Exome-wide Rare Variant Analysis Identifies TUBA4A Mutations Associated with Familial ALS (original) (raw)
. Author manuscript; available in PMC: 2015 Oct 22.
SUMMARY
Exome sequencing is an effective strategy for identifying human disease genes. However, this methodology is difficult in late-onset diseases where limited availability of DNA from informative family members prohibits comprehensive segregation analysis. To overcome this limitation, we performed an exome-wide rare variant burden analysis of 363 index cases with familial ALS (FALS). The results revealed an excess of patient variants within TUBA4A, the gene encoding the Tubulin, Alpha 4A protein. Analysis of a further 272 FALS cases and 5,510 internal controls confirmed the overrepresentation as statistically significant and replicable. Functional analyses revealed that TUBA4A mutants destabilize the microtubule network, diminishing its repolymerization capability. These results further emphasize the role of cytoskeletal defects in ALS and demonstrate the power of gene-based rare variant analyses in situations where causal genes cannot be identified through traditional segregation analysis.
INTRODUCTION
The identification of mutations for human disorders is critical for our understanding of their pathogenesis. Although numerous studies have successfully used exome sequencing to identify novel disease genes, this approach is still limited by the fact that each individual harbors many rare variants, thus requiring DNA from multiple affected individuals from an affected family to be successful. Unfortunately, for many diseases the collection of multiple affected samples from the same family can be difficult. This is especially true for familial amyotrophic lateral sclerosis (FALS), a late-onset, rapidly progressive neurodegenerative disease. As such, there is a need to develop approaches to use individual affected cases to identify disease genes. Exome-wide rare variant analysis has been proposed as a promising alternative strategy (Panoutsopoulou et al., 2013; Stitziel et al., 2011). By this approach, the total number of gene variants seen in cases is compared to that in controls. Threshold minor allele frequency and possibly functional significance filters are implemented to prevent statistical noise from common or neutral variants masking true gene associations. Importantly, although an abundance of variants in cases suggests the presence of pathogenic mutations, an associated variant set will still likely include neutral variants in both cases and controls. Since it does not rely on related samples, we decided to use this approach to identify novel causative genes for FALS.
RESULTS
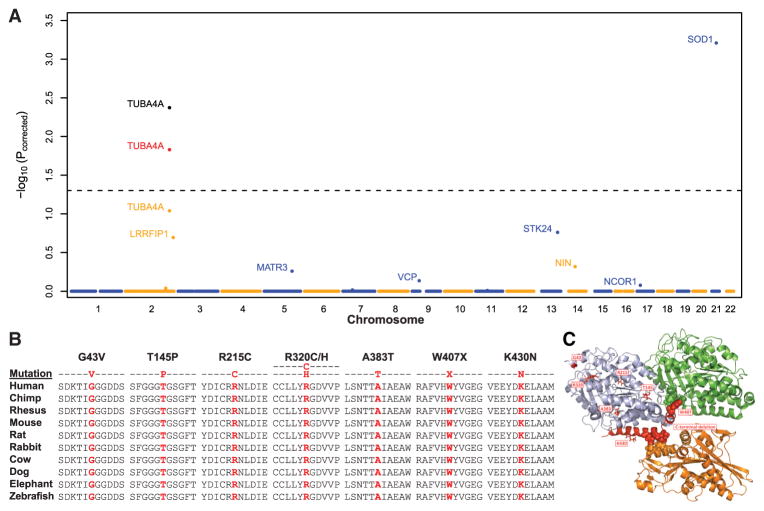
We performed exome sequencing on a discovery cohort of 363 index cases (i.e., one affected sample per family) from six countries (Supplemental Experimental Procedures available online) devoid of mutations/repeat expansions in known ALS genes (SOD1, C9orf72, TARDBP, FUS, PFN1, UBQLN2, OPTN, VCP, and ANG). An average of 3.2 × 109 target bases was sequenced per sample to an average depth of 90.4×. Control exome data included 4,300 European Americans available through the NHLBI’s Exome Variant Server (EVS) (Tennessen et al., 2012) and 31 internally sequenced samples. As proof of principle, we spiked our sample set with nine FALS exomes with known causative mutations (six in SOD1 and three in VCP). The analysis was restricted to the 12,487 genes fulfilling all quality control filters and to variants either predicted to be damaging by PolyPhen-2 or resulting in a stop gain/loss. Variants were also only considered if they exhibited minor allele frequencies less than 0.04%, in keeping with previously reported ALS mutations (Chiò et al., 2012; Kenna et al., 2013) (Supplemental Experimental Procedures). The significance of disease associations following multiple test correction (Pcorrected) was calculated using permutation procedures (Kiezun et al., 2012). Strong signals of disease association were observed for SOD1 (6 cases [1.6%] versus 0 controls, p = 5.5 × 10−8, Pcorrected = 6.2 × 10−4) and VCP (3 cases [0.8%] versus 0 controls, p = 1.1 × 10−4, Pcorrected = 0.73) (Figure 1A and Figure S1). Excluding SOD1, the top hit from our analysis was TUBA4A, encoding the Tubulin, Alpha 4A protein (4 cases [1.1%] versus 0 controls, p = 9.1 × 10−6, Pcorrected = 0.09). Similar significance for TUBA4A was also obtained when using alternative bioinformatic tools to identify putatively damaging variants (SIFT, GERP, MutationTaster, and phyloP). TUBA4A ranked first or second in all tests (Table S1 and Figure S1), while no other gene ranked in the top ten hits for all five tests. Besides TUBA4A, our strategy revealed other strong candidate genes for FALS (Figure 1A; Table S1), including MATR3, responsible for distal myopathy 2 and recently identified as causative for FALS (Johnson et al., 2014). Based on these results, we further investigated TUBA4A as a candidate gene associated with FALS.
