Timing Is Everything: Coordinated Control of Host Shutoff by Influenza A Virus NS1 and PA-X Proteins (original) (raw)
Abstract
Like all viruses, influenza viruses (IAVs) use host translation machinery to decode viral mRNAs. IAVs ensure efficient translation of viral mRNAs through host shutoff, a process whereby viral proteins limit the accumulation of host proteins through subversion of their biogenesis. Despite its small genome, the virus deploys multiple host shutoff mechanisms at different stages of infection, thereby ensuring successful replication while limiting the communication of host antiviral responses. In this Gem, we review recent data on IAV host shutoff proteins, frame the outstanding questions in the field, and propose a temporally coordinated model of IAV host shutoff.
INFLUENZA VIRUS GENE EXPRESSION
Influenza virus (IAV) transcription and genome replication take place in the nucleus. The viral RNA-dependent RNA polymerase (RDRP) that performs these tasks is a trimeric complex composed of polymerase basic protein 2 (PB2), PB1, and polymerase acidic protein (PA) (for an excellent recent review, see reference 1). Following uncoating, each of the eight negative-sense single-stranded RNA L(−)ssRNA] IAV genome segments arrives in the nucleus bound to a single RDRP complex that is poised to begin transcription. Transcription begins when the endonucleolytic PA subunit of RDRP cleaves capped host pre-mRNAs at a location shortly downstream from the 5′ end. These short capped RNA oligonucleotides are used as primers to initiate synthesis of (+)ssRNA complementary to each (−)ssRNA genome segment. This process is commonly known as “cap snatching.” Following the transcription of each genome segment, the RDRP generates a 3′ poly(A) tail by “stuttering” on a short template polyuridine tract. In this way, the RDRP synthesizes viral mRNAs that are structurally similar to host mRNAs, with 5′ m7G caps and 3′ poly(A) tails, thereby protecting them from degradation by host enzymes and permitting efficient export to the cytoplasm and utilization of host translation machinery.
The earliest rounds of transcription, known as primary transcription, are performed solely by the RDRP that entered the nucleus of the newly infected cell bound to viral genome segments. As new viral proteins are synthesized and imported into the nucleus, they participate in viral genome replication, which requires newly synthesized RDRP subunits and nucleoprotein. In the next phase of the replication cycle, known as secondary transcription, the transcription of freshly replicated genomic segments generates large amounts of viral mRNAs. Finally, RDRP becomes fully committed to genome replication and at later stages of the infection cycle, continuous viral protein synthesis is maintained on mRNA templates produced during primary and secondary transcription phases. High rates of viral translation are needed to support robust viral genome replication and rapid assembly of large quantities of progeny virions. For this reason, viral mRNAs must persist for many hours in the cytoplasm, resisting degradation and remaining associated with polysomes. Accordingly, IAV has evolved ways to gain priority access of viral mRNAs to the translation machinery and to defeat host antiviral defenses that work, in part, by arresting translation.
THE DISCOVERY OF PA-X, A NEW HOST SHUTOFF ENDONUCLEASE
To ensure priority access to host translation machinery, viruses utilize host shutoff mechanisms that eliminate competition from cellular transcripts. Classic examples of such mechanisms include the poliovirus proteases 2Apro (2) and 3Cpro (3–5), which cleave essential host translation initiation factors that are dispensable for viral internal ribosome entry site-mediated initiation of translation. Another example is the host shutoff endonucleases of herpesviruses that destroy host mRNAs, thereby ensuring efficient cap-dependent translation of viral mRNAs (6). Until recently, no analogous cytoplasmic shutoff mechanism had been described for IAV, despite the fact that, similar to herpesviruses, IAV proteins are translated in a cap-dependent manner and therefore face direct competition from cellular transcripts for access to translation machinery.
In 2012, a new IAV protein with properties clearly relevant to host shutoff was discovered (7). Inspection of IAV nucleotide sequences that displayed lower-than-expected rates of synonymous substitutions led to the identification of a short open reading frame (dubbed X-ORF) in the PA coding sequence that could be accessed through +1 ribosomal frameshifting at Phe-191. This frameshifting event gives rise to a novel IAV protein called PA-X, which has the same endonuclease domain as PA but lacks the C-terminal region responsible for association with other RDRP subunits. Even though ribosomal frameshifting as a mechanism of extending the coding capacity of small genomes is well documented for many viruses, this was the first time this phenomenon was described for IAV. In vitro assays demonstrated that PA-X strongly inhibited protein production from ectopically coexpressed constructs by causing mRNA degradation. Introduction of single amino acid mutations known to disrupt PA endonuclease activity completely prevented PA-X-mediated mRNA degradation. Because it lacked the C-terminal domain required for binding of RDRP subunits and homing to the nucleus, PA-X was proposed to function as a novel cytosolic host shutoff endonuclease.
