The structure of Rpf2–Rrs1 explains its role in ribosome biogenesis (original) (raw)
Abstract
The assembly of eukaryotic ribosomes is a hierarchical process involving about 200 biogenesis factors and a series of remodeling steps. The 5S RNP consisting of the 5S rRNA, RpL5 and RpL11 is recruited at an early stage, but has to rearrange during maturation of the pre-60S ribosomal subunit. Rpf2 and Rrs1 have been implicated in 5S RNP biogenesis, but their precise role was unclear. Here, we present the crystal structure of the Rpf2–Rrs1 complex from Aspergillus nidulans at 1.5 Å resolution and describe it as Brix domain of Rpf2 completed by Rrs1 to form two anticodon-binding domains with functionally important tails. Fitting the X-ray structure into the cryo-EM density of a previously described pre-60S particle correlates with biochemical data. The heterodimer forms specific contacts with the 5S rRNA, RpL5 and the biogenesis factor Rsa4. The flexible protein tails of Rpf2–Rrs1 localize to the central protuberance. Two helices in the Rrs1 C-terminal tail occupy a strategic position to block the rotation of 25S rRNA and the 5S RNP. Our data provide a structural model for 5S RNP recruitment to the pre-60S particle and explain why removal of Rpf2–Rrs1 is necessary for rearrangements to drive 60S maturation.
INTRODUCTION
Ribosomes are macromolecular machines that translate the genetic information encoded by messenger RNA into proteins. Ribosome biogenesis is a hierarchical process (1–3) that involves the processing and folding of different pre-ribosomal RNAs (pre-rRNAs) coupled with the simultaneous assembly of ribosomal proteins (r-proteins). Eukaryotic ribosome biogenesis starts with the co-transcriptional assembly of a pre-ribosomal subunit (90S) in the nucleolus that later will split into pre-40S and pre-60S subunits. Both follow independent processing and maturation steps before they are exported into the cytoplasm for final assembly.
During maturation of the 60S subunit more than 70 non-ribosomal assembly factors cluster with the different intermediates, conferring directionality and accuracy to the process. These pre-60S subunits define distinct structural neighborhoods, with a specific pre-rRNA/protein composition and intracellular localization. At least 14 assembly factors are required for the 25S rRNA processing. They are involved in the final endonucleolytic cleavage at the C2 site of the internal transcribed spacer 2 (ITS2) and by that, they recruit other molecules into the pre-ribosomes or trigger structural rearrangements (4). According to their function, they can be divided into different groups: RNA-binding proteins including Tif6, Nip7, Rpf2, Rlp24 and Nsa2; GTPases such as Nog1 and Nog2; DEAD box proteins/ATPases like Spb4, Dbp10, Drs1 and Has1 and the scaffolding protein Mak11. Nop2 could be an RNA methyltransferase, while Rrs1 has no predicted function (4). A model for the concerted recruitment of these assembly factors to nascent ribosomes has been established (4). With respect to the formation of the central protuberance a Rpf2 subcomplex has been described containing the ribosome assembly factors Rpf2 and Rrs1, and the components of the 5S RNP, the 5S rRNA, and the r-proteins RpL5 and RpL11 (5). Rpf2 has been annotated as an Imp4 superfamily protein (6–8). Proteins of this superfamily are characterized by the Brix domain fold, and eukaryotic members are known by their essential role in ribosome biogenesis. Rrs1 is an essential protein with unknown fold required for pre-rRNA processing (9). Further studies revealed that Rrs1 is primarily involved in 60S ribosomal subunit assembly (10,11).
The cryo-EM structure of a pre-60S subunit intermediate (the Arx1 particle) showed the presence of numerous assembly factors (12) and an unexpected arrangement of the 5S RNP at the central protuberance (13). The 5S RNP has already acquired its final structure (14), but is rotated by about 180° with respect to its position in the mature 60S subunit. All the neighboring 25S rRNA helices are deformed, but helix 84 (H84) interacts already with RpL11 of the 5S RNP as in the mature 60S subunit. We have recently shown how this interaction site on RpL11 is shielded by Symportin 1 (Syo1) (15), preventing RpL11 from unspecific interactions during nuclear import (16,17) and during assembly of the 5S RNP prior recruitment to the pre-60S particle (15). Several studies point towards an early incorporation of the 5S RNP (5,18) that needs to be rotated into its final position probably simultaneously with the release of assembly factors (13,19). At the pre-60S particle a number of assembly factors are present, and some of them were positioned using our previously determined crystal structures (13,19). However, although the Rpf2 and Rrs1 form a complex (5), which is already present in the pre-60S particle (19), they could not be positioned as the structure was not available.
