Sleeping Beauty Transposition of Chimeric Antigen Receptors Targeting Receptor Tyrosine Kinase-Like Orphan Receptor-1 (ROR1) into Diverse Memory T-Cell Populations (original) (raw)
Abstract
T cells modified with chimeric antigen receptors (CARs) targeting CD19 demonstrated clinical activity against some B-cell malignancies. However, this is often accompanied by a loss of normal CD19+ B cells and humoral immunity. Receptor tyrosine kinase-like orphan receptor-1 (ROR1) is expressed on sub-populations of B-cell malignancies and solid tumors, but not by healthy B cells or normal post-partum tissues. Thus, adoptive transfer of T cells specific for ROR1 has potential to eliminate tumor cells and spare healthy tissues. To test this hypothesis, we developed CARs targeting ROR1 in order to generate T cells specific for malignant cells. Two Sleeping Beauty transposons were constructed with 2nd generation ROR1-specific CARs signaling through CD3ζ and either CD28 (designated ROR1RCD28) or CD137 (designated ROR1RCD137) and were introduced into T cells. We selected for T cells expressing CAR through co-culture with γ-irradiated activating and propagating cells (AaPC), which co-expressed ROR1 and co-stimulatory molecules. Numeric expansion over one month of co-culture on AaPC in presence of soluble interleukin (IL)-2 and IL-21 occurred and resulted in a diverse memory phenotype of CAR+ T cells as measured by non-enzymatic digital array (NanoString) and multi-panel flow cytometry. Such T cells produced interferon-γ and had specific cytotoxic activity against ROR1+ tumors. Moreover, such cells could eliminate ROR1+ tumor xenografts, especially T cells expressing ROR1RCD137. Clinical trials will investigate the ability of ROR1-specific CAR+ T cells to specifically eliminate tumor cells while maintaining normal B-cell repertoire.
Introduction
T cells can be rendered specific for tumor-associated antigens (TAAs) independent of their endogenous T-cell receptor (TCR) via gene transfer of chimeric antigen receptors (CARs) [1]. CARs are constructed from the genes encoding a single-chain variable fragment (scFv) of a TAA-specific monoclonal antibody (mAb), extracellular hinge or scaffold with transmembrane domain, and portions of CD3ζ and CD28 or CD137 (4-1BB) endodomains. Introduction of this chimeric gene generates T cells that proliferate, produce cytokines, and direct cytolysis of tumor cells in a TAA-dependent manner [2]. Infusion of T cells expressing CAR specific for CD19 with either CD3ζ /CD28 or CD3ζ /CD137 can induce complete tumor regressions in subsets of patients with B-lineage lymphomas, acute lymphoblastic leukemia (B-ALL), or chronic lymphocytic leukemia (CLL) [3–10]. In addition to the structure of the CAR, the subset of T cells that serves as a template for bioengineering can impact the anti-tumor effect. For instance, murine immunotherapy models have demonstrated that less differentiated T cells, i.e., naïve (TN), stem-cell memory (TSCM), or central memory (TCM) T cells, can clear tumor cells more effectively than more differentiated T cells, i.e., effector memory (TEM) or terminally-differentiated effector memory RA (TEMRA) T cells [11, 12]. Thus, the specificity of the T cell and the underlying memory phenotype can be important predictors for therapeutic efficacy.
T cells that express CARs specific for CD19 cannot distinguish between neoplastic or normal CD19-bearing B cells. Indeed, the initial clinical data has demonstrated that most patients benefiting from an anti-tumor response have concomitant B-cell depletion. Such B-cell aplasia requires that the patient receive timely intravenous infusions of normal immunoglobulin to alleviate the threat for opportunistic infections [13]. Nevertheless, such treatment cannot eliminate the risk for serious infection, as one recipient of CD19-specific CAR+ T cells died due to opportunistic infection [14]. Thus, targeting a TAA, which is not expressed on normal B cells or other adult tissues, would mitigate the risk for B-cell depletion and potentially improve outcome.
One TAA that may serve as an alternative to CD19-directed T-cell therapy is receptor tyrosine kinase-like orphan receptor-1 (ROR1). ROR1 is principally expressed during embryonic development but its expression attenuates during fetal development and is negligible at term [15]. However, ROR1 is aberrantly expressed by some B-cell malignancies, e.g., lymphomas, CLL, and t(1;19) B-ALL, and by many solid-tumor malignancies, e.g., adrenal, bladder, breast, colon, lung, pancreas, prostate, ovary, skin, testes, uterus, and neuroblastoma [16–21]. ROR1 has also been discovered on ovarian cancer stem cells yielding the potential to eliminate tumor initiating cells by targeting ROR1 [22]. Monoclonal antibodies specific for ROR1 have reacted with some bone marrow adipocytes [23] and hematogones [24] (B-cell precursors dispensable for maintaining peripheral B-cell repertoire), but not on other normal tissues [18, 20]. Non-human primates treated with autologous ROR1-specific T cells had no toxicity from treatment suggesting that targeting ROR1 is a safe approach for humans [25]. Thus, it appears T cells that express CARs specific for ROR1 would not deplete B-cells, but rather target neoplastic B cells or solid tumors.
Clinical trials have not yet evaluated the efficacy or safety of targeting ROR1 using adoptive immunotherapy. In preparation for the human application of genetically modified T cells, we evaluated two 2nd generation CAR species specific for ROR1. Our approach is based on the methodology employed in our first-in-human clinical trials using CD19-specific T cells generated by synchronous electro-transfer of CD19-specific CAR as a Sleeping Beauty (SB) transposon and a hyperactive SB transposase [26, 27]. Following transfection the T cells are co-cultured with irradiated activating and propagating cells (AaPC), which select for T cells that have stable expression of the CAR through direct interactions with AaPC bearing its cognate antigen, e.g., CD19 [28–31]. In this study we generated AaPC that express ROR1 and developed SB transposons to express CARs specific for ROR1 that signal via chimeric CD3ζ/CD28 (designated ROR1RCD28) or chimeric CD3ζ/CD137 (designated ROR1RCD137). We show that T cells expressing either CAR can proliferate in response to cells bearing ROR1 and specifically kill ROR1+ tumor cells. We found that ROR1RCD137+ T cells had more effective anti-tumor activity than ROR1RCD28+ T cells in immune-deficient mice engrafted with ROR1+ tumor cells. A phase I clinical trial (NCT02194374) is open for the infusion of autologous ROR1RCD137 CAR+ T cells in patients with CLL.
Materials and Methods
Ethics statement
All human samples acquired for this study were obtained after written informed consent was granted in accordance with protocols established and approved by the MD Anderson Cancer Center Internal Review Board (IRB). The identities of all samples were kept private. Animals treated in this study were handled in accordance with the strict guidelines established by the MD Anderson Cancer Center Institutional Animal Care and Use Committee (IACUC), which specifically approved this study (Protocol#03-06-04333). Mice were housed in pathogen-free conditions and were monitored daily for welfare-related assessments in accordance to IACUC guidelines. Moribund mice, e.g., ruffled fur, hunched posture, tumor size, were humanely euthanized with inhaled CO2 as per direction of IACUC guidelines. No mice died without euthanasia. All efforts were made to minimize animal suffering and inhaled isoflurane was administered for anesthesia as required.
Chimeric antigen receptors
Cloning of second generation CD19-specific CARs signaling through CD3ζ and CD28 (CD19RCD28) or CD137 (CD19RCD137) have been previously described [32, 33]. Heavy and light chain immunoglobulin sequences from the 4A5 mAb hybridoma were codon optimized and synthesized de novo (GeneArt; Invitrogen, Grand Island, NY) to create the “ROR1R” nucleotide sequence of (i) murine IgGκ signal peptide, (ii) VL, (iii) Whitlow linker (GSTSGSGKPGSGEGSTKG), (iv) VH, and (v) the first 73 amino acids of a modified human IgG4 stalk. ROR1R was amplified by PCR with ROR1RCoOpF (GCTAGCCGCCACCATGGGCTGGTCCTGCATC) and ROR1Rrev (GCTCCTCCCGGGGCTTTGTCTTGGC) primers then sub-cloned into pCR4-TOPO with TOPO TA Cloning Kit (Invitrogen) to generate ROR1R(CoOp)/pCR4-TOPO and sequence was verified with T7 and T13-0 primers (DNA Sequencing Core, MDACC). ROR1R(CoOp)/pCR4-TOPO and CD19RCD28mZ(CoOp)/pEK plasmids were digested with NheI and SmaI and ligated to generate ROR1RCD28mZ(CoOp)/pEK. The ROR1-specific CAR was then transferred into a SB transposon by digestion of CD19RCD28mZ(CoOp)/pSBSO-MCS and ROR1RCD28mZ(CoOp)/pEK with NheI and SpeI to generate ROR1RCD28mZ(CoOp)/pSBSO-MCS. The final ROR1RCD28 SB transposon plasmid was constructed by digesting CD19RCD28mZ(CoOp)/pSBSO-SIM with NheI, XmaI, and Antarctic Phosphatase and ROR1RCD28mZ(CoOp)/pSBSO-MCS with NheI, XmnI, and XmaI to generate ROR1RCD28/pSBSO-SIM plasmid. Similarly, the final ROR1RCD137 transposon plasmid was constructed by digesting CD19R-CD28Tm-41BBCyt-Z(CoOp)/pSBSO-FRA with NheI, XmaI, and Antarctic Phosphatase and ROR1RCD28mZ(CoOp)/pSBSO-MCS with NheI, XmnI, and XmaI to generate ROR1RCD137/pSBSO-FRA plasmid. Identities of final ROR1R plasmids were distinguished from one another with BsrGI and from CD19R plasmids by PmlI (not present). The entire sequence of both plasmids was verified by Sanger Sequencing (DNA Sequencing Core, MDACC).