Inspection of IAV properties in animal infection models confirmed an important role for PA-X in host shutoff; recombinant IAVs with mutations in the PA ORF that inhibited +1 frameshifting at Phe-191 were less effective in suppressing host gene expression than were parental wild-type strains (7). PA-X-deficient viruses displayed higher virulence in mice without significantly affecting viral replication efficiency (7, 8). Further examination of these IAV frameshifting mutants in vitro revealed that PA-X accumulation was strongly linked to host shutoff. Recently, we demonstrated that PA-X expression is required for depletion of bulk poly(A) RNA in the cytoplasm of infected cells and the relocalization of poly(A) binding protein (PABP) to the nucleus at later times postinfection (9). Taken together, these findings strongly support the idea that PA-X is a bona fide host shutoff endonuclease. Nevertheless, the molecular mechanisms whereby PA-X exerts its effects remain largely uncharacterized. For example, it is not known whether PA-X has any RNA sequence or structural specificity. During cap snatching, the cap-binding activity of the PB2 subunit of the viral polymerase determines the positioning of the PA endonuclease domain to cleave pre-mRNAs 10 to 15 nucleotides downstream from the 5′ end (10, 11). It remains to be determined whether PA-X might cleave specific RNA sequences or structural motifs or utilize RNA-binding proteins to guide the selection of cleavage sites. Many outstanding questions remain to be addressed. Does PA-X selectively target mRNAs? Does it discriminate between viral and host mRNAs? If it does, then what is the basis for this discrimination? The X-ORF does not resemble any known protein and yet is strongly conserved among IAV isolates. Desmet et al. demonstrated that the X-ORF potentiates the shutoff function of the N-terminal endonuclease domain, suggesting that it may play a role in target selection (12). The potential effects of the X-ORF on protein-protein interactions, stability, or subcellular localization of PA-X remain to be determined.
Previous studies of other viral shutoff endonucleases may provide some important clues to the function of PA-X. For example, the herpes simplex virus shutoff endonuclease vhs is recruited to mRNAs through protein-protein interactions with eukaryotic translation initiation factor 4H, which influences the position of mRNA cleavage (13). Remarkably, this endonuclease degrades both host and viral mRNAs, thereby sharpening transitions between distinct viral gene expression programs. Another herpesviral host shutoff endonuclease, the Kaposi's sarcoma-associated herpesvirus SOX protein, appears to have much in common with PA-X. Both members of the PD-(D/E)XK nuclease superfamily, SOX and PA-X cause depletion of cytoplasmic poly(A) RNA and nuclear relocalization of PABP. The relocalized PABP in SOX-expressing cells induces aberrant hyperadenylation of nuclear RNAs that prevents their export to the cytoplasm (14). We observed a strong increase in the nuclear poly(A) RNA signal in IAV-infected cells at late times postinfection (9). It is possible that nuclear PABP relocalization could trigger hyperadenylation in the nuclei of IAV-infected cells as well. The mechanism of SOX-mediated mRNA destruction has been partially elucidated; SOX induces the cleavage of mRNAs associated with polysomes, followed by processive 5′-3′ degradation by host exonuclease Xrn1 (15, 16). It is not yet known whether PA-X could function in a similar manner. The discovery of PA-X has rekindled interest in understanding IAV host shutoff. Further characterization of the biogenesis, properties, and mechanism of action of PA-X should firmly integrate this protein into the model of influenza virus host shutoff and help us better understand how host shutoff relates to viral adaptation and pathogenesis.
PROPOSED MODEL OF COORDINATED CONTROL OF HOST SHUTOFF BY INFLUENZA A VIRUS
(i) The early, nuclear phase of host shutoff.