Figure 1. Rare Variant Analysis Identifies TUBA4A Mutations in FALS.
(A) Manhattan plot displaying permutation-based corrected p values generated by a rare variant analysis of FALS. The dotted line represents a Pcorrected = 0.05. The points in yellow, red, and black for TUBA4A denote the p value for the discovery, replication, and joint analyses, respectively. (B) The evolutionary conservation of TUBA4A mutations is displayed. Mutated residues are shown in red. (C) The interactions of bovine TUBA1B and TUBB2B with the rat kinesin KIF5B (Protein Data Bank accession number 4ATX) are shown using the PyMOL Molecular Graphics System (v.1.5.0.5). All relevant residues are identical between bovine TUBA1B and human TUBA4A. Light blue, α-tubulin; green, β-tubulin; orange, kinesin; red, mutant or deleted residues identified in this study; spheres, residues involved in interprotein interactions. As shown, the C-terminal region interacts with both β-tubulin and the kinesin motor domain.
Exome sequencing revealed a total of five nonsynonymous TUBA4A changes, all localized in exon 4 and confirmed by Sanger sequencing (Table S2). None of them were observed in the 4,300 EVS controls and all occurred at highly conserved positions (Figure 1B). The mutations included two missense changes in the same residue (R320C/H) and a nonsense mutation (W407X) removing the last 41 amino acids. This C-terminal region contacts the β-tubulin subunit as well as the motor domain of kinesins and other microtubule-associated proteins (Howes et al., 2014; Liu et al., 2012) (Figure 1C). Additional missense changes included R215C and A383T mutations. PolyPhen-2 predicted four of the five mutations to be deleterious (Table S2). In contrast, only three nonsynonymous changes were observed in the 4,300 EVS controls (V68A, I276V, and V375M), all predicted to be benign. No relatives of the affected patients were available to test segregation.
To further evaluate TUBA4A as a causative ALS gene, we sequenced an independent replication cohort comprising a further 272 index FALS cases and 5,510 internal European American controls for rare damaging exon 4 variants. Reimplementation of the variant filtering strategies employed during the discovery analysis and subsequent testing using Fisher’s exact method revealed a significant excess of mutation carriers among patients (2 cases [0.65%] versus 2 controls [0.04%], p = 1.5 × 10−2). A combined analysis of the discovery and replication cohorts resulted in a statistically significant overabundance of rare variants after multiple test correction (OR = 36 [95% CI: 10–210], p = 4.3 × 10−7, Pcorrected = 4.2 × 10−3). Of the two mutations observed in the replication cohort, a T145P variant segregated with disease within the family, while a K430N variant was not detected in an affected first cousin of the sequenced proband (Figure S1C), suggesting that this is either a neutral polymorphism, as is often observed in rare variant analyses, or the two affected family members are phenocopies. Variants identified in controls included E386K and G365E substitutions but none of the variants detected in FALS patients from either the discovery or replication cohorts. Sequencing of the entire coding region in 1,355 sporadic ALS (SALS) cases also identified a G43V mutation in a single sample (Table S2). This variant was predicted to be benign by PolyPhen-2 and detected within 1 control from the internal replication panel. No TUBA4A mutations were identified in 131 ALS samples (89 FALS and 42 SALS) with known mutations/repeat expansions.
All patients carrying TUBA4A mutations had spinal-onset, classical ALS, with upper and lower motor neuron signs. Two cases also developed a cognitive decline of frontal type, consistent with a diagnosis of frontotemporal dementia (FTD) and another had a first-degree relative with FTD (Figure S1 and Table S3). Screening of 1,053 samples analyzed by the 1000 Genome project and 2,200 African Americans from the EVS revealed the A383T mutation within a single individual of African ancestry. Therefore, with the exception of A383T and G43V, the set of observed patient variants were not detected within a total of 13,023 control samples (Table S4).
