Multiple Interactions of Rad23 Suggest a Mechanism for Ubiquitylated Substrate Delivery Important in Proteolysis (original) (raw)
Abstract
The mechanism underlying the delivery of ubiquitylated substrates to the proteasome is poorly understood. Rad23 is a putative adaptor molecule for this process because it interacts with ubiquitin chains through its ubiquitin-associated motifs (UBA) and with the proteasome through a ubiquitin-like element (UBL). Here, we demonstrate that the UBL motif of Rad23 also binds Ufd2, an E4 enzyme essential for ubiquitin chain assembly onto its substrates. Mutations in the UBL of Rad23 alter its interactions with Ufd2 and the proteasome, and impair its function in the UFD proteolytic pathway. Furthermore, Ufd2 and the proteasome subunit Rpn1 compete for the binding of Rad23, suggesting that Rad23 forms separate complexes with them. Importantly, we also find that the ability of other UBL/UBA proteins to associate with Ufd2 correlates with their differential involvement in the UFD pathway, suggesting that UBL-mediated interactions may contribute to the substrate specificity of these adaptors. We propose that the UBL motif, a protein-protein interaction module, may be used to facilitate coupling between substrate ubiquitylation and delivery, and to ensure the orderly handoff of the substrate from the ubiquitylation machinery to the proteasome.
INTRODUCTION
Ubiquitin (Ub) is best known as a signal that targets proteins for destruction by a multisubunit, ATP-dependent protease termed the proteasome (Baumeister et al., 1998; DeMartino and Slaughter, 1999). The Ub-proteasome system consists of two phases (Pickart, 2001; Weissman, 2001; Schwartz and Hochstrasser, 2003). In the Ub conjugative phase, Ub is activated and transferred to the substrate through several enzymes, including a Ub-activating enzyme (E1), a Ub-conjugating enzyme (E2), and a Ub-protein ligase (E3). For substrates targeted for degradation, successive Ub molecules are added to form a Ub chain on the substrates by E3 or E4 enzymes that are specifically involved in Ub chain elongation. E4 is a recently identified component of the Ub system and required for the synthesis of multi-Ub chains for a subset of substrates (Koegl et al., 1999). Two of the known E4s are Ufd2 (Koegl et al., 1999) and p300, the latter being important for the ubiquitylation of p53 (Grossman et al., 2003). In the second phase, the ubiquitylated substrate is delivered to the 26S proteasome; the substrate is unfolded and processively degraded into small peptides, and Ub is released by specific deubiquitylating enzymes (Verma et al., 2002; Yao and Cohen, 2002). The biochemical mechanisms of the postubiquitylation events are poorly understood.
In addition to its classical role in protein degradation, Ub is also emerging as a nonproteolytic signal for protein transport and processing (Pickart, 2001; Weissman, 2001). The fate of a substrate depends on the number of Ub moieties conjugated (mono-Ub or Ub chains), as well as the lysine linkage of Ub-Ub conjugation. Three types of Ub chains have known functions (Weissman, 2001; Schwartz and Hochstrasser, 2003). K29 and K48 are the major sites used in the formation of Ub chains that target substrates for proteasome-mediated degradation. Multi-Ub chains linked through K63 can be recognized by the proteasome in vitro (Hofmann and Pickart, 2001), but not in vivo (Finley, 2001). Hence, it has been suggested that specific Ub chain-recognition adaptors serve to distinguish between these Ub chains in vivo (Hofmann and Pickart, 2001); and the adaptors that recognize the K29- or K48-linked chain likely play a key role in substrate delivery to the proteasome. The involvement of adaptor molecules in substrate proteolysis is also consistent with the paradigm emerging from regulated proteolysis in Escherichia coli (Levchenko et al., 2003).
A key attribute of the adaptors is likely to be an ability to bind ubiquitylated proteins and the proteasome. Recent studies have found that proteins bearing the UBA domain bind K48-linked Ub chains and ubiquitylated substrates in vivo and in vitro (Bertolaet et al., 2001; Wilkinson et al., 2001; Chen and Madura, 2002; Rao and Sastry, 2002; Raasi and Pickart, 2003). Interestingly, a subset of UBA-containing proteins also harbors a UBL motif at the N terminus. The UBL domain consists of ∼70 amino acid residues and shares ∼20% identity with Ub. The UBL domain of yeast proteins binds the Rpn1 subunit of the proteasome (Elsasser et al., 2002; Saeki et al., 2002b). Three Saccharomyces cerevisiae proteins, Rad23, Dsk2, and Ddi1, contain both the UBA and UBL motifs and are putative adaptors in Ub-mediated proteolysis. The loss of Rad23 leads to the partial stabilization of the substrates of the UFD proteolytic pathway (Lambertson et al., 1999; Rao and Sastry, 2002), and Schizosaccharomyces pombe Rad23 is involved in the degradation of the CDK inhibitor Rum1 (Wilkinson et al., 2001), indicating a positive role for Rad23 in proteolysis. Importantly, the stabilized substrates in the rad23 and dsk2 mutant cells are fully ubiquitylated, suggesting that Rad23 and Dsk2 function at a postubiquitylation, but preproteasome, step (Lambertson et al., 1999; Rao and Sastry, 2002). The UBA motif is essential for the function of Rad23 and Dsk2 in proteolysis (Bertolaet et al., 2001; Rao and Sastry, 2002). Here, we show that the UBL domain of Rad23 binds Ufd2, an E4 component essential for Ub chain assembly onto UFD substrates. Moreover, we demonstrate that UBL-mediated interactions are important for the functioning of Rad23 in proteolysis. Our results suggest a mechanism for ubiquitylated substrate delivery to the proteasome. We further propose that the substrate specificity of Ub-binding proteins may be determined by other protein-protein interaction motifs.
MATERIALS AND METHODS
Yeast Strains and Plasmids
S. cerevisiae PJ69-4A (_MATa lys2-801 ura3-52 trp1-109 his3-_Δ_200 leu2-3112 gal4_Δ _gal80_Δ GAL2-ADE2 LYS2::GAL1-HIS3 met::GAL7-lacZ) was used for yeast two-hybrid assays. Strains MY3589 (MATa ura3-52 leu ade2) and MY3588 (MATa ura3-52 trp1 ade2 his3 dsk2::LEU2) were gifts from M. Rose (Biggins et al., 1996). The strain MY3587F (_MAT_α ura3-52 leu2 his3 ade2 rad23) was published previously (Rao and Sastry, 2002). In strain YHR56 (MATa ura3-52 leu ade2 ddi1::KanR), DDI1 was replaced with a kanamycin-resistant gene. Yeast strain YHR60H (MATa ura3-52 ade2 his3 rad23::TRP1 dsk2::HIS3) was constructed by replacing RAD23 and DSK2 with TRP1 and HIS3, respectively. Strains YHR58 (MAT_α ura3-52 leu2 his3 ade2 rad23 ddi1::KanR) and YHR59 (MATa ura3-52 trp1 ade2 his3 dsk2::LEU2 ddi1::KanR) were constructed by replacing DDI1 with a kanamycin-resistant gene in strains MY3587F and MY3588, respectively. DDI1 was replaced by HIS3 in YHR60 to obtain YHR66 (MATa ura3-52 ade2 rad23::TRP1 dsk2::LEU2 ddi1::HIS3). Strains YHR69 (MAT_α ura3-52 leu2 his3 ade2 rad23 ddi1_Δ_UBL::RAD23 Δ_UBL-KanR), and YHR71 (MATa ura3-52 leu ade2 rad23_Δ_UBL::DDI1 Δ_UBL-KanR) were constructed by swapping the C-terminal ΔUBL domains of RAD23 and DDI1, which are linked to a kanamycin-resistant gene to facilitate the selections, in strains MY3587F and MY3589, respectively. Strains YHR83 (MATa ura3 leu2 his3 trp1 rad23::KanR), and YHR84 (ura3 leu2 his3 trp1 rad23::KanR ufd2::HIS3) were constructed by replacing RAD23 with a kanamycin-resistant gene and UFD2 with HIS3 in strain EGY128 (MATa ura3 leu2 his3 trp1). Strain YHR85 (leu2 his3 trp1 rad23::KanR ufd2::HIS3 ddi1::URA3) was obtained by replacing DDI1 with hisG-URA3-hisG. Cultures were grown in rich (YPD) or in synthetic media containing standard ingredients and either 2% glucose (SD medium) or 2% galactose (SG medium).
