Suppression of PGC-1α is critical for reprogramming oxidative metabolism in renal cell carcinoma (original) (raw)
. Author manuscript; available in PMC: 2016 Jul 7.
Summary
Long believed to be a byproduct of malignant transformation, reprogramming of cellular metabolism is now recognized as a driving force in tumorigenesis. In clear cell renal cell carcinoma (ccRCC) frequent activation of HIF-signaling induces a metabolic switch that promotes tumorigenesis. Here we demonstrate that PGC-1α, a central regulator of energy metabolism, is suppressed in VHL-deficient ccRCC by a HIF/Dec1-dependent mechanism. In VHL wild type cells, PGC-1α suppression leads to decreased expression of the mitochondrial transcription factor Tfam and impaired mitochondrial respiration. Conversely, PGC-1α expression in VHL-deficient cells restores mitochondrial function and induces oxidative stress. ccRCC cells expressing PGC-1α exhibit impaired tumor growth and enhanced sensitivity to cytotoxic therapies. In patients, low levels of PGC-1α expression are associated with poor outcome. These studies demonstrate that suppression of PGC-1α recapitulates key metabolic phenotypes of ccRCC and highlight the potential of targeting PGC-1α expression as a therapeutic modality for the treatment of ccRCC.
Introduction
First observed by Otto Warburg in the early 20th century, metabolic reprogramming is now accepted as an emerging hallmark of cancer (Hanahan and Weinberg, 2011). Constitutive activation of hypoxia inducible factor (HIF) signaling and accumulation of cytosolic lipid droplets indicate profound changes in cellular metabolism take place during ccRCC tumorigenesis. Indeed, the signaling networks that regulate metabolic behavior are frequently altered in ccRCC, leading to the description of this tumor type as a “metabolic disease” (Linehan et al., 2010). Despite the seemingly important role of altered metabolism in ccRCC tumorigenesis, the molecular mechanisms underlying these metabolic shifts are incompletely understood. Clinically, ccRCC is refractory to conventional cytotoxic agents, with therapeutic approaches instead favoring surgery and targeted therapies such as tyrosine kinase inhibitors, mTOR inhibitors, and immunotherapy (Rini et al., 2009). Importantly, the mechanism of resistance to chemo/radiotherapy is poorly understood, and whether altered metabolism influences therapeutic response remains unknown.
Deletions and mutations to the tumor suppressor gene, Von Hippel-Lindau (Vhl), are the most frequent genetic alterations in ccRCC (Cancer Genome Research Atlas, 2013; Gnarra et al., 1994). The Vhl gene encodes an E3 ubiquitin ligase that is essential for oxygen-dependent regulation of HIF-α transcription factors (Kondo et al., 2002; Maxwell et al., 1999). In normal oxygen conditions, HIF-1α and HIF-2α subunits are hydroxylated on key proline residues by oxygen-dependent prolyl hydroxylases (PHDs), bound by VHL, and rapidly degraded by the proteasome (Ivan et al., 2001; Jaakkola et al., 2001). In hypoxia, prolyl hydroxylation is inhibited, resulting in HIF-α stabilization and dimerization with aryl hydrocarbon receptor nuclear transactivator (ARNT) (Wang et al., 1995). Consequently, VHL loss of function results in constitutive activation of HIF-α which promotes tumorigenesis through transcriptional activation of genes involved in angiogenesis, invasion, metastasis, and metabolism (Gordan and Simon, 2007).
Constitutive activation of HIF transcription factors is thought to be a primary driving force of metabolic reprogramming in ccRCC. HIF transcription factors activate a gene expression program that upregulates glycolytic flux while simultaneously inhibiting mitochondrial activity (Fukuda et al., 2007; Papandreou et al., 2006). The ability of HIF to negatively regulate mitochondrial activity fits with the evolutionary need for coupling oxygen consumption in the mitochondria to nutrient and oxygen availability. In ccRCC, mitochondrial content is inversely correlated with tumor grade, indicating that suppression of mitochondrial activity may play an important role in ccRCC progression (Simonnet et al., 2002). Importantly, the mechanism underlying suppression of mitochondrial content in ccRCC, and the consequences thereof remain incompletely understood.
The PPARγ coactivators (PGC) are a family of transcriptional coactivators that are regulated by a wide range of environmental stimuli to coordinate mitochondrial biogenesis and metabolic flux (Puigserver et al., 1998). While the PGC coactivators (consisting of PGC-1α, PGC-1β, and PRC) exhibit some degree of redundancy, PGC-1α knockout mice exhibit multi-tissue defects in mitochondrial metabolism indicating unique functions for PGC-1α that cannot be compensated for by the other family members (Leone et al., 2005; Lin et al., 2004). A growing body of evidence points toward an important role for PGC-1α in cancer, however an important dichotomy exists, with reports of pro and anti-tumorigenic effects of PGC-1α expression in different cancer types (D'Errico et al., 2011; Haq et al., 2013; LeBleu et al., 2014; Lim et al., 2014; Vazquez et al., 2013; Yan et al., 2014). A better understanding of the role of PGC-1α in different tumor types and at different stages of tumorigenesis will be important in determining whether this pathway will be amenable to therapeutic intervention.
Past studies indicate that dynamic interplay between the HIF transcription factors and c-Myc can regulate expression of PGC-1β and mitochondrial biogenesis (Zhang et al., 2007). Additionally, hypoxia has been linked to the regulation of PGC-1α in adipose tissue, where hypoxia inhibits SIRT2-mediated deacetylation of PGC-1α (Krishnan et al., 2012). Conversely, PGC-1α-induced oxygen consumption and ROS production have been reported to stabilize and activate HIFs (O'Hagan et al., 2009; Shoag and Arany, 2010). To this point, it has remained unclear whether PGC-1α expression or activity are modulated in ccRCC, and if so, what effect this regulation would have on the metabolic phenotypes and tumorigenic potential of ccRCC.
Results
Suppression of mitochondrial biogenesis transcription factors in ccRCC
To explore the gene expression changes underlying metabolic alteration in ccRCC, we analyzed previously published microarray datasets from ccRCC cell lines and tumors. Gene set enrichment analysis (GSEA) revealed significant enrichment of hypoxia gene sets and depletion of β-oxidation and TCA cycle gene sets in ccRCC compared to the non-malignant kidney (Figure 1A, Supplemental Figures S1A and S1B). Furthermore, DAVID analysis revealed suppression of genes involved in mitochondrial function in VHL-deficient ccRCC cells in vitro (Figure 1B and Supplemental Figure S1C). We hypothesized that these changes in gene expression might result from suppression of the key transcriptional regulators of mitochondrial biogenesis.
