release into soybean nodule cells as studied with two Bradyrhizobium japonicum mutant strains. Eur. J. Cell Biol. 49, 24–32 [PubMed] [Google Scholar]
- 8.Hohnjec N., Lenz F., Fehlberg V., Vieweg M. F., Baler M. C., Hause B., Küster H. (2009) The signal peptide of the Medicago truncatula modular nodulin MtNOD25 operates as an address label for the specific targeting of proteins to nitrogen-fixing symbiosomes. Mol. Plant Microbe Interact. 22, 63–72 [DOI] [PubMed] [Google Scholar]
- 9.Liu J., Miller S. S., Graham M., Bucciarelli B., Catalano C. M., Sherrier D. J., Samac D. A., Ivashuta S., Fedorova M., Matsumoto P., Gantt J. S., Vance C. P. (2006) Recruitment of novel calcium-binding proteins for root nodule symbiosis in Medicago truncatula. Plant Physiol. 141, 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meckfessel M. H., Blancaflor E. B., Plunkett M., Dong Q., Dickstein R. (2012) Multiple domains in MtENOD8 protein including the signal peptide target it to the symbiosome. Plant Physiol. 159, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day D. A., Whitehead L. F., Hendriks J. H. M., Tyerman S. D. (1995) Nitrogen and carbon exchange across symbiotic membranes from soybean nodules. In: Tikhonovich I. A., Provorov N. A., Romanov V. I., Newton W. E., eds. Nitrogen fixation: fundamentals and applications, pp. 557–564, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 12.Udvardi M. K., Price G. D., Gresshoff P. M., Day D. A. (1988) A dicarboxylate transporter on the peribacteroid membrane of soybean nodules. FEBS Lett. 231, 36–40 [Google Scholar]
- 13.Ouyang L. J., Whelan J., Weaver C. D., Roberts D. M., Day D. A. (1991) Protein phosphorylation stimulates the rate of malate uptake across the peribacteroid membrane of soybean nodules. FEBS Lett. 293, 188–190 [DOI] [PubMed] [Google Scholar]
- 14.Day D. A., Kaiser B. N., Thomson R., Udvardi M. K., Moreau S., Puppo A. (2001) Nutrient transport across symbiotic membranes from legume nodules. Aust. J. Plant Physiol. 28, 667–674 [Google Scholar]
- 15.Niemietz C. M., Tyerman S. D. (2000) Channel-mediated permeation of ammonia gas through the peribacteroid membrane of soybean nodules. FEBS Lett. 465, 110–114 [DOI] [PubMed] [Google Scholar]
- 16.Hwang J. H., Ellingson S. R., Roberts D. M. (2010) Ammonia permeability of the soybean nodulin 26 channel. FEBS Lett. 584, 4339–4343 [DOI] [PubMed] [Google Scholar]
- 17.Tyerman S. D., Whitehead L. F., Day D. A. (1995) A channel-like transporter for NH4+ on the symbiotic interface of N2-fixing plants. Nature 378, 629–632 [Google Scholar]
- 18.Dean R. M., Rivers R. L., Zeidel M. L., Roberts D. M. (1999) Purification and functional reconstitution of soybean nodulin 26. An aquaporin with water and glycerol transport properties. Biochemistry 38, 347–353 [DOI] [PubMed] [Google Scholar]
- 19.Fortin M. G., Morrison N. A., Verma D. P. S. (1987) Nodulin-26, a peribacteroid membrane nodulin is expressed independently of the development of the peribacteroid compartment. Nucleic Acids Res. 15, 813–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masalkar P., Wallace I. S., Hwang J. H., Roberts D. M. (2010) Interaction of cytosolic glutamine synthetase of soybean root nodules with the C-terminal domain of the symbiosome membrane nodulin 26 aquaglyceroporin. J. Biol. Chem. 285, 23880–23888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivers R. L., Dean R. M., Chandy G., Hall J. E., Roberts D. M., Zeidel M. L. (1997) Functional analysis of nodulin 26, an aquaporin in soybean root nodule symbiosomes. J. Biol. Chem. 272, 16256–16261 [DOI] [PubMed] [Google Scholar]
- 22.Castelli J. M. (2006) Characterization of putative transporters maintaining iron homeostasis in symbiotic soybeans. School of biomedical, biomolecular and chemical sciences, faculty of life and physical sciences, p. 212, University of Western Australia, Perth [Google Scholar]