Two-hybrid plasmids containing the full-length of RAD23, DSK2, DDI1, UFD2, RPN1 were gifts from E. Friedberg (University of Texas), S. Fields (University of Washington), S. Jentsch (Max Planck Institute of Biochemistry), and T. Ito (Kanazawa University). The polymerase chain reaction (PCR) products of UFD2 derivatives were verified by DNA sequencing and were fused, in frame, to the 3′ end of the GAL4 fragments in pRS414Gal1 vector, thus creating pRS414Gal-Ufd2 for the overexpression in yeast (Mumberg et al., 1994). To generate the Snc2-Ufd2 fusion, the PCR product of the Snc2 fragment (amino acids 4-93) (Lustgarten and Gerst, 1999) is inserted in-frame to the N terminus of Ufd2 in the pRS414Gal-Ufd2 plasmid. The promoter and RAD23 gene was amplified by PCR to add FLAG epitope at 3′ end of the open reading frame, and cloned to pRS315, a low copy vector. RAD23 was amplified by PCR to incorporate the FLAG epitope and cloned to the 3′ end of the GAL1 promoter in yeast vector pRS425Gal1 for their overexpression. Mutations in the UBL domain were obtained using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). For their expression in E. coli, Rad23-FLAG, Ufd2-myc, and RGS/His6-Rpn1 were cloned into pQE30 (QIAGEN, Valencia, CA), pGEX (Pharmacia Biotech, Piscataway, NJ), and pMAL-TEV (a gift from Dr. R. Yew, University of Texas Health Science Center at San Antonio, San Antonio, TX), respectively. Construction details are available upon request. E. coli BL21 (DE3), M15 were used to express proteins GST-Ufd2-myc, RGS/His6-Rpn1, and His6-Rad23-FLAG.
Pulse-Chase Analysis
Protein degradation was determined by pulse-chase analysis as described previously (Rao et al., 2001). Yeast cells, carrying plasmids that expressed UbV76-Val-βgal from the P_GAL1_ promoter, were grown at 30°C to OD600 of ∼1 in SRG medium with auxotrophic supplements and 2% raffinose and galactose as the carbon source. Cells from a 20-ml culture were harvested by centrifugation, washed with 0.8 ml of SRG, resuspended in 0.4 ml of SRG, and labeled for 5 min with 0.16 mCi of 35S-Express (PerkinElmer Life and Analytical Sciences, Boston, MA), followed by centrifugation and resuspension of cells in glucose-containing SD medium with 4 mM l-methionine and 2 mM l-cysteine. Samples (0.1 ml) were taken at the indicated time points and processed for immunoprecipitation with anti-βgal antibody (Sigma-Aldrich, St. Louis, MO), followed by SDS-PAGE, as described previously (Rao et al., 2001; Rao and Sastry, 2002).
Glutathione S-Transferase (GST) Binding Assays
GST fusion proteins were purified as described previously (Rao and Sastry, 2002) and were dialyzed with 1× phosphate-buffered saline buffer. GST fusion protein or GST alone (∼2 μg) was mixed with yeast extracts in 200 μl of binding buffer (150 mM NaCl, 1% Triton X-100, 10% glycerol, 0.1 mg/ml bovine serum albumin, 50 mM Na-HEPES, pH 7.5) containing 1× protease inhibitor mix (Roche Diagnostics, Indianapolis, IN), and incubated with 10 μl (bed volume) of glutathione-agarose beads (Sigma-Aldrich) for 2 h at 4°C. The beads were washed three times with the binding buffer, followed by SDS-PAGE of the retained proteins, and immunoblotting with a monoclonal anti-myc antibody (Covance, Berkeley, CA).
Coimmunoprecipitation/Immunoblotting Assay
W303-1A (wild-type) cells carrying either two vectors (pRS414Gal and pRS425Gal), or pRS414Gal-Ufd2myc (expressing myc-tagged Ufd2) and pRS425Gal, or pRS414Gal-Ufd2myc and pRS425Gal-Rad23Flag (expressing FLAG-tagged Rad23), or pRS414Gal-Ufd2myc and pRS425Gal harboring the FLAG-tagged rad23 derivatives together, were grown in the galactose-containing SG medium to OD600 of ∼1, followed by preparation of extracts, immunoprecipitation with beads linked to specific antibodies indicated, SDS-8% PAGE, and immunoblotting, separately, with anti-FLAG (Sigma-Aldrich), anti-myc (Covance). To detect their bindings to the proteasome, W303-1A cells containing the FLAG-tagged rad23 alleles were processed similarly. Immunoblots were probed with anti-FLAG, or anti-Rpt5 antibodies (Affinity, Devon, United Kingdom). The blot was incubated with a 1:2000 dilution of the goat anti-mouse horseradish peroxidase conjugate and was developed using enhanced chemiluminescence reagents (Amersham Biosciences, Piscataway, NJ).
Purification of Ufd2, Rpn1, and Rad23 from E. coli
GST fusion of Ufd2-myc was purified as described previously (Rao and Sastry, 2002). RGS/His6-Rad23-FLAG was expressed in M15 strain and purified by two-step affinity columns as follows. Expression was carried out at 30°C by induction with 1 mM isopropyl β-d-thiogalactoside for 4 h. Cells were suspended in lysis buffer (300 mM NaCl, 10 mM imidazole, 0.05% Tween 20, 50 mM NaH2PO4, pH 8) and lysed by sonication. The extract was then centrifuged at 13,500 rpm for 30 min at 4°C. The supernatant was passed over a Ni2+-NTA resin (QIAGEN). After repeated washes, we eluted proteins with elution buffer (300 mM NaCl, 250 mM imidazole, 0.05% Tween 20, 50 mM NaH2PO4, pH 8). Eluent was then mixed with FLAG-agarose beads (Sigma-Aldrich) for 1 h at 4°C. Proteins were eluted with FLAG peptides (Sigma-Aldrich) and subsequently dialyzed with 1× binding buffer (150 mM NaCl, 1% Triton X-100, 5 mM EDTA, 50 mM Na-HEPES, pH 7.5).