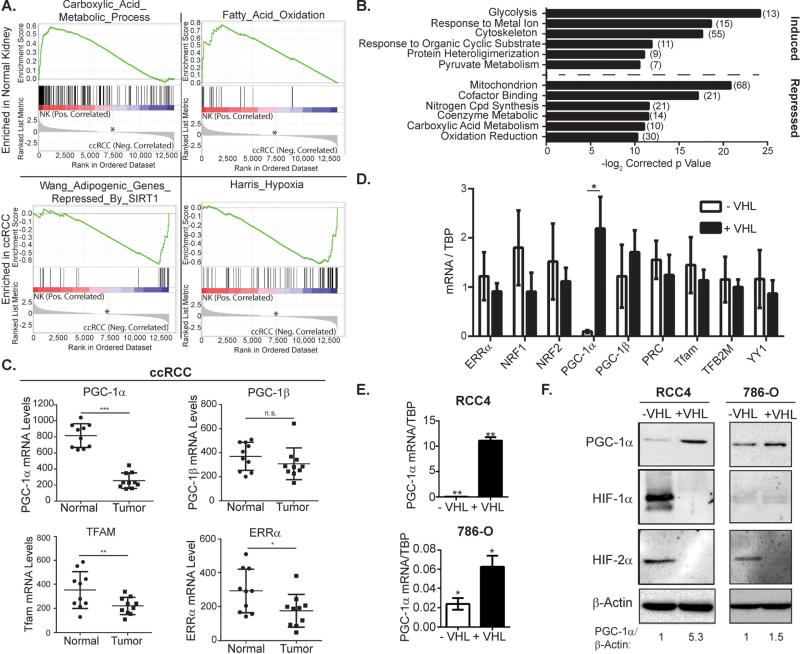
Figure 1. PGC-1α expression is suppressed in VHL-deficient clear cell renal cell carcinoma.
A. GSEA of normal kidney and ccRCC tumors. * zero cross at 7239, (GEO accession:GSE6344) B. DAVID analysis of gene expression in RCC4 cells in VHL-deficiency or hypoxia. (Array data from (Krieg et al., 2010)) C. mRNA expression levels in normal kidney or ccRCC. Data from GSE6344. * p<0.04; ** p<0.03; *** p<0.0001; n.s. p>0.05 by Student's two-tailed t-test. D. mRNA levels in VHL deficient (−VHL) or wild type (+VHL) RCC4 cells. * p<0.0001 by Student's two-tailed t-test. E. mRNA levels in VHL deficient (−VHL) or wild type (+VHL) RCC4 and 786-O cells. * p=0.01; ** p=0.001 by Student's two-tailed t-test. F. PGC-1α protein levels in VHL deficient (−VHL) or wild type (+VHL) RCC4 and 786-O cells. All bar graphs plotted as mean ± SD. See also Figures S1 and S2.
Mitochondrial biogenesis is controlled by a network of nuclear transcription factors and coactivators that include the nuclear respiratory factors (NRFs), the estrogen related receptors (ERRs) and the PGC family of transcriptional coactivators. To determine whether differential expression of these transcriptional regulators could explain the gene expression changes outlined above, we profiled the expression of several genes known to act as regulators of these pathways. To our surprise, PGC-1α, but not PGC-1β was highly suppressed in ccRCC (Figures 1C, S2A-B). Additionally, expression of the mitochondrial transcription factor Tfam (a known target of PGC-1α) was decreased in ccRCC, as was ERRα, a transcription factor through which PGC-1α function is transduced (Mootha et al., 2004) (Figure 1C). Consistent with these data, PGC-1α mRNA and protein were suppressed in several VHL-deficient cell lines (Figure 1D-F, S2C). Given its central role as a regulator of mitochondrial function, we hypothesized that suppression of PGC-1α disrupts mitochondrial content and function in ccRCC.
PGC-1α is essential for mitochondrial homeostasis in renal proximal tubule cells
Studies using genetically engineered mouse models indicate that PGC-1α deficiency results in defective mitochondrial metabolism in multiple tissue types (Leone et al., 2005). Similarly, VHL deficient ccRCC cell lines exhibit HIF-dependent decreases in mitochondrial content and activity (Figure S3A-D). We reasoned that the suppression of PGC-1α in VHL-deficient ccRCC could underlie the mitochondrial deficiency characteristic of this cancer type. Consistent with this hypothesis, PGC-1α knockdown in VHL-wild type malignant (TK10) and non-malignant proximal tubule (HK2) cells decreased expression of the essential mitochondrial transcription factor, Tfam (Figure 2A). Importantly, PGC-1α suppression also functionally impaired mitochondrial oxygen consumption rates (OCR) in both HK2 and TK10 cells indicating that endogenous expression of PGC-1α is essential for maintaining basal mitochondrial respiration in these cells (Figure 2B, S3E-G). Consistent with impaired mitochondrial function in PGC-1α deficient cells, GC-MS analysis revealed significant decreases in TCA cycle metabolite abundance (Figure 2C). These results indicate that PGC-1α expression is essential for mitochondrial function and suggest that suppression of PGC-1α may contribute to the depletion of mitochondrial function observed in ccRCC.
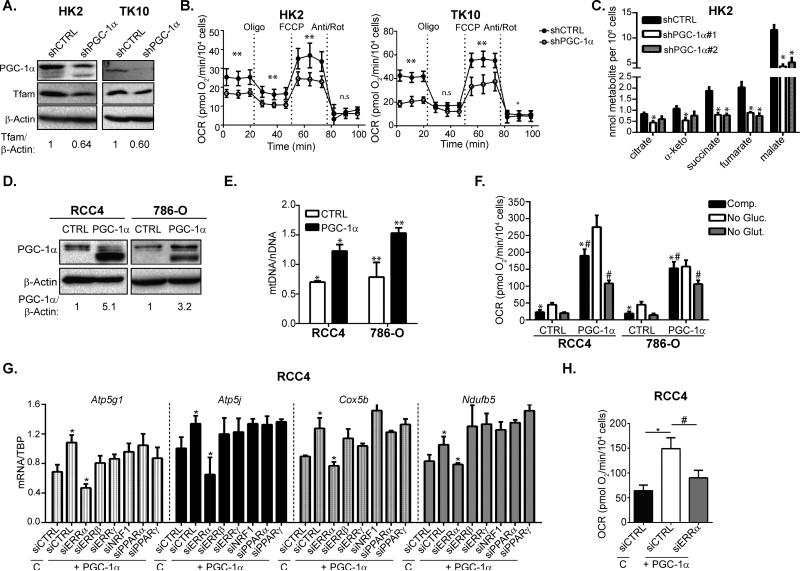
Figure 2. PGC-1α regulates mitochondrial homeostasis and OXPHOS activity in ccRCC.
A. Western Blots of VHL-wild type HK2 and TK10 cells transduced with shCTRL or shPGC-1α. B. Oxygen consumption rates (OCR) in HK2 and TK10 cells transduced with shCTRL or shPGC-1α. Oligomycin (Oligo), FCCP, and Antimycin A/Rotenone (Anti/Rot) were injected at the indicated time points. Data presented as mean + SEM. Student's two-tailed t-test calculated by pooling the 3 measurements for each condition ** p≤0.01;* p<0.05. C. GC-MS quantification of TCA cycle metabolites from shCTRL and shPGC-1α transduced HK2 cells * p≤0.01 by Student's two-tailed t-test. D. Ectopic expression of PGC-1α in VHL-deficient RCC4 and 786-O cells. E. mitochondrial DNA (mtDNA) measurements normalized to nuclear DNA (nDNA) in CTRL or PGC-1α transduced RCC4 and 786-O cells. * p=0.01; ** p<0.03 by Student's two-tailed t-test. F. OCR measurements in CTRL and PGC-1α transduced RCC4 and 786-O cells cultured with the indicated carbon sources. *, # two-tailed Student's t-test p<0.0001. G. qPCR analysis of mitochondrial gene expression in RCC4 cells transduced with control (C) or PGC-1α retrovirus and transfected with the indicated siRNA. * two-tailed Student's t-test p<0.05. H. Basal OCR in CTRL and PGC-1α transduced RCC4 cells transfected with the indicated siRNA. *,# two-tailed Student's t-test p<0.0001. All bar graphs plotted as mean ± SD unless otherwise noted. See also Figures S3.