Tumor cell tissue culture
EL4 cell line was acquired from American Type Culture Collection (Manassas, VA; cat# ATCC TIB-39). NALM-6 cell line was purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (Germany; cat# ACC-128). Kasumi-2 was a gift from Jeffrey Tyner (Oregon Health & Science University) [34]. Clone#9 AaPC (previously referred to as artificial antigen presenting cells; aAPC) was generated though enforced co-expression of truncated CD19, CD64, CD86, and CD137L on K-562 cells genetically modified with lentiviral vectors and was a gift from Carl June (University of Pennsylvania) [35]. This AaPC was further modified using SB system to co-express IL15/IL15Rα fusion protein (designated membrane-bound IL-15; mIL15) and sub-cloned to generate clone#27. Clone#27 was then genetically modified using SB system to express ROR1, and single-cell clones were isolated by FACS based on co-staining of ROR1, CD137L, and IL15 during the sort. The clone#1 AaPC uniformly and stably co-expressed CD19, CD32, CD64, CD86, CD137L, mIL15, and ROR1. These cell lines were maintained in complete media (RPMI, 10% FBS (Hyclone, Logan, UT), and 1x Glutamax-100). Identities of all cell lines were confirmed by STR DNA Fingerprinting at MDACC’s Cancer Center Support Grant (CCSG) supported facility “Characterized Cell Line Core.” All tumor cells were free of mycoplasma and other microbial pathogens.
Numeric expansion of ROR1-specific CAR+ T cells
CAR+ T cells were propagated based on modifying standard operating protocols as previously described [28, 32]. Cryopreserved PBMC, obtained from healthy donors after informed consent, were thawed the day of the electroporation (designated day 0) and rested for 2 hours at 37°C. PBMC for electroporation were spun at 200g for 10 minutes and 2x107 cells were mixed with supercoiled DNA plasmids (5 μg SB11 transposase and 15 μg SB transposon) in Human T cell Nucleofector Solution (cat#VPA-1002, Lonza), added to a cuvette, and electroporated on the U-014 program of Amaxa Nucleofector II (Lonza). Electroporated cells were transferred to a 6-well plate containing phenol-free RPMI, 20% FBS, and 1x Glutamax-100. The following day, electroporated T cells were phenotyped and stimulated with AaPC clone#1 (irradiated using cesium source to 100 Gy) at 1:1 ratio of clone#1 to ROR1-specific CAR+ T cells. Each co-culture was supplemented with IL-21 (cat# AF20021; Peprotech, Rocky Hill, NJ; 30 ng/mL) starting at initiation of culture and every 2–3 days thereafter and with IL-2 (Aldesleukin; Novartis, Switzerland; 50 IU/mL) added every 2–3 days starting at the second stimulation. CAR expression was evaluated weekly to determine the number of AaPC to add to co-cultures every 7 days. If contaminating NK cells reached >10% of the total population, they were depleted from co-cultures with paramagnetic CD56 microbeads (cat#130-050-401, Miltenyi Biotec, Auburn, CA) and LS columns (cat#130-042-401, Miltenyi Biotec). CAR+ T cells were cryopreserved at days 14, 21, 28, and 35 (where applicable). Phenotyping and functional analyses were performed between days 21 to 29. ROR1-specific CAR+ T cells from patients with CLL diagnosis were derived in the same manner, except that tumor cells were depleted the day following electroporation with CD19 microbeads (cat# 130–019–301; Miltenyi) and then stimulated at above. Mock electroporated “No DNA” T cells were stimulated with OKT3-loaded AaPC clone#4 from each donor/patient for negative controls as previously described [36].
Abundance and identity of mRNA molecules by digital profiling
At designated times after co-culture on AaPC and cytokines, T cells were lysed with 160 μL RLT Buffer (Qiagen, Valencia, CA) per 106 cells and lysates were frozen at -80°C. RNA lysates were thawed and immediately analyzed using nCounter Analysis System (NanoString Technologies, Seattle, WA) with “lymphocyte codeset array” (LCA). LCA data was normalized to both spike positive control RNA and housekeeping genes (ACTB, G6PD, OAZ1, POLR1B, POLR2A, RPL27, RPS13, and TBP) as detailed previously [37] and normalized counts were reported. Limit-of-detection (LOD) was calculated from the negative control counts and reported as the mean plus two-times the standard deviation (mean+2xSD) and shown as dashed lines in graphs of mRNA data.
Flow cytometry and Immunohistochemistry
All mAbs were purchased from BD Biosciences (San Jose, CA), except for CCR7 mAb (eBioscience, San Diego, CA), IL-15 mAb (R&D Systems, Minneapolis, MN), Fc mAb used to detect CAR (Invitrogen), and 4A5 mAb used to detect ROR1 (Kipps TJ laboratory, UCSD). Staining was performed as described [38]. Samples for flow cytometry were acquired on FACS Calibur (BD Biosciences) and analyzed with FlowJo software (version 7.6.3). For immunohistochemistry, fresh frozen sections of malignant and normal pancreas were stained with 100 μg/ml of 4A5 mouse-anti-human ROR1 mAb or control mouse IgG2b mAb (isotype), and developed using an anti-mouse secondary antibody conjugated with horse radish peroxidase. Tissues were visualized with diaminobenzidine (DAB) and counterstained with hematoxylin. Slides were evaluated by the study pathologist to identify the tissue or cell-type stained and intensity of staining.
In vitro functional assays
In vitro specific lysis was assessed using a standard 4-hour chromium release assay, as previously described [32]. Expression of cytokines was evaluated by intracellular staining and flow cytometry. CAR+ T cells were incubated with an equal volume and number of target cells for 6 hours at 37°C in the presence of Brefeldin-A (GolgiPlug; BD Biosciences) to block exocytosis and secretion of cytokines. Co-cultures were then (i) stained for surface markers, e.g., CD3 and CAR (Fc-specific antibody), (ii) fixed and permeabilized with BD Cytofix/Cytoperm (cat# 555028, BD Biosciences), (iii) stained for intracellular IFNγ, and (iv) analyzed by flow cytometry.
Mouse experiments
Kasumi-2-_ffLuc_-mKate tumor cells were generated according to a protocol described elsewhere [37]. Transduced Kasumi-2 cells were sorted for uniform mKate expression by FACS to obtain cells with ffLuc activity suitable for non-invasive BLI. Immunocompromised female NSG mice (6–12 weeks of age; NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ; Jackson Laboratory, Bar Harbor, ME) were intravenously (i.v.) injected with 4x104 Kasumi-2-ffLuc-mKate cells (designated day 0), and the following day mice (total n = 17) were mixed together then randomly distributed into 3 groups, which were treated with (i) no treatment (n = 5), (ii) ROR1RCD28+ T cells (n = 5), and (iii) ROR1RCD137+ T cells (n = 5). Mice injected with only T cells served as controls for xenogeneic graft-versus-host-disease (one mouse per T-cell group). T-cell doses (107 total cells per mouse) were administered on days 1, 8, and 15. CAR expression on infused T cells was 96%, 91%, and 90% for ROR1RCD28 and 94%, 62%, and 46% for ROR1RCD137 on days 1, 8, and 15, respectively. IL-2 (60,000 IU) was intraperitoneally administered the day of T-cell dosing and the following day. BLI from tumor ffLuc was monitored twice per week. Log-rank (Mantel-Cox) test was used for statistical analysis between groups of mice (n = 5 per group).
Results
ROR1 is expressed on subsets of tumor cells, but not healthy pancreas
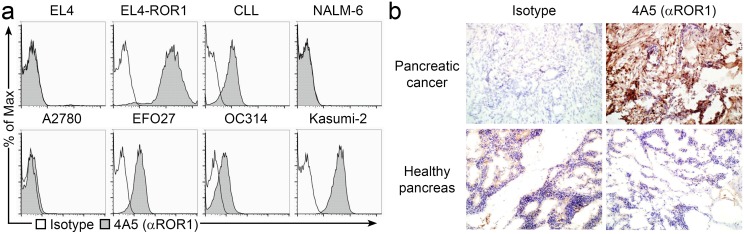
Before constructing CARs from 4A5, a mAb with specificity for human ROR1 [18–20, 24], we sought to validate its staining of tumor cells. The murine T-cell lymphoma cell line EL4 did not express ROR1 but following genetic modification with a ROR1 transposon this cell line showed bright staining for ROR1 (Fig 1a). Consistent with other reports, all of the CLL samples that we tested were ROR1+ as measured by 4A5 mAb staining (representative example is displayed). The t(1;19) B-ALL cell line Kasumi-2 expressed ROR1 whereas the non-t(1;19) B-ALL cell line NALM-6 did not. Eleven of twelve ovarian cancer cell lines tested positive for ROR1 (EFO27 and OC314 are shown as examples) and A2780 was the only ovarian cancer cell line we evaluated that did not express ROR1. Previous reports have identified ROR1 mRNA in the pancreas [39], so we tested 4A5 mAb staining against healthy and malignant pancreatic tissue using immunohistochemistry (Fig 1b). We were unable to detect ROR1 expression in healthy pancreas but ROR1 was widely and strongly expressed by pancreatic cancer tissue. Thus, this staining dataset corroborated our rationale to use the 4A5 mAb to construct CARs specific for ROR1 expressed on tumor cells.
