Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets (original) (raw)
. Author manuscript; available in PMC: 2016 May 21.
Summary
Cells, the basic units of biological structure and function, vary broadly in type and state. Single-cell genomics can characterize cell identity and function, but limitations of ease and scale have prevented its broad application. Here we describe Drop-Seq, a strategy for quickly profiling thousands of individual cells by separating them into nanoliter-sized aqueous droplets, associating a different barcode with each cell’s RNAs, and sequencing them all together. Drop-Seq analyzes mRNA transcripts from thousands of individual cells simultaneously while remembering transcripts’ cell of origin. We analyzed transcriptomes from 44,808 mouse retinal cells and identified 39 transcriptionally distinct cell populations, creating a molecular atlas of gene expression for known retinal cell classes and novel candidate cell subtypes. Drop-Seq will accelerate biological discovery by enabling routine transcriptional profiling at single-cell resolution.
Introduction
Individual cells are the building blocks of tissues, organs, and organisms. Each tissue contains cells of many types, and cells of each type can switch among biological states. In most biological systems, our knowledge of cellular diversity is incomplete; for example, the cell-type complexity of the brain is unknown and widely debated (Luo et al., 2008; Petilla Interneuron Nomenclature et al., 2008). To understand how complex tissues work, it will be important to learn the functional capacities and responses of each cell type.
A major determinant of each cell’s function is its transcriptional program. Recent advances now enable mRNA-seq analysis of individual cells (Tang et al., 2009). However, methods of preparing cells for profiling have been applicable in practice to just hundreds (Hashimshony et al., 2012; Picelli et al., 2013) or (with automation) a few thousand cells (Jaitin et al., 2014), typically after first separating the cells by flow sorting (Shalek et al., 2013) or microfluidics (Shalek et al., 2014) and then amplifying each cell’s transcriptome separately. Fast, scalable approaches are needed to characterize complex tissues with many cell types and states, under diverse conditions and perturbations.
Here we describe Drop-Seq, a method to analyze mRNA expression in thousands of individual cells by encapsulating cells in tiny droplets for parallel analysis. Droplets – nanoliter-scale aqueous compartments formed by precisely combining aqueous and oil flows in a microfluidic device (Thorsen et al., 2001; Umbanhowar, 2000) – have been used as tiny reaction chambers for PCR (Hindson et al., 2011; Vogelstein and Kinzler, 1999) and reverse transcription (Beer et al., 2008). We sought here to use droplets to compartmentalize cells into nanoliter-sized reaction chambers for analysis of all of their RNAs. A basic challenge of using droplets for transcriptomics is to retain a molecular memory of the identity of the cell from which each mRNA transcript was isolated. To accomplish this, we developed a molecular barcoding strategy to remember the cell-of-origin of each mRNA. We critically evaluate Drop-Seq, then use it to profile cell states along the cell cycle. We then applied it to a complex neural tissue, mouse retina, and from 44,808 cell profiles retrieved 39 distinct populations, each corresponding to one or a group of closely related cell types. Our results demonstrate how large-scale single-cell analysis can help deepen our understanding of the biology of complex tissues and cell populations.
Results
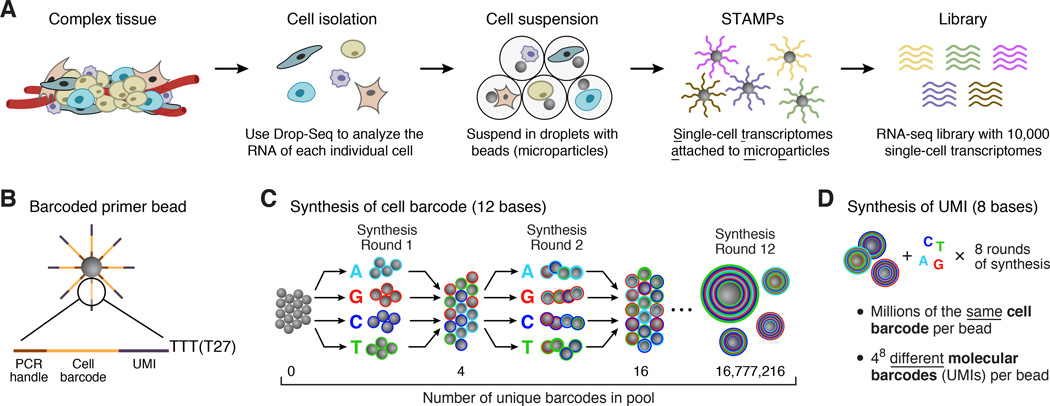
Drop-Seq consists of the following steps (Figure 1A): (1) prepare a single-cell suspension from a tissue; (2) co-encapsulate each cell with a distinctly barcoded microparticle (bead) in a nanoliter-scale droplet; (3) lyse cells after they have been isolated in droplets; (4) capture a cell’s mRNAs on its companion microparticle, forming STAMPs (Single-cell Transcriptomes Attached to Microparticles); (5) reverse-transcribe, amplify, and sequence thousands of STAMPs in one reaction; and (6) use the STAMP barcodes to infer each transcript’s cell of origin.
Figure 1. Molecular barcoding of cellular transcriptomes in droplets.
(A) Drop-Seq barcoding schematic. A complex tissue is dissociated into individual cells, which are then encapsulated in droplets together with microparticles (gray circles) that deliver barcoded primers. Each cell is lysed within a droplet; its mRNAs bind to the primers on its companion microparticle. The mRNAs are reverse-transcribed into cDNAs, generating a set of beads called “single-cell transcriptomes attached to microparticles” (STAMPs). The barcoded STAMPs can then be amplified in pools for high-throughput mRNA-seq to analyze any desired number of individual cells.
(B) Sequence of primers on the microparticle. The primers on all beads contain a common sequence (“PCR handle”) to enable PCR amplification after STAMP formation. Each microparticle contains more than 108 individual primers that share the same “cell barcode” (panel C) but have different unique molecular identifiers (UMIs), enabling mRNA transcripts to be digitally counted (panel D). A 30 bp oligo dT sequence is present at the end of all primer sequences for capture of mRNAs.
(C) Split-and-pool synthesis of the cell barcode. To generate the cell barcode, the pool of microparticles is repeatedly split into four equally sized oligonucleotide synthesis reactions, to which one of the four DNA bases is added, and then pooled together after each cycle, in a total of 12 split-pool cycles. The barcode synthesized on any individual bead reflects that bead’s unique path through the series of synthesis reactions. The result is a pool of microparticles, each possessing one of 412 (16,777,216) possible sequences on its entire complement of primers (see also Figure S1).
(D) Synthesis of a unique molecular identifier (UMI). Following the completion of the “split-and-pool” synthesis cycles, all microparticles are together subjected to eight rounds of degenerate synthesis with all four DNA bases available during each cycle, such that each individual primer receives one of 48 (65,536) possible sequences (UMIs).
A split-pool synthesis approach to generate large numbers of distinctly barcoded beads
To deliver large numbers of distinctly barcoded primer molecules into individual droplets, we use microparticles (beads). We synthesized oligonucleotide primers directly on beads (from 5’ to 3’, yielding free 3’ ends available for enzymatic priming). Each oligonucleotide is composed of four parts (Figure 1B): (1) a constant sequence (identical on all primers and beads) for use as a priming site for downstream PCR and sequencing; (2) a “cell barcode” (identical across all the primers on the surface of any one bead, but different from the cell barcodes on other beads); (3) a Unique Molecular Identifier (UMI) (different on each primer, to identify PCR duplicates) (Kivioja et al., 2012); and (4) an oligo-dT sequence for capturing polyadenylated mRNAs and priming reverse transcription.
To efficiently generate massive numbers of beads, each with a distinct barcode, we developed a “split-and-pool” DNA synthesis strategy (Figure 1C). A pool of millions of microparticles is divided into four equally sized groups; a different DNA base (A, G, C, or T) is then added to each. All microparticles are then re-pooled, mixed, and re-split at random into another four groups, and then a different DNA base (A, G, C, or T) is added to each of the four new groups. After 12 cycles of split-and-pool DNA synthesis, the primers on any given microparticle possess the same one of 412 = 16,777,216 possible 12-bp barcodes, but different microparticles have different sequences (Figure 1C). The entire microparticle pool then undergoes eight rounds of degenerate oligonucleotide synthesis to generate the UMI on each oligo (Figure 1D); finally, an oligo-dT sequence (T30) is synthesized on the 3’ end of all oligos on all beads.