To further explore the role of PGC-1α suppression in ccRCC metabolism, we tested whether restored PGC-1α expression in VHL-deficient cells was sufficient to rescue mitochondrial function (Figure 2D). Consistent with increased mitochondrial content, PGC-1α expression in VHL-deficient RCC4 and 786-O cells increased mitochondrial DNA content and CMXRos staining (Figure 2E, S3H). PGC-1α expression also functionally increased mitochondrial respiration in both RCC4 and 786-O, an effect that was significantly blunted by withdrawal of exogenous glutamine (Figure 2F). Increased mitochondrial respiration was also partially sensitive to the CPT-1α inhibitor, etomoxir, indicating that PGC-1α stimulation of OCR occurred at least in part due to increases in β-oxidation (Figure S3I-J). These results provide evidence that restored PGC-1α expression is sufficient to restore mitochondrial content and function in VHL-deficient ccRCC.
PGC-1α function is transduced through interaction with a wide range of nuclear transcription factors and we next sought to identify the transcription factor(s) acting in concert with PGC-1α to stimulate mitochondrial biogenesis in ccRCC. Using siRNA to transiently suppress expression of several transcription factors known to interact with PGC-1α, we determined that ERRα was required for PGC-1α induced mitochondrial gene expression in VHL-deficient RCC4 cells (Figure 2G, S3K). Furthermore, ERRα suppression also abrogated PGC-1α increased mitochondrial respiration rate, indicating its essential role as a co-regulator of PGC-1α in regulating mitochondrial function (Figure 2H). These results indicate that ERRα is required for PGC-1α mediated induction of mitochondrial biogenesis in ccRCC.
PGC-1α induces oxidative stress and sensitizes ccRCC cells to cytotoxic therapy
Reactive oxygen species are potent byproducts of electron transport chain activity and play a complex role in tumorigenesis. Malignant cells often exhibit elevated steady state levels of ROS rendering them exquisitely sensitive to further increases in ROS production from the mitochondria (Trachootham et al., 2009). We reasoned that the elevated mitochondrial respiratory activity observed upon expression of PGC-1α would likely be accompanied by an increase in ROS production. Furthermore, VHL-deficient ccRCC cell lines exhibit HIF-dependent decreases in both mitochondrial respiration and ROS production, indicating that the two processes are functionally linked (Figures S3C and S4A). In support of this hypothesis, RCC4 and 786-O cells ectopically expressing PGC-1α contained significantly elevated levels of hydroxyl and superoxide radicals as measured by DCF-DA and MitoSOX staining respectively (Figures 3A-B, S4B). These results were surprising given the known ability of PGC-1α to coordinate expression of ROS detoxification enzymes with enhanced mitochondrial biogenesis (St-Pierre et al., 2006). Indeed, PGC-1α expression in VHL-deficient cells did increase the expression of several antioxidant genes (Figure 3C). To determine whether the antioxidant response was sufficient to detoxify the observed increases in ROS, we next stained for 8-hydroxyguanosine, an oxidized derivative of guanosine that is commonly used as a biomarker of oxidative stress. Strikingly, both RCC4 and 786-O cells transduced with PGC-1α exhibited significantly elevated levels of 8oxoG compared to control, indicating that PGC-1α expression was sufficient to induce oxidative stress in VHL-deficient ccRCC cells (Figure 3D and S4C). Furthermore, PGC-1α expression increased γH2AX levels in RCC4 cells, indicating the presence of an active DNA damage response (Figure 3E). These results suggest that PGC-1α induced mitochondrial activity is tightly associated with oxidative stress in VHL-deficient cells.
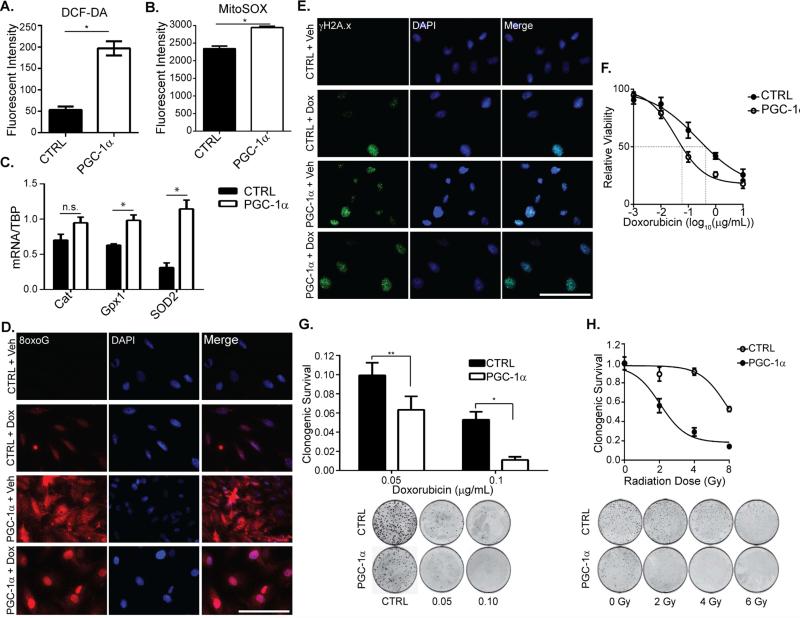
Figure 3. PGC-1α increases oxidative stress in ccRCC and enhances sensitivity to cytotoxic therapies.
A. Mean DCF-DA staining of control (CTRL) or PGC-1α transduced RCC4 cells. * p<0.001 by Student's two-tailed t-test. _B._ Mean MitoSOX staining intensity in control or PGC-1α transduced RCC4 cells. * p<0.0002 by Student's two-tailed t-test. _C._ mRNA levels of the indicated genes in CTRL and PGC-1α transduced RCC4 cells. * p<0.025; n.s. p>0.05 by Student's two-tailed t-test. D. 8-hydroxyguanosine (8oxoG) immunofluorescent (IF) staining of CTRL and PGC-1α transduced RCC4 cells treated with vehicle (Veh) or 0.1 g/mL doxorubicin (Dox) for 48hrs. Scale bar, 100 μm. E. Phosphorylated histone H2A.x (Ser 139) (γH2AX) IF staining of CTRL and PGC-1α transduced RCC4 cells treated with Veh or Dox for 48 hrs. Scale bar 100 μm. F. XTT assay of CTRL and PGC-1α transduced RCC4 cells treated with the indicated dose of doxorubicin for 48hrs. IC50 for doxorubicin killing indicated by dashed line (CTRL = 0.23 μM, PGC-1α = 0.04 μM). G. Clonogenic survival of CTRL and PGC-1α transduced RCC4 cells treated with the indicated concentration of doxorubicin. * p<0.02; ** p=0.01 by Student's two-tailed t-test. H. Clonogenic survival of CTRL and PGC-1α transduced RCC4 cells treated with 0, 2, 4, or 6 Gy of ionizing radiation. All bar graphs plotted as mean ± SD. See also Figure S4.
ccRCC is resistant to cytotoxic chemotherapy and radiotherapy and we hypothesized that PGC-1α would sensitize ccRCC cells to cytotoxic therapies. To test this hypothesis, we exposed RCC4 cells to doxorubicin and found that ectopic expression of PGC-1α sensitized ccRCC cells to doxorubicin treatment (Figures 3F). Consistent with this result, RCC4 cells transduced with PGC-1α exhibited a significant decrease in clonogenic survival compared to control (Figure 3G). Similarly, PGC-1α expression dramatically sensitized RCC4 cells to ionizing radiation as measured using the clonogenic survival assay (Figure 3H). These results indicate that therapeutic induction of PGC-1α expression may be an unrecognized mechanism to enhance the efficacy of chemo- and radiotherapy in the treatment of ccRCC.