- 23.Krusell L., Krause K., Ott T., Desbrosses G., Krämer U., Sato S., Nakamura Y., Tabata S., James E. K., Sandal N., Stougaard J., Kawaguchi M., Miyamoto A., Suganuma N., Udvardi M. K. (2005) The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17, 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Vier K. D., Day D. A., Guerinot M. L. (1996) Bacterial iron acquisition in the soybean-Bradyrhizobium japonicum symbiosis. Plant Physiol. 111, 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreau S., Meyer J. M., Puppo A. (1995) Uptake of iron by symbiosomes and bacteroids from soybean nodules. FEBS Lett. 361, 225–228 [DOI] [PubMed] [Google Scholar]
- 26.Kaiser B. N., Moreau S., Castelli J., Thomson R., Lambert A., Bogliolo S., Puppo A., Day D. A. (2003) The soybean NRAMP homolog, GmDMT1, is a symbiotic divalent metal transporter capable of ferrous iron transport. Plant J. 35, 295–304 [DOI] [PubMed] [Google Scholar]
- 27.Moreau S., Day D. A., Puppo A. (1998) Ferrous iron is transported across the peribacteroid membrane of soybean nodules. Planta 207, 83–87 [Google Scholar]
- 28.Blumwald E., Fortin M. G., Rea P. A., Verma D. P. S., Poole R. J. (1985) Presence of host plasma membrane type H+-ATPase in the membrane envelope enclosing the bacteroids in soybean root nodules. Plant Physiol. 78, 655–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fedorova E., Thomson R., Whitehead L. F., Maudoux O., Udvardi M. K., Day D. A. (1999) Localization of H+-ATPases in soybean root nodules. Planta 209, 25–32 [DOI] [PubMed] [Google Scholar]
- 30.Udvardi M. K., Day D. A. (1989) Electrogenic ATPase activity on the peribacteroid membrane of soybean (Glycine max L.) root nodules. Plant Physiol. 90, 982–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panter S., Thomson R., De Bruxelles G., Laver D., Trevaskis B., Udvardi M. (2000) Identification with proteomics of novel proteins associated with the peribacteroid membrane of soybean root nodules. Mol. Plant Microbe Interact. 13, 325–333 [DOI] [PubMed] [Google Scholar]
- 32.Winzer T., Bairl A., Linder M., Linder D., Werner D., Müller P. (1999) A novel 53-kDa nodulin of the symbiosome membrane of soybean nodules, controlled by Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 12, 218–226 [DOI] [PubMed] [Google Scholar]
- 33.Saalbach G., Erik P., Wienkoop S. (2002) Characterization by proteomics of peribacteroid space and peribacteroid membrane preparations from pea (Pisum sativum) symbiosomes. Proteomics 2, 325–337 [DOI] [PubMed] [Google Scholar]
- 34.Wienkoop S., Saalbach G. (2003) Proteome analysis. Novel proteins identified at the peribacteroid membrane from Lotus japonicus root nodules. Plant Physiol. 131, 1080–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bestel-Corre G., Dumas-Gaudot E., Poinsot V., Dieu M., Dierick J.-F., van Tuinen D., Remacle J., Gianinazzi-Pearson V., Gianinazzi S. (2002) Proteome analysis and identification of symbiosome-related proteins from Medicago truncatula Gaertn. by two-dimensional electrophoresis and mass spectrometry. Electrophoresis 23, 122–137 [DOI] [PubMed] [Google Scholar]
- 36.Catalano C. M., Lane W. S., Sherrier D. J. (2004) Biochemical characterization of symbiosome membrane proteins from Medicago truncatula root nodules. Electrophoresis 25, 519–531 [DOI] [PubMed] [Google Scholar]
- 37.Unkovich M., Herridge D. F., Peoples M. B., Cadisch G., Boddey B., Giller K., Alves B., Chalk P. (2008) Measuring plant-associated nitrogen fixation in agricultural systems, ACIAR, Adelaide [Google Scholar]
- 38.Day D. A., Price G. D., Udvardi M. K. (1989) Membrane interface of the Bradyrhizobium japonicum-Glycine max symbiosis: peribacteroid units from soybean nodules. Aust. J. Plant Physiol. 16, 69–84 [Google Scholar]