TUBA4A Mutants Display Altered Incorporation into Microtubules
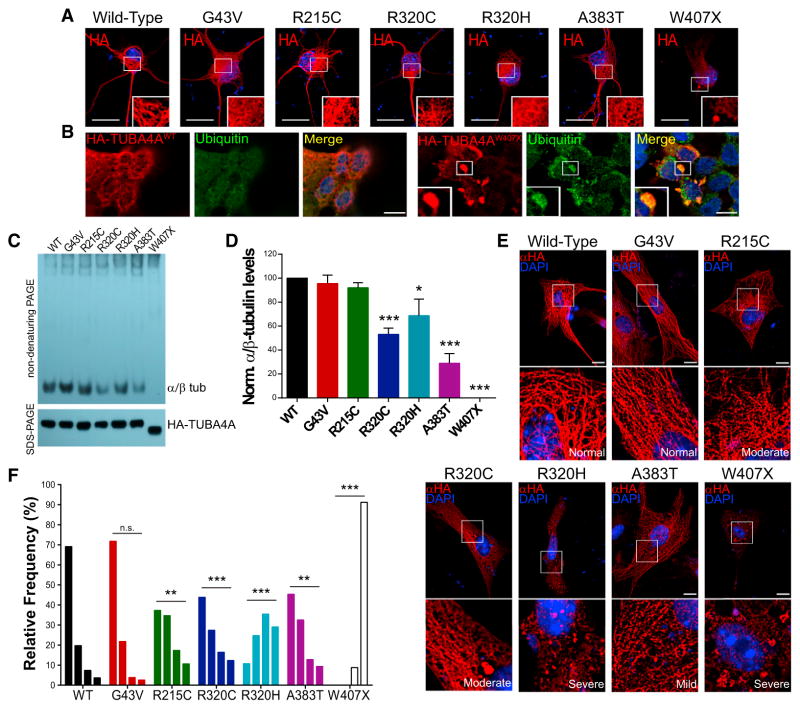
Since several causative ALS proteins form insoluble inclusions in postmortem brain tissues and in cell culture, we investigated the ability of TUBA4A mutants to aggregate by expressing hemagglutinin (HA)-tagged TUBA4A constructs in primary motor neurons (PMNs) and HEK293 cells. Interestingly, the W407X mutant did not incorporate into the microtubule network and formed small ubiquitinated cytoplasmic inclusions in ~40% of transfected PMNs and ~85% of transfected HEK293 cells (Figures 2A and 2B; Figure S2). Immunohistochemistry of brain and spinal cord tissue from SALS cases (without mutations) yielded a clear staining of the perikarya and neuropil region in both the spinal cord and motor cortex; however, no TUBA4A aggregates were identified (Figure S2). No coaggregation was observed in cells coexpressing an aggregation-prone TDP-43 C-terminal fragment and wild-type TUBA4A (Figure S2). While the other TUBA4A mutants formed cytoplasmic inclusions in 10%–30% of transfected HEK293 cells (data not shown), no aggregation was observed in PMNs. However, subtle alterations in their cytoplasmic distribution were observed, such as a more diffuse staining compared to the wild-type protein, which was mainly incorporated into the microtubule network (Figure 2A). We thus investigated the ability of mutant TUBA4A to efficiently form microtubules using a cell-free system to quantify its incorporation into α-/β-tubulin dimers (Cleveland et al., 1978; Jaglin et al., 2009; Tian et al., 2010). Each mutant was subjected to in vitro translation in rabbit reticulocyte lysate that contains all components needed to form tubulin dimers. The products were then analyzed by nondenaturing gel electrophoresis to measure incorporation of the translated recombinant protein into tubulin dimers. Interestingly, TUBA4AW407X yielded no discernible dimers, while the A383T and both R320 mutants displayed significantly lower levels of assembly relative to the wild-type protein (Figures 2C and 2D). The other mutants did not differ from the control, save for a small but reproducible migration difference for TUBA4AR215C. In parallel, we tested the incorporation of mutant TUBA4A into microtubules in cultured cells. We chose primary astroglial cells, as they have a large cytoplasm and a relatively long cell cycle that result in an extensive and stable microtubule network. We performed an unbiased analysis of the cytoskeletal incorporation of HA-tagged TUBA4A constructs. The ratio between the fluorescence intensities of the filamentous versus overall staining, ranging from 1 to 0, was used to categorize each cell as normal (1–0.75), mild (0.75–0.5), moderate (0.5–0.25), or severe (0.25–0) (Figures 2E and 2F and Figure S3). The wild-type HA-tagged TUBA4A protein yielded nearly 70% of cells displaying a high level of microtubule incorporation. In contrast, several mutants displayed an altered incorporation into microtubules. In particular, the W407X yielded the most dramatic effect with an absence of tagged protein in the microtubule network and the presence of multiple aggregate-like inclusions. The R320C/H, A383T, and R215C mutants all displayed a significantly different distribution relative to the wild-type, but no differences were detected for the G43V protein.
Figure 2. Mutant TUBA4A Alters Microtubule Polymerization.