HIF-α dependent suppression of PGC-1α in VHL-deficiency and Hypoxia
To further assess the potential of PGC-1α induction as a therapeutic modality in ccRCC, we sought to characterize the molecular mechanisms underlying suppression of PGC-1α in ccRCC. The tumor suppressor function of VHL is tightly linked with its ability to regulate stability of HIF-1α and HIF-2α. To determine whether PGC-1α suppression in VHL-deficient ccRCC cells required HIF-α expression, we used shRNAs to target expression of either HIF-1α or HIF-2α. Indicating an essential role for HIFs in mediating PGC-1α suppression, knockdown of either HIF-1α or HIF-2α resulted in increased PGC-1α expression in VHL-deficient RCC4 and 786-O cells indicating that HIF-α expression is required for suppression of PGC-1α (Figure 4A-B, S5A). We further probed the role of HIF transcriptional regulatory activity in PGC-1α suppression by assessing whether the obligate HIF-α binding partner, ARNT was required for PGC-1α suppression. Consistent with the results from HIF-1α and HIF-2α knockdown, suppression of ARNT resulted in increased levels of PGC-1α protein and mRNA in VHL-deficient cells (Figure 4C-D, S5B). These results demonstrate that a functional HIF-α/ARNT transcriptional complex is required for suppression of PGC-1α in VHL-deficient ccRCC.
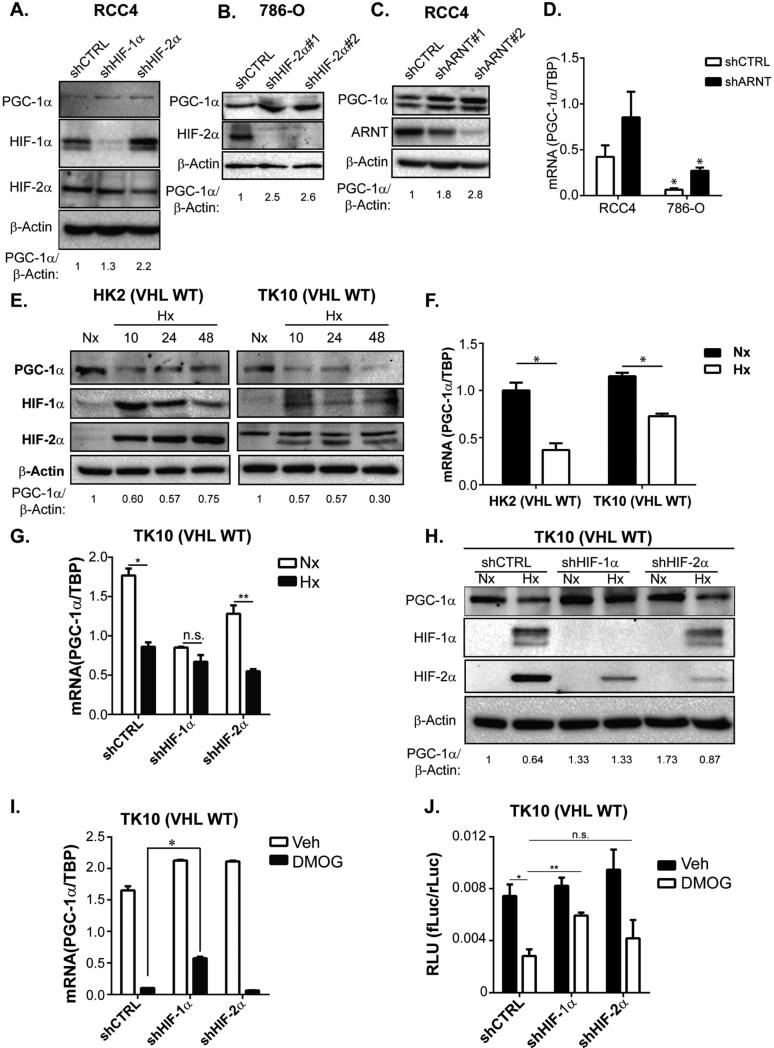
Figure 4. Regulation of PGC-1α expression by HIF-1α and HIF-2α.
A. Western Blots of VHL deficient RCC4 cells transduced with shCTRL, shHIF-1α, or shHIF-2α B. Western Blots of VHL-deficient 786-O cells transduced with shCTRL or shHIF-2α. C. Western Blots of RCC4 cells transduced with shCTRL or shARNT. D. PGC-1α mRNA levels in RCC4 and 786-O cells transduced with shCTRL or shARNT. * p<0.05 by Student's two-tailed t-test. _E._ Western Blots of VHL-wild type HK2 and TK10 cells cultured in normoxia (21% O2, Nx) or hypoxia (0.5% O2, Hx) for indicated time (hrs). _F._ PGC-1α mRNA levels in HK2 and TK10 cells cultured in normoxia (21% O2, Nx) or hypoxia (0.5% O2, Hx) for 24hrs. * p<0.01 by Student's two-tailed t-test. G. PGC-1α mRNA levels in TK10 cells transduced with shCTRL, shHIF-1α or shHIF-2α shRNA constructs and cultured in normoxia (21% O2, Nx) or hypoxia (0.5% O2, Hx) for 24hrs. * p<0.0005; ** p<0.001; n.s. p>0.06 by Student's two-tailed t-test. H. Western Blots of TK10 cells transduced with shCTRL, shHIF-1α, or shHIF-2α and cultured in normoxia (21% O2, Nx) or hypoxia (0.5% O2, Hx) for 24 hrs. I. PGC-1α mRNA levels in TK10 cells transduced with shCTRL, shHIF-1α or shHIF-2α shRNA constructs and treated with 1 mM DMOG or vehicle control (CTRL) for 24hrs. * p<0.001 by Student's two-tailed t-test. J. Ppargc1a promoter reporter activity (firefly luciferase/CMV-renilla luciferase activity (RLU)) in TK10 cells transduced with control, shCTRL, shHIF-1α, or shHIF-2α and cultured in vehicle (Veh) or 1 mM DMOG for 18hrs. Two-tailed Student's t-test results: * shCTRL Veh vs. shCTRL DMOG: p<0.05; ** shCTRL DMOG vs. shHIF-1α DMOG: p<0.005; n.s. shCTRL DMOG vs. shHIF-2α DMOG: p=0.23. All bar graphs plotted as mean ± SD. See also Figure S5.