Here we present the X-ray structure of the Aspergillus nidulans Rpf2–Rrs1 complex and dissect the interactions with components of the 5S RNP. The structure shows how Rrs1 completes the Brix domain of Rpf2 and creates a double anticodon-binding domain. Docking of the complex into a cryo-EM density of the previously described pre-60S particle (13) provides detailed information on interaction of Rpf2–Rrs1 with the 5S RNP, the assembly factor Rsa4 and the 25S rRNA. Two additional α-helices at the Rrs1 C-terminal tail are perfectly positioned to block the rearrangement of the 25S rRNA. The Rpf2–Rrs1 complex therefore seems to prevent premature rearrangements and to serve as a tether for recruiting the 5S RNP to the pre-60S.
MATERIALS AND METHODS
Cloning
The genes encoding Rpf2, Rrs1, Syo1, RpL5 and RpL11 from Chaetomium thermophilum were cloned from cDNA (_ct_Syo1, _ct_RpL5 and _ct_RpL11 (16); _ct_Rpf2 and _ct_Rrs1, kindly provided by M. Thoms, Hurt lab), whereas Rpf2 and Rrs1 from Aspergillus nidulans (Rpf2 and Rrs1) were purchased from Eurofins (MWG Operon). All clones were verified by DNA sequencing. For co-expression, Rpf2 homologs were cloned into pET-modified vectors (20) carrying an N-terminal strep tag (kanr), whereas Rrs1 homologs with addition of a C-terminal (His)6-tag were cloned into pET-16b (ampr) (Invitrogen). For pull-down purposes, the protein variants were cloned into pET-modified vectors containing (His)6-GST tags (for full list of constructs see Supplementary Table S1).
Protein expression and purification
Co-expression of Rpf2 and Rrs1-(His)6 was carried out in E. coli BL21 (DE3) Rosetta 2 cells. Cells were grown in auto-induction medium (21) at 30°C for 8–10 hours under rigorous shaking. Expression of Se-Met labeled proteins was carried out as previously described (22).
Cell pellets were resuspended in lysis buffer (20 mM HEPES pH 7.5, 250 mM NaCl, 20 mM MgCl2, 20 mM KCl) and lysed with an M-110 L Microfluidizer (Microfluidics). The lysate was clarified by centrifugation and the supernatant loaded onto a HisTrap HP column (GE Healthcare). The column was washed with lysis buffer containing 40 mM imidazole. The protein was eluted in lysis buffer containing 500 mM imidazole. The protein was then loaded onto a Resource S column (GE Healthcare). The flow-through containing the protein complex was concentrated and subjected to size-exclusion chromatography (SEC) using a S75 26/60 column (GE Healthcare) in SEC buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1% (v/v) glycerol). (His)6-GST-_ct_RpL5 and (His)6-GST-_ct_RpL11 proteins were purified by affinity chromatography in two steps using a HisTrap HP column (GE Healthcare) followed by a SP sepharose (GE Healthcare). (His)6-_ct_Rpf2, _ct_Rrs1-(His)6, the _ct_Syo1-RpL5-(His)6-RpL11 complex and the _ct_Syo1-RpL5-(His)6-RpL11–5S rRNA complex were purified as described (15,16).
Preparation of RNA
A synthetic DNA template encoding the 5S rRNA-head (5′ ACGTACGACCATACCCAGTGGAAAGCACGGCATCCCGTCCGCTCTGCCCTAGTTAAGCCACTGAGGGCCgtgaGGTGTTGACG 3′) or 5S rRNA-tail (5′ GAGGGTTAGTAGTTGGGTCGGTGACGACCAGCGAATCCCCTC3′) followed by the hammerhead ribozyme sequence were purchased from Eurofins (MWG Operon). Preparation of 5S rRNA was done as described (15,16). 5S rRNA-head and 5S rRNA-tail constructs fused to hammerhead ribozymes were designed by the method as described (23).
In vitro pull-down assays
Purified (His)6-GST-_ct_RpL11 or (His)6-GST-_ct_RpL5 were loaded on a GSTrap HP column (GE Healthcare). Excess of protein was removed by washing with 10 column volumes of lysis buffer containing 500 mM NaCl and 5 column volumes of binding buffer (20 mM HEPES-Na pH7.5, 200 mM NaCl, 5 mM KCl, 5 mM MgCl2). Purified (His)6-_ct_Rpf2 or _ct_Rrs1-(His)6 were hence added onto the column and incubated for 45 min. The column was washed with 10 volumes of SEC buffer containing 200 mM NaCl. The protein was eluted with three volumes of elution buffer (50 mM Tris-HCl pH 8.0, 200 mM NaCl, 30 mM reduced glutathione).