(A) PMNs transfected with HA-tagged TUBA4A constructs. (B) HEK293 cells expressing wild-type or mutant HA-TUBA4A were immunostained for ubiquitin. Ubiquitin-positive aggregates were visible only in the HA-TUBA4AW407X-transfected cells. (C) Nondenaturing (top) and SDS-gel electrophoresis (bottom) of HA-TUBA4A constructs after in vitro translation. (D) The intensity of the bands representing the α-/β-tubulin dimers was quantified. Bars represent mean and SEM (one-way ANOVA and Dunnett’s post hoc test, n = 6, *p < 0.05, ***p < 0.001). (E) Representative images of primary astroglial cells transfected with the HA-TUBA4A constructs. (F) The ratio between HA-TUBA4A protein distributions in filaments versus its overall levels was measured and distributed into four categories from mainly filamentous to mainly diffuse. The frequency of each phenotype was compared between the wild-type and mutant proteins (Kolmogorov-Smirnov test; n = 75–90 cells per condition from 3 experiments, **p < 0.01, ***p < 0.001). From left to right, bars represent the relative frequencies of the normal, mild, moderate, and severe phenotypes. Red, HA; green, ubiquitin; blue, DAPI. White boxes indicate the regions enlarged in the insets. Scale bars, 10 μm.
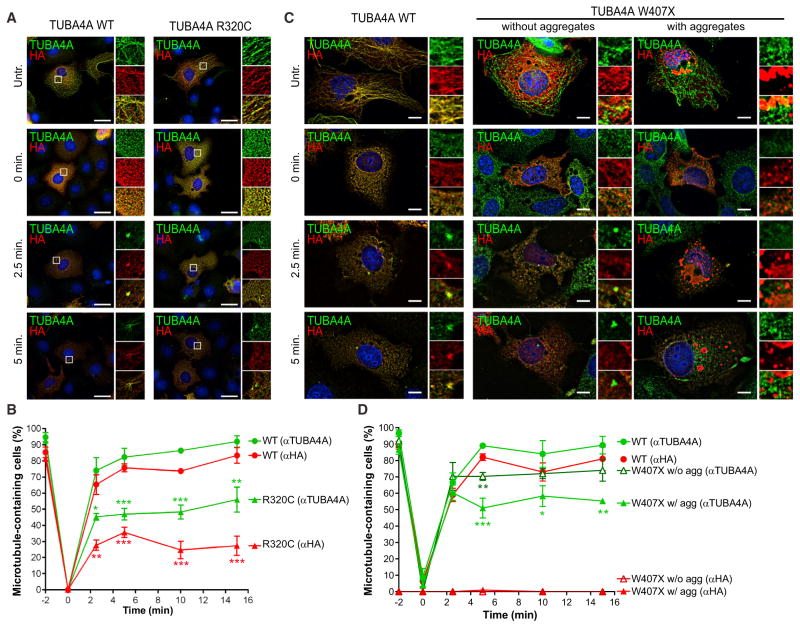
TUBA4A Mutants Disrupt Microtubule Dynamics and Stability through a Dominant-Negative Mechanism
We next determined whether TUBA4A mutants affected microtubule dynamics by testing the ability of transfected COS7 cells to recover after transient microtubule depolymerization (Figure 3) (Gilissen et al., 2011; Tian et al., 2008, 2010). For this assay, the wild-type, R320C, and W407X proteins were compared, as these mutants displayed an intermediate and severe phenotype, respectively, in the previous experiments. While ~80% of cells transfected with TUBA4AWT formed centrosomes positive for the HA-tagged TUBA4A protein within 5 min of nocodazole removal, only ~20% of TUBA4AR320C-transfected cells formed new microtubules containing the mutant protein at 15 min. The expression of mutant TUBA4A also impaired the ability of the endogenous protein to form microtubules, as only 50% of the cells contained TUBA4A-positive microtubules after 15 min, compared to ~90% in wild-type-transfected cells (Figures 3A and 3B). Similar results were also observed in cells containing TUBA4AW407X-positive aggregates. On the contrary, cells expressing the TUBA4AW407X mutant without the presence of aggregates did not display any defect in the rate of microtubule repolymerization (Figures 3C and 3D). These results suggest that mutant TUBA4A by incorporating into the microtubules may disrupt their dynamics through a dominant-negative mechanism.
Figure 3. Mutant TUBA4A Alters Microtubule Dynamics.
(A and C) Representative images of nocodazole-treated COS7 cells expressing wild-type and mutant TUBA4A (A, TUBA4AR320C; C, TUBA4AW407X). Cells were allowed to recover in nocodazole-free medium for 2.5, 5, 10, or 15 min prior to fixation. Red, HA; green, TUBA4A; blue, DAPI. White boxes indicate the regions enlarged in the insets. Scale bars, 10 μm. (B and D) The percentage of cells containing microtubules positive for the HA-tagged or the endogenous TUBA4A protein was quantified from more than 100 cells per condition from 3 independent experiments and compared between wild-type and mutant TUBA4A (B, TUBA4AR320C; D, TUBA4AW407X). Points represent mean and SEM (one-way ANOVA and Dunnett’s post hoc test; *p < 0.05, **p < 0.01, ***p < 0.001).