While prevalent in ccRCC, VHL-loss of function mutations are rare in most tumor types. In contrast, hypoxia is a nearly ubiquitous characteristic of solid tumors. To determine whether HIF-dependent regulation of PGC-1α is intact in VHL-wild type cells exposed to low oxygen tensions, we compared expression of PGC-1α protein and mRNA in cell lines cultured in normoxia or hypoxia. Consistent with our results in VHL-deficient cells, hypoxia suppressed PGC-1α protein and mRNA levels in both non-transformed HK2 proximal tubule cells and in VHL-wild type TK10 ccRCC cells (Figure 4E-F). When compared to normoxia counterparts, knockdown of HIF-1α blunted the decrease in PGC-1α mRNA and protein in cells exposed to hypoxia, indicating that this effect was HIF-dependent (Figures 4G-H, S5C). Similarly, HIF-α stabilization induced by treatment with the prolyl hydroxylase inhibitor, dimethyloxaloylglycine (DMOG), resulted in HIF-1α-dependent suppression of PGC-1α mRNA (Figure 4I). Treatment with DMOG also resulted in HIF-1α-dependent inhibition of Ppargc1a promoter activity, indicating that diminished expression of PGC-1α likely occurs at least in part through transcriptional repression (Figure 4J). These results demonstrate that PGC-1α transcription is inhibited in a HIF-dependent manner in ccRCC.
Dec1 is necessary and sufficient for transcriptional repression of PGC-1α
While hypoxia is associated with widespread changes in gene expression, including both induction and repression of numerous genes, HIF-1α and HIF-2α function exclusively as transcriptional activators of their direct target genes (Schodel et al., 2011). We thus hypothesized that a transcriptional repressor must function as an intermediate in PGC-1α suppression. The basic helix-loop-helix transcription factor Dec1 (Stra13/Bhlhe40) is a hypoxia inducible gene that acts as a transcriptional repressor (Sun and Taneja, 2000). Interestingly, Dec1 has previously been reported to suppress PPARγ2 during adipocyte differentiation and PGC-1α during myogenesis (Hsiao et al., 2009; Yun et al., 2002). We thus hypothesized that Dec1 acts downstream of HIF-stabilization to functionally suppress PGC-1α in ccRCC.
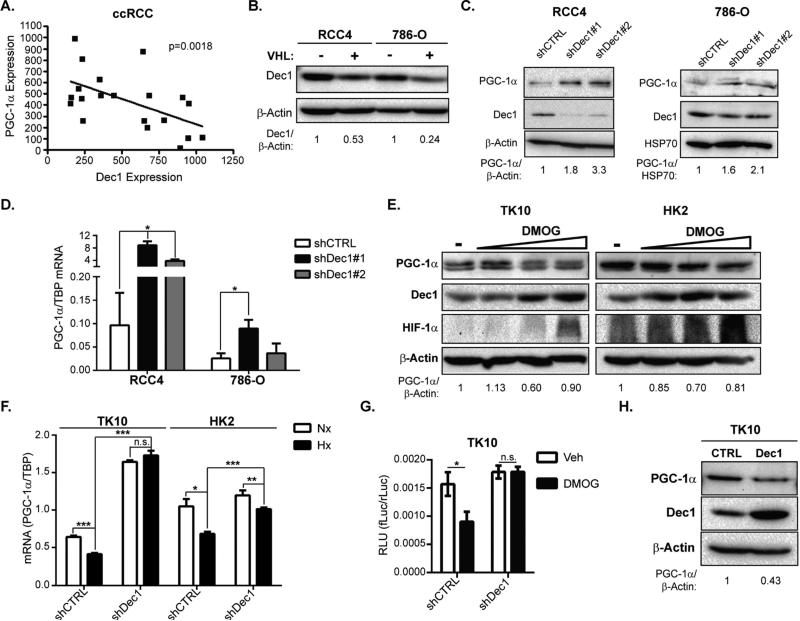
In support of this hypothesis, Dec1 expression was inversely correlated with PGC-1α in ccRCC and normal kidney samples (Figure 5A). Dec1 protein levels were elevated in VHL-deficient ccRCC cell lines and suppression of Dec1 in these cells increased PGC-1α mRNA and protein levels (Figure 5B-D). In VHL-wild type cells, DMOG treatment increased Dec1 protein levels, while decreasing PGC-1α protein levels (Figure 5E). Consistent with an essential role for Dec1 as an effector of HIF-mediated PGC-1α suppression, genetic inhibition Dec1 abrogated suppression of PGC-1α expression in hypoxia (Figure 5F). Additionally, in VHL-wild type TK10 cells, inhibition of Ppargc1a promoter activity by DMOG treatment required expression of Dec1 (Figure 5G). Phylogenetic analysis of the Ppargc1a promoter revealed two highly conserved putative Dec1 binding sites, indicating the evolutionary importance of Dec1-mediated regulation of PGC-1α (Supplemental Figure S5D-F). To examine whether Dec1 could suppress PGC-1α in the absence of HIF-activation, we ectopically expressed Dec1 in TK10 cells, and found that Dec1 expression was sufficient to suppress PGC-1α (Figure 5H). Together, these results indicate that Dec1 is necessary and sufficient for suppression of PGC-1α in ccRCC and provide a mechanistic link between HIF, the predominant oncogenic driver of ccRCC, and PGC-1α.
Figure 5. Dec1 is necessary and sufficient for transcriptional suppression of PGC-1α.
A. mRNA expression of PGC-1α and Dec1 in renal cortex and ccRCC tumors. GEO Accession # GSE6344. Pearson correlation co-efficient ρ = −0.55, two-tailed t-test p=0.0018 B. Western Blots of VHL deficient (−VHL) or wild type (+VHL) RCC4 and 786-O cells. C. Western Blots of RCC4 and 786-O cells transduced with shCTRL or shDec1. D. PGC-1α mRNA levels in RCC4 and 786-O cells transduced with shCTRL or shDec1. * p=0.01 by Student's two-tailed t-test shDec1 vs. shCTRL. E. Western Blots of VHL-wild type HK2 and TK10 cells treated with vehicle or 0.1, 0.5, or 1 mM DMOG for 24 hrs. F. PGC-1α mRNA levels in TK10 and HK2 cells transduced with shCTRL or shDec1 and cultured in normoxia (Nx) or hypoxia (Hx) for 24hrs. * p=0.017; ** p < 0.05; *** p≤0.0001.; n.s. p>0.05 by Student's two-tailed t-test. G. Ppargc1a promoter reporter activity (firefly luciferase/CMV-renilla luciferase activity (RLU)) in vehicle (Veh) and DMOG (1 mM) treated TK10 cells transduced with shCTRL, or shDec1. * p<0.02; n.s. p>0.05 by Student's two-tailed t-test. H. Western Blots of TK10 cells ectopically expressing Dec1. Bar graphs plotted as mean ± SD. See also Figure S5.