To confirm that we could distinguish RNAs based on attached barcodes, we reverse-transcribed a pool of synthetic RNAs onto 11 microparticles and sequenced the resulting cDNAs (Figure S1A and Extended Experimental Procedures); 11 microparticle barcodes each constituted 3.5% – 14% of the resulting sequencing reads, whereas the next-most-abundant 12-mer constituted only 0.06% (Figure S1A). These results suggested that the microparticle-of-origin for most cDNAs can be recognized by sequencing. We also found that each bead contained more than 108 barcoded primer sites and that the sequence complexity of the barcodes approached theoretical limits (Figures S1B and S1C, Extended Experimental Procedures).
Microfluidics device for co-encapsulating cells with beads
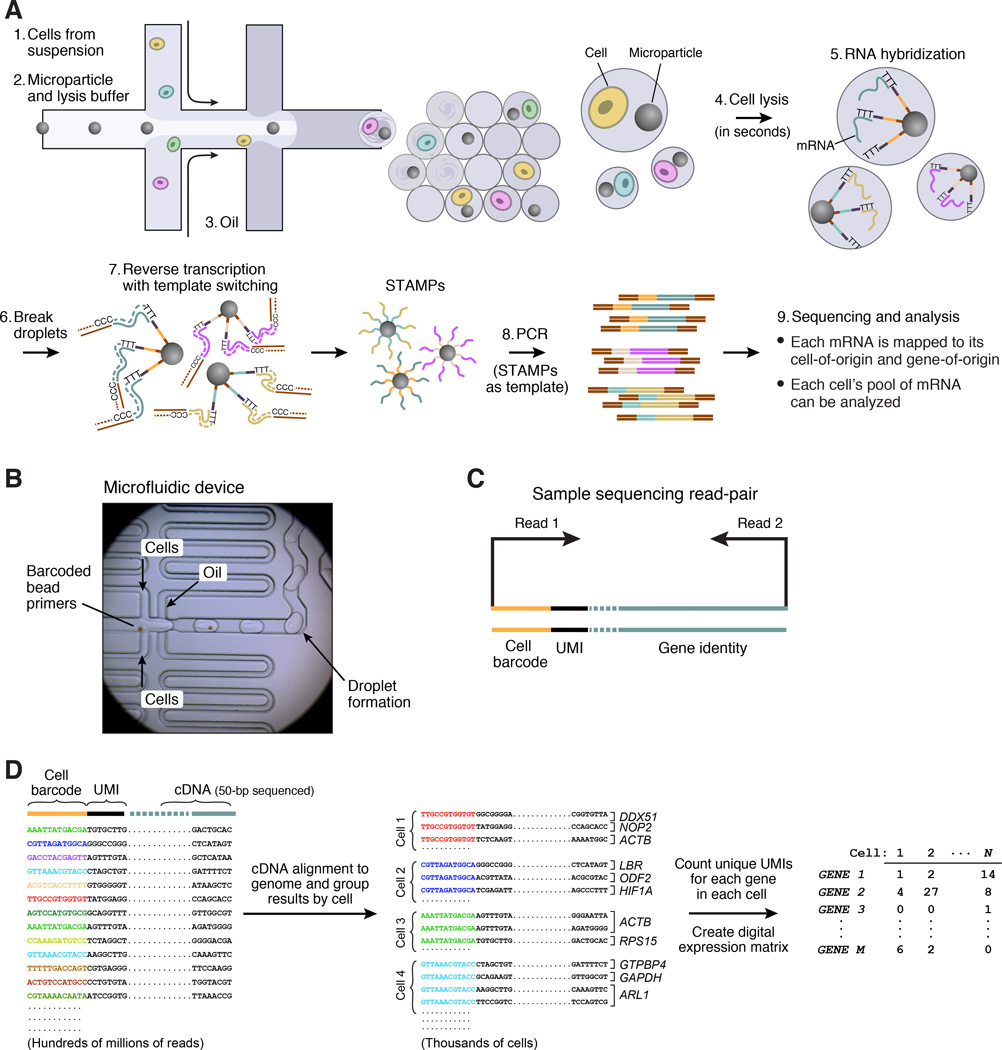
We designed a microfluidic “co-flow” device (Utada et al., 2007) to co-encapsulate cells with barcoded microparticles (Figures 2A, S2 and DataFile 1). This device quickly co-flows two aqueous solutions across an oil channel to form more than 100,000 nanoliter-sized droplets per minute. One flow contains the barcoded microparticles suspended in a lysis buffer; the other flow contains a cell suspension (Figure 2A, left, Figure 2B). The number of droplets created greatly exceeds the number of beads or cells injected, so that a droplet will generally contain zero or one cells, and zero or one beads. Millions of nanoliter-sized droplets are generated per hour, of which thousands contain both a bead and a cell (Movie S1). STAMPs are produced in the subset of droplets that contain both a bead and a cell.
Figure 2. Extraction and processing of single-cell transcriptomes by Drop-Seq.
(A) Schematic of single-cell mRNA-Seq library preparation with Drop-Seq. A custom-designed microfluidic device joins two aqueous flows before their compartmentalization into discrete droplets. One flow contains cells, and the other flow contains barcoded primer beads suspended in a lysis buffer. Immediately following droplet formation, the cell is lysed and releases its mRNAs, which then hybridize to the primers on the microparticle surface. The droplets are broken by adding a reagent to destabilize the oil-water interface (Experimental Procedures), and the microparticles collected and washed. The mRNAs are then reverse-transcribed in bulk, forming STAMPs, and template switching is used to introduce a PCR handle downstream of the synthesized cDNA (Zhu et al., 2001).
(B) Microfluidic device used in Drop-Seq. Beads (brown in image), suspended in a lysis agent, enter the device from the central channel; cells enter from the top and bottom. Laminar flow prevents mixing of the two aqueous inputs prior to droplet formation (see also Movie S1). Schematics of the device design and how it is operated can be found in Figure S2.
(C) Molecular elements of a Drop-Seq sequencing library. The first read yields the cell barcode and UMI. The second, paired read interrogates sequence from the cDNA (50 bp is typically sequenced); this sequence is then aligned to the genome to determine a transcript’s gene of origin.
(D) In silico reconstruction of thousands of single-cell transcriptomes. Millions of paired-end reads are generated from a Drop-Seq library on a high-throughput sequencer. The reads are first aligned to a reference genome to identify the gene-of-origin of the cDNA. Next, reads are organized by their cell barcodes, and individual UMIs are counted for each gene in each cell (Extended Experimental Procedures). The result, shown at far right, is a “digital expression matrix” in which each column corresponds to a cell, each row corresponds to a gene, and each entry is the integer number of transcripts detected from that gene, in that cell.
Sequencing and analysis of many STAMPs in a single reaction
To efficiently process thousands of STAMPs at once, we break droplets, collect the mRNA-bound microparticles, and reverse-transcribe the mRNAs (from the microparticle-attached primers) together in one reaction, forming covalent, stable STAMPs (Figure 2A, step 7, and Experimental Procedures). A scientist can then select any desired number of STAMPs for the preparation of 3’-end digital expression libraries (Figure 2C, Experimental Procedures). We sequence the resulting molecules from each end (Figure 2C) using high-capacity parallel sequencing. We digitally count the number of mRNA transcripts of each gene ascertained in each cell, using the UMIs to avoid double-counting sequence reads that arose from the same mRNA transcript. We thereby create a matrix of digital gene-expression measurements (one measurement per gene per cell) for further analysis (Figure 2D, Experimental Procedures).