Electrophoretic mobility shift assay (EMSA)
EMSAs were carried out as described (15). To a constant amount of RNA, increasing molar ratios of protein (0.5x, 1x, 1.5x fold) were added and the salt concentration was adjusted to maintain similar buffer conditions. The samples were incubated for 15 min at 25°C and were analyzed by Tris-borate agarose electrophoresis in 0.5x Tris-borate buffer for 25 min at 20 mA at room temperature and ethidium bromide staining.
Size exclusion chromatography
The _ct_Syo1–RpL5–RpL11 complex in the presence or absence of 5S rRNA was incubated in stoichiometric amounts or with a 1:4 or 1:10 ratio of either purified _ct_Rpf2 or _ct_Rrs1. After incubation the samples were concentrated and subjected to SEC (S200, GE Healthcare) in SEC buffer.
Crystallization
Crystallization for native and Se-Met labeled Rpf2 (21–262)–Rrs1 (10–113) complexes were performed by automated crystallization at 18°C by sitting drop vapor diffusion methodology mixing equal volumes (200 nl) of protein complex (10 mg/ml) and crystallization buffer (JCSG core suites I-IV). Crystals of the native complex appeared within two days while crystals of the Se-Met labeled complex appeared within five days. The best diffracting crystal (resolution of 1.5 Å) was obtained for Se-Met labeled protein and grew in 0.2 M KCl and 20% (w/v) PEG 3350. Crystals were cryo-protected by soaking in mother liquor containing either 15% (v/v) ethylene glycol or 20% (v/v) glycerol and directly flash-cooled in liquid nitrogen.
Data collection and structure determination
Automatic characterization and data collection were performed at the newly established high-throughput MASSIF-1 screening beamline ID30A-1 at the European Synchrotron Radiation Facility (ESRF) in Grenoble/France. Initially, more than 70 native crystals were measured providing us with several data sets of the complex in different space groups. Single wavelength anomalous dispersion (SAD) data sets from 40 Se-Met derived crystals were collected close to the Se-edge at 0.968 Å. Data sets were processed with the XDS package (24). The initial model of the Rpf2–Rrs1 complex was obtained from a single SAD data set using the AutoSol and Autobuild programs of the PHENIX suite (25). Model building and refinement was performed with the COOT and PHENIX suites (25,26). Data collection and refinement statistics are summarized in Table 1. The model quality was analyzed with PROCHECK and MOLPROBITY (27,28). Figures were prepared using program PyMOL (Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC; http://www.pymol.org).
Table 1. Data collection and refinement statistics.
| Resolution range (Å) | 42.45–1.52 (1.57–1.52) |
|---|---|
| Space group | C 2 2 21 |
| Unit cell (Å, °) | 49.2 84.1 194.3 90 90 90 |
| Unique reflections | 61799 (5385) |
| Multiplicity | 4.8 (2.4) |
| Completeness (%) | 98.0 (84.1) |
| Mean I/sigma(I) | 11.6 (0.7) |
| Wilson B-factor (Å2) | 18.9 |
| _R_pim (%) | 4.3 (95.7) |
| CC* | 0.999 (0.675) |
| Anomalous signal+ | 2.23 |
| FOM before DM+ | 0.43 |
| _R_work (%) | 14.6 (33.0) |
| _R_free (%) | 19.1 (36.9) |
| Number of non-hydrogen atoms | 3020 |
| macromolecules | 2595 |
| water | 425 |
| RMSD (bonds, Å) | 0.012 |
| RMSD (angles, °) | 1.32 |
| Ramachandran plot (%) | |
| favored | 99 |
| outliers | 0.3 |
| Average B-factor (Å2) | 29.1 |
| macromolecules | 27.2 |
| solvent | 41.0 |
Docking of the Rpf2–Rrs1 complex into the cryo-EM density
The Rpf2–Rrs1 complex was docked as rigid body into the cryo-EM density of the pre-60S particle (13) using the program CHIMERA (29). The Rpf2–Rrs1 complex was manually pre-oriented according to its elongated structure, previous proximity studies (5) and biochemical data (this study) into the unassigned electron density next to the 5S RNP. Real space refinement was applied at the resolution cut-off of the cryo-EM study at 8.7 Å. The selected position corresponds to the highest real space correlation (0.88) and at the same time reveals excellent charge and shape complementarity. In contrast, positioning the Rpf2–Rrs1 complex rotated by 180° according to its internal pseudo-two-fold symmetry, resulted in a correlation of only 0.76 and in severe clashes with the 5S RNP.