- 39.Millar A. H., Heazlewood J. L. (2003) Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis. Plant Physiol. 131, 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seigneurin-Berny D., Rolland N., Garin J., Joyard J. (1999) Differential extraction of hydrophobic proteins from chloroplast envelope membranes: a subcellular-specific proteomic approach to identify rare intrinsic membrane proteins. Plant J. 19, 217–228 [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., Roberts D. M. (1995) Expression of soybean nodulin 26 in transgenic tobacco. Targetting to the vacuolar membrane and effects of floral and seed development. Mol. Biol.Cell 6, 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee C. P., Eubel H., Millar A. H. (2010) Diurnal changes in mitochondrial function reveal daily optimization of light and dark respiratory metabolism in Arabidopsis. Mol, Cell. Proteomics 9, 2125–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., Hyten D. L., Song Q., Thelen J. J., Cheng J., Xu D., Hellsten U., May G. D., Yu Y., Sakurai T., Umezawa T., Bhattacharyya M. K., Sandhu D., Valliyodan B., Lindquist E., Peto M., Grant D., Shu S., Goodstein D., Barry K., Futrell-Griggs M., Abernathy B., Du J., Tian Z., Zhu L., Gill N., Joshi T., Libault M., Sethuraman A., Zhang X. C., Shinozaki K., Nguyen H. T., Wing R. A., Cregan P., Specht J., Grimwood J., Rokhsar D., Stacey G., Shoemaker R. C., Jackson S. A. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 [DOI] [PubMed] [Google Scholar]
- 44.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 45.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 46.Kall L., Storey J. D., Noble W. S. (2008) Nonparametric estimation of posterior error probabilities associated with peptides identified by tandem mass spectrometry. Bioinformatics 24, i42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thimm O., Blasing O., Gibon Y., Nagel A., Meyer S., Kruger P., Selbig J., Muller L. A., Rhee S. Y., Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939 [DOI] [PubMed] [Google Scholar]
- 48.Hirokawa T., Boon-Chieng S., Mitaku S. (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379 [DOI] [PubMed] [Google Scholar]
- 49.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 50.von Heijne G. (1992) Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225, 487–494 [DOI] [PubMed] [Google Scholar]
- 51.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 52.Small I., Peeters N., Legeai F., Lurin C. (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4, 1581–1590 [DOI] [PubMed] [Google Scholar]
- 53.Chou K. C., Shen H. B. (2010) Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5, e11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoglund A., Donnes P., Blum T., Adolph H. W., Kohlbacher O. (2006) MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics 22, 1158–1165 [DOI] [PubMed] [Google Scholar]
- 55.Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 56.Fankhauser N., Maser P. (2005) Identification of GPI anchor attachment signals by a Kohonen self-organizing map. Bioinformatics 21, 1846–1852 [DOI] [PubMed] [Google Scholar]
- 57.Mohammadi-Dehcheshmeh M., Ebrahimie E., Tyerman S. D., Kaiser B. N. (2014) A novel method based on combination of semi-in vitro and in vivo conditions in _Agrobacterium rhizogenes_-mediated hairy root transformation of Glycine species. In Vitro Cell. Dev. Biol. Plant 50, 282–291 [Google Scholar]
- 58.Weaver C. D., Shomer N. H., Louis C. F., Roberts D. M. (1994) Nodulin 26, a nodule-specific symbiosome membrane protein from soybean, is an ion channel. J. Biol. Chem. 269, 17858–17862 [PubMed] [Google Scholar]
- 59.Napier R. M., Fowke L. C., Hawes C., Lewis M., Pelham H. R. (1992) Immunological evidence that plants use both HDEL and KDEL for targeting proteins to the endoplasmic reticulum. J. Cell Sci. 102 (Pt 2), 261–271 [DOI] [PubMed] [Google Scholar]
- 60.Lefebvre B., Timmers T., Mbengue M., Moreau S., Herve C., Toth K., Bittencourt-Silvestre J., Klaus D., Deslandes L., Godiard L., Murray J. D., Udvardi M. K., Raffaele S., Mongrand S., Cullimore J., Gamas P., Niebel A., Ott T. (2010) A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 107, 2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andreev I. M., Dubrovo P. N., Krylova V. V., Izmailov S. F. (1999) Functional identification of ATP-driven Ca2+ pump in the peribacteroid membrane of broad bean root nodules. FEBS Lett. 447, 49–52 [DOI] [PubMed] [Google Scholar]