The single cell accuracy and sensitivity of Drop-Seq libraries
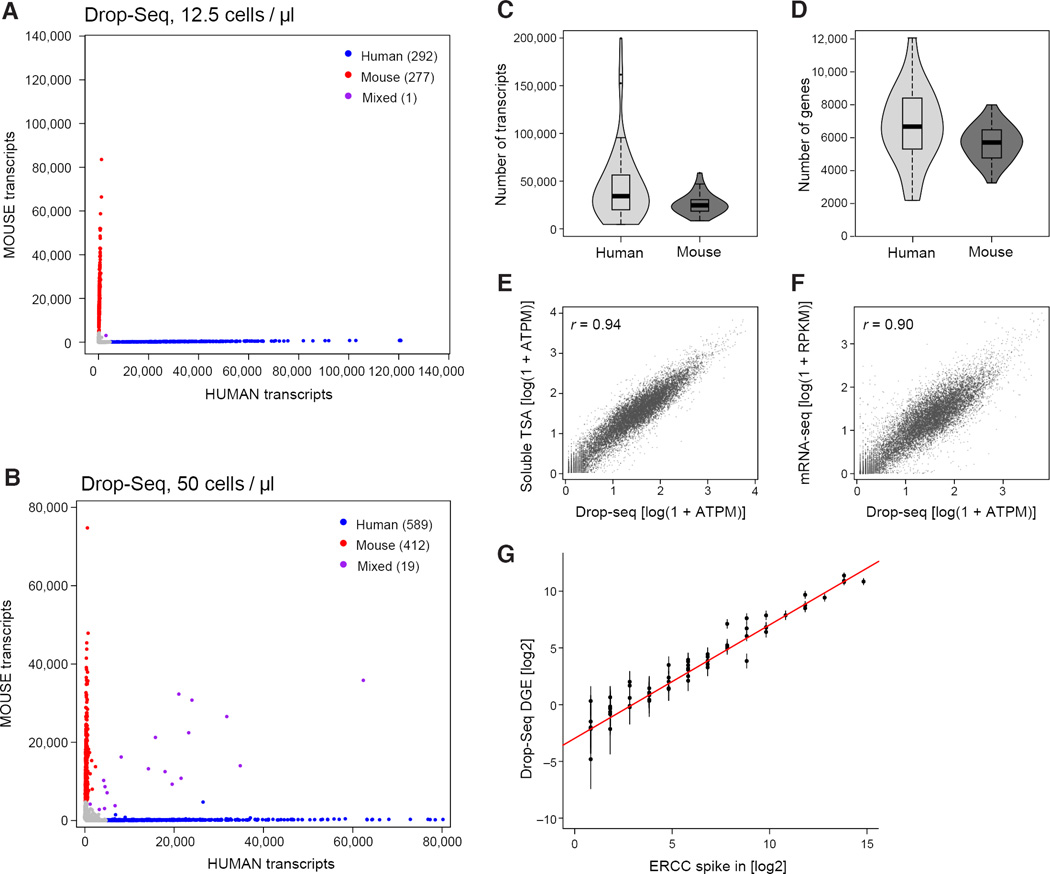
To measure the accuracy with which Drop-Seq remembers the cell-of-origin of each mRNA, we analyzed mixtures of cultured human (HEK) and mouse (3T3) cells, scoring the numbers of human and mouse transcripts that associated with each cell barcode (Figure 3A, 3B, S3A). We found that the individual STAMPs created by Drop-Seq were highly organism-specific (Figure 3A, 3B), indicating high single-cell integrity of the libraries. At saturating levels of sequence coverage, we detected an average of 44,295 mRNA transcripts from 6,722 genes in HEK cells and 26,044 transcripts from 5,663 genes in 3T3 cells (Figures 3C and 3D).
Figure 3. Critical evaluation of Drop-Seq using species-mixing experiments.
(A,B) Drop-Seq analysis of mixutres of mouse and human cells. Mixtures of human (HEK) and mouse (3T3) cells were analyzed by Drop-Seq at the concentrations shown. The scatter plot shows the number of human and mouse transcripts associating to each STAMP. Blue dots indicate STAMPs that were designated from these data as human-specifiic (average of 99% human transcripts); red dots indicate STAMPs that were mouse-specific (average 99%). At the lower cell concentration, one STAMP barcode (of 570) associated with a mixture of human and mouse transcripts (panel A, purple). At the higher cell concentration, about 1.9% of STAMP barcodes associated with mouse-human mixtures (panel B). Data for other cell concentrations and a different single-cell analysis platform are in Figures S3B and S3C.
(C,D) Sensitivity analysis of Drop-Seq at high read-depth. Violin plots show the distribution of the number of transcripts (C, scored by UMIs) and genes (D) detected per cell for 54 HEK (human) STAMPs (blue) and 28 3T3 (mouse) STAMPs (green) that were sequenced to a mean read depth of 737,240 high-quality aligned reads per cell.
(E,F) Correlation between gene expression measurements in Drop-Seq and non-single-cell RNA-seq methods. Comparison of Drop-Seq gene expression measurements (averaged across 550 STAMPs) to measurements from bulk RNA analyzed by: (E) an in-solution template switch amplification (TSA) procedure similar to Smart-Seq2 (Picelli et al., 2013) (Extended Experimental Procedures); and (F) Illumina TruSeq mRNA-Seq. All comparisons involve RNA derived from the same cell culture flask (3T3 cells). All expression counts were converted to average transcripts per million (ATPM) and plotted as log (1+ATPM).
(G) Quantitation of Drop-Seq capture efficiency by ERCC spike-ins. Drop-Seq was performed with ERCC control synthetic RNA at an estimated concentration of 100,000 ERCC RNA molecules per droplet. 84 beads were sequenced at a mean depth of 2.4 million reads, aligned to the ERCC reference sequences, and UMIs counted for each ERCC species, after applying a stringent down-correction for potential sequencing errors (Table S1 and Extended Experimental Procedures). For each ERCC RNA species above an average concentration of one molecule per droplet, the predicted number of molecules per droplet was plotted in log space (x-axis), versus the actual number of molecules detected per droplet by Drop-Seq, also in log space (y-axis). The intercept of a regression line, constrained to have a slope of 1 and fitted to the seven highest points, was used to estimate a conversion factor (0.128). A second estimation, using the average number of detected transcripts divided by the number of ERCC molecules used (100,000), yielded a conversion factor of 0.125.
To understand how Drop-Seq libraries compare to other single-cell methods, we used three quality metrics: (i) the frequency of cell-cell doublets; (ii) single-cell purity; and (iii) transcript capture rates.
Cell doublets
One potential mode of failure in any single-cell method involves cells that stick together or happen to otherwise be co-isolated for library preparation. In Drop-Seq, across four conditions spanning 12.5 cells/µL to 100 cells/µL, the fraction of species-mixed STAMPs correlated with cell concentration (Figure 3A, 3B, S3B; Experimental Procedures), with cell doublet estimates ranging from 0.36% to 11.3% for the various cell concentrations tested (under the assumption that human-mouse doublets account for half of all doublets). This reflects the greater chance at higher cell concentrations that a droplet could encapsulate multiple cells. By comparison, previous studies that used FACS (Jaitin et al., 2014) or a commercial microfluidics platform (Shalek et al., 2014) to isolate single cells reported doublet rates of 2.3% and 11% respectively, based upon examining microscopy images of captured cells. In analyzing the above mouse-human cell suspension mixture in a commercial microfluidics system (Fluidigm C1), we found that 30% of the resulting libraries in that experiment were species-mixed (Figure S3C); about one-third of these doublets were visible in the microscopy images.
Single-cell impurity
Species-mixing experiments enabled us to measure single-cell purity across thousands of libraries prepared at different cell concentrations. We found that purity was strongly related to cell concentration, ranging from 98.8% at 12.5 cells / µL to 90.4% at 100 cells / µL (Figure S3B). The largest source of single-cell impurity appeared to be ambient RNA that is present in the cell suspension (a first step of almost all single-cell methods) and presumably results from cells that are damaged during preparation (Figure S3D). We measured a mean single-cell purity of 95.8% for the same cell mixtures in the Fluidigm C1 system (Figure S3C), similar to Drop-Seq at 50 cells / µL.
Conversion efficiency
The use of synthetic RNA “spike-in” controls at known concentrations, together with UMIs to avoid double-counting, allows estimation of capture rates for digital single-cell expression technologies (Brennecke et al., 2013; Islam et al., 2014). We identified evidence that PCR and sequencing errors inflate the numbers of apparently unique UMIs (Table S1 and Extended Experimental Procedures), so we developed a more conservative estimation method than has been used in earlier studies (Islam et al., 2014); in our approach, we collapse similar UMI sequences into a single count. Using this approach we calculated a capture rate of 12.8% for Drop-Seq (Figure 3G). We corroborated this estimate by making independent digital expression measurements (on bulk RNA from 50,000 HEK cells) on 10 genes using droplet digital PCR (ddPCR) (Hindson et al., 2011), calculating an average conversion efficiency of 10.7% (Figures S4A, S4B, and S4C).