RESULTS
Analysis of Rpf2 and Rrs1 interaction with the 5S RNP complex
We have previously shown that Syo1 chaperones 5S RNP assembly in ribosome biogenesis and that 5S RNP formation can proceed in presence of Syo1 (15). However, at which point Syo1 is released was not clear. We reasoned that other biogenesis factors might be involved in Syo1 release before or upon docking of the 5S RNP to the pre-60S subunit. As Rpf2 and Rrs1 have been implicated in 5S RNP biogenesis, we tested whether Rpf2 has an effect on the Syo1/5S RNP complex (15) by size exclusion chromatography (SEC) using proteins from the thermophilic fungus Chaetomium thermophilum (Figure 1A) (30). The Syo1–RpL5–RpL11 complex does not dissociate in the presence of increasing amounts of Rpf2 (1:1 to 1:10). Instead, we observed degradation of Rpf2 with a shorter fragment being incorporated into the Syo1–RpL5–RpL11 complex (confirmed by mass spectrometry) (Supplementary Figure S1A). We repeated the experiment with this truncated Rpf2 variant (29–308) (Figure 1A), and again observed the formation of a stoichiometric complex (Figure 1B). These data show that the truncated Rpf2 variant binds to the Syo1 complex in the absence of the 5S rRNA and Rpf2 does not release Syo1.
Figure 1.
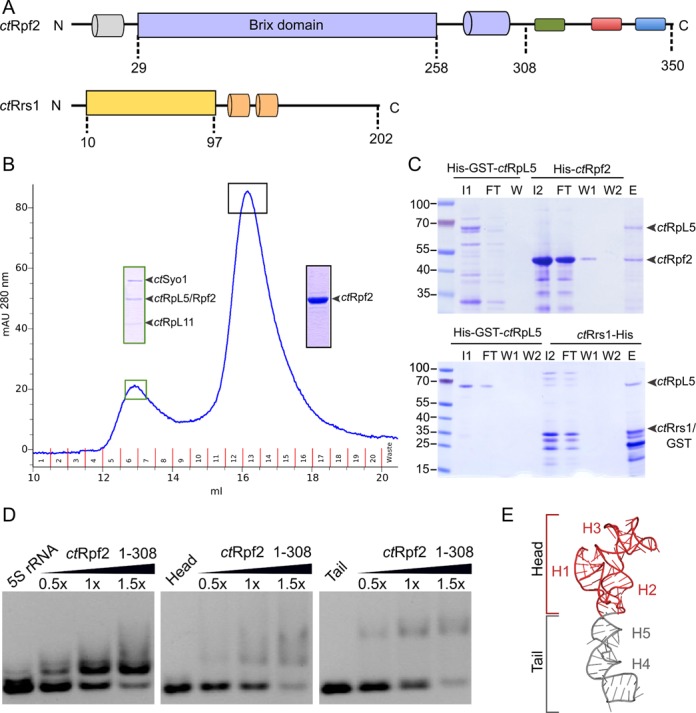
Biochemical characterization of Rpf2. (A) Domain architecture of _ct_Rpf2 and _ct_Rrs1. _Ct_Rpf2 consists of an N-terminal helix (grey), a Brix domain (29–258) (blue), a highly conserved region (green), and two highly charged regions (acidic: red, basic: blue). The core of _ct_Rrs1 (10–97) is shown in yellow and two additional helices in the C-terminal tail are represented in orange. (B) SEC elution profile for the interaction of the _ct_Syo1-RpL5-RpL11 complex with _ct_Rpf2 (29–308) (used in a 1:10 ratio). The insertions correspond to SDS PAGE lanes of the respective peak fractions (complex: green boxes; _ct_Rpf2 (29–308): black boxes). (C) Pull-down assays of _ct_Rpf2 and _ct_Rrs1 with _ct_RpL5. (His)6-GST tagged variants of _ct_RpL5 (lane I1) were immobilized on a GSTrap column and incubated with _ct_Rpf2 (lane I2, top panel) or _ct_Rrs1 (lane I2, bottom panel). SDS-PAGE samples are labeled FT: flow-through, W: wash fractions, and E: elution. (D) RNA binding assay for _ct_Rpf2 (1–308) using the full-length 5S rRNA (left panel) and different variants (middle and right panels). The protein/RNA ratio is given at the top. (E) The 5S rRNA head (red) is defined for helices H1 to H3, while the tail (grey) corresponds to helices H4 and H5.