RGS/His6-tagged Rpn1 was linked to myelin basic protein (MBP) with a TEV protease cleavage site. Its expression was induced with 0.5 mM isopropyl β-d-thiogalactoside for 3 h at 30°C. Cells were collected after centrifugation and dissolved in MBP-suspension buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Tween 20). After the sonication, the lysate was centrifuged at 13,500 rpm for 30 min at 4°C. The supernatant was incubated with amylose beads for 1 h at 4°C. We washed the resin with MBP-suspension buffer and eluted the proteins with the TEV protease in 1× reaction buffer (Invitrogen, Carlsbad, CA) for 10 h at 4°C. Eluent was dialyzed with lysis buffer (300 mM NaCl, 10 mM imidazole, 0.05% Tween 20, 50 mM NaH2PO4, pH 8) and incubated with Ni2+-NTA beads for 1 h at 4°C. After extensive washing, we eluted proteins with elution buffer (300 mM NaCl, 250 mM imidazole, 0.05% Tween 20, 50 mM NaH2PO4, pH 8) and subsequently dialyzed with 1× binding buffer (150 mM NaCl, 1% Triton X-100, 5 mM EDTA, 50 mM Na-HEPES, pH 7.5).
RESULTS
Phenotypic Analysis of Yeast Cells Lacking One or More UBL/UBA Genes
To understand their physiological roles, we have constructed various yeast mutants lacking one or multiple UBL/UBA containing genes in the MY3589 background. All mutants are viable at 30°C (Figure 1). One of the major functions of the proteasome is protein quality control in response to various stresses (Gottesman et al., 1997). Hence, the mutants were tested under a number of stress conditions. None of the mutants is sensitive to the treatment with dithiothreitol, or the alkylating agent methyl methanesulfonate (our unpublished data). Cells lacking RAD23 are known to be UV sensitive (Watkins et al., 1993), but mutations in DSK2 and/or DDI1 do not lead to UV sensitivity and do not enhance the UV sensitivity of rad23 mutants (Bertolaet et al., 2001; our unpublished data), indicating that only RAD23 has a DNA repair function. Cells lacking both RAD23 and DSK2 have been shown to be inviable at 37°C (Biggins et al., 1996). As expected, the triple mutant dies at 37°C, but the rad23, ddi1, dsk2 ddi1, and rad23 ddi1 mutants are not sensitive to elevated temperature (our unpublished data).
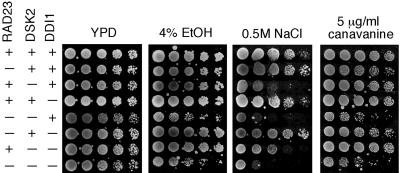
Figure 1.
Phenotypes associated with yeast cells lacking UBL/UBA-containing genes. Mutants display stress-sensitive phenotypes. Isogenic yeast strains were grown to an optical density A600 of ∼1.2, and fourfold dilutions were spotted onto various media indicated above each panel. The plates were incubated at 30°C for 2-5 d. The strain backgrounds are labeled on the left.
Incorporation of canavanine, an amino acid analog, into proteins leads to their misfolding. Many mutants of the Ub-proteasome system are sensitive to canavanine. Loss of RAD23 was previously shown to render cells moderately sensitive to canavanine (Lambertson et al., 1999). Interestingly, we found that although the loss of DSK2 and/or DDI1 did not engender sensitivity to canavanine, cells became extremely sensitive to canavanine when the loss of RAD23 is combined with the dsk2 mutation (Figure 1). Whereas single mutants showed little sensitivity to ethanol, rad23 dsk2 and rad23 dsk2 ddi1 mutants were markedly sensitive to ethanol (Figure 1). Furthermore, we found that the loss of DSK2 conferred sensitivity to salt, and the rad23 mutation exacerbated the salt sensitivity (Figure 1). These results suggest that RAD23 and DSK2 provide overlapping functions in responding to stress conditions.
Rad23 Interacts with Ufd2
To understand the function of Rad23 in proteolysis, we tested whether Rad23 interacts with other proteins involved in the Ub-proteasome pathways. Using the yeast two-hybrid assay, we found that Rad23 interacts with Ufd2 in vivo but not with Cdc48, Doa4, or Ubr1 (Figure 2; our unpublished data). While this study was in progress, a complex containing both Ufd2 and Rad23 was detected by a proteomic analysis of biochemical interactions in yeast (Ho et al., 2002). Ufd2 is an E4 enzyme essential for Ub chain assembly onto UFD substrates (Koegl et al., 1999). Ufd2 is linked to stress tolerance in yeast and the human Ufd2 homologue is a candidate neuroblastoma tumor suppressor (Koegl et al., 1999; Krona et al., 2003). The genetic dissection of the proteolytic pathway that degrades the model substrate UbV76-V-βgal fusion led to the discovery of Ub fusion degradation (UFD) pathway (Johnson et al., 1995). Two recently identified physiological UFD substrate are UBB(+1), a mutant Ub carrying a 19-amino acid C-terminal extension that often accumulates in affected neurons in Alzheimer's disease (Lindsten et al., 2002), and Ataxin-3, a polyQ-containing protein (Matsumoto et al., 2004).
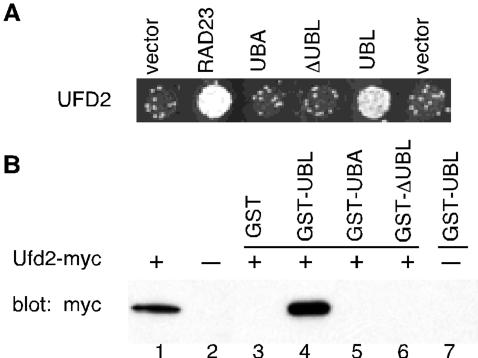
Figure 2.
Interactions between Ufd2 and Rad23. (A) Two-hybrid interactions. Yeast cells were cotransformed with plasmids encoding Gal4 DNA-binding domain fused to Ufd2 protein indicated on the left with Gal4 activation domain fused to Rad23 or portions of Rad23, as indicated above the panel. Growth on -his plate is indicative of protein-protein interaction. (B) GST pull-down assays. The mixtures containing glutathione beads, various GST fusion proteins, and yeast extracts with overexpressed myc-tagged Ufd2 were incubated at 4°C for 2 h. Lanes 1 and 2 contain 5% input extracts with or without tagged Ufd2, respectively. The bound proteins were eluted, fractioned by SDS-PAGE, and immunoblotted with antibody to myc. Ufd2 and Rad23 have also been detected in a complex by a whole-genome approach (Ho et al., 2002).