Fig 1. Evaluation of ROR1 expression on tumor cells, pancreatic cancer, and healthy pancreas.
(a) Flow cytometry plots of samples stained with 4A5 mouse-anti-human ROR1 mAb (filled histograms) or matched isotype control (open histograms). Cell types tested were: EL4 murine T-cell lymphoma cell line, EL4 cells genetically modified to express ROR1 (EL4-ROR1), PBMC from patient with CLL diagnosis, non-t(1;19) B-ALL cell line NALM-6, t(1;19) B-ALL cell line Kasumi-2, and ovarian cancer cell lines A2780, EFO27, and OC314. (b) Fresh frozen sections of malignant and normal pancreas were stained with 4A5 mAb or isotype mAb, and developed using an anti-mouse secondary antibody conjugated with horse radish peroxidase. Tissues were visualized with diaminobenzidine and counterstained with hematoxylin. Slides were evaluated by the study pathologist to identify the tissue or cell-type stained and intensity of staining.
Sleeping Beauty transposition and co-culture on ROR1+ AaPC with cytokines generates ROR1-specific T cells
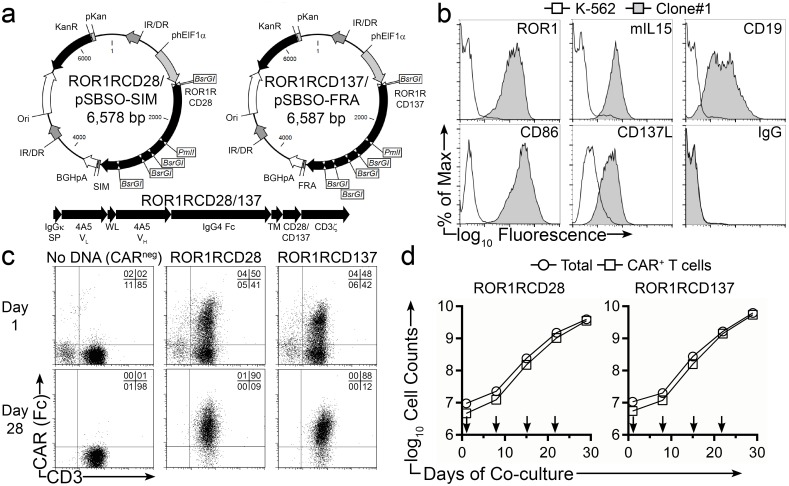
SB transposition followed by co-culture with AaPC, IL-2, and IL-21 was employed to propagate T cells expressing a 2nd generation CD19-specific CAR that activates via CD3ζ/CD28 endodomain (designated CD19RCD28) and these T cells have been infused into patients with B-cell malignancies [28–33, 40]. This strategy was adapted to target ROR1 by replacing the CD19-specific scFv sequence in the CAR with the scFv sequence derived from 4A5 mAb. Two SB transposons were constructed for side-by-side comparison of 2nd generation ROR1-specific CARs that differ in signaling via CD28 (ROR1RCD28) or CD137 (ROR1RCD137) (Fig 2a). The remaining portions of the CARs, e.g., murine IgGκ signal peptide, Whitlow linker, modified human IgG4-Fc stalk, human CD28 transmembrane, and human CD3ζ intracellular domains (with three ITAM activation signals) were identical between the two constructs. We used AaPC (designated clone#1) to achieve the selective expansion of CAR+ T cells. These feeder cells were derived from K-562 cells genetically modified to co-express ROR1, CD19, CD86, CD137L, and a membrane-bound IL-15/IL-15-Receptor-α (IL15Rα) fusion protein (mIL15) (Fig 2b). Peripheral blood mononuclear cells (PBMC) from healthy donors were co-electroporated with DNA plasmids expressing SB11 transposase and either ROR1RCD28 or ROR1RCD137 transposons. The following day, expression of CARs (from both integrated and episomal transgenes) was detected in ROR1RCD28+ and ROR1RCD137+ T cells at 41% ± 6% and 41% ± 8% (mean ± SD; n = 3), respectively, as evidenced by co-staining for Fc (the IgG4-Fc extracellular stalk of CAR) and CD3 (Fig 2c top). Sham-electroporated “No DNA” T cells were co-cultured with γ-irradiated OKT3-loaded clone#4 AaPC, IL-2, and IL-2 and served as negative controls [36]. Co-cultures of CAR+ T cells and γ-irradiated clone#1 AaPC were then initiated in the presence of exogenous IL-2 and IL-21 in parallel to the “No DNA” co-cultures. Recursive stimulations of AaPC, IL-2, and IL-21 were performed every 7 days for a total of four stimulations, and IL-2 and IL-21 were replaced every 2–3 days during co-culture. At day 28 of co-culture, CAR was expressed in T cells at 90% ± 3% and 79% ± 11% (mean ± SD; n = 3) for ROR1RCD28 and ROR1RCD137, respectively (Fig 2c bottom). These two CAR-expressing T cell populations appear to proliferate at similar rates, as noted by the total numbers of cells counted at the end of culture (p = 0.66; Two-way ANOVA), and in number of CAR+ T cells generated (p = 0.74). Total cell numbers closely coincided with CAR+ T-cell counts for both ROR1RCD28 and ROR1RCD137, resulting in production of at least 109 CAR+ T cells (Fig 2d). In similar studies, CLL patient-derived PBMC, composed of >91% CD19+ malignant cells, were electroporated with SB11 and ROR1-specific CAR SB transposons. The following day, cultures were depleted of malignant cells with CD19 paramagnetic microbeads and stimulated weekly with clone#1 AaPC in the presence of IL-2 and IL-21, as was performed for healthy donor PBMC. This resulted in >160-fold expansion of CAR+ T cells with 92% and 80% expressing ROR1RCD28 and ROR1RCD137, respectively. This indicates that autologous ROR1-specific T cells can be generated from recipients with advanced CLL. Thus, SB transposition and co-culture on AaPC clone#1 with IL-2/-21 generated clinically-appealing quantities of ROR1-specific T cells with high frequencies of CAR expression.
Fig 2. Sustained numeric expansion of ROR1-specific CAR+ T cells upon clone#1 AaPC and cytokines.
(a) DNA plasmid vector maps for ROR1RCD28/pSBSO-SIM and ROR1RCD137/pSBSO-FRA. Abbreviations are IR/DR: Sleeping Beauty Inverted Repeat/Direct Repeat, phElF1α: Human Elongation Factor-1-α region hybrid promoter, ROR1RCD28: Human codon-optimized ROR1-specific scFv:Fc:CD28:CD3ζ CAR, ROR1RCD137: Human codon-optimized ROR1-specific scFv:Fc:CD137:CD3ζ CAR, SIM: “SIM” PCR tracking oligonucleotides, FRA: “FRA” PCR tracking oligonucleotides, BGHpA; bovine growth hormone polyadenylation sequence, Ori: minimal E.coli origin of replication, KanR: Bacterial selection gene encoding Kanamycin resistance, pKan: prokaryotic Kanamycin promoter. Digestion with BsrGI enzyme can distinguish the two plasmids, which have high degrees of similarity, and with PmlI enzyme can distinguish them from CD19RCD28 plasmid, which does not have PmlI site. The entire plasmid sequences were verified by Sanger-based sequencing techniques. (b) Parental K-562 (open histograms) and clone#1 AaPC (shaded histograms) were stained for CD19, CD86, CD137L, ROR1, and IL15 (membrane-bound IL-15; mIL15) where isotype (IgG) was used a negative control. (c) Expression CAR in ROR1RCD28 (middle) and ROR1RCD137 (right) T cells the day following electroporation (top panels) and after 28 days of co-culture on clone#1 AaPC (bottom panels) where “no DNA” T cells (left) were used as negative controls. T cells were marked by CD3 staining and CAR+ cells were detected with Fc-specific antibody. Quadrant frequencies are displayed in upper right corners. (d) Proliferation kinetics of total cells (circles) and CAR+ T cells (squares) on clone#1 AaPC over 28 days of co-culture. ROR1RCD28 displayed on the left and ROR1RCD137 shown on the right. Arrows represent addition of γ-irradiated clone#1 AaPC. Data are representative of 3 donors expanded in 3 independent experiments. Expansion data can be found in S1 Dataset, Fig 2 tab.