- 62.Roth L. E., Stacey G. (1989) Bacterium release into host cells of nitrogen-fixing soybean nodules: the symbiosome membrane comes from three sources. Eur. J. Cell Biol. 49, 13–23 [PubMed] [Google Scholar]
- 63.Atkins C. A., Smith P., Storer P. J. (1997) Re-examination of the intracellular localization of de novo purine synthesis in cowpea nodules. Plant Physiol. 113, 127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen T., Zelechowska M., Foster V., Bergmann H., Verma D. P. (1985) Primary structure of the soybean nodulin-35 gene encoding uricase II localized in the peroxisomes of uninfected cells of nodules. Proc. Natl. Acad. Sci. U.S.A. 82, 5040–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Libault M., Farmer A., Brechenmacher L., Drnevich J., Langley R. J., Bilgin D. D., Radwan O., Neece D. J., Clough S. J., May G. D., Stacey G. (2010) Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 152, 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Severin A. J., Woody J. L., Bolon Y. T., Joseph B., Diers B. W., Farmer A. D., Muehlbauer G. J., Nelson R. T., Grant D., Specht J. E., Graham M. A., Cannon S. B., May G. D., Vance C. P., Shoemaker R. C. (2010) RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 10, 160 doi: 10.1186/1471-2229-10-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith P. M., Atkins C. A. (2002) Purine biosynthesis. Big in cell division, even bigger in nitrogen assimilation. Plant Physiol. 128, 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller S. S., Driscoll B. T., Gregerson R. G., Gantt J. S., Vance C. P. (1998) Alfalfa malate dehydrogenase (MDH): molecular cloning and characterization of five different forms reveals a unique nodule-enhanced MDH. Plant J. 15, 173–184 [DOI] [PubMed] [Google Scholar]
- 69.Palmieri F., Pierri C. L., De Grassi A., Nunes-Nesi A., Fernie A. R. (2011) Evolution, structure, and function of mitochondrial carriers: a review with new insights. Plant J. 66, 161–181 [DOI] [PubMed] [Google Scholar]
- 70.Ott T., van Dongen J. T., Gunther C., Krusell L., Desbrosses G., Vigeolas H., Bock V., Czechowski T., Geigenberger P., Udvardi M. K. (2005) Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. Biol. 15, 531–535 [DOI] [PubMed] [Google Scholar]
- 71.Kamauchi S., Wadahama H., Iwasaki K., Nakamoto Y., Nishizawa K., Ishimoto M., Kawada T., Urade R. (2008) Molecular cloning and characterization of two soybean protein disulfide isomerases as molecular chaperones for seed storage proteins. FEBS J. 275, 2644–2658 [DOI] [PubMed] [Google Scholar]
- 72.Schaller A., Stintzi A., Graff L. (2012) Subtilases – versatile tools for protein turnover, plant development, and interactions with the environment. Physiol. Plant. 145, 52–66 [DOI] [PubMed] [Google Scholar]
- 73.Takeda N., Kistner C., Kosuta S., Winzer T., Pitzschke A., Groth M., Sato S., Kaneko T., Tabata S., Parniske M. (2007) Proteases in plant root symbiosis. Phytochemistry 68, 111–121 [DOI] [PubMed] [Google Scholar]
- 74.Takeda N., Sato S., Asamizu E., Tabata S., Parniske M. (2009) Apoplastic plant subtilases support arbuscular mycorrhiza development in Lotus japonicus. Plant J. 58, 766–777 [DOI] [PubMed] [Google Scholar]
- 75.Rawlings N. D., Waller M., Barrett A. J., Bateman A. (2014) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 42, D503–D509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Von Groll U., Berger D., Altmann T. (2002) The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14, 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Olivares J. E., Díaz-Camino C., Estrada-Navarrete G., Alvarado-Affantranger X., Rodríguez-Kessler M., Zamudio F. Z., Olamendi-Portugal T., Márquez Y., Servín L. E., Sánchez F. (2011) Nodulin 41, a novel late nodulin of common bean with peptidase activity. BMC Plant Biol. 11, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xia Y., Suzuki H., Borevitz J., Blount J., Guo Z., Patel K., Dixon R. A., Lamb C. (2004) An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 23, 980–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van de Velde W., Zehirov G., Szatmari A., Debreczeny M., Ishihara H., Kevei Z., Farkas A., Mikulass K., Nagy A., Tiricz H., Satiat-Jeunemaitre B., Alunni B., Bourge M., Kucho K., Abe M., Kereszt A., Maroti G., Uchiumi T., Kondorosi E., Mergaert P. (2010) Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126 [DOI] [PubMed] [Google Scholar]