To further evaluate how the digital transcriptomes ascertained by Drop-Seq related to the underlying mRNA content of cells, we compared Drop-Seq log-expression measurements to those made by a commonly used in-solution amplification process, finding strong correlation (r = 0.94, Figure 3E), though Drop-Seq ascertained GC-rich transcripts at a lower rate (Figure S4D). We also compared Drop-Seq single-cell log-expression measurements with measurements from bulk mRNA-seq, observing a correlation of _r_=0.90 (Figures 3F, S4E, and S4F).
Cell states: Drop-Seq analysis of the cell cycle
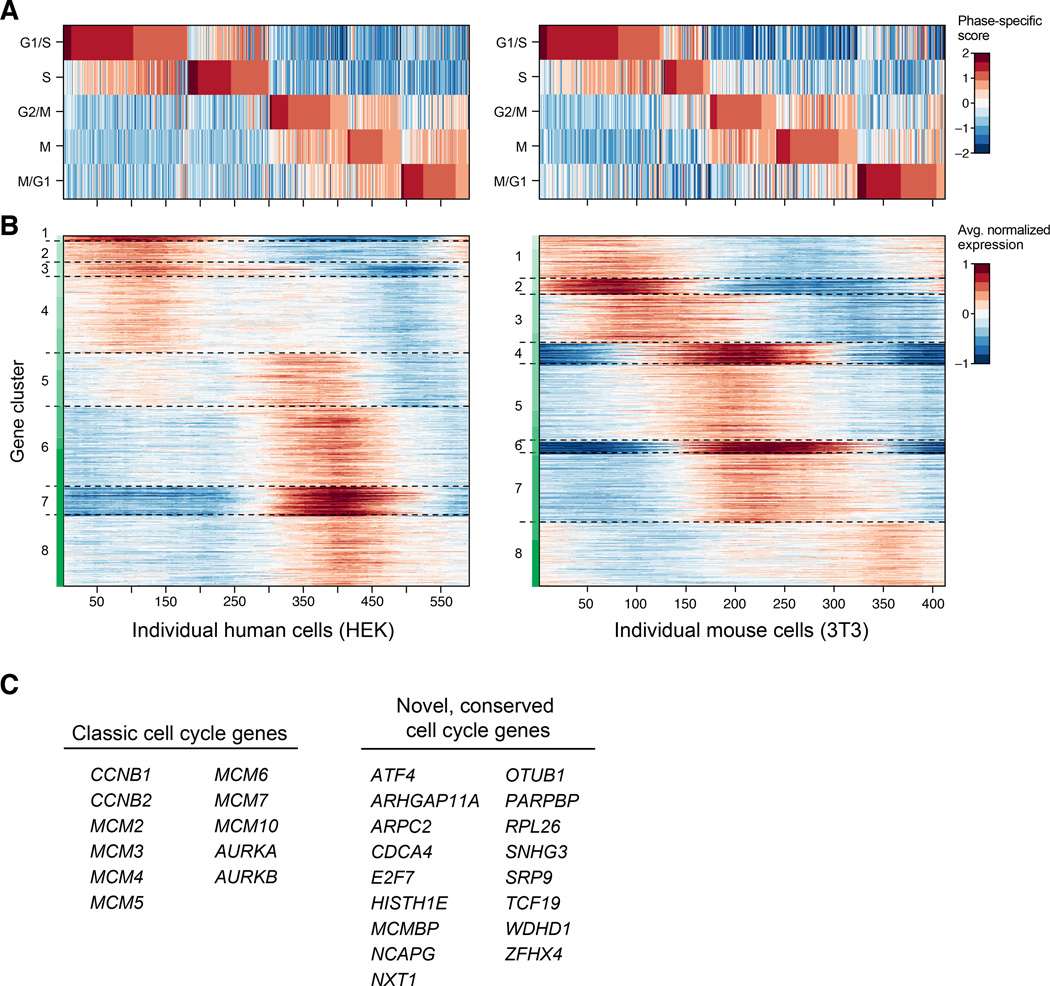
To evaluate the visibility of cell states in Drop-Seq, we first examined cell-to-cell variation among the 589 HEK and 412 3T3 STAMPs shown in Figure 3B. Both cultures consisted of asynchronously dividing cells; principal components analysis (PCA) of the single-cell expression profiles showed the top principal components to be dominated by genes with roles in protein synthesis, growth, DNA replication, and other aspects of the cell cycle. We inferred the cell-cycle phase of each of the 1,001 cells by scoring for gene sets (signatures) reflecting five phases of the cell cycle previously characterized in chemically synchronized cells (G1/S, S, G2/M, M, and M/G1) (Figure 4A, Table S2) (Whitfield et al., 2002). We identified 544 human and 668 mouse genes with expression patterns that varied along the cell cycle (at a false discovery rate of 5%; Experimental Procedures) (Figure 4B), including 200 orthologous gene pairs (p < 10−65 by hypergeometric test). Of these orthologous gene pairs, most (82.5%) have been previously annotated as related to the cell cycle in at least one species; among the other 17.5%, we found some that would be expected to show cell cycle variation (e.g. E2F7 and PARPBP) and many that to our knowledge were not previously connected to the cell cycle (Figure 4C and Table S3). Single-cell analysis at this scale enabled characterization of cell-cycle gene expression without chemical synchronization and at high temporal resolution.
Figure 4. Cell-cycle analysis of HEK and 3T3 cells analyzed by Drop-Seq.
(A) Cell-cycle state of 589 HEK cells (left) and 412 3T3 cells (right) measured by Drop-Seq. Cells were assessed for their progression through the cell cycle by comparison of each cell’s global pattern of gene expression with gene sets known to be enriched in one of five phases of the cycle (horizontal rows). A phase-specific score was calculated for each cell across these five phases (Extended Experimental Procedures), and the cells ordered by their phase scores.
(B) Discovery of cell cycle regulated genes. Heat map showing the average normalized expression of 544 human and 668 mouse genes found to be regulated by the cell cycle. Maximal and minimal expression was calculated for each gene across a sliding window of the ordered cells, and compared with shuffled cells to obtain a false discovery rate (FDR) (Experimental Procedures). The plotted genes (FDR threshold of 5%) were then clustered by k-means analysis to identify sets of genes with similar expression patterns. Cluster boundaries are represented by dashed gray lines.
(C) Representative cell cycle regulated genes discovered by Drop-Seq. Selected genes that were found to be cell cycle regulated in both the HEK and 3T3 cell sets. Left, genes that are well-known to be cell cycle regulated. Right, some genes identified in this analysis that were not previously known to be associated with the cell cycle (Experimental Procedures). A complete list of cell cycle regulated genes can be found in Table S2.