Electroporated and propagated CAR+ T cells have a transcriptional profile consistent with diverse memory phenotypes and potential for effector functions
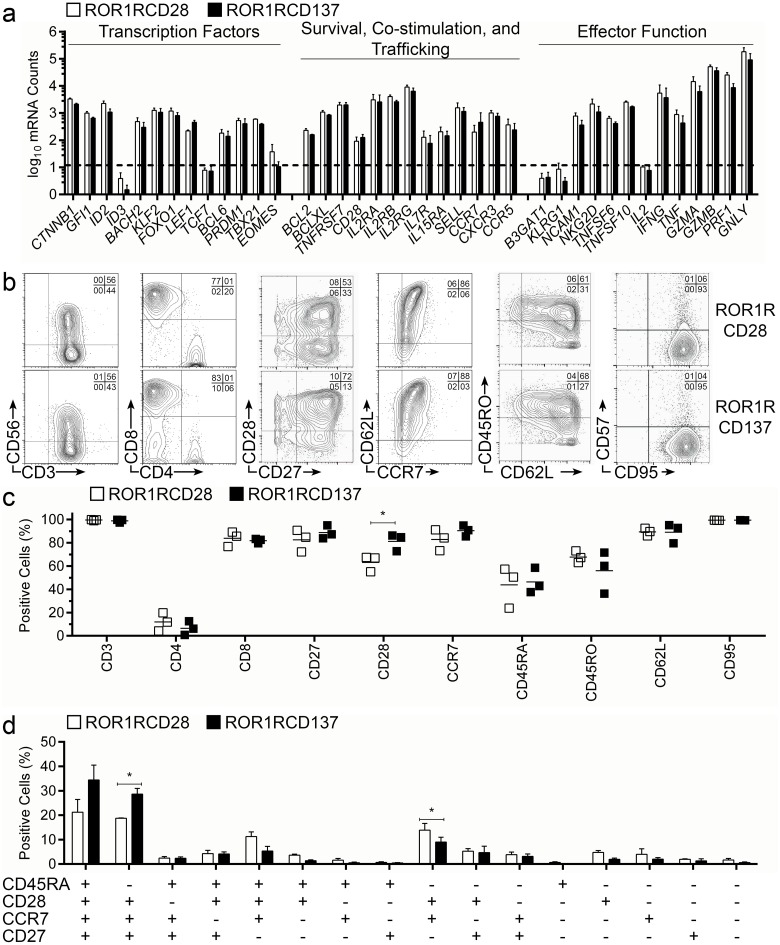
T-cell memory phenotype can be used to predict proliferation, survival, and effector potential of an infused CAR+ T-cell product. We used non-enzymatic digital multiplex array of mRNA transcripts (NanoString) to assess the transcriptional profile of CAR+ T cells following numeric expansion with AaPC and cytokines. Transcription factors associated with less differentiated T-cell phenotype, e.g., CTNNB1 (β-Catenin), GFI1 (Growth Factor Independence-1), ID2 (Inhibitor of DNA Binding-2), BACH2 (BTB and CNC Homology-2), KLF2 (Kruppel-like Factor-2), FOXO1 (Forkhead Box-O1), and LEF1 (Lymphoid Enhancer Binding Factor-1), were expressed by populations of ROR1-specific T cells concurrently with transcription factors associated with T cells in later memory stages, including BCL6 (B-cell Lymohoma-6), PRDM1 (BLIMP-1), and TBX21 (T-bet), suggesting that the outgrowth of CAR+ T cells was heterogeneous in memory gene regulation (Fig 3a left). Quantities of transcription factors ID3 (Inhibitor of DNA Binding-3; reciprocal and upstream to ID2), TCF7 (T-cell Factor-1; TCF1; reciprocal to and upstream of LEF1), and EOMES (Eomesodermin; similar in function to T-bet) were at or below the limit-of-detection, indicating that coordination through some of the gene regulation cascades had occurred during or prior to expansion on AaPC. Thus, CAR+ T cells had a continuum of memory transcription factor expression. Genes associated with intrinsic memory T-cell survival, e.g., BCL2 (B-cell Lymphoma-2) and BCLXL (BCL2-related protein long isoform), and receptors for cytokines responsible for extrinsic memory T-cell survival and proliferation, e.g., IL2RA (IL-2-Receptor-α; CD25), IL2RB (IL-2-Receptor-β; CD122), IL2RG (IL-2-Receptor-γ; CD132), IL7R (IL-7-Receptor-α; CD127), and IL15RA (IL-15-Receptor-α), were detected in CAR+ T cells suggesting that persistence and survival of these cells following adoptive transfer is plausible (Fig 3a middle). Memory-associated T-cell trafficking molecules, e.g., SELL (L-Selectin; CD62L), CCR7 (CCR7), CXCR3 (CXCR3), and CCR5 (CCR5), and lymphocyte co-stimulatory molecules, e.g., TNFRSF7 (CD27) and CD28 (CD28), were detected in ROR1RCD28+ and ROR1RCD137+ T cells, indicating that the CAR+ T cells may have been derived from or developed into T-cell memory cells. Importantly, genes associated with exhaustion and terminal differentiation, including B3GAT1 (Beta-1,3-Glucuronyltransferase-1; CD57) and KLRG1 (KLRG1) were not expressed above the limit-of-detection by the CAR+ T cells (Fig 3a right). Genes encoding surface receptors leading to killer function, e.g., NCAM1 (Neural cell adhesion molecule-1; CD56) and NKG2D (NKG2D), cytokines with pro-inflammatory properties, e.g., IFNG (Interferon-γ) and TNF (Tumor Necrosis Factor), and molecules responsible for directing cell death and/or cytolysis, e.g., TNFSF6 (Fas-ligand), TNFSF10 (TRAIL), GZMA (Granzyme-A), GZMB (Granzyme-B), PRF1 (Perforin-1), and GNLY (Granulysin), were expressed by CAR+ T cells suggesting that these cells could have effector function in addition to their potential for memory formation. In aggregate, CAR+ T cells which emerge after recurrent additions of AaPC and IL-2/IL-21 appear to contain sub-populations with desirable gene expression predictive of therapeutic efficacy after adoptive transfer.
Fig 3. Lymphocyte transcriptional profile and memory markers expressed on CAR+ T-cell surface.
After 29 days of expansion on clone#1 AaPC/IL-2/IL-21, ROR1RCD28 and ROR1RCD137 cells were (i) lysed for mRNA expression analysis using bar-codes or (ii) phenotyped for T-cell surface markers by flow cytometry. (a) RNA lysates were profiled for expression of a selected group of lymphocyte genes with non-enzymatic digital multiplex array of mRNA transcripts (NanoString) where transcription factors are shown on the left, genes associated with survival, co-stimulation, and trafficking are shown on the middle, and genes associated with effector function are shown on the right. Dashed line represents the limit-of-detection calculated by mean + 2xSD of negative controls. (b) Flow cytometry of ROR1RCD28 and ROR1RCD137 T cells showing co-staining for CD3 and CD56, CD4 and CD8, CD28 and CD27, CD62L and CCR7, CD45RO and CD62L, or CD95 and CD57 in cells gated for CAR expression based staining with Fc-specific antibody. ROR1RCD28+ T cells are shown in the top panels and ROR1RCD137 T cells are displayed in the bottom panels. One of 3 representative donors is displayed and quadrant frequencies are shown in the upper right corners. (c) Cumulative frequencies of cells staining positive for each memory marker within an extended memory phenotype panel. (d) Multi-parameter flow cytometry was used to determine frequencies of cells staining positive for combinations of CD45RA, CD27, CD28, and CCR7. For (c) and (d) ROR1RCD28 are in open shapes/bars and ROR1RCD137 are in closed shapes/bars and lines displayed in (c) are means (n = 3) and in (d) are mean ± SD (n = 3). Student’s two-tailed t-tests were used for statistical analysis between the two groups. *p<0.05 Expression and phenotype data can be found in S1 Dataset, Fig 3 tab.
CAR+ T cells express surface markers consistent with T-cell memory
Flow cytometry was used to validate the NanoString panel, examine the surface expression of canonical T-cell markers, and determine the frequencies of memory populations with multi-parameter staining. CAR+ T-cell populations expressed CD56 (NCAM1), although CD3negCD56+ NK cells were absent from final cultures (Fig 3b, first panels). A preponderance of CD8+ T cells was present in the cultures relative to CD4+ T cells (Fig 3b, second panels), and >99% of cells expressed CD3. High frequencies of CAR+ T cells were found co-expressing CD28 and CD27 co-stimulatory molecules (Fig 3b, third panels), which are characteristics of T-cell memory. CD27, in particular, has been correlated to durable regressions resulting from CD8+ T-cell treatments [41]. The stemness of CD8+ memory cells has been linked to expression of L-selectin [12], which was detected in CAR+ T cell’s mRNA (SELL) and as protein (CD62L), and was concurrently expressed with the associated lymphoid homing marker CCR7 (Fig 3b, fourth panels). Thus, such CAR+ T cells with long-lived potential could migrate to secondary lymphoid structures harboring ROR1+ leukemias. Few CD45RO+CD62Lneg cells were detected, which are generally associated with TEM or late-stage effector memory cells, and could be contrasted to the larger abundance of CD45ROnegCD62L+ cells usually associated with less differentiated T cells (Fig 3b, fifth panels). Virtually all cells expressed CD95, a marker for activation and differentiation beyond TN lineages, which was expected on T cells cultured ex vivo, but the same T cells did not express CD57, a marker of exhaustion (Fig 3b, sixth panels). In aggregate, the staining of these markers was consistent amongst donors (Fig 3c). Multi-parameter flow cytometry revealed that most electroporated/propagated T cells belonged to less-differentiated memory phenotype primarily composed of TSCM (CD45RA+CD27+CD28+CCR7+) and TCM (CD45RAnegCD27+CD28+CCR7+) (Fig 3d) [42, 43]. There were appreciable frequencies of CD45RA+CD27negCD28+CCR7+ and CD45RAnegCD27negCD28+CCR7+, which we were not able to define as a specific memory subset, and almost no TEMRA cells (CD45RA+CD27negCD28negCCR7neg). In aggregate, the surface phenotype of ROR1-specific CAR+ T cells corroborated the mRNA digital barcoding profiling data (NanoString) and indicated that these cells have desirable characteristics for fighting ROR1+ malignancies.