PGC-1α expression suppresses ccRCC tumor growth
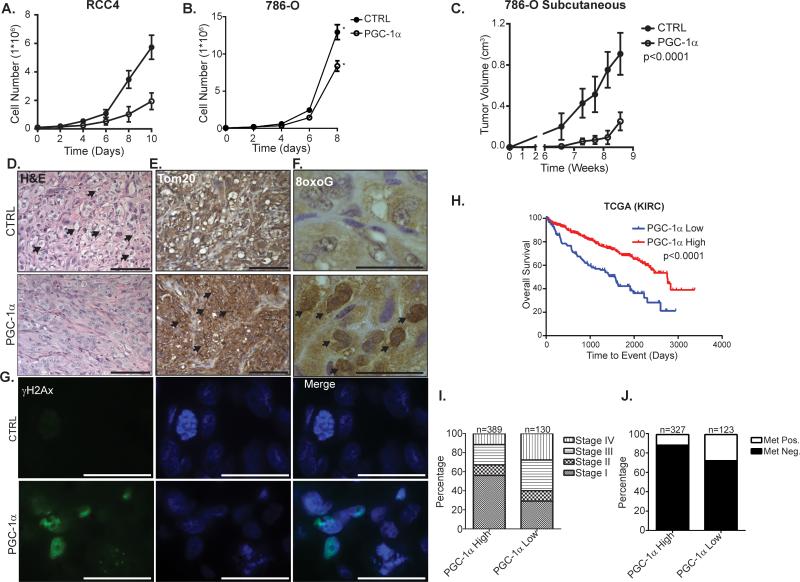
Given our finding that ectopic PGC-1α expression stimulates oxidative stress in ccRCC cells, we hypothesized that restored expression of PGC-1α could suppress ccRCC growth. Supporting this hypothesis PGC-1α expression inhibited growth of two VHL-deficient ccRCC cell lines in vitro (Figure 6A-B). Consistent with the anti-proliferative effects of PGC-1α expression in vitro, subcutaneous growth of 786-O tumors was significantly impaired by expression of PGC-1α (Figures 6C, S6). Furthermore, PGC-1α transduced 786-O tumors were characterized by an apparent reduction in the clear cell morphology (Figure 6D). Consistent with our in vitro data indicating that PGC-1α expression is sufficient to stimulate increased mitochondrial content, IHC analysis of 786-O tumors revealed increased expression of the mitochondrial protein, Tom20 in tumors expressing PGC-1α (Figures 6E). Corroborating our in vitro data that PGC-1α expression induces oxidative stress in ccRCC cells, PGC-1α transduced tumors exhibited elevated levels of 8oxoG and γH2AX (Figures 6F-G). These results indicate that increased expression of PGC-1α inhibits ccRCC growth and highlight the potential of PGC-1α induction as a therapeutic modality in the treatment of ccRCC.
Figure 6. PGC-1α suppresses ccRCC growth.
A. in vitro growth curves of CTRL or PGC-1α transduced RCC4 cells. B. in vitro growth curves of CTRL or PGC-1α transduced 786-O cells. * p<0.05 by Student's two-tailed t-test. C. Subcutaneous tumor volume of CTRL or PGC-1α transduced 786-O cells. Mixed effects model p<0.0001 between CTRL vs. PGC-1α. D. H&E stained CTRL or PGC-1α transduced 786-O subcutaneous tumors. Arrows denote cells with evident clear cell morphology. Scale bar, 100 μm. E. Tom20 immunohistochemical (IHC) staining of CTRL or PGC-1α transduced 786-O subcutaneous tumors. Arrows denote cells with strong perinuclear Tom20 staining. Scale bar, 100 μm. F. 8oxoG IHC staining of CTRL or PGC-1α transduced 786-O subcutaneous tumors. Arrows denote nuclei with strongly positive 8oxoG staining. Scale bar, 25 μm G. γH2Ax IF staining of CTRL or PGC-1α transduced 786-O subcutaneous tumors. Nuclei were counterstained with DAPI. Scale bar 25 μm. H. Kaplan-Meier Curve for ccRCC patients segregated by low (bottom quartile) or high expression of PGC-1α. Log-rank test p<0.0001. I. Percentage of patients with AJCC stage I-IV disease, segregated by PGC-1α expression as in panel H. J. Percentage of patients with detectable metastases. Patients segregated by PGC-1α expression as in H. All bar graphs presented as mean ± S.D. See also Figure S6.
To assess the impact of PGC-1α expression in clinical cases of ccRCC, we next analyzed RNA sequencing data from over 500 patients with ccRCC from the Cancer Genome Atlas (TCGA) to further explore the role of PGC-1α in ccRCC tumorigenesis. These analyses revealed that low expression of PGC-1α was significantly correlated with worse overall survival than patients whose tumors expressed relatively high levels of PGC-1α (Figure 6H). Consistent with these findings, patients harboring tumors with low expression of PGC-1α were significantly more likely to have advanced stage tumors and metastatic disease (Figure 6I-J). Together, these results indicate that low expression of PGC-1α may be a prognostic factor in ccRCC.
Discussion
Our data indicate that PGC-1α is suppressed in _VHL_-deficient ccRCC through HIF-dependent induction of the transcriptional repressor, Dec1. In VHL-wild type proximal tubule cells, PGC-1α suppression disrupts mitochondrial respiration. Conversely, rescued expression of PGC-1α in VHL-deficient ccRCC cell lines restores mitochondrial function in an ERRα dependent manner. Importantly, the increases in mitochondrial respiration upon expression of PGC-1α are associated with induction of oxidative stress in ccRCC cell lines. In addition to stimulating oxidative stress, PGC-1α expression also sensitizes VHL-deficient ccRCC lines to cytotoxic therapies and inhibits in vitro and in vivo growth, highlighting the potential of targeting this pathway in the treatment of ccRCC. In clinical ccRCC datasets, PGC-1α expression is decreased compared to normal renal cortex, and low levels of PGC-1α expression are associated with poor outcome, further indicating that PGC-1α can suppress ccRCC tumorigenesis.
PGC-1α has been reported to have pro- or anti-tumorigenic function in various tumors, but the reasons for this heterogeneous response remain unclear. One possibility is that PGC-1α interacts with different tissue specific transcription factors, thus driving distinct genetic programs in different cancer types. However, increased mitochondrial biogenesis appears to be a ubiquitous response to PGC-1α expression regardless of the cancer type or outcome of PGC-1α expression. Given this, we postulate that the outcome of PGC-1α expression on tumor growth is likely dictated at least in part by whether the genetic landscape or tumor microenvironment are permissive to increased mitochondrial activity. For example, LeBleu, et al. report increased PGC-1α in circulating tumor cells (CTCs) relative to the primary orthotopic tumor. One could reasonably expect the metabolic demands of CTCs in the oxygen rich circulatory system to be different from a hypoxic cancer cell. In the case of ccRCC, the increased production of ROS upon expression of PGC-1α is inadequately detoxified, leading to oxidative stress. Whether the antioxidant response dictates outcome to enhanced mitochondrial biogenesis downstream of PGC-1α activation will require further investigation.
PGC-1α expression is induced in a subset of melanomas where its expression is regulated by MITF (Haq et al., 2013; Vazquez et al., 2013). Interestingly, hypoxia negatively regulates MITF expression through induction of Dec1, the same transcriptional repressor implicated in our studies as a repressor of PGC-1α (Feige et al., 2011). These results suggest that the tumor microenvironment could play a meaningful role in dictating tumor response to PGC-1α expression. Future studies will be needed to fully understand the mechanisms underlying these differences in tumor response to PGC-1α, and the clinical implications thereof.
Here we demonstrate that restored expression of PGC-1α increases mitochondrial content, ROS production, and oxidative damage, with the ultimate effect of impaired tumor growth and enhanced therapeutic response. ccRCC tumors are typically refractory to cytotoxic chemotherapy and radiotherapy, but the induction of PGC-1α may present an opportunity to enhance efficacy and improve the treatment of ccRCC. Despite the apparent difficulties associated with targeting transcription factors, several groups have conducted high throughput screens that have identified both transcriptional and post-translational activators of PGC-1α (Arany et al., 2008; Lee et al., 2014). In the future, it will be important to determine whether the pharmacological activation of PGC-1α, either through increasing PGC-1α expression or activating the existing pool of endogenous PGC-1α, is a viable therapeutic modality for the treatment of ccRCC.