To test whether release might depend on the 5S rRNA, we performed the binding experiment with Rpf2 (29–308) now in the presence of the 5S rRNA (Supplementary Figure S1B). Using the reconstituted Syo1/5S RNP complex we observed that addition of Rpf2 (29–308) leads again to incorporation of Rpf2 to the Syo1/5S RNP complex. Therefore, Rpf2 does not release Syo1 also in the presence of the 5S rRNA. Next, we tested whether Rrs1 interacts with the Syo1 complex using the same approach. However, SEC experiments with SDS-PAGE analysis of peak fractions followed by mass spectrometry indicate that Rrs1 alone does not bind to the Syo1 complex or that the interaction is not stable in size exclusion chromatography (Supplementary Figure S1C).
Analysis of Rpf2 and Rrs1 interaction with RpL5 and RpL11 by in vitro pull-down assays
In order to understand to which component of the 5S RNP Rpf2 binds, we performed in vitro pull-down assays with different combinations of the protein components. Glutathione S-transferase (GST)-tagged variants of RpL5 and RpL11 were used as baits together with (His)6-tagged versions of Rpf2 and Rrs1 as preys. In contrast to previous studies (5,31), we did not observe an interaction between Rpf2 or Rrs1 with RpL11 (Supplementary Figure S1D). This discrepancy could be due to the absence of washing and elution steps in pull-down assays used in previous studies (5,32). RpL5 seems to interact with both assembly factors in a one to one stoichiometry (Figure 1C). Taken together, our data show that RpL5 interacts with both assembly factors and suggests that RpL5 plays a central role in the previously described Rpf2 subcomplex (5,11).
Analysis of 5S rRNA binding by EMSAs
Since Rpf2 binds to the Syo1/5S RNP complex, we tested whether Rpf2 variants bind directly to the 5S rRNA using electrophoretic mobility shift assays (EMSAs, Figure 1D and Supplementary Figure S1E). These data show that Rpf2 directly binds the 5S rRNA. While full-length Rpf2 also binds non-specifically to the 4.5S RNA of E. coli SRP, a C-terminal truncation variant (Rpf2 1–308, as derived from sequence alignments and limited proteolysis), still binding the 5S rRNA, does not. Unspecific binding is therefore due to a charged region at the Rpf2 C-terminus.
The 5S rRNA can be described as two parts connected by a three-way junction: a ‘head’ (helices 1 to 3) and a ‘tail’ region (helices 4 and 5) (Figure 1E). In order to analyze the contribution of both parts to Rpf2 binding, we produced them separately and analyzed their ability to bind Rpf2. Our data show that Rpf2 contacts both parts of the 5S rRNA with similar efficiency, and indicate that Rpf2 might bind at the three-way junction between helices H2 and H5. This position would also allow to contact RpL5 in both the Rpf2 subcomplex as well as the Syo1/5S RNP complex.
Structure of the Rpf2–Rrs1 complex
Having shown that Rpf2 binds to the 5S rRNA, we set-out to determine the structure of the Rpf2–Rrs1 complex. A number of variants of the Chaetomium thermophilum Rpf2–Rrs1 complex derived from limited proteolysis and mass spectrometry were used for crystallization trials. However, as these complexes did not crystallize or the crystals diffracted poorly, we screened for other sources for structural studies and finally obtained crystals of the Aspergillus nidulans Rpf2–Rrs1 complex. The crystal structure of Rpf2 (residues 23–253) in complex with Rrs1 (residues 18–102) was determined at 1.5 Å resolution by the single anomalous dispersion (SAD) method (Figure 2A and Table 1) at the newly established high-throughput MASSIF-1 beamline at the ESRF (Grenoble/France). The Rpf2–Rrs1 complex crystallized in the orthorhombic space group C2221 with unit cell parameters of a = 49.2, b = 84.1 and c = 194.3 Å and contains one heterodimer in the asymmetric unit. A structural similarity search (33) for Rpf2 reveals a putative non-ribosomal factor from Archaea (PDB entry 2CXH, RMSD of 3.6 Å for 160 residues) and the Imp4-like protein Mil (PDB entry 1W94, RMSD of 2.9 Å for 139 residues) as closest structural neighbors. Residue numbers in the text refer to the A. nidulans proteins (for yeast, C. thermophilum, or H. sapiens see Supplementary Figure S2A and B). While this study was written up for publication, another structure of the Rpf2–Rrs1 complex was published at 2.35 Å resolution (34).
Figure 2.