ROR1-specific CAR+ T cells produce IFNγ in response to ROR1+ tumor cells
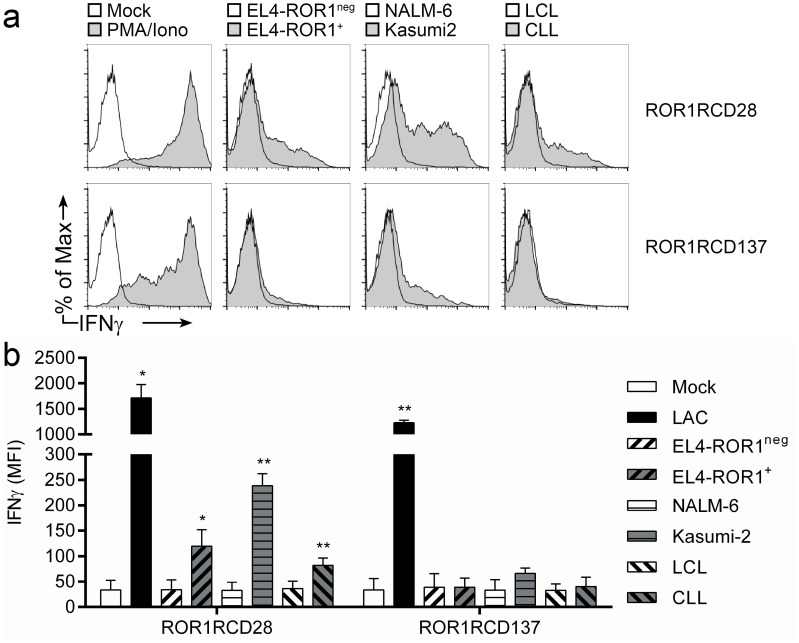
We monitored for the production of IFNγ by intracellular cytokine staining of healthy donor CAR+ T cells following 6 hour co-culture with target cells. Both ROR1RCD28+ and ROR1RCD137+ T cells produced significant quantities of IFNγ when treated with PMA/Ionomycin (non-specific mitogenic stimulus; positive control) relative to mock-activated (media only; negative control) cultures (Fig 4). ROR1RCD28+ T cells stained brightly for IFNγ when co-cultured with ROR1+ tumor targets EL4-ROR1, Kasumi-2, and CLL cells, but did not produce IFNγ, as evidenced by similar staining of unchallenged cultures (mock; media only) to co-cultures with ROR1neg cells from EBV-transformed healthy donor B-cell lymphoblastoid cell lines (LCL), NALM-6, and parental EL4 cells (Fig 4a top). IFNγ production by ROR1RCD137+ T cells slightly increased only in response to Kasumi-2 cells, demonstrating that there were differences in the effector cytokine production of the two CAR populations to certain tumor cells (Fig 4a bottom). In summary, effector function of CAR+ T cells, as manifested by IFNγ expression, was restricted to ROR1+ tumor cells and ROR1RCD28+ T cells produced more IFNγ than did ROR1RCD137+ T cells in response to ROR1.
Fig 4. IFNγ production by ROR1-specific CAR+ T cells in response to ROR1+ targets.
At day 29 of co-culture with AaPC/IL2/IL-21, CAR+ T cells were co-cultured for 6 hours at 37°C with tumor targets or non-specific mitogenic stimuli (PMA/Iono) then analyzed for expression of IFNγ in CAR+ T cells (gated based on Fc expression). Brefeldin-A (GolgiPlug) was added to T cells to block IFNγ secretion. (a) Representative flow cytometry plots where ROR1RCD28 cultures are on the top and ROR1RCD137 cultures are on the bottom. Percentage of max (y-axes) normalized total cell numbers in each sample for consistency between samples so that IFNγ fluorescent intensity (x-axes) could be compared between conditions. (b) Cumulative mean fluorescence intensities (MFI) of IFNγ staining from co-cultures where mean ± SD (n = 3 donors) is shown. Student’s paired, 1-tailed t-test for statistical analysis between target co-culture and T cells alone (Mock; media only). *p<0.05 and **p<0.01 IFNγ data can be found in S1 Dataset, Fig 4 tab.
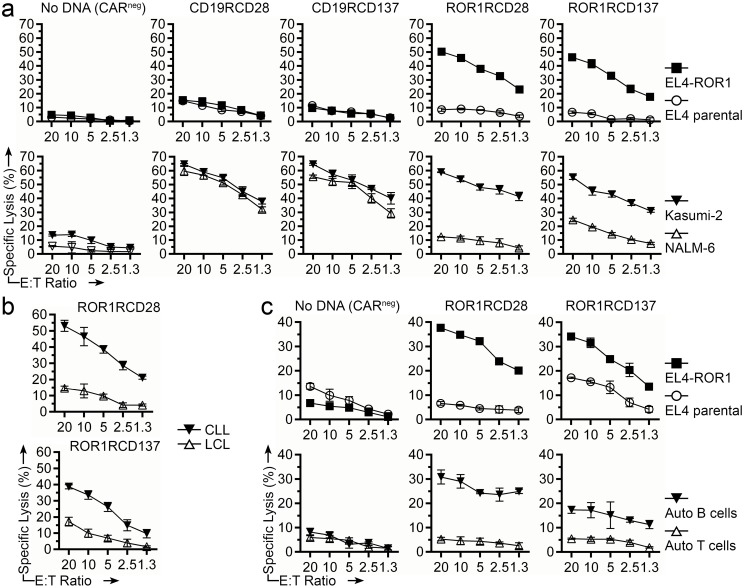
ROR1-specific killing by CAR+ T cells in vitro
The killing of tumor cells by genetically modified T cells is another measurement of re-directed specificity. Four-hour chromium release assay was used to assess the specific lysis of tumor-cell lines and primary tumor cells. ROR1RCD28+ and ROR1RCD137+ T cells derived from healthy donor PBMC efficiently lysed EL4-ROR1+ and Kasumi-2 but showed minimal lysis of EL4 parental (ROR1neg) and NALM-6 cells (Fig 5a). Furthermore, autologous CD19RCD28+ and CD19RCD137+ T cells displayed minimal lysis of EL4 and EL4-ROR1 but lysed both Kasumi-2 and NALM-6 (both CD19+) suggesting that the ROR1RCD28+ and ROR1RCD137+ T cells were more discriminant in their targeting of these B-ALL cell lines (Fig 5a bottom). Autologous CARneg T cells (No DNA) showed no lysis of these 4 cell lines indicating that the CAR responses were specific. ROR1RCD28+ and ROR1RCD137+ T cells generated from healthy donor PBMC also lysed allogeneic CLL patient B cells (ROR1+), but spared allogeneic LCL (ROR1neg) derived from healthy donor B-cells (Fig 5b). Similarly, ROR1RCD28+ and ROR1RCD137+ T cells generated from PBMC of a patient with CLL were able to lyse EL4-ROR1 and autologous tumor cells (ROR1+; isolated with CD19-specific magnetic beads), while autologous T cells and parental EL4 (both ROR1neg) cells were spared (Fig 5c). In summary, ROR1-specific CAR+ T cells demonstrated effective in vitro specific lysis of ROR1+ tumor cells in both autologous and allogeneic settings.
Fig 5. Specific cytolysis of ROR1+ tumor cells by CAR+ T cells.
Four-hour chromium release assay was used to assess specific lysis by T cells at decreasing effector to target (E:T) ratios. (a) No DNA (CARneg), CD19RCD28+, CD19RCD137+, ROR1RCD28+, or ROR1RCD137+ T cells generated from healthy donor PBMC were challenged with EL4-ROR1+ cell line (closed squares), EL4 parental (ROR1neg) cell line (open circles), Kasumi-2 cell line (closed inverted triangles), or NALM-6 cell line (open triangles). (b) CAR+ T cells generated from healthy donor PBMC were challenged with allogeneic CLL cells (closed inverted triangles), healthy donor allogeneic B-cell LCL (open triangles). (c) No DNA (CARneg), ROR1RCD28+, or ROR1RCD137+ T cells generated from CLL patient PBMC were challenged with EL4-ROR1+ cell line (closed squares), EL4 parental (ROR1neg) cell line (open circles), autologous B cells (closed inverted triangles), or autologous T cells (open triangles). Data are mean ± SD of triplicate measurements in CRA and are representative of four donors from four independent experiments. Lysis data can be found in S1 Dataset, Fig 5 tab.