- 80.Kondorosi E., Mergaert P., Kereszt A. (2013) A paradigm for endosymbiotic life: cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu. Rev. Microbiol. 67, 611–628 [DOI] [PubMed] [Google Scholar]
- 81.Cheon C. I., Lee N. G., Siddique A. B., Bal A. K., Verma D. P. (1993) Roles of plant homologs of Rab1p and Rab7p in the biogenesis of the peribacteroid membrane, a subcellular compartment formed de novo during root nodule symbiosis. EMBO J. 12, 4125–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cui Y., Zhao Q., Gao C., Ding Y., Zeng Y., Ueda T., Nakano A., Jiang L. (2014) Activation of the Rab7 GTPase by the MON1-CCZ1 complex is essential for PVC-to-vacuole trafficking and plant growth in Arabidopsis. Plant Cell 26, 2080–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Limpens E., Lvanov S., Van Esse W., Voets G., Fedorova E., Bisseling T. (2009) Medicago N2-Fixing symbiosomes acquire the endocytic identity marker Rab7 but delay the acquisition of vacuolar identity. Plant Cell 21, 2811–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ivanov S., Fedorova E. E., Limpens E., De Mita S., Genre A., Bonfante P., Bisseling T. (2012) Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc. Natl. Acad. Sci. U.S.A. 109, 8316–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zheng H., Camacho L., Wee E., Batoko H., Legen J., Leaver C. J., Malho R., Hussey P. J., Moore I. (2005) A Rab-E GTPase mutant acts downstream of the Rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell 17, 2020–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Speth E. B., Imboden L., Hauck P., He S. Y. (2009) Subcellular localization and functional analysis of the Arabidopsis GTPase RabE. Plant Physiol. 149, 1824–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moore I., Diefenthal T., Zarsky V., Schell J., Palme K. (1997) A homolog of the mammalian GTPase Rab2 is present in Arabidopsis and is expressed predominantly in pollen grains and seedlings. Proc. Natl. Acad. Sci. U.S.A. 94, 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Catalano C. M., Czymmek K. J., Gann J. G., Sherrier D. J. (2007) Medicago truncatula syntaxin SYP132 defines the symbiosome membrane and infection droplet membrane in root nodules. Planta 225, 541–550 [DOI] [PubMed] [Google Scholar]
- 89.Bassham D. C., Brandizzi F., Otegui M. S., Sanderfoot A. A. (2008) The secretory system of Arabidopsis. Arabidopsis Book 6, e0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kalde M., Nuhse T. S., Findlay K., Peck S. C. (2007) The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc. Natl. Acad. Sci. U.S.A. 104, 11850–11855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pumplin N., Zhang X., Noar R. D., Harrison M. J. (2012) Polar localization of a symbiosis-specific phosphate transporter is mediated by a transient reorientation of secretion. Proc. Natl. Acad. Sci. U.S.A. 109, E665–E672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wallace I. S., Choi W. G., Roberts D. M. (2006) The structure, function, and regulation of the nodulin 26-like intrinsic protein family of plant aquaglyceroporins. Biochim. Biophys. Acta 1758, 1165–1175 [DOI] [PubMed] [Google Scholar]