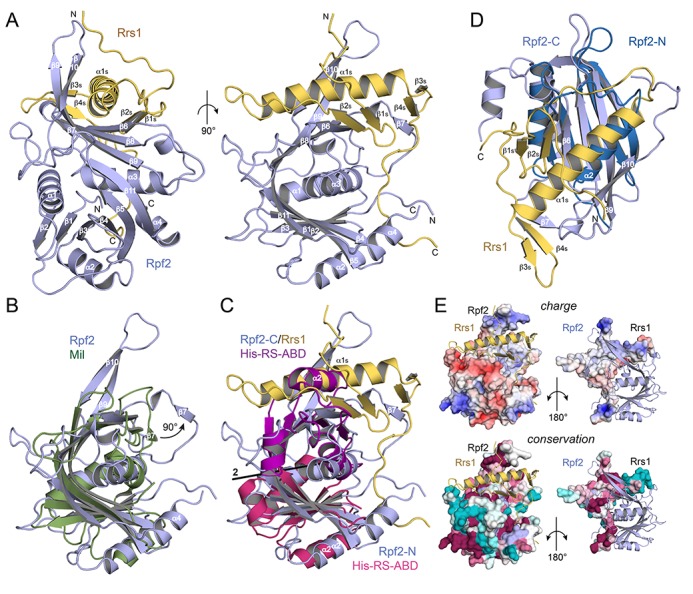
The structure of the Rpf2–Rrs1 core complex. (A) Overall structure of the complex. The Rpf2 Brix domain is shown in blue and Rrs1 in yellow. Rrs1 complements the fold of the Brix domain and its tails are stabilized by the β9–β10 hairpin and the termini of the Rpf2 core. (B) Structural comparison of the Rpf2 (blue) and archaeal Mil (green) Brix domains. Rpf2 is extended by the β9–β10 hairpin and helix α4. Strand β7 of Rpf2 is detached from the β-sheet as indicated. (C) Superposition of the ABD of a class II aminoacyl-tRNA synthetase (His-RS) with Rpf2-N and Rpf2-C, respectively. Rpf2-N and Rpf2-C are related by pseudo-twofold symmetry. (D) Superposition of Rpf2-N and Rpf2-C/Rrs1 showing their structural similarity and the equivalence of helices α2 and the N-terminal part of α1s. The C-terminal part of helix α1s is accommodated in the β-sheet extension formed by the two β-hairpins of Rrs1. (E) Electrostatic surface potentials (top; red: negative, blue: positive, contour at ±5kT) and surface conservation (bottom; magenta: conserved, cyan: variable) of Rpf2 and Rrs1 in views onto the interface. Rrs1 forms a cross-like structure spanning Rpf2 (right panels). The termini of Rrs1 (top and bottom of the cross) are positively charged and bind to highly conserved surfaces of Rpf2. The most highly basic and conserved patch of Rpf2 (bottom left in both left panels) is a major binding site for 5S rRNA.
Our structure comprises the folded core of the Rpf2–Rrs1 complex lacking the N-terminal and elongated C-terminal tails of Rpf2 (Rpf2-Ct, residues 254 to 331) and Rrs1 (Rrs1-Ct, residues 103 to 218) (Figure 2A). Rpf2 shows the fold of the Brix (biogenesis of ribosomes in Xenopus, AF319877) domain family (35) that has been described for the archaeal Imp4-like Mil protein (36) as a saddle-shaped six-stranded antiparallel β-sheet (β3β4β5β8β9β10) extended at both sides by two parallel β-strands (β1β2 and β6β7) and flanked by three α-helices (α1α2α3). Rpf2 is also extended at the C-terminus by helix α4 and strand β10 is split by a glycine-proline insertion (G229 and P230) into strands β10 and β11. Strands β9 and β10 form an extended β-hairpin not present in Mil (Figure 2B) and strand β7 is detached (about 90° rotation) from the β-sheet and creates a binding platform for the Rrs1 protein. The Brix family belongs to the anticodon-binding domain (ABD) fold (37) and resembles a duplicated ABD lacking the C-terminal helix of class IIa aminoacyl-tRNA synthetases (38,39) (Figure 2C). Gene duplication of an ABD was suggested as ancestral event in Brix domain evolution (36). The internal duplication creates two similar subdomains, Rpf2-N (residues Leu23 to Pro122) and Rpf2-C (residues Gly141 to Gln253) (Figure 2D), with an RMSD of 2.9 Å and Rpf2-C having only one N-terminal helix. Rpf2-N and Rpf2-C are related by pseudo-symmetry through a twofold rotation axis in between helices α1 and α3 (Figure 2C).