We reasoned that the Ufd2-Rad23 interaction is probably physiologically relevant because we and others have previously demonstrated the importance of RAD23 in the yeast UFD pathway (Lambertson et al., 1999; Rao and Sastry, 2002). Moreover, yeast cells lacking UFD2, or RAD23 and DSK2, are sensitive to ethanol (Koegl et al., 1999) (Figure 1), suggesting that these genes function in the same pathway. To determine the region of Rad23 involved in Ufd2 interaction, derivatives of Rad23 containing various functional domains (Figure 2A) were tested in the two-hybrid assay for interaction with Ufd2. Interestingly, we found that the UBL motif is sufficient for interaction with Ufd2 (Figure 2). We used GST pull-down assays to further examine the binding between Ufd2 and Rad23. For this purpose, Rad23ΔUBL, Rad23UBL, and Rad23UBA2 were separately fused to the C terminus of GST. These proteins were expressed in E. coli and purified using glutathione beads (Rao and Sastry, 2002). Purified proteins were then mixed with extracts from yeast cells expressing myc-tagged Ufd2 in the presence of glutathione beads. Ufd2-Rad23 binding was assayed by immunoblotting with monoclonal anti-myc antibody to determine the amount of Ufd2 associated with the GST fusions. Consistent with the two-hybrid results (Figure 2A), the biochemical analysis showed a specific association of Ufd2 with UBL-portion of Rad23 (Figure 2B). Thus, we identified a potentially novel function of the UBL motif in proteolysis. Because the Ub-chain binding domains (UBA) of Rad23 are not required for the binding of Ufd2 in vivo and in vitro (Figure 2), the Ufd2-Rad23 interaction is likely not mediated by Ub molecules (also see Figure 5C). Ufd2 was previously shown to bind Ub (Koegl et al., 1999) and may use the same domain to bind the UBL motif.
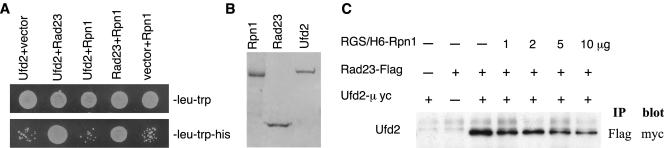
Figure 5.
Interactions among Rad23, Ufd2, and Rpn1. (A) Two-hybrid interactions. Two-hybrid analysis was performed as in Figure 2A. The bait and prey are indicated above the panels. Growth on -leu-trp-his plate (bottom) is indicative of protein-protein interaction. Top, control plate (-leu-trp), indicating that equal amounts of cells were plated. (B) Purification of Rpn1, Rad23, and Ufd2 from E. coli. A Coomassie Blue-stained gel shows purified RGS/His6-tagged Rpn1, FLAG, and His6 double-tagged Rad23, and GST-linked Ufd2-myc. (C) Increasing amount of Rpn1 reduce the Ufd2-Rad23 interaction. Purified GST-Ufd2-myc (1 μg) and Rad23-FLAG (1 μg) were mixed in various combinations as indicated with increasing amount of RGS/His6-Rpn1 in 200 μl of binding buffer (50 mM Na-HEPES, pH 7.5, 150 mM NaCl, 5 mM EDTA, 2% Triton X-100, 0.2 mg/ml bovine serum albumin) containing 1× protease inhibitor mix (Roche Diagnostics), and incubated with 10 μl (bed volume) of FLAG-agarose beads (Sigma-Aldrich) for 2 h at 4°C. The beads were washed four times with the binding buffer, followed by SDS-PAGE of the retained proteins and immunoblotting with a monoclonal anti-myc antibody.
Mutations in Rad23 Alter Its Binding to Ufd2
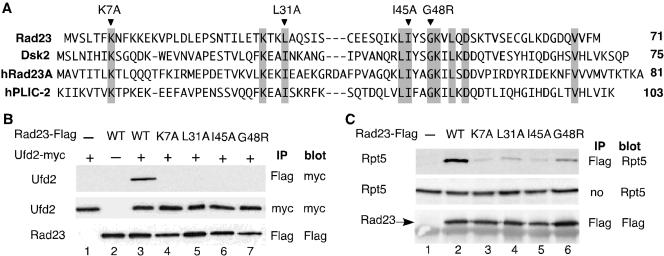
To our knowledge, previous functional analysis of the UBL motif relied on the UBL-deletions. To establish the significance of the UBL domain in mediating Ufd2-Rad23 binding, we looked for mutations in Rad23 that would alter its binding to Ufd2. We compared sequences of UBL domains of Rad23 and Dsk2 (Figure 3A) to identify conserved amino acid residues among them. We constructed four variants of the UBL domain of Rad23, each containing a mutation in one of the highly conserved residues K7, L31, I45, or G48 (Figure 3A). Note that the I75A mutation, corresponding to the I45A mutation in yeast Rad23, was made in the UBL element of a human Dsk2 homologue hPLIC-2 and does not affect the structure of the UBL domain as determined by NMR spectroscopy (Walters et al., 2002).
Figure 3.
Mutations in Rad23 alter its binding to Ufd2 and the proteasome. (A) Sequence alignment of the UBL element of Rad23 and Dsk2 from yeast and human. hRad23A represents human Rad23, and hPLIC-2 is human Dsk2 homologue. Highly conserved residues are indicated by the gray boxes. Mutations constructed are indicated above the sequences. (B) Coimmunoprecipitation analysis of interactions between Ufd2 and Rad23 derivatives. Proteins were extracted from cells expressing Ufd2 and Rad23 derivatives and immunoprecipitated with beads coupled to various antibodies as indicated. Immunoprecipitates were separated on SDS-PAGE, transferred to polyvinylidene difluoride membrane, and then probed with antibodies. The identity of the bands is indicated on the left. Mutations in Rad23 are indicated above the panels. The antibodies used for immunoprecipitation (IP) and Western blot (blot) are indicated on the right of the panels. (C) Coimmunoprecipitation analysis of interactions between the proteasome and Rad23 derivatives. The experiments were performed similarly as in Figure 3B, except we used the antibody against Rpt5 (Affinity), a subunit of the proteasome, to determine the amount of the proteasome.
These mutations were introduced into full-length, FLAG epitope-linked Rad23 expressed from the Gal1 promoter, which facilitates the detection of the gene products. UFD2 was fused to the myc epitope and expressed from the Gal1 promoter as well. Plasmids carrying RAD23 mutant alleles were cotransformed with the plasmid bearing UFD2 into yeast cells. FLAG-tagged wild-type RAD23 was used as a positive control. K7A, L31A, I45A, and G48R mutations abolished the Ufd2-Rad23 interaction in the coimmunoprecipitation assay (Figure 3B). Note that these mutations did not affect the levels of Rad23 and Ufd2 (Figure 3B). Similarly, we examined the effect of these mutations on the Rad23-proteasome interaction. Interestingly, the binding of Rad23 to the proteasome was detectable, but significantly reduced, in these mutants (Figure 3C), suggesting that Ufd2 and the proteasome might bind overlapping sites in the UBL domain of Rad23.