Cell types: Drop-Seq analysis of the retina
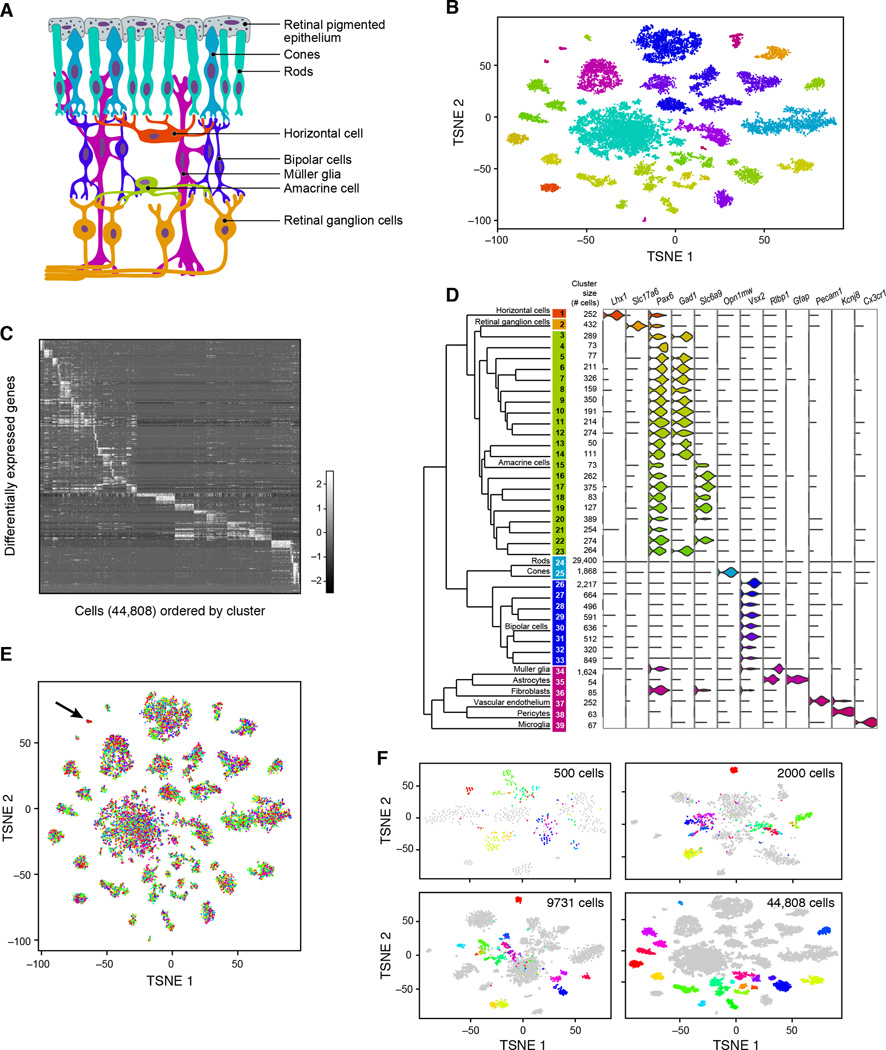
We selected the retina as the first tissue to study with Drop-Seq because decades of work has generated molecular information about many retinal cell types (Masland, 2012; Sanes and Zipursky, 2010), allowing us to relate our RNA-seq data to prior classification. The retina contains five neuronal classes—retinal ganglion, bipolar, horizontal, photoreceptor, and amacrine—each defined by morphological, physiological, and molecular criteria (Figure 5A). Most of the classes are divisible into discrete types – a total currently estimated at about 100 – but well under half of these types possess known, distinguishing molecular markers.
Figure 5. Ab initio reconstruction of retinal cell types from 44,808 single-cell transcription profiles prepared by Drop-Seq.
(A) Schematic representation of major cell classes in the retina. Photoreceptors (rods or cones) detect light and pass information to bipolar cells, which in turn contact retinal ganglion cells that extend axons into other CNS tissues. Amacrine and horizontal cells are retinal interneurons; Müller glia act as support cells for surrounding neurons.
(B) Clustering of 44,808 Drop-Seq single-cell expression profiles into 39 retinal cell populations. The plot shows a two-dimensional representation (tSNE) of global gene expression relationships among 44,808 cells; clusters are colored by cell class, according to Figure 5A.
(C) Differentially expressed genes across 39 retinal cell populations. In this heat map, rows correspond to individual genes found to be selectively upregulated in individual clusters (p < 0.01, Bonferroni corrected); columns are individual cells, ordered by cluster (1–39). Clusters with > 1,000 cells were downsampled to 1,000 cells to prevent them from dominating the plot.
(D) Gene expression similarity relationships among 39 inferred cell populations. Average expression across all detected genes was calculated for each of 39 cell clusters, and the relative (Euclidean) distances between gene-expression patterns for the 39 clusters are represented by a dendrogram. The branches of the dendrogram were annotated by examining the differential expression of known markers for retina cell classes and types. Twelve examples are shown at right, using violin plots to represent the distribution of expression within the clusters. Violin plots for additional genes are in Figure S6A.
(E) Representation of experimental replicates in each cell population. tSNE plot from Figure 2B, with each cell now colored by experimental replicate (for visual clarity, the central rod cluster was downsampled to 10,000 cells). Each of the 7 replicates contributes to all 39 cell populations. Cluster 36 (arrow), in which these replicates are unevenly represented, expressed markers of fibroblasts, which are not native to the retina and are presumably a dissection artifact (see also Figure S6B).
(F) Trajectory of amacrine clustering as a function of number of cells analyzed. Three different downsampled datasets were generated: (1) 500, (2) 2,000, or (3) 9,731 cells (Extended Experimental Procedures). Cells identified as amacrines (clusters 3–23) in the full analysis are here colored by their cluster identities in that analysis. Analyses of smaller numbers of cells incompletely distinguished these subpopulations from one another.
We sequenced 49,300 STAMPs prepared from 14-day-old mouse retinas (STAMPs were collected in seven batches over four days). We performed principal components analysis on the 13,155 largest libraries (Figure S5, Table S3), then reduced the 32 statistically significant PCs (Experimental Procedures) to two dimensions using t-Distributed Stochastic Neighbor Embedding (tSNE) (Amir el et al., 2013; van der Maaten and Hinton, 2008). We projected the remaining 36,145 cells in the data into the tSNE analysis. We then combined a density clustering approach with post hoc differential expression analysis to divide 44,808 cells among 39 transcriptionally distinct clusters (Extended Experimental Procedures) ranging from 50 to 29,400 cells (Figures 5B and 5C). Finally, we organized the 39 cell populations into larger categories (classes) by building a dendrogram of similarity relationships among the 39 cell populations (Figure 5D, left).
The cell populations inferred from this analysis were readily matched to the known retinal cell types, including all five neuronal cell classes, based on the specific expression of known markers for these cell types (Figure 5D, right, and Figure S6A). Additional clusters corresponded to astrocytes (associated with retinal ganglion cell axons exiting the retina), resident microglia, endothelial cells (from intra-retinal vasculature), pericytes, and fibroblasts (Figure 5D). The relative abundances of the major cell classes in our data agreed with earlier estimates from microscopy (Jeon et al., 1998) (Table 1).
Replication and cumulative power of Drop-Seq data
Replication across experimental sessions enables the construction of cumulatively powerful datasets – but only if data are replicable and comparable. The retinal STAMPs were generated on four different days (weeks apart), utilizing different litters and multiple runs in several sessions, for a total of seven replicates. One of the runs was performed at a particularly low cell concentration (15 cells/µL) and thus high purity, to evaluate whether results were artifacts of cell-cell doublets or single-cell impurity. We found that all 39 clusters contained cells from every experiment. One cluster (arrow in Figure 5E; star in Figure S6B), which drew disproportionately from two replicates, expressed markers of fibroblasts, a nonretinal cell type that is present in tissue surrounding the retina, and hence likely represents imprecise dissection.
We examined how the classification of cells (based on their patterns of gene expression) evolved as a function of the numbers of cells in analysis. We used 500, 2,000, or 9,731 cells from our dataset, and asked how (for example) cells identified as amacrines in the full dataset clustered in analyses of smaller numbers of cells (Figure 5F). As the number of cells in the data increased, distinctions between related clusters become clearer, stronger, and finer in resolution, with the result that a greater number of rare amacrine cell sub-populations (each representing 0.1–0.9% of the cells in the experiment) could ultimately be distinguished from one another (Figure 5F).