- 93.Tomatsu H., Takano J., Takahashi H., Watanabe-Takahashi A., Shibagaki N., Fujiwara T. (2007) An Arabidopsis thaliana high-affinity molybdate transporter required for efficient uptake of molybdate from soil. Proc. Natl. Acad. Sci. U.S.A. 104, 18807–18812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Léran S., Varala K., Boyer J. C., Chiurazzi M., Crawford N., Daniel-Vedele F., David L., Dickstein R., Fernandez E., Forde B., Gassmann W., Geiger D., Gojon A., Gong J. M., Halkier B. A., Harris J. M., Hedrich R., Limami A. M., Rentsch D., Seo M., Tsay Y. F., Zhang M., Coruzzi G., Lacombe B. (2014) A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 19, 5–9 [DOI] [PubMed] [Google Scholar]
- 95.Karim S., Holmstrom K. O., Mandal A., Dahl P., Hohmann S., Brader G., Palva E. T., Pirhonen M. (2007) AtPTR3, a wound-induced peptide transporter needed for defence against virulent bacterial pathogens in Arabidopsis. Planta 225, 1431–1445 [DOI] [PubMed] [Google Scholar]
- 96.Song W., Steiner H. Y., Zhang L., Naider F., Stacey G., Becker J. M. (1996) Cloning of a second Arabidopsis peptide transport gene. Plant Physiol. 110, 171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Komarova N. Y., Thor K., Gubler A., Meier S., Dietrich D., Weichert A., Suter Grotemeyer M., Tegeder M., Rentsch D. (2008) AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiol. 148, 856–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Williams L. E., Millar A. J. (2001) Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 659–688 [DOI] [PubMed] [Google Scholar]
- 99.Tsay Y. F., Chiu C. C., Tsai C. B., Ho C. H., Hsu P. K. (2007) Nitrate transporters and peptide transporters. FEBS Lett. 581, 2290–2300 [DOI] [PubMed] [Google Scholar]
- 100.Krouk G., Lacombe B., Bielach A., Perrine-Walker F., Malinska K., Mounier E., Hoyerova K., Tillard P., Leon S., Ljung K., Zazimalova E., Benkova E., Nacry P., Gojon A. (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18, 927–937 [DOI] [PubMed] [Google Scholar]
- 101.Beeckman T., Friml J. (2010) Nitrate contra auxin: nutrient sensing by roots. Dev. Cell 18, 877–878 [DOI] [PubMed] [Google Scholar]
- 102.Morere-Le Paven M. C., Viau L., Hamon A., Vandecasteele C., Pellizzaro A., Bourdin C., Laffont C., Lapied B., Lepetit M., Frugier F., Legros C., Limami A. M. (2011) Characterization of a dual-affinity nitrate transporter MtNRT1.3 in the model legume Medicago truncatula. J. Exp. Biol. 62, 5595–5605 [DOI] [PubMed] [Google Scholar]
- 103.Rosendahl L., Jochimsen B. U. (1995) In vitro indole-3-acetic acid uptake in symbiosomes from soybean (Glycine max L.) root nodules. Symbiosis 19, 99–110 [Google Scholar]
- 104.Nour-Eldin H. H., Andersen T. G., Burow M., Madsen S. R., Jorgensen M. E., Olsen C. E., Dreyer I., Hedrich R., Geiger D., Halkier B. A. (2012) NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488, 531–534 [DOI] [PubMed] [Google Scholar]
- 105.Jeong J., Suh S., Guan C., Tsay Y. F., Moran N., Oh C. J., An C. S., Demchenko K. N., Pawlowski K., Lee Y. (2004) A nodule-specific dicarboxylate transporter from alder is a member of the peptide transporter family. Plant Physiol. 134, 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Day D. A., Poole P. S., Tyerman S. D., Rosendahl L. (2001) Ammonia and amino acid transport across symbiotic membranes in nitrogen-fixing legume nodules. Cell. Mol. Life Sci. 58, 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Streeter J., Wong P. P. (1988) Inhibition of legume nodule formation and N2 fixation by nitrate. Critic. Rev. Plant Sci. 7, 1–23 [Google Scholar]
- 108.Harris J. M., Dickstein R. (2010) Control of root architecture and nodulation by the LATD/NIP transporter. Plant Signal. Behav. 5, 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bright L. J., Liang Y., Mitchell D. M., Harris J. M. (2005) The LATD gene of Medicago truncatula is required for both nodule and root development. Mol. Plant Microbe Interact. 18, 521–532 [DOI] [PubMed] [Google Scholar]
- 110.Yendrek C. R., Lee Y. C., Morris V., Liang Y., Pislariu C. I., Burkart G., Meckfessel M. H., Salehin M., Kessler H., Wessler H., Lloyd M., Lutton H., Teillet A., Sherrier D. J., Journet E. P., Harris J. M., Dickstein R. (2010) A putative transporter is essential for integrating nutrient and hormone signaling with lateral root growth and nodule development in Medicago truncatula. Plant J. 62, 100–112 [DOI] [PubMed] [Google Scholar]