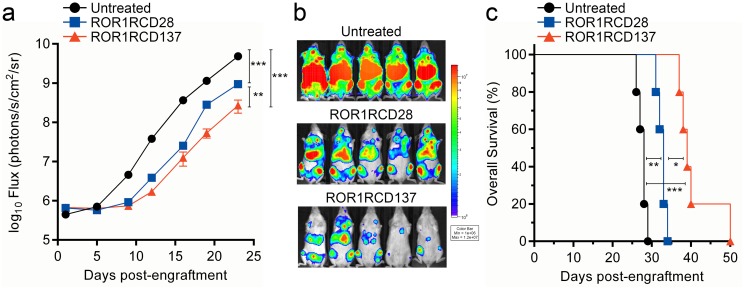
In vivo clearance of leukemia by ROR1-specific CAR+ T cells
In order to evaluate the anti-tumor activity of ROR1-specific CAR+ T cells in vivo, a xenograft model of leukemia was established in immunocompromised mice and treated with ROR1-specific CAR+ T cells. As Kasumi-2 cells were sensitive to ROR1-specific T-cell lysis and were not lysed by autologous CARneg T cells (Fig 5), they were genetically modified with lentivirus particles to introduce mKate red fluorescence protein for sorting transduced cells and enhanced firefly luciferase (ffLuc) [44] for serial non-invasive bioluminescence imaging (BLI) of tumor burden in vivo. NOD.scid.γc -/- (NSG) mice were used because they lack functional adaptive immune systems and can therefore accept human tumor xenografts. Healthy, 6–12 week old female NSG mice were engrafted with Kasumi-2-_ffLuc_-mKate and distributed into 3 treatment groups (n = 5 per group): (i) no treatment, (ii) ROR1RCD28+ T-cell treated, and (iii) ROR1RCD137+ T-cell treated, where T cells were given at days 1, 8, and 15 post-tumor cell engraftment (Fig 6). Untreated mice had consistent log10-fold increases in bioluminescence flux from their tumors (Fig 6a black circles and 6b top) and succumbed to disease after an average of 27 days following engraftment (Fig 6c black circles). ROR1RCD28+ T cells were able to diminish tumor burden compared to untreated mice as measured by tumor BLI flux (Fig 6a blue squares and 6b middle) and prolonged survival compared to untreated mice (Fig 6c blue squares). ROR1RCD137+ T cells eliminated tumor burden above both untreated mice and ROR1RCD28+ T cell-treated mice (Fig 6c red triangles and 6b bottom), and were able to increase survival compared to both untreated mice and ROR1RCD28+ T cell-treated (Fig 6c red triangles). In summary, ROR1-specific CAR+ T cells can efficiently treat ROR1+ leukemia in mice which supports testing of these T cells in the clinic as an investigational therapy for patients with ROR1+ malignancies.
Fig 6. In vivo tumor clearance by ROR1-specific CAR+ T cells.
NSG mice were intravenously (i.v.) injected with Kasumi-2-_ffLuc_-mKate cells and were treated with three i.v. doses of T cells to assess the ability of ROR1-specific T cells to manage disease. High-dose IL-2 was added intraperitoneally the day of injection and the following day. (a) Non-invasive BLI reported as flux was serially measured in untreated (black circles), ROR1RCD28+ T cell-treated (blue squares), and ROR1RCD137+ T cell-treated (red triangles) mice. Data are mean ± SEM (n = 5). Two-way ANOVA was used for statistical analysis. (b) Representative BLI images at day +23 post-tumor cell engraftment. (c) Overall survival of mice. *p<0.05, **p<0.01, ***p<0.001 BLI and survival data can be found in Supporting Information, Fig 6 tab.
Discussion
This work reveals the pre-clinical basis to support our current “first-in-human” Phase I clinical trial of CAR+ T cells for investigational therapy of ROR1+ malignancies. We demonstrate that SB transposition achieved stable CAR expression in T cells and, in concert with co-culture on clone#1 AaPC, resulted in heterogeneous outgrowth of T-cell memory populations with ROR1-restricted anti-tumor activity. Currently, this process takes 28 days to achieve clinical numbers of T cells. Potentially shortening the production time for cell growth may (i) simplify the overall process, (ii) preserve more of the minimally differentiated subsets, and (iii) could take advantage of in vivo proliferation of CAR+ T cells thereby augmenting therapeutic efficacy. As SB transposition was used to express ROR1-specific CARs in naïve or minimally-differentiated T-cell populations present in quiescent PBMC, as has been done with other SB-based CAR studies [31, 38], the prospect of giving T cells earlier in the proliferative cycle is of great interest to our group. Homeostatic cytokines, e.g., IL-15, can promote the survival of less differentiated T cells, and trans-presentation of IL-15 by IL15Rα has higher signaling potency than IL-15 alone [45, 46]. Thus, we chose to introduce mIL15 (IL-15/IL-15Rα fusion protein) rather than IL-15 tethered to a hinge/Fc stalk, which is the moiety expressed on AaPC used to generate CAR+ T cells expressing CD19RCD28 [47], on the clone#1 AaPC. The AaPC feeder platform, which is amenable to genetic introduction of TAAs (e.g., ROR1) and molecules that promote T-cell survival (e.g., mIL15), in concert with SB transposition can support the propagation of populations of T cells predicted to have sustained engraftment, i.e., TSCM and CD62L+ TCM populations [12, 48], while enforcing CAR expression on T cells through recursive antigen exposure; therefore, they are attractive platforms for the generation of ROR1-specific T cells for cancer immunotherapy.
The choice of signaling motif within a 2nd generation CAR endodomain may impact the therapeutic activity of infused T cells. CD19-specific CARs activating T cells via chimeric CD28 or CD137 endodomains led to objective clinical responses [3–7]. We designed CARs that used the same extracellular structure in order to reveal differences arising between the endodomains. The most notable differences were that ROR1RCD137+ T cells displayed enhanced in vivo tumor clearance (Fig 6) despite showing reduced IFNγ production in vitro relative to ROR1RCD28+ T cells (Fig 4). The decrease in IFNγ-response with CARs signaling through CD137 relative to those signaling through CD28 is consistent with independent studies comparing ROR1-specific CARs [25, 39, 49, 50]. These studies introduced CARs derived from different ROR1-specific antibodies (2A2 and R12) into TCM cells with lentivirus and showed that CD137 signaling resulted in reduced cytokine production and increased anti-tumor activity in vivo [50]. CARs with short extracellular spacers, especially those without Fc stalks that could be bound by cells expressing CD64 (high-affinity Fc receptor), were optimal for other ROR1 CARs (2A2 and R12 moieties) [49]. Future studies from our group will evaluate whether truncation or replacement of the Fc stalk in CARs constructed from 4A5 mAb, introduced into T cells with SB, and cultured on AaPC will produce similar benefits. An inverse correlation between in vitro effector function (cytokine production and cytolysis) and in vivo tumor clearance by CD8+ tumor-specific T cells has also been established for adoptive immunotherapy of melanoma, further supporting the notion that ex vivo culture conditions and memory phenotype of infused T cells are useful parameters to correlate with therapeutic efficacy [11]. A side-by-side comparison of the two CARs using a competitive repopulation experiment in a clinical trial is likely needed to reveal which CAR design is superior for a given tumor and perhaps even a particular patient.
A major advantage of targeting ROR1 over the current T-cell therapies targeting CD19 is that recipients would not deplete B cells and develop hypogammaglobulinemia, thereby mitigating the risk for impaired humoral immunity [32, 51]. ROR1 was originally identified on the surface of CLL cells with absent expression on normal tissues, including hematopoietic cells [18, 24]. Subsequently, ROR1 was described on some acute leukemias and solid tumors [19, 20, 34]. The 4A5 ROR1-specific mAb (used in this study to construct the CAR) did not detect ROR1 in healthy tissues by either immunoblot or immunohistochemistry (Fig 1b) [18, 20]. The only healthy cells to be stained with 4A5 mAb are hematogones, which are rare pre-B-cells that do not appear to be obligate precursors to mature B cells [24]. There has been some discordance between the mRNA transcript data and protein staining data, which may be explained by off targeting of ROR2 gene, which is expressed after birth in many tissues, and is highly homologous to ROR1. Given that ROR1-specific T cells propagated in this study have potential for long-term engraftment due to their memory T-cell phenotype, it is plausible that patients treated with these genetically modified T cells could experience prolonged anti-tumor effects with potential for elimination of minimal disease or relapsed ROR1+ malignancies. As a control for adverse events, conditional suicide genes, e.g., inducible Caspase9, could be co-expressed with CAR in order to eliminate T cells in vivo as needed [52].