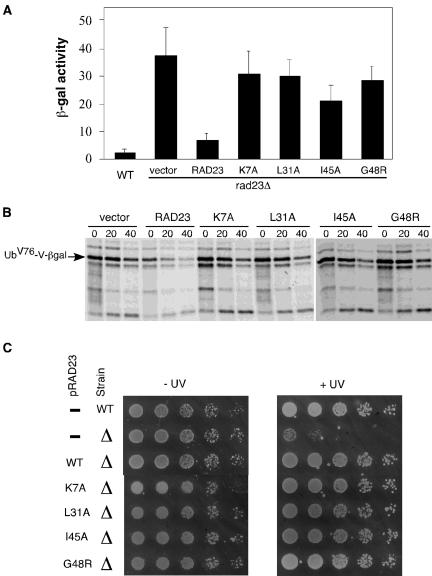
Mutations in Rad23 Impair the Degradation of a UFD Substrate, but Maintain Its Nonproteolytic Role in DNA Repair
To determine their effects on proteolysis in vivo, the UBL mutations (K7A, L31A, I45A, and G48R) were introduced into full-length Rad23 under the control of its own promoter on a low copy plasmid to avoid potential artifacts caused by overexpression. These plasmids were cotransformed with a plasmid expressing the UFD substrate, UbV76-V-βgal (Johnson et al., 1995), to _rad23_Δ cells. We used the LacZ assay to gauge the effects of the UBL mutations on the intracellular concentration of the UFD substrate. As shown in Figure 4A, _rad23_Δ cells had much higher levels of β-gal activity than wild-type cells. As expected, the expression of RAD23 in the _rad23_Δ cells restored the low levels of β-gal activity. Interestingly, higher levels of β-gal activity were maintained in _rad23_Δ cells expressing the RAD23 mutants (Figure 4A), suggesting the UBL element is important in fulfilling the proteolytic roles of Rad23 in the UFD pathway. We also carried out pulse-chase assays to measure the stability of UbV76-V-βgal in these cells. As shown in Figure 4B, UbV76-V-βgal is stabilized in _rad23_Δ cells. As expected, the expression of RAD23 in the _rad23_Δ cells rendered UbV76-V-βgal unstable. We found that the UFD substrate is stabilized in _rad23_Δ cells expressing the RAD23 mutants (Figure 4B), suggesting that these four amino acid residues (K7A, L31A, I45A, and G48R) are essential for the proteolytic function of RAD23. These results indicate that the UFD pathway is impaired in Rad23 mutants defective for Ufd2 and the proteasome interactions.
Figure 4.
The UFD proteolytic pathway, but not the nucleotide excision repair (NER) pathway, is impaired in rad23 mutant backgrounds. (A) Levels of β-gal activity in wild-type or _rad23_Δ cells carried a plasmid bearing UbV76-V-βgal, a UFD substrate, and a low-copy plasmid with or without expressing RAD23 derivatives from the RAD23 promoter as indicated. The experiments were done at least three times, and the average values with SD are shown. (B) Pulse-chase analysis of UbV76-V-βgal in _rad23_Δ cells with or without RAD23 and its derivatives as indicated. Yeast cells were grown to the exponential phase and pulse labeled with 35S for 5 min in vivo, and then cold Met/Cys mix was added to start the chase. We took samples at 20-min intervals and processed them for immunoprecipitation with antibodies against β-galactosidase, followed by SDS-PAGE and phosphorimaging analysis, as described in Rao and Sastry (2002). (C) Rad23 mutants maintain a fully functional nucleotide excision repair pathway. Yeast cultures were grown to an optical density A600 of ∼0.8, and fivefold dilutions were spotted onto YPD media. One set of cells was exposed to 20 J/m2 UV radiation. The plates were incubated at 30°C for 3 d. The RAD23 alleles on the plasmids and the status of RAD23 in the strains are labeled on the left.
Rad23 is also known to function as a DNA damage recognition factor in nucleotide excision repair (Watkins et al., 1993; Gillette et al., 2001). Neither Dsk2 nor Ddi1 has a DNA repair function (Bertolaet et al., 2001; our unpublished data). Genetic and biochemical studies have shown the proteolytic function of Rad23 is separable from its DNA repair role (Bertolaet et al., 2001; Gillette et al., 2001). Specifically, the UBA motif is essential for its proteolytic role but not required for its involvement in DNA repair. However, the UBL motif is important for the nonproteolytic functioning of Rad23 in DNA repair. UBL promotes efficient repair through interaction with the 19S regulatory complex of the proteasome. We examined the UV sensitivity of _rad23_Δ cells expressing the RAD23 alleles. As expected, the expression of RAD23 in the _rad23_Δ cells restored the resistance to UV radiation. Interestingly, UV resistance was restored in _rad23_Δ cells expressing the RAD23 mutants (Figure 4C), suggesting that the reduced proteasome binding of the Rad23 mutants is still sufficient in fulfilling the nonproteolytic roles of Rad23 in the DNA repair. Because the Ufd2-Rad23 interaction is eliminated in these mutants, these results with UV resistance indicate that the Ufd2-Rad23 association is not required for DNA repair.
Rpn1 Competes with Ufd2 for the Binding of Rad23
The UBL motif of Rad23 was previously shown to bind proteasome subunit Rpn1. Do Rpn1 and Ufd2 bind to Rad23 together or separately? We tested the possibility that Ufd2, Rad23, and Rpn1 form a trimeric complex. However, we failed to detect the interaction between Ufd2 and Rpn1 by two-hybrid assays or coimmunoprecipitations (Figure 5A; our unpublished data), suggesting that Ufd2 and Rpn1 likely form separate complexes with Rad23. To examine their interactions more directly, we expressed and purified the epitope-tagged Rad23, Rpn1, and Ufd2 separately from E. coli (Figure 5B). Purified proteins were mixed in various combinations (Rad23-Rpn1, Ufd2-Rad23, Ufd2-Rpn1, Ufd2-Rad23-Rpn1), and coimmunoprecipitation experiments were performed. We found that Rad23 directly interacts with Ufd2 or Rpn1, and Ufd2 does not interact with Rpn1 in the presence or absence of Rad23 (Figure 5C; our unpublished data).
Using coimmunoprecipitation assays, we also tested whether Rpn1 influences Ufd2-Rad23 association. Increasing amounts of Rpn1 were added to the mixture containing Ufd2-myc and Rad23-FLAG, and the immunoprecipitates were probed with monoclonal antibody against myc to determine the amount of Ufd2 retained in the FLAG beads. We found that the Ufd2-Rad23 binding was reduced with increasing amount of Rpn1 (Figure 5C). These results indicate that Ufd2 directly binds Rad23 and competes with Rpn1 for Rad23-association, which is consistent with the mutation analysis (Figure 3) that the Rad23 binding sites for Ufd2 and Rpn1 overlap.
Differential Involvement of UBL/UBA Proteins in the UFD Pathway
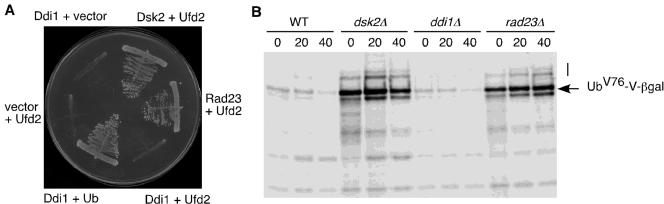
Similar to Rad23, the other two yeast UBL/UBA proteins, Dsk2 and Ddi1, interact with Ub chains and the proteasome (Elsasser et al., 2002; Saeki et al., 2002a,b). Do Dsk2 and Ddi1 interact with Ufd2? Yeast two-hybrid and coimmunoprecipitation assays showed that Dsk2, but not Ddi1, interacts with Ufd2 (Figure 6A; our unpublished data). We have previously demonstrated that both Rad23 and Dsk2 are important for the degradation of the UFD substrate UbV76-V-βgal (Rao and Sastry, 2002). Is DDI1 involved in the UFD pathway? We examined the degradation of UFD substrate in cells lacking DDI1. We carried out pulse-chase assays with UbV76-V-βgal. The UFD pathway is impaired in cells lacking RAD23 or DSK2, as indicated by stabilization of UbV76-V-βgal (Figure 6B). In contrast, deletion of DDI1 did not stabilize the UFD substrate (Figure 6B). Our data suggest that different UBL motifs have distinct activities and may be a key in determining the functions of the UBL/UBA-containing proteins.