- 111.Bagchi R., Salehin M., Adeyemo O. S., Salazar C., Shulaev V., Sherrier D. J., Dickstein R. (2012) Functional assessment of the Medicago truncatula NIP/LATD protein demonstrates that it is a high-affinity nitrate transporter. Plant Physiol. 160, 906–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Prell J., White J. P., Bourdes A., Bunnewell S., Bongaerts R. J., Poole P. S. (2009) Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acids. Proc. Natl. Acad. Sci. U.S.A. 106, 12477–12482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kang J., Park J., Choi H., Burla B., Kretzschmar T., Lee Y., Martinoia E. (2011) Plant ABC Transporters. Arabidopsis Book 9, e0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Verrier P. J., Bird D., Burla B., Dassa E., Forestier C., Geisler M., Klein M., Kolukisaoglu U., Lee Y., Martinoia E., Murphy A., Rea P. A., Samuels L., Schulz B., Spalding E. J., Yazaki K., Theodoulou F. L. (2008) Plant ABC proteins – a unified nomenclature and updated inventory. Trends Plant Sci. 13, 151–159 [DOI] [PubMed] [Google Scholar]
- 115.Kim S., Yamaoka Y., Ono H., Kim H., Shim D., Maeshima M., Martinoia E., Cahoon E. B., Nishida I., Lee Y. (2013) AtABCA9 transporter supplies fatty acids for lipid synthesis to the endoplasmic reticulum. Proceedings of the National Academy of Sciences of the United States of America 11 Proc. Natl. Acad. Sci. U.S.A. 0, 773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vanneste S., Friml J. (2009) Auxin: a trigger for change in plant development. Cell 136, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 117.Lee M., Choi Y., Burla B., Kim Y. Y., Jeon B., Maeshima M., Yoo J. Y., Martinoia E., Lee Y. (2008) The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat. Cell Biol. 10, 1217–1223 [DOI] [PubMed] [Google Scholar]
- 118.Kretzschmar T., Kohlen W., Sasse J., Borghi L., Schlegel M., Bachelier J. B., Reinhardt D., Bours R., Bouwmeester H. J., Martinoia E. (2011) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483, 341–344 [DOI] [PubMed] [Google Scholar]
- 119.Panikashvili D., Savaldi-Goldstein S., Mandel T., Yifhar T., Franke R. B., Hofer R., Schreiber L., Chory J., Aharoni A. (2007) The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiol. 145, 1345–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Le Hir R., Sorin C., Chakraborti D., Moritz T., Schaller H., Tellier F., Robert S., Morin H., Bako L., Bellini C. (2013) ABCG9, ABCG11 and ABCG14 ABC transporters are required for vascular development in Arabidopsis. Plant J. 76, 811–824 [DOI] [PubMed] [Google Scholar]
- 121.Bird D., Beisson F., Brigham A., Shin J., Greer S., Jetter R., Kunst L., Wu X., Yephremov A., Samuels L. (2007) Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 52, 485–498 [DOI] [PubMed] [Google Scholar]
- 122.Lee M., Lee K., Lee J., Noh E. W., Lee Y. (2005) AtPDR12 contributes to lead resistance in Arabidopsis. Plant Physiol. 138, 827–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim D. Y., Bovet L., Maeshima M., Martinoia E., Lee Y. (2007) The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 50, 207–218 [DOI] [PubMed] [Google Scholar]
- 124.Xi J., Xu P., Xiang C. B. (2012) Loss of AtPDR11, a plasma membrane-localized ABC transporter, confers paraquat tolerance in Arabidopsis thaliana. Plant J. 69, 782–791 [DOI] [PubMed] [Google Scholar]
- 125.Haney C. H., Long S. R. (2010) Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Simon-Plas F., Perraki A., Bayer E., Gerbeau-Pissot P., Mongrand S. (2011) An update on plant membrane rafts. Curr. Opin. Plant Biol. 14, 642–649 [DOI] [PubMed] [Google Scholar]