In summary, our data support the clinical application of ROR1-specific CAR+ T cells and a Phase I clinical trial is open to patients with CLL as of July 2014 (NCT02194374). Clinical-grade DNA plasmids coding for ROR1RCD137 and SB11 have been produced and a master-cell bank of AaPC clone#1 has been generated. Furthermore, regulatory documents and materials are in place to undertake the clinical trial. This will be the first time ROR1 on CLL has been targeted by T cells; therefore, the primary endpoint will be to determine toxicity and maximum tolerated T-cell dose. Future trials can then assess the clinical impact of targeting ROR1 on solid tumors.
Supporting Information
S1 Dataset. Raw data as Excel spreadsheet.
(Fig 2 tab) Cell counts at designated times as measured by trypan blue exclusion. (Fig 3 tab) Normalized mRNA counts from NanoString array of T cells at day 29 of co-culture (top), surface phenotype of CAR+ T cells (middle), and multiparameter memory phenotype of T cells (bottom). (Fig 4 tab) MFI of IFNγ staining of CAR+ T cells following 6 hour co-culture with target cells. (Fig 5 tab) 4-hour chromium release assay of T cells co-cultured with target cells. (Fig 6 tab) BLI flux kinetics of Kasumi2-ffLuc-mKate cells following challenge with CAR+ T cells (top) and days of mouse euthanasia (bottom).
(XLSX)
Acknowledgments
We thank Dr. Carl June and colleagues (University of Pennsylvania) for help generating K-562-derived AaPC clone #9 and Dr. Perry Hackett (University of Minnesota) for his assistance with the SB system. The flow cytometry core lab (MDACC) assisted with FACS. We thank Dr. Eric Tran (NCI Surgery Branch) for editing this document. Drew C. Deniger was an American Legion Auxiliary Fellow in Cancer Research (UT-GSBS-Houston), Andrew Sowell-Wade Huggins Scholar in Cancer Research (Cancer Answers Foundation), and a Teal Pre-doctoral Scholar (Department of Defense Ovarian Cancer Research Program) during this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by funding from: Cancer Center Core Grant (CA16672); RO1 (CA124782, CA120956, CA141303; CA141303); R33 (CA116127); P01 (CA148600); S10RR026916; SPORE (CA83639); Albert J. Ward Foundation; Burroughs Wellcome Fund; Cancer Prevention and Research Institute of Texas; CLL Global Research Foundation; Department of Defense; Estate of Noelan L. Bibler; Gillson Longenbaugh Foundation; Harry T. Mangurian, Jr., Fund for Leukemia Immunotherapy; Fund for Leukemia Immunotherapy; Institute of Personalized Cancer Therapy; Leukemia and Lymphoma Society SCOR; Lymphoma Research Foundation; Miller Foundation; Mr. Herb Simons; Mr. and Mrs. Joe H. Scales; Mr. Thomas Scott; MDACC Moon Shot; National Foundation for Cancer Research; Pediatric Cancer Research Foundation; Production Assistance for Cellular Therapies (PACT); TeamConnor; Thomas Scott; William Lawrence and Blanche Hughes Children's Foundation.
References
- 1.Jena B, Dotti G, Cooper LJ. Redirecting T-cell specificity by introducing a tumor-specific chimeric antigen receptor. Blood. 2010;116(7):1035–44. Epub 2010/05/05. doi: blood-2010-01-043737 [pii] 10.1182/blood-2010-01-043737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.June CH. Principles of adoptive T cell cancer therapy. J Clin Invest. 2007;117(5):1204–12. Epub 2007/05/04. 10.1172/JCI31446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368(16):1509–18. Epub 2013/03/27. 10.1056/NEJMoa1215134 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brentjens RJ, Riviere I, Park JH, Davila ML, Wang X, Stefanski J, et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood. 2011;118(18):4817–28. Epub 2011/08/19. 10.1182/blood-2011-04-348540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kochenderfer JN, Dudley ME, Feldman SA, Wilson WH, Spaner DE, Maric I, et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood. 2012;119(12):2709–20. Epub 2011/12/14. 10.1182/blood-2011-10-384388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalos M, Levine BL, Porter DL, Katz S, Grupp SA, Bagg A, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3(95):95ra73. Epub 2011/08/13. doi: 3/95/95ra73 [pii] 10.1126/scitranslmed.3002842 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365(8):725–33. Epub 2011/08/13. 10.1056/NEJMoa1103849 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz CR, Micklethwaite KP, Savoldo B, Ramos CA, Lam S, Ku S, et al. Infusion of donor-derived CD19-redirected virus-specific T cells for B-cell malignancies relapsed after allogeneic stem cell transplant: a phase 1 study. Blood. 2013;122(17):2965–73. Epub 2013/09/14. 10.1182/blood-2013-06-506741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371(16):1507–17. Epub 2014/10/16. 10.1056/NEJMoa1407222 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015;385(9967):517–28. 10.1016/S0140-6736(14)61403-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gattinoni L, Klebanoff CA, Palmer DC, Wrzesinski C, Kerstann K, Yu Z, et al. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells. J Clin Invest. 2005;115(6):1616–26. Epub 2005/06/03. 10.1172/JCI24480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graef P, Buchholz VR, Stemberger C, Flossdorf M, Henkel L, Schiemann M, et al. Serial transfer of single-cell-derived immunocompetence reveals stemness of CD8(+) central memory T cells. Immunity. 2014;41(1):116–26. Epub 2014/07/19. 10.1016/j.immuni.2014.05.018 . [DOI] [PubMed] [Google Scholar]
- 13.Jolles S, Sewell WA, Misbah SA. Clinical uses of intravenous immunoglobulin. Clin Exp Immunol. 2005;142(1):1–11. 10.1111/j.1365-2249.2005.02834.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brentjens R, Yeh R, Bernal Y, Riviere I, Sadelain M. Treatment of chronic lymphocytic leukemia with genetically targeted autologous T cells: case report of an unforeseen adverse event in a phase I clinical trial. Mol Ther. 2010;18(4):666–8. Epub 2010/04/02. 10.1038/mt.2010.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nomi M, Oishi I, Kani S, Suzuki H, Matsuda T, Yoda A, et al. Loss of mRor1 enhances the heart and skeletal abnormalities in mRor2-deficient mice: redundant and pleiotropic functions of mRor1 and mRor2 receptor tyrosine kinases. Mol Cell Biol. 2001;21(24):8329–35. Epub 2001/11/20. 10.1128/MCB.21.24.8329-8335.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baskar S, Kwong KY, Hofer T, Levy JM, Kennedy MG, Lee E, et al. Unique cell surface expression of receptor tyrosine kinase ROR1 in human B-cell chronic lymphocytic leukemia. Clin Cancer Res. 2008;14(2):396–404. Epub 2008/01/29. 10.1158/1078-0432.CCR-07-1823 . [DOI] [PubMed] [Google Scholar]
- 17.Daneshmanesh AH, Mikaelsson E, Jeddi-Tehrani M, Bayat AA, Ghods R, Ostadkarampour M, et al. Ror1, a cell surface receptor tyrosine kinase is expressed in chronic lymphocytic leukemia and may serve as a putative target for therapy. Int J Cancer. 2008;123(5):1190–5. Epub 2008/06/12. 10.1002/ijc.23587 . [DOI] [PubMed] [Google Scholar]
- 18.Fukuda T, Chen L, Endo T, Tang L, Lu D, Castro JE, et al. Antisera induced by infusions of autologous Ad-CD154-leukemia B cells identify ROR1 as an oncofetal antigen and receptor for Wnt5a. Proc Natl Acad Sci U S A. 2008;105(8):3047–52. Epub 2008/02/22. 10.1073/pnas.0712148105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang S, Chen L, Cui B, Chuang HY, Yu J, Wang-Rodriguez J, et al. ROR1 is expressed in human breast cancer and associated with enhanced tumor-cell growth. PLoS One. 2012;7(3):e31127 Epub 2012/03/10. 10.1371/journal.pone.0031127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, Chen L, Wang-Rodriguez J, Zhang L, Cui B, Frankel W, et al. The onco-embryonic antigen ROR1 is expressed by a variety of human cancers. Am J Pathol. 2012;181(6):1903–10. Epub 2012/10/09. 10.1016/j.ajpath.2012.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rebagay G, Yan S, Liu C, Cheung NK. ROR1 and ROR2 in Human 1Malignancies: Potentials for Targeted Therapy. Front Oncol. 2012;2:34 Epub 2012/06/02. 10.3389/fonc.2012.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S, Cui B, Lai H, Liu G, Ghia EM, Widhopf GF 2nd, et al. Ovarian cancer stem cells express ROR1, which can be targeted for anti-cancer-stem-cell therapy. Proc Natl Acad Sci U S A. 2014;111(48):17266–71. 10.1073/pnas.1419599111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dave H, Anver MR, Butcher DO, Brown P, Khan J, Wayne AS, et al. Restricted cell surface expression of receptor tyrosine kinase ROR1 in pediatric B-lineage acute lymphoblastic leukemia suggests targetability with therapeutic monoclonal antibodies. PLoS One. 2012;7(12):e52655 Epub 2013/01/04. 