Experimental Procedures
Cell Culture
Cells were cultured in DMEM + 10% complete or charcoal-stripped FBS where indicated. Human PGC-1α cDNA was ectopically expressed by retroviral transduction using the pLPC vector. All shRNA constructs other than shHIF-1α (pSIREN) were in pLKO.1 vector and were purchased from Open Biosystems. Cell viability was assessed by XTT assay as described previously (Turcotte et al., 2008). For clonogenic assays, cells were pre-treated for 48 hours with the indicated dose of doxorubicin, or irradiated with the indicated dose of ionizing radiation, cultured for 10 days and stained with crystal violet to visualize colonies. Clonogenic survival was determined by dividing the number of surviving colonies by the number of cells plated. For siRNA experiments, ON-TARGETplus siRNA SMARTpools containing 4 siRNAs were purchased from Dharmacon. siRNAs were transfected into cells using Dharmafect1 transfection reagent (Dharmacon) and cells were collected for gene expression analysis or extracellular flux analysis 48hrs post-transfection. All cell lines used in these studies have been previously described elsewhere (Bear et al., 1987; Maxwell et al., 1999; Ryan et al., 1994).
Extracellular Flux Analysis
OCR/ECAR measurements were performed using XF96 extracellular flux analyzer (Seahorse Biosciences). Experiments were conducted in DMEM with 5% FBS, 25 mM glucose, 2 mM glutamine, and 1 mM pyruvate, but without NaHCO3. Inhibitors were injected to final concentrations of: 2.5 μg/mL oligomycin, 5 μM FCCP, 2.5 μM antimycin A, and 1 μM rotenone, 100 μM etomoxir. All chemicals were purchased from Sigma-Aldrich.
GC/MS Metabolomics
Cell pellets were extracted with 0.45 ml water/ methanol (1:1 volume, containing 20 μM L-norvaline) and 0.225 ml chloroform containing 4 μg/ml heptadecanoic acid as described in detail before (Ratnikov et al., 2015; Scott et al., 2011). Polar metabolites extracted into the water/ methanol phase were derivatized and analyzed by GC-MS.
Luciferase Assays
A 1.8 kb region of the proximal Ppargc1a promoter was cloned from HK2 cells into the pGL3-basic reporter plasmid (Promega). TK10 cells were transiently transfected with this construct along with CMV-Renilla reporter to monitor transfection efficiency. Firefly and renilla luciferase levels were quantified with Bright-Glo and Renilla luciferase assays (Promega).
Microarray and RNA Sequencing Analysis
GSE6344 and GSE15641 were selected for analysis from the GEO database due to the representation of both normal renal cortex and ccRCC specimens within each dataset (Gumz et al., 2007; Jones et al., 2005; Tun et al., 2010). For GSE15641, data from non-ccRCC subtypes of kidney cancer were excluded from our analysis. Datasets were analyzed using the GSEA module in the GenePattern software suite (Reich et al., 2006; Subramanian et al., 2005). Microarrays comparing expression changes in VHL-deficient and wild type RCC4 cells and after culture of RCC4+VHL cells in hypoxia have been previously described (Krieg et al., 2010). Differentially expressed genes above or below a fold change threshold (>1.5 fold or <-1.5 fold) in both datasets were analyzed using the DAVID functional annotation tool (Huang da et al., 2009a; Huang da et al., 2009b).
Ppargc1a expression levels were extracted from Level 3 RNA sequencing data from the KIRC TCGA dataset using the “normalized_count” parameter (http://tcga-data.nci.nih.gov/tcga/) and matched by TCGA barcode with corresponding clinical information including time to death or last follow up, American Joint Committee on Cancer tumor staging, and the presence or absence of metastatic disease. To segregate patients into high or low expression of PGC-1α, the median expression from all patients in the dataset was calculated and patients were segregated into the PGC-1α low group if their PGC-1α expression in the tumor was in the bottom quartile of expression. The remainder of patients were included in the PGC-1α high group.
Microscopy and Immunohistochemistry
For fluorescent labeling of mitochondria, live cells were incubated with 100 nM CMXROS (Molecular Probes) in buffer containing 100 mM KCl to depolarize the mitochondrial membrane for 30 minutes at 37 °C. Antibodies used for immunohistochemical (IHC) and immunofluorescence experiments: 8-hydroxyguanosine (Abcam), γH2AX (Millipore), and Tom20 (Santa Cruz). For IHC experiments, primary antibody labeling was detected after incubation with the appropriate secondary antibody by incubation with 3, 3'-diaminobenzidine (DAB, Vector Labs). For IF experiments, nuclei were counterstained for 5 minutes with 4',6-diamidino-2-phenylindole (DAPI). Cells were counterstained with hematoxylin.
Quantitative Real Time-PCR
mRNA was extracted from cells using Trizol (Invitrogen) and reverse transcribed using Superscript II reverse transcriptase (Invitrogen). Relative mRNA levels were calculated using the standard curve methodology. For mitochondrial DNA quantification, total DNA was isolated from cultured cells using phenol/chloroform/isoamyl alcohol (Invitrogen). Primers were designed to amplify the 16S rRNA gene from the mtDNA genome and were normalized to the nuclear gene β2-microglobulin. A complete list of primer sets can be found in Table S1.
ROS Measurements
Live cells were stained with either DCF-DA or MitoSOX stains according to manufacturer recommended protocol. Briefly, cells were incubated with 10 μM DCF-DA (Molecular Probes) or 5 μM MitoSOX Red (Life Technologies) for 30 min at 37 °C. After PBS wash, cells were analyzed by flow cytometry using an LSRII flow cytometer (Becton Dickenson).
Western Blotting
Western Blot images were acquired using the ChemiDoc CCD imaging system (BioRad). The following antibodies were used for Western Blotting: ARNT (Novus), β-Actin (Sigma), Dec1 (Novus), HIF-1α (BD), HIF-2α (Novus), HSP70 (Sigma), PGC-1α (Sigma and Santa Cruz), Tfam (Sigma). Densitometry analysis was performed using ImageJ software and relative values were calculated by normalizing to the control group from each experiment (Schneider et al., 2012).
Xenograft Tumor Growth
5 × 106 CTRL or PGC-1α transduced 786-O cells were injected into the flanks of NOD/SCID mice (n=5 for control group, n=8 for PGC-1α group). Tumor size was measured weekly, and volume was calculated according to the following formula: volume = (width_2×_length)÷2.
The tumor volumes of the mice in both treatment groups (CTRL and PGC-1α) followed a log normal distribution and thus the data was log transformed to render the data normally distributed and to analyze the data in a mixed effect model. The data was analyzed in a mixed effects model to accommodate the repeated measure of tumor volume over time. The effect of treatment was included as fixed effects in the model and day was included as random effect. The variance-covariance matrix was assumed to have a compound symmetry structure. In the final model treatment effects (CTRL vs. PGC-1α) was significant (treatment p < 0.0001).
All animal experiments were conducted in accordance with standard IACUC protocols.