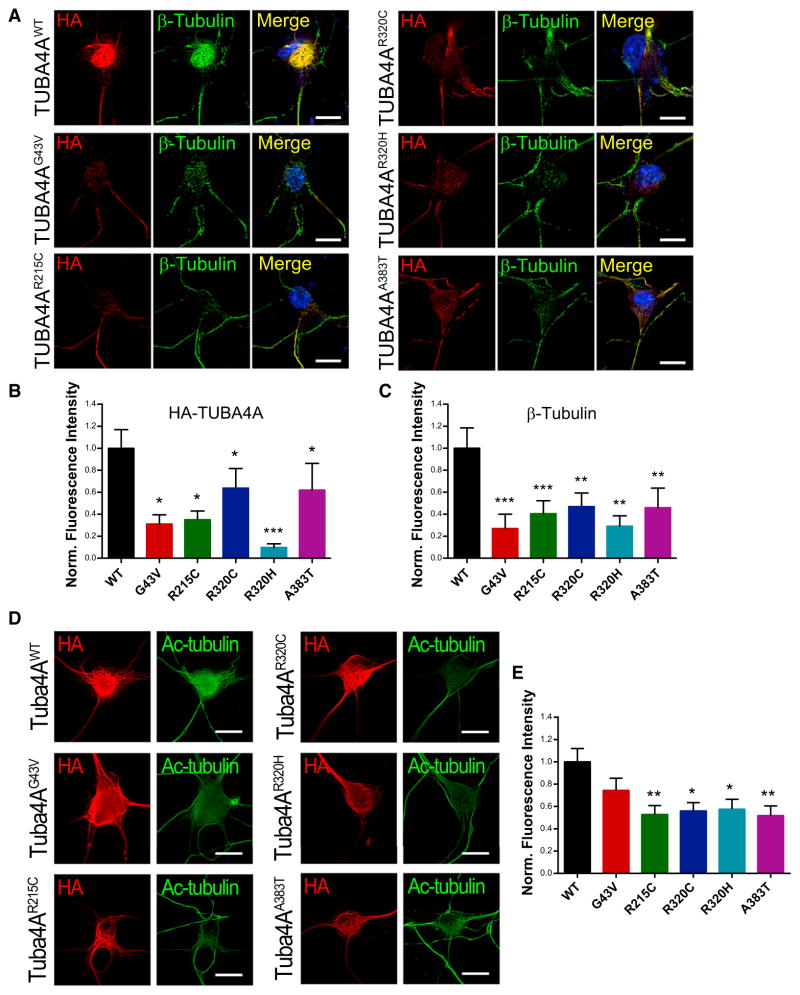
To pursue this hypothesis, we investigated whether TUBA4A mutants would influence the overall stability of the microtubule network. PMNs expressing wild-type and mutant TUBA4A constructs were permeabilized with 0.1% Triton X-100 to extract soluble proteins before fixation and immunostained for HA-TUBA4A and β-tubulin. Under these conditions, the cytoskeleton was largely preserved in wild-type-transfected neurons, whereas each of the missense mutants resulted in decreased HA-TUBA4A and β-tubulin immunofluorescence (Figures 4A–4C). Furthermore, the levels of α-tubulin acetylation, which have been shown to correlate with increased microtubule stability (Matsuyama et al., 2002; Piperno et al., 1987), were significantly reduced in mutant TUBA4A-expressing PMNs (Figures 4D and 4E). Of note, the G43V mutation, which is uniquely localized to the protein N terminus, did not impair tubulin acetylation, suggesting that defective binding of the acetyl-transferase αTAT1 to α-tubulin C-terminal tail (Howes et al., 2014; Panoutsopoulou et al., 2013; Stitziel et al., 2011), where TUBA4A mutations are clustered, may be responsible for this effect. Together, these results further suggest that missense mutants of TUBA4A weaken the microtubule network by a dominant-negative mechanism.
Figure 4. Mutant TUBA4A Destabilizes the Microtubule Network.
(A) PMNs transfected with wild-type or mutant TUBA4A constructs were permeabilzed with 0.1% Triton X-100 for 5 min prior to fixation. (B and C) The fluorescence intensity of (B) HA-TUBA4A and (C) β-tubulin was quantified and compared. Bars represent mean and SEM (Kruskal-Wallis test, n = 23–34 cells per condition from 3 independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001). (D and E) Acetylated tubulin (green) levels were measured in PMNs expressing TUBA4A mutants (red). Representative images are shown (D). Bars represent mean and SEM (E, one-way ANOVA and Dunnett’s post hoc test, n = 30–45 cells per condition from 3 independent experiments, *p < 0.05, **p < 0.01). Scale bars, 10 μm.