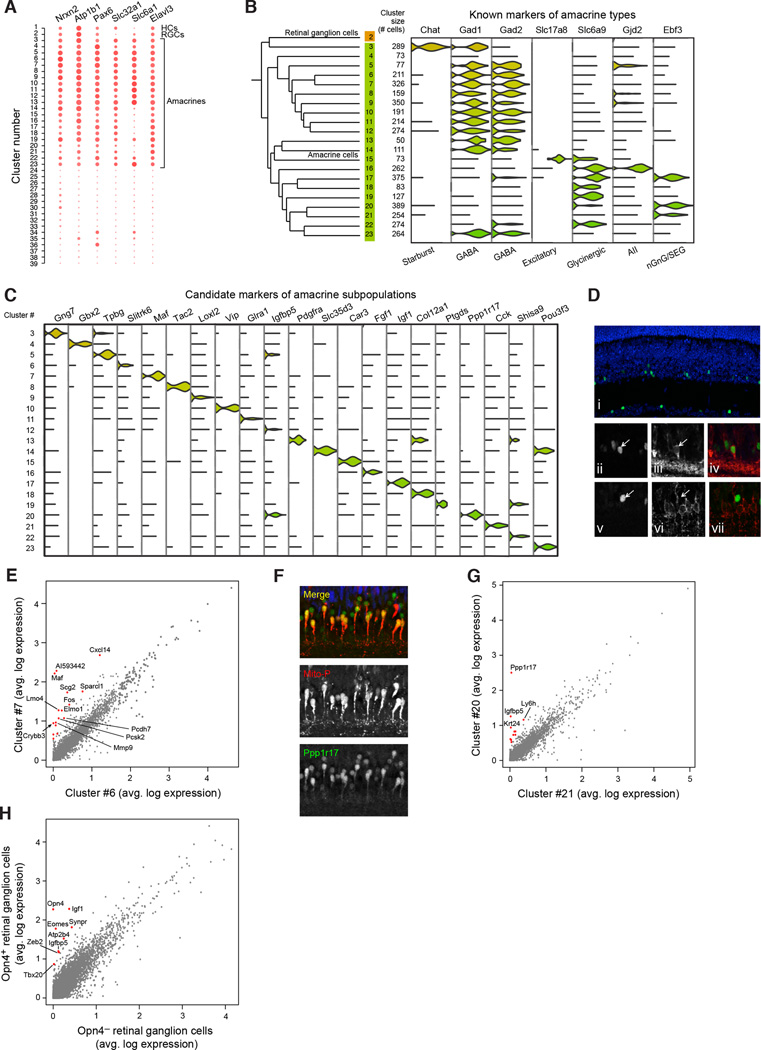
Profiles of amacrine cell types
To characterize distinctions among closely related cell populations, we focused on the 21 clusters of amacrines. Amacrines are the most morphologically diverse neuronal class (Masland, 2012), but the majority of types lack defining molecular markers. Most amacrine cells are inhibitory, utilizing either GABA or glycine as a neurotransmitter. Excitatory amacrine cells that release glutamate have also been identified (Haverkamp and Wassle, 2004). Another amacrine cell population expresses no GABAergic, glycinergic or glutamatergic markers; its neurotransmitter is unidentified (nGnG amacrines) (Kay et al., 2011).
We first identified markers that were most universally expressed by amacrines relative to other cell classes (Figure 6A). We then assessed the expression of known glycinergic and GABAergic markers; their mutually exclusive expression is a fundamental distinction among amacrines. Of the 21 amacrine clusters, 12 were identifiable as GABAergic (Gad1 and/or _Gad2_-positive) and 5 others were glycinergic (glycine transporter _Slc6a9_-positive) (Figure 6B). An additional cell population was identified as excitatory by its expression of a glutamate transporter, Slc17a8 (Figure 6B). The remaining three clusters (clusters 4, 20, and 21) had low levels of GABAergic, glycinergic, and glutamatergic markers; these likely include nGnG amacrines.
Figure 6. Finer-scale expression distinctions among amacrine cells, cones and retinal ganglion cells.
(A) Pan-amacrine markers. The expression levels of the six genes identified (Nrxn2 Atp1b1 Pax6 Slc32a1 Slc6a1 Elavl3) are represented as dot plots across all 39 clusters; larger dots indicate broader expression within the cluster; deeper red denotes a higher expression level.
(B) Identification of known amacrine types among clusters. The twenty-one amacrine clusters consisted of twelve GABAergic, five glycinergic, one glutamatergic and three non-GABAergic non-glycinergic clusters. Starburst amacrines were identified in cluster 3 by their expression of Chat; excitatory amacrines by expression of Slc17a8; A-II amacrines by their expression of Gjd2; and SEG amacrine neurons by their expression of Ebf3.
(C) Nomination of novel candidate markers of amacrine subpopulations. Each cluster was screened for genes differentially expressed in that cluster relative to all other amacrine clusters (p<0.01, Bonferroni corrected) (McDavid et al., 2013), and filtered for those with highest relative enrichment. Expression of a single candidate marker for each cluster is shown across all amacrines.
(D) Validation of MAF as a marker for a GABAergic amacrine population. Staining of a fixed adult retina from wild-type mice for MAF (panels i, ii, v, and green staining in iv and vii), GAD1 (panels iii and iv, red staining), and SLC6A9 (panels vi and vii, red staining), demonstrating co-localization of MAF with GAD1, but not SLC6A9.
(E) Differential expression of cluster 7 (MAF+) with nearest neighboring amacrine cluster (#6). Average gene expression was compared between cells in clusters 6 and 7; sixteen genes (red dots) were identified with >2.8-fold enrichment in cluster 7 (p<10−9).
(F) Validation of PPP1R17 as a marker for an amacrine subpopulation. Staining of a fixed adult retina from Mito-P mice, which express CFP in both nGnG amacrines and type 1 bipolars (Kay et al., 2011). Overlapping labeling by PPP1R17 antibody (green) and Mito-P CFP (red) supports Drop-Seq identification of Ppp1r17 expression in the nGnG amacrine neurons. 85% of CFP+ cells were PPP1R17+ and 50% of the PPP1R17+ cells were CFP-, suggesting a second amacrine type expressing this marker. Blue staining is for VSX2, a marker of bipolar neurons.
(G) Differential expression of cluster 20 (PPP1R17+) with nearest neighboring amacrine cluster (#21). Average gene expression was compared between cells in clusters 20 and 21; twelve genes (red dots) were identified with >2.8-fold enrichment in cluster 20 (p<10−9).
(H) Differential expression of melanopsin-positive and negative RGCs. Average expression was compared between _Opn4_-positive and –negative RGCs in cluster 2. Seven genes were identified as enriched in _Opn4_-positive cells (red dots, >2-fold, p<10−9).
Among the glycinergic and GABAergic clusters, we found many amacrine types with known markers. A-II amacrine neurons appeared to correspond to the most divergent glycinergic cluster (Figure 6B, cluster 16), as this was the only cluster to strongly express the Gjd2 gene encoding the gap junction protein connexin 36 (Feigenspan et al., 2001). Ebf3, a transcription factor found in SEG glycinergic as well as nGnG amacrines, was specific to clusters 17 and 20. Starburst amacrine neurons (SACs), the only retinal cells that use acetylcholine as a co-transmitter, were identifiable as cluster 3 by their expression of the cholinergic marker Chat (Figure 6B). Unlike other GABAergic cells, SACs expressed Gad1 but not Gad2, as previously observed in rabbit (Famiglietti and Sundquist, 2010).
We then identified selectively expressed markers for each of the 21 amacrine cell populations (Figure 6C and Table S4). We validated two of the markers immunohistochemically. First, we co-stained retinal sections with antibodies to the transcription factor MAF, the top marker of cluster 7, plus antibodies to either GAD1 or SLC6A9, markers of GABAergic and glycinergic transmission, respectively. As predicted by the Drop-Seq analysis, MAF was found in a small subset of amacrine cells that were GABAergic and not glycinergic (Figure 6D). Cluster 7 had numerous genes that were enriched relative to its nearest neighbor, cluster 6 (Figure 6E, 16 genes > 2.8-fold enrichment, p<10−9), including _Crybb3_, which belongs to the crystallin family of proteins that are known to be directly upregulated by _Maf_ (Yang and Cvekl, 2005), and another, the protease _Mmp9_, which accepts crystallins as substrates (Descamps et al., 2005). Second, we stained sections with antibodies to PPP1R17 (Figure 6F). Cluster 20 shows weak, infrequent glycine transporter expression and is one of only two clusters (with cluster 21) that express _Neurod6_, a marker of nGnG neurons (Kay et al., 2011). We used a transgenic strain (MitoP) that has been shown to express cyan fluorescent protein (CFP) specifically in nGnG amacrines (Kay et al., 2011). PPP1R17 stained 85% of all CFP-positive amacrines in the MitoP line, validating this as a marker of nGnG cells (Figure 6F). PPP1R17 was one of several markers that distinguished Cluster 20 from its closest neighbor, Cluster 21 (Figure 6G; 12 genes > 2.8-fold enrichment, p<10−9). The differences between Clusters 20 and 21 suggest a hitherto unsuspected level of heterogeneity among nGnG amacrines.