Supplementary Material
1
2
Acknowledgements
The authors would like to thank Adam Krieg for generating the RCC4 VHL microarray datasets used for DAVID analysis in Figure 1B (Krieg, et al. 2010). This work was funded by National Institutes of Health grants CA67166 and CA116685. E.L.L. was supported by US National Cancer Institute Training Grant CA121940. C.W. was supported by a training grant from the Canadian Institutes of Health and Research. C.M.T. was supported by RSNA(Radiological Society of North America) Resident Research Grants 1018 and 1111. C.K.D. and J.T.C. were supported by the following grants: NIH CA125618, CA106520 and the Department of Defense W81XWH-12-1-0148 (J.T.C). A.J.G. was supported by grants from NIH (CA 67166 and 88480), the Silicon Valley Foundation, and the Skippy Frank Foundation. The Cancer Metabolism facility at SBMRI is supported by NCI award number 5P30CA030199.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
E.L.L. and A.J.G. designed all experiments, wrote and revised the manuscript, and shared oversight of this project. E.L.L. performed and analyzed data for most experiments. C.W. provided critical assistance with in vivo tumor growth studies and revised manuscript. C.M.T. performed cloning of PGC-1α constructs. C.K.D. and J.T.C. assisted with bioinformatics analysis, including analysis of Ppargc1a expression in ccRCC GEO datasets. D.A.S. and A.D.R. assisted in experimental design and execution of GC-MS analysis of TCA cycle metabolite abundance.
References
- Arany Z, Wagner BK, Ma Y, Chinsomboon J, Laznik D, Spiegelman BM. Gene expression-based screening identifies microtubule inhibitors as inducers of PGC-1alpha and oxidative phosphorylation. Proc Natl Acad Sci U S A. 2008;105:4721–4726. doi: 10.1073/pnas.0800979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear A, Clayman RV, Elbers J, Limas C, Wang N, Stone K, Gebhard R, Prigge W, Palmer J. Characterization of two human cell lines (TK-10, TK-164) of renal cell cancer. Cancer Res. 1987;47:3856–3862. [PubMed] [Google Scholar]
- Cancer Genome Research Atlas Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Errico I, Salvatore L, Murzilli S, Lo Sasso G, Latorre D, Martelli N, Egorova AV, Polishuck R, Madeyski-Bengtson K, Lelliott C, et al. Peroxisome proliferator-activated receptor-gamma coactivator 1-alpha (PGC1alpha) is a metabolic regulator of intestinal epithelial cell fate. Proc Natl Acad Sci U S A. 2011;108:6603–6608. doi: 10.1073/pnas.1016354108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige E, Yokoyama S, Levy C, Khaled M, Igras V, Lin RJ, Lee S, Widlund HR, Granter SR, Kung AL, Fisher DE. Hypoxia-induced transcriptional repression of the melanoma-associated oncogene MITF. Proc Natl Acad Sci U S A. 2011;108:E924–933. doi: 10.1073/pnas.1106351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, Latif F, Liu S, Chen F, Duh FM, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- Gordan JD, Simon MC. Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr Opin Genet Dev. 2007;17:71–77. doi: 10.1016/j.gde.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, Frederick DT, Hurley AD, Nellore A, Kung AL, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao SP, Huang KM, Chang HY, Chen SL. P/CAF rescues the Bhlhe40-mediated repression of MyoD transactivation. Biochem J. 2009;422:343–352. doi: 10.1042/BJ20090072. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- Krieg AJ, Rankin EB, Chan D, Razorenova O, Fernandez S, Giaccia AJ. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30:344–353. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan J, Danzer C, Simka T, Ukropec J, Walter KM, Kumpf S, Mirtschink P, Ukropcova B, Gasperikova D, Pedrazzini T, Krek W. Dietary obesity-associated Hif1alpha activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD+ system. Genes Dev. 2012;26:259–270. doi: 10.1101/gad.180406.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu VS, O'Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014 doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Dominy JE, Choi YJ, Jurczak M, Tolliday N, Camporez JP, Chim H, Lim JH, Ruan HB, Yang X, et al. Cyclin D1-Cdk4 controls glucose metabolism independently of cell cycle progression. Nature. 2014;510:547–551. doi: 10.1038/nature13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Luo C, Vazquez F, Puigserver P. Targeting mitochondrial oxidative metabolism in melanoma causes metabolic compensation through glucose and glutamine utilization. Cancer Res. 2014;74:3535–3545. doi: 10.1158/0008-5472.CAN-13-2893-T. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol. 2010;7:277–285. doi: 10.1038/nrurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan KA, Cocchiglia S, Zhdanov AV, Tambuwala MM, Cummins EP, Monfared M, Agbor TA, Garvey JF, Papkovsky DB, Taylor CT, Allan BB. PGC-1alpha is coupled to HIF-1alpha-dependent gene expression by increasing mitochondrial oxygen consumption in skeletal muscle cells. Proc Natl Acad Sci U S A. 2009;106:2188–2193. doi: 10.1073/pnas.0808801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Ratnikov B, Aza-Blanc P, Ronai ZA, Smith JW, Osterman AL, Scott DA. Glutamate and asparagine cataplerosis underlie glutamine addiction in melanoma. Oncotarget. 2015;5 doi: 10.18632/oncotarget.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373:1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- Ryan MJ, Johnson G, Kirk J, Fuerstenberg SM, Zager RA, Torok-Storb B. HK-2: an immortalized proximal tubule epithelial cell line from normal adult human kidney. Kidney Int. 1994;45:48–57. doi: 10.1038/ki.1994.6. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117:e207–217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DA, Richardson AD, Filipp FV, Knutzen CA, Chiang GG, Ronai ZA, Osterman AL, Smith JW. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. J Biol Chem. 2011;286:42626–42634. doi: 10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoag J, Arany Z. Regulation of hypoxia-inducible genes by PGC-1 alpha. Arterioscler Thromb Vasc Biol. 2010;30:662–666. doi: 10.1161/ATVBAHA.108.181636. [DOI] [PubMed] [Google Scholar]
- Simonnet H, Alazard N, Pfeiffer K, Gallou C, Beroud C, Demont J, Bouvier R, Schagger H, Godinot C. Low mitochondrial respiratory chain content correlates with tumor aggressiveness in renal cell carcinoma. Carcinogenesis. 2002;23:759–768. doi: 10.1093/carcin/23.5.759. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Taneja R. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Proc Natl Acad Sci U S A. 2000;97:4058–4063. doi: 10.1073/pnas.070526297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- Tun HW, Marlow LA, von Roemeling CA, Cooper SJ, Kreinest P, Wu K, Luxon BA, Sinha M, Anastasiadis PZ, Copland JA. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. PLoS One. 2010;5:e10696. doi: 10.1371/journal.pone.0010696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte S, Chan DA, Sutphin PD, Hay MP, Denny WA, Giaccia AJ. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell. 2008;14:90–102. doi: 10.1016/j.ccr.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K, Clish CB, Granter SR, Widlund HR, Spiegelman BM, Puigserver P. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Gingras MC, Dunlop EA, Nouet Y, Dupuy F, Jalali Z, Possik E, Coull BJ, Kharitidi D, Dydensborg AB, et al. The tumor suppressor folliculin regulates AMPK-dependent metabolic transformation. J Clin Invest. 2014;124:2640–2650. doi: 10.1172/JCI71749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun Z, Maecker HL, Johnson RS, Giaccia AJ. Inhibition of PPAR gamma 2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev Cell. 2002;2:331–341. doi: 10.1016/s1534-5807(02)00131-4. [DOI] [PubMed] [Google Scholar]
- Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2