- 127.Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Lu F., Marchler G. H., Mullokandov M., Omelchenko M. V., Robertson C. L., Song J. S., Thanki N., Yamashita R. A., Zhang D., Zhang N., Zheng C., Bryant S. H. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mongrand S., Morel J., Laroche J., Claverol S., Carde J. P., Hartmann M. A., Bonneu M., Simon-Plas F., Lessire R., Bessoule J. J. (2004) Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279, 36277–36286 [DOI] [PubMed] [Google Scholar]
- 129.Lubkowitz M. (2006) The OPT family functions in long-distance peptide and metal transport in plants. Genet. Eng. 27, 35–55 [DOI] [PubMed] [Google Scholar]
- 130.Conte S. S., Walker E. L. (2012) Genetic and biochemical approaches for studying the yellow stripe-like transporter family in plants. Curr. Top. Membr. 69, 295–322 [DOI] [PubMed] [Google Scholar]
- 131.Hofstetter S. S., Dudnik A., Widmer H., Dudler R. (2013) Arabidopsis YELLOW STRIPE-LIKE7 (YSL7) and YSL8 transporters mediate uptake of Pseudomonas virulence factor syringolin A into plant cells. Mol. PlantMicrobe Interact. 26, 1302–1311 [DOI] [PubMed] [Google Scholar]
- 132.Brear E. M., Day D. A., Smith P. M. (2013) Iron: an essential micronutrient for the legume-rhizobium symbiosis. Front. Plant Sci. 4, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Clarke V. C., Loughlin P. C., Day D. A., Smith P. M. C. (2014) Transport processes of the legume symbiosome membrane. Front. Plant Sci. 5, 699 doi: 10.3389/fpls.2014.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Song W. Y., Choi K. S., Kim do Y., Geisler M., Park J., Vincenzetti V., Schellenberg M., Kim S. H., Lim Y. P., Noh E. W., Lee Y., Martinoia E. (2010) Arabidopsis PCR2 is a zinc exporter involved in both zinc extrusion and long-distance zinc transport. Plant Cell 22, 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Song W. Y., Martinoia E., Lee J., Kim D., Kim D. Y., Vogt E., Shim D., Choi K. S., Hwang I., Lee Y. (2004) A novel family of cys-rich membrane proteins mediates cadmium resistance in Arabidopsis. Plant Physiol. 135, 1027–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Moreau S., Thompson R. M., Kaiser B. N., Trevaskis B., Guerinot M. L., Udvardi M. K., Day D. A. (2002) GmZIP1 encodes a symbiosome-specific zinc transporter in soybean. J. Biol. Chem. 277, 4738–4746 [DOI] [PubMed] [Google Scholar]
- 137.Nakagawa Y., Katagiri T., Shinozaki K., Qi Z., Tatsumi H., Furuichi T., Kishigami A., Sokabe M., Kojima I., Sato S., Kato T., Tabata S., Iida K., Terashima A., Nakano M., Ikeda M., Yamanaka T., Iida H. (2007) Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc. Natl. Acad. Sci. U.S.A. 104, 3639–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Song W. Y., Hortensteiner S., Tomioka R., Lee Y., Martinoia E. (2011) Common functions or only phylogenetically related? The large family of PLAC8 motif-containing/PCR genes. Mol. Cell 31, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Frary A., Nesbitt T. C., Grandillo S., Knaap E., Cong B., Liu J., Meller J., Elber R., Alpert K. B., Tanksley S. D. (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88 [DOI] [PubMed] [Google Scholar]
- 140.Guo M., Rupe M. A., Dieter J. A., Zou J., Spielbauer D., Duncan K. E., Howard R. J., Hou Z., Simmons C. R. (2010) Cell Number Regulator1 affects plant and organ size in maize: implications for crop yield enhancement and heterosis. Plant Cell 22, 1057–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Song WY, Choi KS, Alexis de A, Martinoia E, Lee Y.Brassica juncea plant cadmium resistance 1 protein (BjPCR1) facilitates the radial transport of calcium in the root. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 19808–19813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Libault M., Stacey G. (2010) Evolution of FW2.2-like (FWL) and PLAC8 genes in eukaryotes. Plant Signal. Behav. 5, 1226–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Roulin A., Auer P. L., Libault M., Schlueter J., Farmer A., May G., Stacey G., Doerge R. W., Jackson S. A. (2012) The fate of duplicated genes in a polyploid plant genome. Plant J. 73, 143–153 [DOI] [PubMed] [Google Scholar]
- 144.Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data