Figure 6.
DDI1 is not involved in the UFD pathway. (A) Ddi1 does not interact with Ufd2. Yeast two-hybrid assays were performed as described in Figure 2A. Cells containing the indicated plasmids were streaked onto -his plates. Positive controls Ddi + Ub and Rad23 + Ufd2 and negative controls Ddi1+ vector and vector + Ufd2 are included. (B) UFD pathway is not affected by the lack of DDI1 gene. Pulse-chase analysis of UbV76-V-βgal in isogenic yeast strains MY3589 (wt), MY3588 (_dsk2_Δ), YHR56 (_ddi1_Δ), and MY3587F (_rad23_Δ) was performed as described in Figure 4B. An arrow indicates UbV76-V-βgal. A line indicates the “ladder” of multiubiquitylated UbV76-V-βgal.
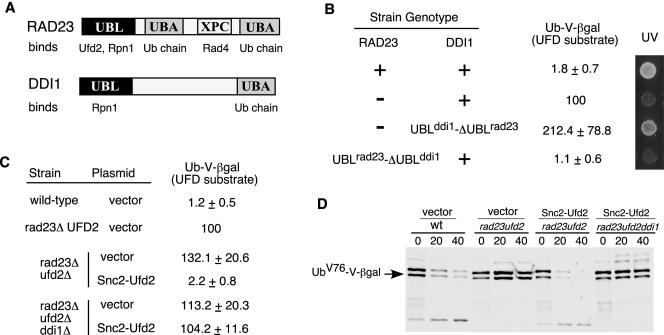
Why do Rad23 and Ddi1 have distinct roles in the UFD pathway (Figure 6B) and DNA repair (Bertolaet et al., 2001)? It is likely due to their different affinities for other proteins. Structurally, Rad23 has one unique XPC domain, which binds Rad4 and is important for its function in DNA repair, and two UBA domains with much stronger affinity for ubiquitylated proteins (Figure 7A). To identify the key determinant for their distinct functions, we swapped the domains between Rad23 and Ddi1 (Figure 7B). We replaced the C-terminal region of Ddi1 with the ΔUBL fragment of Rad23 by integration at the genomic DDI1 locus in rad23 mutant background. We found that the UBLddi1-ΔUBLrad23 fusion restored UV resistance but not the degradation of the UFD substrate in rad23 mutant cells (Figure 7B). Thus, the C-terminal fragment of Rad23 is the critical determinant for the differential involvements of Rad23 and Ddi1 in DNA repair, but not for their distinct roles in proteolysis. Similarly, we linked the UBL motif of Rad23 to the C-terminal fragment of Ddi1 at the normal genomic locus of RAD23. Interestingly, the UBLrad23-ΔUBLddi1 fusion promotes the degradation of the UFD substrate but cells are UV sensitive (Figure 7B). The results indicate that the UBL domain is the critical element that differentiates the proteolytic functions of Rad23 and Ddi1.
Figure 7.
UBL-mediated Ufd2 interaction is important for the functioning of Rad23. (A) Domain organization of Rad23 and Ddi1. Proteins that bind to each domain are indicated. (B) Domain swapping between Rad23 and Ddi1. The strain genotypes are labeled on the left. UV response was done as in Figure 4C and shown on the right. Levels of βgal activity in yeast cells carried a plasmid bearing the UFD substrate, UbV76-V-βgal, were determined as described in Figure 4A. We arbitrarily set the βgal levels of _rad23_Δ cells as 100. Average values, presented as percentages of βgal activity in _rad23_Δ mutants, and standard deviations are shown in the middle of the panel. (C) Expression of Snc2-Ufd2 fusion restores the degradation of UFD substrates in cells lacking RAD23. βgal activity in yeast cells coexpressing UbV76-V-βgal with either vector alone or the UFD2 alleles expressed from the _GAL1_-promoter. The strain genotypes and the UFD2 alleles on the plasmids are labeled on the left. (D) Pulse chase analysis of UbV76-V-βgal in isogenic yeast strains, used in Figure 7C, was performed as described in Figure 4B. An arrow indicates UbV76-V-βgal. Genotypes of various mutant strains and the plasmids used are indicated above the panel.
Biological Significance of the Ufd2-Rad23 Interaction in Proteolysis
Clearly, the UBL-mediated interactions are important for the functioning of Rad23 in proteolysis (Figures 4 and 7B). However, it is unclear whether the Ufd2-Rad23 interaction is important in proteolysis because the mutations in RAD23 affected the bindings of Rad23 to both Ufd2 and the proteasome (Figure 3). The loss-of function studies could be difficult to interpret because mutations may have multiple effects. Therefore, we took a “gain-of-function” approach to examine the significance of the UBL-mediated Ufd2 interaction. We hypothesized that an engineered Ddi1-interacting Ufd2 could act through Ddi1 and bypass the requirement of Rad23 in the UFD pathway. In this setting, Ddi1 maintains its normal activities and gains the ability to bind the Ufd2 variant.
Ddi1 was shown to bind Snc2, a regulator of the secretory pathway (Lustgarten and Gerst, 1999). We fused the Snc2 fragment (amino acids 4-93) (Lustgarten and Gerst, 1999) to the N terminus of Ufd2. Note that Rad23 binds to the N-terminal region of Ufd2 (our unpublished data). We used the LacZ assay to determine the intracellular concentration of the UFD substrate, UbV76-V-βgal. As expected, wild-type cells had very low levels of βgal activity because the substrates were rapidly degraded (Figure 7C). Higher levels of βgal activity were maintained in _rad23_Δ and _rad23_Δ _ufd2_Δ cells. We noticed that _rad23_Δ _ufd2_Δ cells had ∼30% higher levels of βgal activity than _rad23_Δ cells. This is expected because the UFD pathway is abolished in _ufd2_Δ cells lacking the E4 component, but it remains partially active in _rad23_Δ cells. Dsk2, the other UBL/UBA protein, can still promote the substrate proteolysis, albeit inefficiently in cells lacking RAD23 (Rao and Sastry, 2002). Interestingly, the expression of the Snc2-Ufd2 fusion, but not the wild-type Ufd2, in _rad23_Δ _ufd2_Δ cells restored the low levels of βgal activity to nearly the same in wild-type cells (Figure 7C; our unpublished data). The expression of the Snc2-Ufd2 fusion failed to restore the low levels of βgal activity if DDI1 is deleted (Figure 7C), suggesting that DDI1 is required for the functioning of the Snc2-Ufd2 chimera. We also carried out pulse-chase assays to directly determine the stability of UbV76-V-βgal in these cells. UbV76-V-βgal is stabilized in _rad23_Δ _ufd2_Δ cells (Figure 7D). Consistent with steady-state measurements, the expression of the Snc2-Ufd2 fusion in the _rad23_Δ _ufd2_Δ cells rendered UbV76-V-βgal unstable. Furthermore, the functioning of the Snc2-Ufd2 fusion requires intact DDI1 (Figure 7D). The results indicate that the Snc2-Ufd2 fusion efficiently bypassed the requirement of RAD23 for the degradation of UFD substrates and further suggest that the Ufd2-binding activity is important for the functioning of UBL/UBA proteins (e.g., Rad23) in proteolysis.