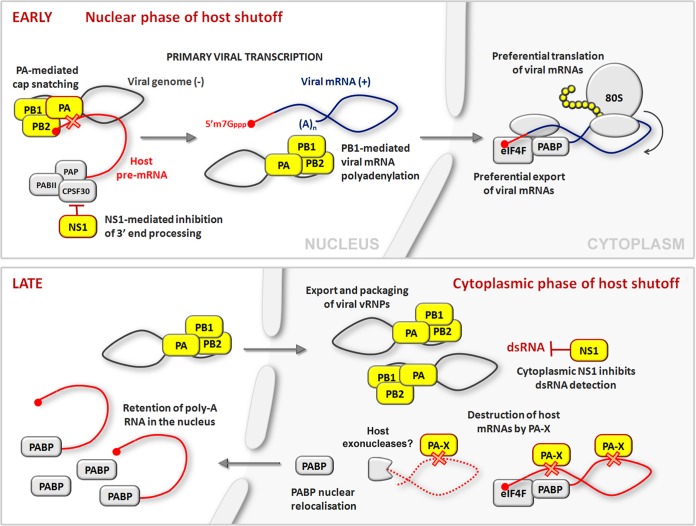
Host shutoff is observed in IAV-infected cells long before PA-X is made, at the earliest stages of infection, coincident with the transcription of viral messages (Fig. 1). At this stage, nascent host transcripts are targeted through at least two mechanisms that act in the nucleus. The first mechanism, cap snatching by viral RDRP, destroys host pre-mRNAs to supply capped primers for viral transcription. The second mechanism involves viral nonstructural protein 1 (NS1), which is capable of binding and inhibiting cellular cleavage and polyadenylation factor 30 (CPSF30) (17). CPSF30 is essential for 3′-end processing and subsequent export of cellular mRNAs, and its inhibition by NS1 effectively blocks polyadenylation and nuclear export of host transcripts. Acting together, these two shutoff mechanisms eliminate competition from nascent host transcripts for nuclear export and available translation factors. In addition, these same mechanisms ensure that host cells are unable to mount antiviral responses that require new gene expression, such as type I interferon production or expression of interferon-stimulated genes. Notably, CPSF30 binding is characteristic of human IAVs and is absent from most animal-adapted strains. Meanwhile, viral mRNAs remain impervious to NS1 because their poly(A) tails are generated by the RDRP, not host machinery. Additional mechanisms of host shutoff, including the destruction of host cytoplasmic mRNAs by PA-X, must play more important roles in these strains.
FIG 1.
A temporal model of influenza A virus host shutoff. See the text for details. dsRNA, double-stranded RNA; eIF4F, eukaryotic translation initiation factor 4F.
(ii) The late, cytoplasmic phase of host shutoff.
As infection progresses, the focus of host shutoff turns away from the nucleus and toward the cytoplasm. This makes some intuitive sense, because all viral mRNAs have been synthesized and transported to the cytoplasm by this time, so the challenge remaining is to maintain priority access to the translation machinery. NS1 accumulates in the nucleus early in infection but is later found in the cytoplasm as well, where it is retasked with ensuring the efficient translation of viral gene products by (i) blocking PKR-mediated sensing of viral double-stranded RNA and translation arrest (18, 19) and (ii) supporting the assembly of translation initiation complexes in the 5′-untranslated regions of viral mRNAs (reviewed in reference 20). Meanwhile, low-efficiency ribosomal frameshifting on mRNA transcribed from IAV genome segment 3 allows slow accumulation of PA-X during infection, likely limiting its effects until later stages. As infection progresses to later stages, continued robust translation of a limited pool of viral mRNAs is likely assisted by PA-X, which, by further depleting host mRNA pools, ensures priority access to the translation machinery. Accumulation of massive amounts of viral materials at later times postinfection provides ample pathogen-associated molecular patterns that could be recognized by cytoplasmic receptors. Concerted action of multiple host shutoff mechanisms ensures that the host cell is unable to initiate an effective antiviral program. Indeed, after nuclear export of viral progeny nucleoproteins, many infected cells have severely depleted levels of total poly(A) RNA in the cytoplasm. Ultimately, IAV triggers apoptosis of the host cell and could block new protein synthesis completely during last few hours of the replication cycle.
SUMMARY
By creating PA-X, the virus effectively repurposes the catalytic activity of the PA subunit to provide an important auxiliary host shutoff mechanism that peaks at later stages of the replication cycle. Although PA-X is not absolutely required for viral replication, it is clearly relevant to IAV pathogenesis in in vivo infection models. Unlike NS1-mediated blockade of CPSF function, which is one of the hallmarks of human adaptation, PA-X is remarkably conserved among all IAV strains. This indicates that PA-X may be essential for IAV biology in its natural host. At the same time, it is clear that the virus has evolved to temper the expression of this potent host shutoff factor, only allowing it to accumulate late in infection. This clearly suggests that PA-X catalytic activity, if left unchecked, may be toxic to the host cell, limiting the assembly and spread of viral progeny.