Rrs1 has a ββαββ topology (secondary structures for Rrs1 are subsequently denoted with an ‘s’) and the C-terminal region traverses the entire Rpf2 surface (Figure 2A and E right panels). Strands β1s and β2s and also β3s and β4s form β-hairpins arranged perpendicular to the C-terminal part of the long helix α1s, which comprises six turns (Figure 2D). While the N-terminus of Rrs1 is close to the tip of the Rpf2 β9–β10 hairpin, the region connecting to the β1s–β2s hairpin crosses over helix α1s and the C-terminal part forms a long proline-rich loop (residues Pro89 to Pro102) that stretches over the Rpf2 surface and locates near to the N- and C-termini of Rpf2.
Taken together, the Rpf2–Rrs1 structure reveals the first Brix domain in complex with a binding partner that together form two ABDs by domain complementation.
Rrs1 completes the Rpf2-C subdomain
Rpf2 and Rrs1 share a very peculiar interface. It is best described as domain complementation necessary to create the second ABD fold (β1α1β2α2β3β4β5). Compared to Rpf2-N, Rpf2-C misses one α-helix (α2) and one β-strand (β2), which are replaced in trans by helix α1s and strand β2s (Figure 2A and D). Together with strand β1s of the β1s–β2s hairpin, the concave β-sheet of Rpf2-C is extended by β-augmentation at strand β6 (to finally six strands). The N-terminal part of helix α1s replaces helix α2 and binds to the β-sheet as in classical ABDs, whereas the Rrs1-specific three last helical turns of helix α1s are accommodated in the augmented β-sheet to create a tight and extended interface of 2.580 Å2. A three-stranded β-sheet formed by the detached strand β7 and the β3s–β4s hairpin contributes to the binding surface for helix α1s. This interface is completed by the β7–β8 loop, forming one helical turn, and an Rpf2-C specific extension in respect to the Brix domain as seen for the archaeal Imp4-protein Mil (Figure 2B) (36). The extension corresponds to the long β9–β10 hairpin of Rpf2, formerly described to include a σ70-like sequence involved in RNA interaction (40). Structure analysis does neither comply with this annotation of a σ70-type peptide nor with its implication in RNA binding. The β9–β10 hairpin contributes to the interaction surface for Rrs1 by wrapping around helix α1s and extending to the N-terminus of Rrs1. Due to these close contacts between Rpf2 and Rrs1 and the fold-complementation the complex does not dissociate in vitro.
The Rpf2–Rrs1 interface
The analysis of surface charge distribution and conservation shows that the Rpf2–Rrs1 interface includes highly conserved surface regions (Figure 2E and Supplementary Figure S2A and B). However, the most prominent, conserved and positively charged surface on Rpf2 (the β3–β4 loop) is involved in RNA interactions (see below), whereas the negatively charged surface patch is not involved in any interactions. The N- and C-termini of Rrs1 are bound by polar and mostly basic residues (Figure 2E top panels and Figure 3), while the amphipathic helix α1s is fixed to the mixed β-sheet of Rpf2–Rrs1 by hydrophobic interactions, which complete the hydrophobic core of the complex. Despite this elaborate interface between Rpf2 and Rrs1, both proteins adopt a stable fold when they are expressed and purified independently.
Figure 3.
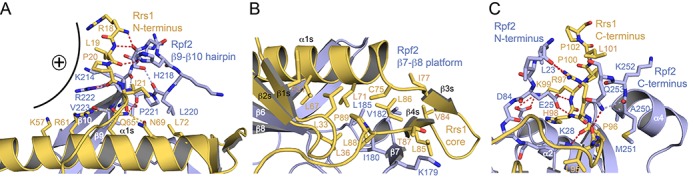
The Rpf2–Rrs1 interface. (A) The N-terminus of Rrs1 together with the β9–β10 hairpin of Rpf2 close like a belt around helix α1s of Rrs1. Hydrogen bonds are shown as dashed lines (red: Rpf2–Rrs1; blue: Rpf2–Rpf2; yellow: Rrs1–Rrs1). The interaction creates a highly basic patch denoted with a plus sign. (B) The detachment of strand β7 of Rpf2 creates a hydrophobic platform that accommodates the β1s–β2s and β3s–β4s hairpins and the C-terminal part of helix α1s. (C) The termini of the Rpf2 core form a clamp that holds the C-terminal tail of the proline-rich loop of Rrs1. Central interactions involve Glu25 of the Rpf2 N-terminus and Arg97 of the Rrs1 C-terminus. Hydrogen-bonding coloring is as in (A).