Supervised analysis reveals additional diversity
Our unsupervised analysis grouped cells into 39 transcriptionally distinct populations, but morphological and functional criteria suggest that there are ~100 retinal cell types. We asked whether supervised analysis could reveal multiple types within individual clusters. For example, retinal ganglion cells (RGCs), which consist of about 30 types (Sanes and Masland, 2015), formed a single cluster in our analysis, perhaps because it is a rare cell population (1%, Table 1). Five RGC types, called intrinsically photosensitive RGCs (ipRGCs), express Opn4, the gene encoding the photopigment melanopsin. Opn4+ RGCs (26/432) expressed nine genes at levels two-fold higher than _Opn4_- RGCs (p < 109, Figure 6H), including Tbr2/Eomes, known to be a selective marker for this population (Sweeney et al., 2014). This result reveals additional heterogeneity that may also emerge ab initio as analyses expand to include more cells.
Discussion
Ascertaining transcriptional variation across individual cells is a valuable way of learning about complex tissues and functional responses, but single-cell analysis has been limited by the time and cost of preparing libraries from many individual cells. A scientist employing Drop-Seq can prepare 10,000 single-cell libraries for sequencing in twelve hours, for about 6.5 cents per cell (Table S5), representing a >100-fold improvement in both time and cost relative to existing methods. A Drop-Seq setup can be constructed quickly and inexpensively in a standard biology lab using readily available equipment (Figure S2B and Extended Experimental Procedures). We hope that ease, speed, and low cost facilitate exuberant experimentation, careful replication, and many cycles of experiments, analyses, ideas, and more experiments.
In validating Drop-Seq, we developed stringent species-mixing experiments to measure single-cell purity and cell doublet rates in our libraries. In another article in this issue, Klein et al. (Klein, 2015) describe a droplet-based approach to single-cell RNA-seq, and also use species-mixing experiments to evaluate it. Our results indicate that all methods of isolating single cells from a cell suspension, including Drop-Seq, fluorescence activated cell sorting (FACS) and microfluidics, are vulnerable to impurities, and highlight the value of performing species mixing experiments to assess single-cell approaches. In our retina analysis, even relatively impure libraries generated in “ultra-high-throughput” modes (100 cells per µL, allowing the processing of 10,000 cells per hour at ~10% doublet and impurity rates) appeared to yield a robust and biologically validated cell classification, but other tissues or applications may require using Drop-Seq in purer modes.
Unsupervised computational analysis of Drop-Seq data identified 39 transcriptionally distinct retinal cell populations, many representing specific subtypes of the major retinal cell classes (Figures 5 and 6). It is a particular strength of the retina that establishing correspondence between cluster and type was in many cases straightforward; an important direction will be to identify cell types and states in other parts of the brain—as well as in other tissues—about which less is currently known.
We see many applications of Drop-Seq, beyond the identification of cell types and cell states. Genome-scale genetic studies are identifying many genes whose variation contributes to disease risk, but biology has lacked similarly high-throughput ways of connecting these genes to specific cell populations and unique functional responses. Drop-Seq could be used to provide initial insights into how these genes function in the diverse cell types composing each tissue. In addition, coupling Drop-Seq to perturbations — such as small molecules, mutations, pathogens, or other stimuli —could generate an information-rich, multi-dimensional readout of the influence of perturbations on many kinds of cells.
The functional implications of a gene’s expression are a product not just of that gene’s intrinsic properties, but also of the entire cell-level context in which the gene is expressed. We hope Drop-Seq enables the abundant and routine discovery of such relationships in many areas of biology.
Experimental Procedures
Device design and fabrication
Microfluidic devices were designed using AutoCAD software (Autodesk, Inc.), and the components tested using COMSOL Multiphysics (COMSOL Inc.). Full details are described in Extended Experimental Procedures.
Barcoded microparticle synthesis
Bead functionalization and reverse-direction phosphoramidite synthesis were performed by Chemgenes Corp (Wilmington, MA). “Split-and-pool” cycles were accomplished by removing the dry resin from each column, hand mixing, and weighing out four equal portions before returning the resin for an additional cycle of synthesis. Full details are described in Extended Experimental Procedures.
Drop-Seq procedure
Monodisperse droplets ~1 nL in size were generated using the microfluidic device described in Extended Experimental Procedures, in which barcoded microparticles, suspended in lysis buffer, were flowed at a rate equal to that of a single-cell suspension, so that resulting droplets were composed of an equal amount of each component. As soon as droplet generation was complete, droplets were broken with perfluorooctanol in 30 mL of 6x SSC. The addition of a large aqueous volume to the droplets reduces hybridization events after droplet breakage, because DNA base pairing follows second-order kinetics (Britten and Kohne, 1968; Wetmur and Davidson, 1968). The beads were then washed and resuspended in a reverse transcriptase mix, followed by a treatment with exonuclease I to remove unextended primers. The beads were then washed, counted, aliquoted into PCR tubes, and PCR amplified. The PCR reactions were purified and pooled, and the amplified cDNA quantified on a BioAnalyzer High Sensitivity Chip (Agilent). The cDNA was fragmented and amplified for sequencing with the Nextera XT DNA sample prep kit (Illumina) using custom primers that enabled the specific amplification of only the 3’ ends (Table S6). The libraries were purified, quantified, and then sequenced on the Illumina NextSeq 500. All details regarding reaction conditions, primers used, and sequencing specifications can be found in the Extended Experimental Procedures.
Cell cycle analysis of HEK and 3T3 cells
Gene sets reflecting five phases of the HeLa cell cycle (G1/S, S, G2/M, M and M/G1) were taken from Whitfield et al. (Whitfield et al., 2002) with some modification (Extended Experimental Procedures and Table S2). A phase-specific score was generated for each cell, across all five phases, using averaged normalized expression levels (log2(TPM+1) of the genes in each set. Cells were then ordered along the cell cycle by comparing the patterns of these five phase scores per cell. To identify cell cycle-regulated genes, we used a sliding window approach, and identified windows of maximal and minimal average expression, both for ordered cells, and for shuffled cells, to evaluate the false-discovery rate. Full details may be found in Extended Experimental Procedures.
Principal components and clustering analysis of retina data
The clustering algorithm for the retinal cell data was implemented and performed using Seurat, a recently developed R package for single-cell analysis (Satija et al., 2015). Principal components analysis (PCA) was first performed on a 13,155-cell “training set” of the 49,300-cell dataset, using single-cell libraries in which transcripts from > 900 genes were detected. We found this approach was more effective in discovering structures corresponding to rare cell types than performing PCA on the full dataset, which was dominated by numerous, tiny rod photoreceptors (Extended Experimental Procedures). Thirty-two statistically significant PCs were identified using a permutation test and independently confirmed using a modified resampling procedure (Chung and Storey, 2014). We projected individual cells within the training set based on their PC scores onto a single two-dimensional map using t-Distributed Stochastic Neighbor Embedding (t-SNE) (van der Maaten and Hinton, 2008). The remaining 36,145 single-cell libraries (< 900 genes detected) were next projected on this t-SNE map, based on their representation within the PC-subspace of the training set (Berman et al., 2014; Shekhar et al., 2014). This approach mitigates the impact of noisy variation in the lower complexity libraries due to gene dropouts. It was also reliable in the sense that when we withheld from the t-SNE all cells from a given cluster and then tried to project them, these withheld cells were not spuriously assigned to another cluster by the projection (Table S7). Point clouds on the t-SNE map represent candidate cell types; density clustering (Ester et al., 1996) identified these regions. Differential expression testing (McDavid et al., 2013) was then used to confirm that clusters were distinct from each other. Hierarchical clustering based on Euclidean distance and complete linkage was used to build a tree relating the clusters. We noted expression of several rod-specific genes, such as Rho and Nrl, in every cell cluster, an observation that has been made in another retinal cell gene expression study (Siegert et al., 2012) and likely arises from solubilization of these high-abundance transcripts during cell suspension preparation. Additional information regarding retinal cell data analysis can be found in the Extended Experimental Procedures.
Data availability
Both raw and analyzed data have been deposited at Gene Expression Omnibus Accession GSE63473.