DISCUSSION
In summary, we have found that Rad23 directly interacts with Ufd2, a U box containing Ub chain elongation factor (E4). Interestingly, this interaction is mediated by the UBL motif, which is known for its affinity toward the proteasome. We found that Ufd2 and proteasome subunit Rpn1 compete for the Rad23 binding. Mutations in highly conserved residues in the UBL element of Rad23 significantly alter its affinities for Ufd2 and the proteasome. Importantly, these mutants impair the function of Rad23 in proteolysis, but not its proteasome-dependent, but proteolysis-independent, role in DNA repair (Figure 4, B and C). The significance of the UBL-mediated Ufd2 interaction in proteolysis is further underscored by the differential involvement of other yeast UBL/UBA-containing proteins, Dsk2 and Ddi1, in the UFD proteolytic pathway. We found that both Rad23 and Dsk2 bind Ufd2 and are important for the UFD pathway, whereas Ddi1 does not bind Ufd2 and is not involved in the UFD pathway. Furthermore, using a gain-of-function Ufd2 variant, we demonstrated that the Ufd2-binding activity is important in fulfilling the proteolytic functions of UBL/UBA proteins. The results suggest that the UBL motif may be a key determinant for the substrate selectivity of UBL/UBA-containing adaptor molecules.
Ubiquitylated species are easily detectable in cell extracts, suggesting many proteins are ubiquitylated. Increasing numbers of proteins have been shown to bind Ub and/or Ub-chains. How do these Ub-binding proteins achieve their substrate specificity? Most of these proteins, including UBA-containing proteins, do not have the UBL motif. It is likely that other protein-protein binding modules may be used to facilitate the functions of these Ub-binding proteins.
Yeast and mammalian proteins use their UBL domains to bind the proteasome. We have uncovered an additional, important function of the UBL domain in Ub-mediated proteolysis. Because the UBL motif is found in many proteins, including Bag1, an important regulator of apoptosis (Takayama et al., 1996), and Parkin, a crucial protein involved in Parkinson's disease (Gasser, 1998), our results suggest that these proteins might use their UBL elements to bind proteins other than the proteasome. To our knowledge, previous studies of UBL-containing proteins used the deletions of the UBL element and the resulting phenotypes were attributed to the loss of the proteasome binding. In light of our results, at least some of the phenotypes may be due to the loss of other UBL-mediated interactions.
Studies from our laboratory and other groups implicated the involvement of Rad23 and Dsk2 in other stress response pathways (Biggins et al., 1996). Rad23 and Dsk2 may bind Ufd2 or other ubiquitylation components in fulfilling these functions. In fact, Rad23 or Dsk2 has been shown to bind several E3/E4 proteins, including E6AP (Kleijnen et al., 2000) and p300 (Zhu et al., 2001), which are involved in the degradation of p53 tumor suppressor (Grossman et al., 2003), and APC/C (Seeger et al., 2003), which is important for cell cycle regulation. It will be important to analyze the function and significance of these Rad23-containing complexes. It also would be of interest to determine whether Rad23 and Dsk2 are required for the degradation of Ataxin-3 and UBB (+1), two known substrates of Ufd2 (Lindsten et al., 2002; Matsumoto et al., 2004).
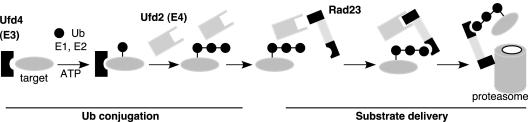
Previous studies have implicated Rad23 and Dsk2 acting as the adaptor molecules in the control of postubiquitylation events. Our results suggest that UBL/UBA adaptor proteins recognize multiubiquitylated substrates and deliver them to the proteasome, likely through a network that involves the components of ubiquitylation machinery (e.g., Ufd2) and proteasome subunits (e.g., Rpn1). We propose a model (Figure 8) in which Rad23 forms various complexes to facilitate substrate delivery to the proteasome. After the attachment of one or two Ub molecules onto the substrate mediated by Ufd4 (E3) and other required components, Ufd2 further assembles the Ub chain that is necessary to target the substrate to the proteasome (Koegl et al., 1999). Rad23 is recruited to the ubiquitylation machinery through its binding to Ufd2, and in turn, the interaction facilitates the recognition of the ubiquitylated substrate by the UBA domain of Rad23 and/or enhances the stability of the substrate-Rad23 complex. These interactions couple the ubiquitylation step to the substrate delivery process directly. Then the UBL domain of Rad23 has to be freed from Ufd2 because the UBL motif is needed to bind proteasome subunit Rpn1. The sequential interactions mediated by the UBL motif likely ensure an orderly handoff of the ubiquitylated substrate from the ubiquitylation machinery to the proteasome. The disassembly of Ufd2-Rad23 complex, and the assembly of Rad23-proteasome complex, may be facilitated by other proteins. The Rad23-Rpn1 interaction brings the ubiquitylated substrate to the proteasome. The final transfer of the substrate to the proteasome may require additional proteins (Glickman et al., 1998; Elsasser et al., 2002; Lam et al., 2002; Rao and Sastry, 2002; Raasi and Pickart, 2003).
Figure 8.
Model for substrate delivery to the proteasome mediated by Rad23. The Ub-conjugation phase is depicted based on published evidence. Through interactions mediated by the UBL and UBA domains in Rad23 with Ufd2, Ub chains, and Rpn1, Rad23 recognizes the substrate and facilitates substrate transfer to the proteasome. See text for details. Although Dsk2 is not shown here, we suspect that it functions similarly.
Acknowledgments
We are grateful to M. Rose, R. Yew, M. Gaczynska, S. Lee, J. Gerst, S. Fields, E. Friedberg, S. Jentsch, T. Ito, and the Yeast Resource Center for strains and plasmids. We thank Y. Rao, D. Sharp, and P. Sung for helpful discussions; and Peter Wu and Xiaolin Qin for preliminary data. We appreciate the helpful comments by an anonymous reviewer. This work was supported by grants to H.R. from San Antonio Cancer Institute, San Antonio Area Foundation, University of Texas Health Science Center Howard Hughes Medical Institute Research Resources Program, and UT Health Science Center Institutional Research grant.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-11-0835. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-11-0835.