10.1371/journal.pone.0052655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broome HE, Rassenti LZ, Wang HY, Meyer LM, Kipps TJ. ROR1 is expressed on hematogones (non-neoplastic human B-lymphocyte precursors) and a minority of precursor-B acute lymphoblastic leukemia. Leuk Res. 2011;35(10):1390–4. Epub 2011/08/05. 10.1016/j.leukres.2011.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger C, Sommermeyer D, Hudecek M, Berger M, Balakrishnan A, Paszkiewicz PJ, et al. Safety of targeting ROR1 in primates with chimeric antigen receptor-modified T cells. Cancer Immunol Res. 2015;3(2):206–16. 10.1158/2326-6066.CIR-14-0163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hackett PB, Largaespada DA, Cooper LJ. A transposon and transposase system for human application. Mol Ther. 2010;18(4):674–83. Epub 2010/01/28. doi: mt20102 [pii] 10.1038/mt.2010.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hackett PB, Largaespada DA, Switzer KC, Cooper LJ. Evaluating risks of insertional mutagenesis by DNA transposons in gene therapy. Transl Res. 2013;161(4):265–83. Epub 2013/01/15. 10.1016/j.trsl.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huls MH, Figliola MJ, Dawson MJ, Olivares S, Kebriaei P, Shpall EJ, et al. Clinical application of Sleeping Beauty and artificial antigen presenting cells to genetically modify T cells from peripheral and umbilical cord blood. J Vis Exp. 2013;(72):e50070 Epub 2013/02/15. 10.3791/50070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maiti SN, Huls H, Singh H, Dawson M, Figliola M, Olivares S, et al. Sleeping beauty system to redirect T-cell specificity for human applications. J Immunother. 2013;36(2):112–23. Epub 2013/02/05. 10.1097/CJI.0b013e3182811ce9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh H, Figliola MJ, Dawson MJ, Olivares S, Zhang L, Yang G, et al. Manufacture of Clinical-Grade CD19-Specific T Cells Stably Expressing Chimeric Antigen Receptor Using Sleeping Beauty System and Artificial Antigen Presenting Cells. PLoS One. 2013;8(5):e64138 Epub 2013/06/07. 10.1371/journal.pone.0064138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh H, Huls H, Kebriaei P, Cooper LJ. A new approach to gene therapy using Sleeping Beauty to genetically modify clinical-grade T cells to target CD19. Immunol Rev. 2014;257(1):181–90. Epub 2013/12/18. 10.1111/imr.12137 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh H, Figliola MJ, Dawson MJ, Huls H, Olivares S, Switzer K, et al. Reprogramming CD19-Specific T Cells with IL-21 Signaling Can Improve Adoptive Immunotherapy of B-Lineage Malignancies. Cancer Res. 2011;71(10):3516–27. Epub 2011/05/12. doi: 0008-5472.CAN-10-3843 [pii] 10.1158/0008-5472.CAN-10-3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh H, Manuri PR, Olivares S, Dara N, Dawson MJ, Huls H, et al. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;68(8):2961–71. Epub 2008/04/17. doi: 68/8/2961 [pii] 10.1158/0008-5472.CAN-07-5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bicocca VT, Chang BH, Masouleh BK, Muschen M, Loriaux MM, Druker BJ, et al. Crosstalk between ROR1 and the Pre-B cell receptor promotes survival of t(1;19) acute lymphoblastic leukemia. Cancer Cell. 2012;22(5):656–67. Epub 2012/11/17. 10.1016/j.ccr.2012.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suhoski MM, Golovina TN, Aqui NA, Tai VC, Varela-Rohena A, Milone MC, et al. Engineering artificial antigen-presenting cells to express a diverse array of co-stimulatory molecules. Mol Ther. 2007;15(5):981–8. Epub 2007/03/22. 10.1038/mt.sj.6300134 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Connor CM, Sheppard S, Hartline CA, Huls H, Johnson M, Palla SL, et al. Adoptive T-cell therapy improves treatment of canine non-Hodgkin lymphoma post chemotherapy. Sci Rep. 2012;2:249 Epub 2012/02/23. 10.1038/srep00249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deniger DC, Maiti SN, Mi T, Switzer KC, Ramachandran V, Hurton LV, et al. Activating and propagating polyclonal gamma delta T cells with broad specificity for malignancies. Clin Cancer Res. 2014;20(22):5708–19. 10.1158/1078-0432.CCR-13-3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deniger DC, Switzer K, Mi T, Maiti S, Hurton L, Singh H, et al. Bispecific T-cells expressing polyclonal repertoire of endogenous gammadelta T-cell receptors and introduced CD19-specific chimeric antigen receptor. Mol Ther. 2013;21(3):638–47. Epub 2013/01/09. 10.1038/mt.2012.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hudecek M, Schmitt TM, Baskar S, Lupo-Stanghellini MT, Nishida T, Yamamoto TN, et al. The B-cell tumor-associated antigen ROR1 can be targeted with T cells modified to express a ROR1-specific chimeric antigen receptor. Blood. 2010;116(22):4532–41. Epub 2010/08/13. 10.1182/blood-2010-05-283309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kebriaei P, Huls H, Jena B, Munsell M, Jackson R, Lee DA, et al. Infusing CD19-directed T cells to augment disease control in patients undergoing autologous hematopoietic stem-cell transplantation for advanced B-lymphoid malignancies. Hum Gene Ther. 2012;23(5):444–50. Epub 2011/11/24. 10.1089/hum.2011.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17(13):4550–7. Epub 2011/04/19. 10.1158/1078-0432.CCR-11-0116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Mitri D, Azevedo RI, Henson SM, Libri V, Riddell NE, Macaulay R, et al. Reversible senescence in human CD4+CD45RA+CD27- memory T cells. J Immunol. 2011;187(5):2093–100. Epub 2011/07/27. 10.4049/jimmunol.1100978 . [DOI] [PubMed] [Google Scholar]
- 43.Klebanoff CA, Gattinoni L, Restifo NP. CD8+ T-cell memory in tumor immunology and immunotherapy. Immunol Rev. 2006;211:214–24. Epub 2006/07/11. 10.1111/j.0105-2896.2006.00391.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rabinovich BA, Ye Y, Etto T, Chen JQ, Levitsky HI, Overwijk WW, et al. Visualizing fewer than 10 mouse T cells with an enhanced firefly luciferase in immunocompetent mouse models of cancer. Proc Natl Acad Sci U S A. 2008;105(38):14342–6. Epub 2008/09/17. 10.1073/pnas.0804105105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stonier SW, Schluns KS. Trans-presentation: a novel mechanism regulating IL-15 delivery and responses. Immunol Lett. 2010;127(2):85–92. Epub 2009/10/13. 10.1016/j.imlet.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stonier SW, Ma LJ, Castillo EF, Schluns KS. Dendritic cells drive memory CD8 T-cell homeostasis via IL-15 transpresentation. Blood. 2008;112(12):4546–54. Epub 2008/09/25. 10.1182/blood-2008-05-156307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manuri PV, Wilson MH, Maiti SN, Mi T, Singh H, Olivares S, et al. piggyBac transposon/transposase system to generate CD19-specific T cells for the treatment of B-lineage malignancies. Hum Gene Ther. 2010;21(4):427–37. Epub 2009/11/13. 10.1089/hum.2009.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17(10):1290–7. Epub 2011/09/20. 10.1038/nm.2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hudecek M, Sommermeyer D, Kosasih PL, Silva-Benedict A, Liu L, Rader C, et al. The nonsignaling extracellular spacer domain of chimeric antigen receptors is decisive for in vivo antitumor activity. Cancer Immunol Res. 2015;3(2):125–35. 10.1158/2326-6066.CIR-14-0127 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hudecek M, Lupo-Stanghellini MT, Kosasih PL, Sommermeyer D, Jensen MC, Rader C, et al. Receptor affinity and extracellular domain modifications affect tumor recognition by ROR1-specific chimeric antigen receptor T cells. Clin Cancer Res. 2013;19(12):3153–64. 10.1158/1078-0432.CCR-13-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kochenderfer JN, Wilson WH, Janik JE, Dudley ME, Stetler-Stevenson M, Feldman SA, et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood. 2010;116(20):4099–102. Epub 2010/07/30. doi: blood-2010-04-281931 [pii] 10.1182/blood-2010-04-281931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Straathof KC, Pule MA, Yotnda P, Dotti G, Vanin EF, Brenner MK, et al. An inducible caspase 9 safety switch for T-cell therapy. Blood. 2005;105(11):4247–54. Epub 2005/02/25. 10.1182/blood-2004-11-4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1 Dataset. Raw data as Excel spreadsheet.
(Fig 2 tab) Cell counts at designated times as measured by trypan blue exclusion. (Fig 3 tab) Normalized mRNA counts from NanoString array of T cells at day 29 of co-culture (top), surface phenotype of CAR+ T cells (middle), and multiparameter memory phenotype of T cells (bottom). (Fig 4 tab) MFI of IFNγ staining of CAR+ T cells following 6 hour co-culture with target cells. (Fig 5 tab) 4-hour chromium release assay of T cells co-cultured with target cells. (Fig 6 tab) BLI flux kinetics of Kasumi2-ffLuc-mKate cells following challenge with CAR+ T cells (top) and days of mouse euthanasia (bottom).
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.