DISCUSSION
To summarize, we have used an unbiased case-control exome-sequencing study to identify a statistically significant and replicable excess of rare damaging TUBA4A variants in FALS. Disease-associated variants affect conserved residues and in most cases are absent in over 13,000 controls. The positive identification of samples with mutations in the SOD1 and VCP genes, as well as in the newly discovered ALS gene MATR3 within the initial cohort, further proves the validity of our approach. Though our burden approach demonstrated the importance of a specific class of TUBA4A variants, a limitation is that it is not informative as to the effects of individual variants. Thus, not all reported patient mutations may be pathogenically relevant or act with equal penetrance. An example of this is the K430N alteration not segregating in an affected first cousin. However, our results from biological characterization provided strong evidence to support a deleterious effect of most variants. Specifically, FALS-associated variants (1) inefficiently form α-/β- tubulin dimers in vitro,(2) display decreased incorporation into microtubules in cultured cells, and (3) inhibit microtubule network assembly and reduce structural stability. Based on these data, TUBA4A mutants appear to disrupt microtubule dynamics and stability through a dominant-negative mechanism. Interestingly, the truncation mutant TUBA4AW407X is deficient in forming dimers or incorporating into the microtubule network, suggesting that it may have a decreased ability to act as a dominant negative. This is illustrated by our observation that in cells expressing the W407X mutant without aggregates, there is no obvious defect in the rate of microtubule repolymerization. However, since TUBA4AW407X shows aggregation propensities analogous to other ALS-associated mutant proteins, its deleterious effect may be through a different mechanism, such as trapping tubulin-binding proteins into aggregates or by overburdening the ubiquitin proteasome system.
Mutations in at least seven other tubulin family members (TUBA1A [Keays et al., 2007; Kumar et al., 2010; Poirier et al., 2007], TUBA8 [Abdollahi et al., 2009], TUBB2B [Jaglin et al., 2009], TUBB3 [Poirier et al., 2010; Tischfield et al., 2010], TUBB5 [Breuss et al., 2012], TUBB4A [Hersheson et al., 2013; Tian et al., 2008], and TUBG1 [Poirier et al., 2013]) have been described to cause several neurodevelopmental and neurodegenerative disorders. Moreover, the progressive motor neuronopathy (pmn) mutant mouse, a commonly used model for human motor neuron disease, bears a mutation in the Tubulin-specific Chaperone E (TBCE) gene (Bömmel et al., 2002; Martin et al., 2002). Most mutant tubulin proteins show similar functional defects compared to TUBA4A mutants, such as impaired dimerization and microtubule incorporation and yet cause very different phenotypes. Interestingly, results from the BrainSpan Atlas and in developing mouse brain tissue show that most α- and β-tubulin subunits responsible for developmental defects are highly expressed during brain development and decrease with age, whereas TUBA4A levels increase dramatically (>50-fold) with age (Figure S4). Furthermore, similar to TUBA4A, expression of TUBB4A also increases over time and mutations in this gene are responsible for a disease with an onset age of 15–37 years (torsion dystonia type 4) (Hersheson et al., 2013). These observations may partially explain the difference between the neurodevelopmental and late-onset symptoms associated to the different tubulin mutations.
TUBA4A is ubiquitously expressed in human tissues with its highest expression in brain (Rustici et al., 2013). Although a downregulation of α-tubulin subunit genes has been reported in motor neurons of SALS patients (Jiang et al., 2005), no specific role has been described for TUBA4A in motor neurons so far. TUBA4A joins other ALS-associated genes encoding for cytoskeletal proteins, such as PFN1 (Wu et al., 2012), DCTN1 (Puls et al., 2003), PRPH (Gros-Louis et al., 2004), and NEFH (Al-Chalabi et al., 1999), thus strengthening the hypothesis that alterations affecting the cytoskeleton architecture and dynamics have a major role in ALS pathogenesis.
EXPERIMENTAL PROCEDURES
Exome Sequencing and Rare Variant Analysis
Ethical approval for this study was received by the IRBs of the participating institutions. Exome sequencing reads were aligned to a human reference (hg19) using Burrows-Wheeler Aligner and processed using the Genome Analysis ToolKit. Rare variant analyses were performed by logistic regression of case-control status with respect to the aggregated count of minor alleles in a given gene window. For each p value from the observed data set analysis, Pcorrected was calculated as the proportion of permuted data sets where an association of equal or greater significance was observed across any gene.
Motor Neuron Culture and Transfection
PMNs were cultured from embryonic day 12.5 embryos and transfected at 2 days in vitro, as previously described (Fallini et al., 2010).
Quantification of Microtubule Incorporation
Using ImageJ, a region of interest (ROI) was selected for each HA-TUBA4A-positive cell and the signal was thresholded to remove background noise. A circularity filter (circularity index ≥ 0.2) was then applied to remove granular staining. The ratio between the fluorescence intensities of the filtered cytoskeleton versus the thresholded ROI yielded an incorporation index ranging from 0 (no incorporation) to 1 (complete microtubule incorporation).
Supplementary Material
1
2
Footnotes
CONSORTIA
The members of the Slagen Consortium are Sandra D’Alfonso, Letizia Mazzini, Giacomo P. Comi, Roberto Del Bo, Mauro Ceroni, Stella Gagliardi, Giorgia Querin, and Cinzia Bertolin.