References
- Baumeister, W., Walz, J., Zühl, F., and Seemüller, E. (1998). The proteasome: paradigm of a self-compartmentalizing protease. Cell 92, 367-380. [DOI] [PubMed] [Google Scholar]
- Bertolaet, B.L., Clarke, D.J., Wolff, M., Watson, M.H., Henze, M., Divita, G., and Reed, S.I. (2001). UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 8, 417-422. [DOI] [PubMed] [Google Scholar]
- Biggins, S., Ivanovska, I., and Rose, M.D. (1996). Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J. Cell Biol. 133, 1331-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., and Madura, K. (2002). Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 22, 4902-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMartino, G.N., and Slaughter, C.A. (1999). The proteasome, a novel protease regulated by multiple mechanisms. J. Biol. Chem. 274, 22123-22126. [DOI] [PubMed] [Google Scholar]
- Elsasser, S., Gali, R.R., Schwickart, M., Larsen, C.N., Leggett, D.S., Muller, B., Feng, M.T., Tubing, F., Dittmar, G.A., and Finley, D. (2002). Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4, 725-730. [DOI] [PubMed] [Google Scholar]
- Finley, D. (2001). Signal transduction. An alternative to destruction. Nature 412, 283, 285-286. [DOI] [PubMed] [Google Scholar]
- Gasser, T. (1998). Genetics of Parkinson's disease. Clin. Genet 54, 259-265. [DOI] [PubMed] [Google Scholar]
- Gillette, T.G., Huang, W., Russell, S.J., Reed, S.H., Johnston, S.A., and Friedberg, E.C. (2001). The 19S complex of the proteasome regulates nucleotide excision repair in yeast. Genes Dev. 15, 1528-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, M.H., Rubin, D.M., Fried, V.A., and Finley, D. (1998). The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 18, 3149-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman, S., Wickner, S., and Maurizi, M.R. (1997). Protein quality control: triage by chaperones and proteases. Genes Dev. 11, 815-823. [DOI] [PubMed] [Google Scholar]
- Grossman, S.R., Deato, M.E., Brignone, C., Chan, H.M., Kung, A.L., Tagami, H., Nakatani, Y., and Livingston, D.M. (2003). Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300, 342-344. [DOI] [PubMed] [Google Scholar]
- Ho, Y., et al. (2002). Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180-183. [DOI] [PubMed] [Google Scholar]
- Hofmann, R.M., and Pickart, C.M. (2001). In vitro assembly and recognition of Lys-63 polyubiquitin chains. J. Biol. Chem. 276, 27936-27943. [DOI] [PubMed] [Google Scholar]
- Johnson, E.S., Ma, P.C.M., Ota, I.M., and Varshavsky, A. (1995). A proteolytic pathway that recognizes ubiquitin as a degradation Signal. J. Biol. Chem. 270, 17442-17456. [DOI] [PubMed] [Google Scholar]
- Kleijnen, M.F., Shih, A.H., Zhou, P., Kumar, S., Soccio, R.E., Kedersha, N.L., Gill, G., and Howley, P.M. (2000). The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell. 6, 409-419. [DOI] [PubMed] [Google Scholar]
- Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H.D., Mayer, T.U., and Jentsch, S. (1999). A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96, 635-644. [DOI] [PubMed] [Google Scholar]
- Krona, C., Ejeskar, K., Abel, F., Kogner, P., Bjelke, J., Bjork, E., Sjoberg, R.M., and Martinsson, T. (2003). Screening for gene mutations in a 500 kb neuroblastoma tumor suppressor candidate region in chromosome 1p; mutation and stage-specific expression in UBE4B/UFD2. Oncogene 22, 2343-2351. [DOI] [PubMed] [Google Scholar]
- Lam, Y.A., Lawson, T.G., Velayutham, M., Zweier, J.L., and Pickart, C.M. (2002). A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416, 763-767. [DOI] [PubMed] [Google Scholar]
- Lambertson, D., Chen, L., and Madura, K. (1999). Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics 153, 69-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko, I., Grant, R.A., Wah, D.A., Sauer, R.T., and Baker, T.A. (2003). Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag. Mol. Cell. 12, 365-372. [DOI] [PubMed] [Google Scholar]
- Lindsten, K., de Vrij, F.M., Verhoef, L.G., Fischer, D.F., van Leeuwen, F.W., Hol, E.M., Masucci, M.G., and Dantuma, N.P. (2002). Mutant ubiquitin found in neurodegenerative disorders is a ubiquitin fusion degradation substrate that blocks proteasomal degradation. J. Cell Biol. 157, 417-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustgarten, V., and Gerst, J.E. (1999). Yeast VSM1 encodes a v-SNARE binding protein that may act as a negative regulator of constitutive exocytosis. Mol. Cell. Biol. 19, 4480-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, M., Yada, M., Hatakeyama, S., Ishimoto, H., Tanimura, T., Tsuji, S., Kakizuka, A., Kitagawa, M., and Nakayama, K.I. (2004). Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J. 23, 659-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg, D., Muller, R., and Funk, M. (1994). Regulatable promoters of _Saccharomyces cerevisiae_—comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22, 5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart, C.M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503-533. [DOI] [PubMed] [Google Scholar]
- Raasi, S., and Pickart, C.M. (2003). Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J. Biol. Chem. 278, 8951-8959. [DOI] [PubMed] [Google Scholar]
- Rao, H., and Sastry, A. (2002). Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J. Biol. Chem. 277, 11691-11695. [DOI] [PubMed] [Google Scholar]
- Rao, H., Uhlmann, F., Nasmyth, K., and Varshavsky, A. (2001). Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature 410, 955-959. [DOI] [PubMed] [Google Scholar]
- Saeki, Y., Saitoh, A., Toh-e, A., and Yokosawa, H. (2002a). Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem. Biophys. Res. Commun. 293, 986-992. [DOI] [PubMed] [Google Scholar]
- Saeki, Y., Sone, T., Toh-e, A., and Yokosawa, H. (2002b). Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem. Biophys. Res. Commun. 296, 813-819. [DOI] [PubMed] [Google Scholar]
- Schwartz, D.C., and Hochstrasser, M. (2003). A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem. Sci. 28, 321-328. [DOI] [PubMed] [Google Scholar]
- Seeger, M., Hartmann-Petersen, R., Wilkinson, C.R., Wallace, M., Samejima, I., Taylor, M.S., and Gordon, C. (2003). Interaction of the anaphase-promoting complex/cyclosome and proteasome protein complexes with multiubiquitin chain-binding proteins. J. Biol. Chem. 278, 16791-16796. [DOI] [PubMed] [Google Scholar]
- Takayama, S., Kochel, K., Irie, S., Inazawa, J., Abe, T., Sato, T., Druck, T., Huebner, K., and Reed, J.C. (1996). Cloning of cDNAs encoding the human BAG1 protein and localization of the human BAG1 gene to chromosome 9p12. Genomics 35, 494-498. [DOI] [PubMed] [Google Scholar]
- Verma, R., Aravind, L., Oania, R., McDonald, W.H., Yates, J.R., 3rd, Koonin, E.V., and Deshaies, R.J. (2002). Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611-615. [DOI] [PubMed] [Google Scholar]
- Walters, K.J., Kleijnen, M.F., Goh, A.M., Wagner, G., and Howley, P.M. (2002). Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 41, 1767-1777. [DOI] [PubMed] [Google Scholar]
- Watkins, J.F., Sung, P., Prakash, L., and Prakash, S. (1993). The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol. Cell. Biol. 13, 7757-7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman, A.M. (2001). Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell. Biol. 2, 169-178. [DOI] [PubMed] [Google Scholar]
- Wilkinson, C.R.M., Seeger, M., Hartmann-Petersen, R., Stone, M., Wallace, M., Semple, C., and Gordon, C. (2001). Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 3, 939-943. [DOI] [PubMed] [Google Scholar]
- Yao, T., and Cohen, R.E. (2002). A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403-407. [DOI] [PubMed] [Google Scholar]
- Zhu, Q., Wani, G., Wani, M.A., and Wani, A.A. (2001). Human homologue of yeast Rad23 protein A interacts with p300/cyclic AMP-responsive element binding (CREB)-binding protein to down-regulate transcriptional activity of p53. Cancer Res. 61, 64-70. [PubMed] [Google Scholar]