ACKNOWLEDGMENTS
We thank members of the McCormick lab for critical reading of the manuscript. We apologize to all of our colleagues whose work could not be cited because of length restrictions.
Work in the McCormick laboratory relevant to this article was supported by Canadian Institutes of Health Research operating grant MOP-136817.
REFERENCES
- 1.Eisfeld AJ, Neumann G, Kawaoka Y. 2015. At the centre: influenza A virus ribonucleoproteins. Nat Rev Microbiol 13:28–41. doi: 10.1038/nrmicro3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ventoso I, MacMillan SE, Hershey JW, Carrasco L. 1998. Poliovirus 2A proteinase cleaves directly the eIF-4G subunit of eIF-4F complex. FEBS Lett 435:79–83. doi: 10.1016/S0014-5793(98)01027-8. [DOI] [PubMed] [Google Scholar]
- 3.Joachims M, Van Breugel PC, Lloyd RE. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol 73:718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuyumcu-Martinez NM, Joachims M, Lloyd RE. 2002. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J Virol 76:2062–2074. doi: 10.1128/jvi.76.5.2062-2074.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE. 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol Cell Biol 24:1779–1790. doi: 10.1128/MCB.24.4.1779-1790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elgadi MM, Hayes CE, Smiley JR. 1999. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J Virol 73:7153–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao H, Sun Y, Hu J, Qi L, Wang J, Xiong X, Wang Y, He Q, Lin Y, Kong W, Seng LG, Sun H, Pu J, Chang KC, Liu X, Liu J. 2015. The contribution of PA-X to the virulence of pandemic 2009 H1N1 and highly pathogenic H5N1 avian influenza viruses. Sci Rep 5:8262. doi: 10.1038/srep08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khaperskyy DA, Emara MM, Johnston BP, Anderson P, Hatchette TF, McCormick C. 2014. Influenza A virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog 10:e1004217. doi: 10.1371/journal.ppat.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plotch SJ, Bouloy M, Krug RM. 1979. Transfer of 5′-terminal cap of globin mRNA to influenza viral complementary RNA during transcription in vitro. Proc Natl Acad Sci U S A 76:1618–1622. doi: 10.1073/pnas.76.4.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plotch SJ, Bouloy M, Ulmanen I, Krug RM. 1981. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 12.Desmet EA, Bussey KA, Stone R, Takimoto T. 2013. Identification of the N-terminal domain of the influenza virus PA responsible for the suppression of host protein synthesis. J Virol 87:3108–3118. doi: 10.1128/JVI.02826-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng P, Everly DNJ, Read GS. 2001. mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor. J Virol 75:10272–10280. doi: 10.1128/JVI.75.21.10272-10280.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar GR, Glaunsinger BA. 2010. Nuclear import of cytoplasmic poly(A) binding protein restricts gene expression via hyperadenylation and nuclear retention of mRNA. Mol Cell Biol 30:4996–5008. doi: 10.1128/MCB.00600-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Covarrubias S, Gaglia MM, Kumar GR, Wong W, Jackson AO, Glaunsinger BA. 2011. Coordinated destruction of cellular messages in translation complexes by the gammaherpesvirus host shutoff factor and the mammalian exonuclease Xrn1. PLoS Pathog 7:e1002339. doi: 10.1371/journal.ppat.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaglia MM, Covarrubias S, Wong W, Glaunsinger BA. 2012. A common strategy for host RNA degradation by divergent viruses. J Virol 86:9527–9530. doi: 10.1128/JVI.01230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell 1:991–1000. doi: 10.1016/S1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 18.Lu Y, Wambach M, Katze MG, Krug RM. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 19.Tan SL, Katze MG. 1998. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J Interferon Cytokine Res 18:757–766. doi: 10.1089/jir.1998.18.757. [DOI] [PubMed] [Google Scholar]
- 20.Yángüez E, Nieto A. 2011. So similar, yet so different: selective translation of capped and polyadenylated viral mRNAs in the influenza virus infected cell. Virus Res 156:1–12. doi: 10.1016/j.virusres.2010.12.016. [DOI] [PubMed] [Google Scholar]