AUTHOR CONTRIBUTIONS
Sample collection, preparation, and clinical evaluation were performed by B.N.S., N.T., A.S.G., S.T., K.P.K., P.K., C. Tiloca, C. Troakes, S.A.S., A.K., D.C., V.P., B.C., J.B., F.B., A.L.t.A., P.C.S., D.M.Y., R.L.M., M.P., J.E.P., J.M.B., M.S., W.v.R., F.P.D., G.L., S.D.U., S.C., C. Cereda, L.C., G.S., K.E.M., K.L.W., G.A.N., I.P.B., P.A.D., C.S.L., G.A.R., O.H., J.H.V., L.v.B., A.A.C., H.P., P.J.S., M.R.T., K.T., F.T., A.G.R., J.D.G., C.G., A.R., R.H.B., V.S., C.E.S., J.E.L., and the SLAGEN Consortium. Experiments and data analysis were performed by B.N.S., N.T., C.F., A.S.G., S.T., J.K., E.L.S., K.P.K., P.K., J.W.M., C. Tiloca, E.W.D., C. Troakes, C. Colombrita, S.A.S., E.A.L., A.K., D.C., B.C., P.C.S., S.A., W.v.R., J.H.V., F.T., A.G.R., Z.W., C.G., A.R., V.S., C.E.S., and J.E.L. Scientific planning and direction were performed by B.N.S., N.T., C.F., A.S.G., S.T., J.K., E.L.S., K.P.K., P.K., C.V., Z.W., C.G., A.R., R.H.B., V.S., C.E.S., and J.E.L. The manuscript was prepared by B.N.S., N.T., C.F., A.S.G., S.T., E.L.S., A.R., V.S., C.E.S., and J.E.L.
References
- Abdollahi MR, Morrison E, Sirey T, Molnár Z, Hayward BE, Carr IM, Springell K, Woods CG, Ahmed M, Hattingh L, et al. Mutation of the variant α-tubulin TUBA8 results in polymicrogyria with optic nerve hypoplasia. Am J Hum Genet. 2009;85:737–744. doi: 10.1016/j.ajhg.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Chalabi A, Andersen PM, Nilsson P, Chioza B, Andersson JL, Russ C, Shaw CE, Powell JF, Leigh PN. Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Hum Mol Genet. 1999;8:157–164. doi: 10.1093/hmg/8.2.157. [DOI] [PubMed] [Google Scholar]
- Bömmel H, Xie G, Rossoll W, Wiese S, Jablonka S, Boehm T, Sendtner M. Missense mutation in the tubulin-specific chaperone E (Tbce) gene in the mouse mutant progressive motor neuronopathy, a model of human motoneuron disease. J Cell Biol. 2002;159:563–569. doi: 10.1083/jcb.200208001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuss M, Heng JIT, Poirier K, Tian G, Jaglin XH, Qu Z, Braun A, Gstrein T, Ngo L, Haas M, et al. Mutations in the β-tubulin gene TUBB5 cause microcephaly with structural brain abnormalities. Cell Rep. 2012;2:1554–1562. doi: 10.1016/j.celrep.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Calvo A, Mazzini L, Cantello R, Mora G, Moglia C, Corrado L, D’Alfonso S, Majounie E, Renton A, et al. Extensive genetics of ALS: a population-based study in Italy. Neurology. 2012;79:1983–1989. doi: 10.1212/WNL.0b013e3182735d36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Kirschner MW, Cowan NJ. Isolation of separate mRNAs for alpha- and beta-tubulin and characterization of the corresponding in vitro translation products. Cell. 1978;15:1021–1031. doi: 10.1016/0092-8674(78)90286-6. [DOI] [PubMed] [Google Scholar]
- Fallini C, Bassell GJ, Rossoll W. High-efficiency transfection of cultured primary motor neurons to study protein localization, trafficking, and function. Mol Neurodegener. 2010;5:17. doi: 10.1186/1750-1326-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hoischen A, Brunner HG, Veltman JA. Unlocking Mendelian disease using exome sequencing. Genome Biol. 2011;12:228. doi: 10.1186/gb-2011-12-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros-Louis F, Larivière R, Gowing G, Laurent S, Camu W, Bouchard JP, Meininger V, Rouleau GA, Julien JP. A frameshift deletion in peripherin gene associated with amyotrophic lateral sclerosis. J Biol Chem. 2004;279:45951–45956. doi: 10.1074/jbc.M408139200. [DOI] [PubMed] [Google Scholar]
- Hersheson J, Mencacci NE, Davis M, MacDonald N, Trabzuni D, Ryten M, Pittman A, Paudel R, Kara E, Fawcett K, et al. Mutations in the autoregulatory domain of β-tubulin 4a cause hereditary dystonia. Ann Neurol. 2013;73:546–553. doi: 10.1002/ana.23832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes SC, Alushin GM, Shida T, Nachury MV, Nogales E. Effects of tubulin acetylation and tubulin acetyltransferase binding on microtubule structure. Mol Biol Cell. 2014;25:257–266. doi: 10.1091/mbc.E13-07-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglin XH, Poirier K, Saillour Y, Buhler E, Tian G, Bahi-Buisson N, Fallet-Bianco C, Phan-Dinh-Tuy F, Kong XP, Bomont P, et al. Mutations in the β-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat Genet. 2009;41:746–752. doi: 10.1038/ng.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YM, Yamamoto M, Kobayashi Y, Yoshihara T, Liang Y, Terao S, Takeuchi H, Ishigaki S, Katsuno M, Adachi H, et al. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2005;57:236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- Johnson JO, Pioro EP, Boehringer A, Chia R, Feit H, Renton AE, Pliner HA, Abramzon Y, Marangi G, Winborn BJ, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17:664–666. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keays DA, Tian G, Poirier K, Huang GJ, Siebold C, Cleak J, Oliver PL, Fray M, Harvey RJ, Molnár Z, et al. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 2007;128:45–57. doi: 10.1016/j.cell.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna KP, McLaughlin RL, Byrne S, Elamin M, Heverin M, Kenny EM, Cormican P, Morris DW, Donaghy CG, Bradley DG, Hardiman O. Delineating the genetic heterogeneity of ALS using targeted high-throughput sequencing. J Med Genet. 2013;50:776–783. doi: 10.1136/jmedgenet-2013-101795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiezun A, Garimella K, Do R, Stitziel NO, Neale BM, McLaren PJ, Gupta N, Sklar P, Sullivan PF, Moran JL, et al. Exome sequencing and the genetic basis of complex traits. Nat Genet. 2012;44:623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, Pilz DT, Babatz TD, Cushion TD, Harvey K, Topf M, Yates L, Robb S, Uyanik G, Mancini GMS, et al. TUBA1A mutations cause wide spectrum lissencephaly (smooth brain) and suggest that multiple neuronal migration pathways converge on alpha tubulins. Hum Mol Genet. 2010;19:2817–2827. doi: 10.1093/hmg/ddq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JS, Schubert CR, Fu X, Fourniol FJ, Jaiswal JK, Houdusse A, Stultz CM, Moores CA, Walsh CA. Molecular basis for specific regulation of neuronal kinesin-3 motors by doublecortin family proteins. Mol Cell. 2012;47:707–721. doi: 10.1016/j.molcel.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N, Jaubert J, Gounon P, Salido E, Haase G, Szatanik M, Guénet JL. A missense mutation in Tbce causes progressive motor neuronopathy in mice. Nat Genet. 2002;32:443–447. doi: 10.1038/ng1016. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, Osada H, Komatsu Y, Nishino N, Khochbin S, et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panoutsopoulou K, Tachmazidou I, Zeggini E. In search of low-frequency and rare variants affecting complex traits. Hum Mol Genet. 2013;22(R1):R16–R21. doi: 10.1093/hmg/ddt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K, Keays DA, Francis F, Saillour Y, Bahi N, Manouvrier S, Fallet-Bianco C, Pasquier L, Toutain A, Tuy FPD, et al. Large spectrum of lissencephaly and pachygyria phenotypes resulting from de novo missense mutations in tubulin alpha 1A (TUBA1A) Hum Mutat. 2007;28:1055–1064. doi: 10.1002/humu.20572. [DOI] [PubMed] [Google Scholar]
- Poirier K, Saillour Y, Bahi-Buisson N, Jaglin XH, Fallet-Bianco C, Nabbout R, Castelnau-Ptakhine L, Roubertie A, Attie-Bitach T, Desguerre I, et al. Mutations in the neuronal β-tubulin subunit TUBB3 result in malformation of cortical development and neuronal migration defects. Hum Mol Genet. 2010;19:4462–4473. doi: 10.1093/hmg/ddq377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K, Lebrun N, Broix L, Tian G, Saillour Y, Boscheron C, Parrini E, Valence S, Pierre BS, Oger M, et al. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat Genet. 2013;45:639–647. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls I, Jonnakuty C, LaMonte BH, Holzbaur ELF, Tokito M, Mann E, Floeter MK, Bidus K, Drayna D, Oh SJ, et al. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- Rustici G, Kolesnikov N, Brandizi M, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Ison J, Keays M, et al. ArrayExpress update—trends in database growth and links to data analysis tools. Nucleic Acids Res. 2013;41(Database issue):D987–D990. doi: 10.1093/nar/gks1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitziel NO, Kiezun A, Sunyaev S. Computational and statistical approaches to analyzing variants identified by exome sequencing. Genome Biol. 2011;12:227. doi: 10.1186/gb-2011-12-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Kong XP, Jaglin XH, Chelly J, Keays D, Cowan NJ. A pachygyria-causing alpha-tubulin mutation results in inefficient cycling with CCT and a deficient interaction with TBCB. Mol Biol Cell. 2008;19:1152–1161. doi: 10.1091/mbc.E07-09-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Jaglin XH, Keays DA, Francis F, Chelly J, Cowan NJ. Disease-associated mutations in TUBA1A result in a spectrum of defects in the tubulin folding and heterodimer assembly pathway. Hum Mol Genet. 2010;19:3599–3613. doi: 10.1093/hmg/ddq276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield MA, Baris HN, Wu C, Rudolph G, Van Maldergem L, He W, Chan WM, Andrews C, Demer JL, Robertson RL, et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Fallini C, Ticozzi N, Keagle PJ, Sapp PC, Piotrowska K, Lowe P, Koppers M, McKenna-Yasek D, Baron DM, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2