Supplementary Material:
Figure S1
Table S4
Movie S1
DataFile 1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Supplemental Information
Table S2
Table S3
Acknowledgements
This work was supported by the Stanley Center for Psychiatric Research (to SM), the MGH Psychiatry Residency Research Program and Stanley-MGH Fellowship in Psychiatric Neuroscience (to EZM), a Stewart Trust Fellows Award (to SM), a grant from the Simons Foundation to the Simons Center for the Social Brain at MIT (to AR, SM and DW), an NHGRI CEGS P50 HG006193 (to AR), the Klarman Cell Observatory (to AR and AB), NIMH grant U01MH105960 (to SM, AR and JRS), NIMH grant R25MH094612 (to EM), NIH F32 HD075541 (to RS). AR is an investigator of the Howard Hughes Medical Institute. Microfluidic device fabrication was performed at the Harvard Center for Nanoscale Systems (CNS), a member of the National Nanotechnology Infrastructure Network (National Science Foundation award no. ECS-0335765), with support from the National Science Foundation (DMR-1310266) and the Harvard Materials Research Science and Engineering Center (DMR-1420570). We thank Christina Usher and Leslie Gaffney for contributions to the manuscript figures and Chris Patil for helpful comments on the manuscript. We thank Connie Cepko for helpful conversations about the retina data; Beth Stevens for advice on retinal dissociations; and Assaf Rotem and Huidan Zhang for advice on microfluidics design and fabrication.
Footnotes
Author Contributions
E.Z.M. developed the barcoding and molecular biology analysis, advised by S.A.M. A.B. designed and fabricated the microfluidic devices, advised by D.A.W. and A.R. E.Z.M. and M.G. developed Drop-Seq experimental protocols and performed the Drop-Seq experiments in S.A.M.’s lab. J.N. developed the methods and software for obtaining digital gene expression measurements for each cell, advised by E.Z.M. and S.A.M. J.N., E.Z.M. and S.A.M. performed the analyses of species-mixing experiments. I.T. performed the cell cycle analysis. A.R.B. prepared the retinal cell suspensions. R.S., K.S., and A.R. developed and performed the retinal cell type clustering analyses with contribution from N.K. E.Z.M., R.S., K.S., and J.R.S. interpreted the retina expression data. E.M.M. and J.R.S. performed the immunohistochemistry experiments. J.J.T. and A.K.S. performed the Fluidigm C1 experiments. E.Z.M., S.A.M., A.R., A.B., and A.K.S. conceived the study and key ways that Drop-Seq works together as an integrated system. E.Z.M. and S.A.M. wrote the manuscript with contributions from all authors.
References
- Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe'er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nature biotechnology. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer NR, Wheeler EK, Lee-Houghton L, Watkins N, Nasarabadi S, Hebert N, Leung P, Arnold DW, Bailey CG, Colston BW. On-chip single-copy real-time reverse-transcription PCR in isolated picoliter droplets. Analytical chemistry. 2008;80:1854–1858. doi: 10.1021/ac800048k. [DOI] [PubMed] [Google Scholar]
- Berman GJ, Choi DM, Bialek W, Shaevitz JW. Mapping the stereotyped behaviour of freely moving fruit flies. Journal of the Royal Society, Interface / the Royal Society. 2014:11. doi: 10.1098/rsif.2014.0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke P, Anders S, Kim JK, Kolodziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nature methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- Britten RJ, Kohne DE. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968;161:529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Chung NC, Storey JD. Statistical Significance of Variables Driving Systematic Variation in High-Dimensional Data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descamps FJ, Martens E, Proost P, Starckx S, Van den Steen PE, Van Damme J, Opdenakker G. Gelatinase B/matrix metalloproteinase-9 provokes cataract by cleaving lens betaB1 crystallin. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2005;19:29–35. doi: 10.1096/fj.04-1837com. [DOI] [PubMed] [Google Scholar]
- Ester M, Kriegel HP, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. Menlo Park, Calif: AAAI Press; 1996. [Google Scholar]
- Famiglietti EV, Sundquist SJ. Development of excitatory and inhibitory neurotransmitters in transitory cholinergic neurons, starburst amacrine cells, and GABAergic amacrine cells of rabbit retina, with implications for previsual and visual development of retinal ganglion cells. Visual neuroscience. 2010;27:19–42. doi: 10.1017/S0952523810000052. [DOI] [PubMed] [Google Scholar]
- Feigenspan A, Teubner B, Willecke K, Weiler R. Expression of neuronal connexin36 in AII amacrine cells of the mammalian retina. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:230–239. doi: 10.1523/JNEUROSCI.21-01-00230.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell reports. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Haverkamp S, Wassle H. Characterization of an amacrine cell type of the mammalian retina immunoreactive for vesicular glutamate transporter 3. The Journal of comparative neurology. 2004;468:251–263. doi: 10.1002/cne.10962. [DOI] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Analytical chemistry. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lonnerberg P, Linnarsson S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nature methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1998;18:8936–8946. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, Voinescu PE, Chu MW, Sanes JR. Neurod6 expression defines new retinal amacrine cell subtypes and regulates their fate. Nature neuroscience. 2011;14:965–972. doi: 10.1038/nn.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivioja T, Vaharautio A, Karlsson K, Bonke M, Enge M, Linnarsson S, Taipale J. Counting absolute numbers of molecules using unique molecular identifiers. Nature methods. 2012;9:72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single cell transcriptomics and its application to embryonic stem cells. Cell PRESS. 2015 doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH. The neuronal organization of the retina. Neuron. 2012;76:266–280. doi: 10.1016/j.neuron.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, Roederer M, Gottardo R. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics. 2013;29:461–467. doi: 10.1093/bioinformatics/bts714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petilla Interneuron Nomenclature G, Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, Buzsaki G, Cauli B, Defelipe J, et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nature reviews Neuroscience. 2008;9:557–568. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nature methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Masland RH. The Types of Retinal Ganglion Cells: Current Status and Implications for Neuronal Classification. Annual review of neuroscience. 2015 doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Zipursky SL. Design principles of insect and vertebrate visual systems. Neuron. 2010;66:15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature biotechnology. 2015 doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Brodin P, Davis MM, Chakraborty AK. Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding (ACCENSE) Proceedings of the National Academy of Sciences of the United States of America. 2014;111:202–207. doi: 10.1073/pnas.1321405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert S, Cabuy E, Scherf BG, Kohler H, Panda S, Le YZ, Fehling HJ, Gaidatzis D, Stadler MB, Roska B. Transcriptional code and disease map for adult retinal cell types. Nature neuroscience. 2012;15:487–495. doi: 10.1038/nn.3032. S481–482. [DOI] [PubMed] [Google Scholar]
- Sweeney NT, Tierney H, Feldheim DA. Tbr2 is required to generate a neural circuit mediating the pupillary light reflex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:5447–5453. doi: 10.1523/JNEUROSCI.0035-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nature methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Thorsen T, Roberts RW, Arnold FH, Quake SR. Dynamic pattern formation in a vesicle-generating microfluidic device. Physical review letters. 2001;86:4163–4166. doi: 10.1103/PhysRevLett.86.4163. [DOI] [PubMed] [Google Scholar]
- Umbanhowar PBPV, Weitz DA. Monodisperse Emulsion Generation via Drop Break Off in a Coflowing Stream. Langmuir. 2000;16:347–351. [Google Scholar]
- Utada AS, Fernandez-Nieves A, Stone HA, Weitz DA. Dripping to jetting transitions in coflowing liquid streams. Physical review letters. 2007;99:094502. doi: 10.1103/PhysRevLett.99.094502. [DOI] [PubMed] [Google Scholar]
- van der Maaten L, Hinton G. Visualizing Data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- Vogelstein B, Kinzler KW. Digital PCR. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmur JG, Davidson N. Kinetics of renaturation of DNA. Journal of molecular biology. 1968;31:349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Molecular biology of the cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Cvekl A. Tissue-specific regulation of the mouse alphaA-crystallin gene in lens via recruitment of Pax6 and c-Maf to its promoter. Journal of molecular biology. 2005;351:453–469. doi: 10.1016/j.jmb.2005.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. BioTechniques. 2001;30:892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Table S4
Movie S1
DataFile 1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Supplemental Information
Table S2
Table S3
Data Availability Statement
Both raw and analyzed data have been deposited at Gene Expression Omnibus Accession GSE63473.