Having described the completion of the ABD fold as a main feature of the Rpf2–Rrs1 interface, there are three Rrs1 regions, which further stabilize Rrs1 on the Rpf2 surface. The N-terminus of Rrs1 is tied to the tip of the Rpf2 β9–β10 hairpin that together close like a belt around the center of helix α1s (Figure 3A). The main chain of the Rrs1 N-terminal region (residues Arg18 to Ile21) is fixed to either main or side chains of the Rpf2 hairpin (residues His218 and Arg222). This interface is completed by residues from five turns of helix α1s (residues Lys57 to Leu72) with especially Gln65 being positioned by Arg61, Asn69 and the main chain of Rpf2 Pro221. Interestingly, the Rpf2–Rrs1 interface creates a highly positively charged outer surface on one side, which seems to play a role in the interaction with the pre-60S ribosomal subunit (see below). The Rrs1 ‘core’ region comprising the two β-hairpins and the three C-terminal turns of helix α1s (residues 64 to 75) is placed on an interaction platform created by Rpf2 (Figure 3B). This involves the detached strand β7 (residues 179 to 181) and the following β7–β8 loop including a single-turn helix (residues 182 to 186). This interface is purely hydrophobic and forms the hydrophobic core of the Rpf2–Rrs1 complex. Finally, the Rrs1 C-terminus contains a long proline-rich stretch that crawls along the surface of Rpf2, with residues Leu95 to Pro102 being clamped between the N- and C-termini of Rpf2 by hydrophilic interactions (Figure 3C). The C-terminus of Rrs1 (His98 and Lys99) is hydrogen-bonded to Rpf2 Glu25 by main chain interactions, Rrs1 Lys99 forms a salt bridge with Rpf2 Asp84, and Arg97 is tethered to Leu23 at the Rpf2 N-terminus. Rrs1 Leu95 interaction with Rpf2 Gln253 completes the clamp.
Docking of the Rpf2–Rrs1 complex to the pre-60S particle
Rpf2 and Rrs1 have been implicated in the recruitment of the 5S RNP into the pre-ribosome (4,5,18) The structure of a pre-60S particle from baker's yeast including the Rpf2–Rrs1 complex has been recently determined by cryo-EM at a resolution of 8.7 Å (13). However, due to the lack of an Rpf2–Rrs1 crystal structure these two assembly factors could not be modeled in the electron density next to the 5S RNP at the ribosome. We can now unambiguously place and orient the Rpf2–Rrs1 complex (real space correlation of 0.88) in the unassigned electron density. It localizes in close contact with the 5S RNP, binding to the 5S rRNA, RpL5 and the ribosome biogenesis factor Rsa4 (Figure 4A). The position of Rpf2–Rrs1 complex at the pre-60S particle correlates with data from our pull-down experiments and EMSAs. The crystal structure fits as rigid body without any necessary adjustments. This docking differs from the model presented by Asano et al. (34), which was based on previous biochemical data only (5).
Figure 4.
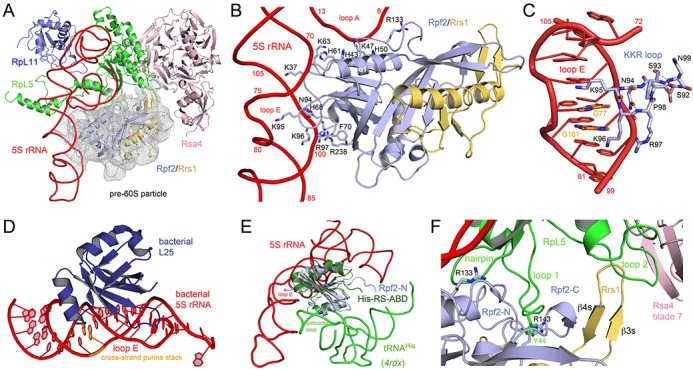
Binding of the Rpf2–Rrs1 complex to the pre-60S particle. (A) The Rpf2–Rrs1 complex perfectly fits unassigned density (2 σ) of a cryo-EM reconstruction of a pre-60S particle. The complex locates to the 5S RNA three-way junction and also binds RpL5 and the ribosome assembly factor Rsa4. Panels A to C and F are approximately in the same orientation. (B) The interaction of Rpf2-N with loops A and E of 5S rRNA. Residues involved in RNA contacts are given as sticks. The β3–β4 loop of Rpf2-N including the conserved 95-KKR motif snugly binds into loop E. (C) Close-up of the KKR loop interaction with the distorted major groove of loop E. Two conserved guanine bases involved in a cross-strand purine stack (orange) seem to be specifically read-out by Rpf2. The KKR-loop forms a β-turn capped by Asn94 and stabilized by hydrogen bonds. (D) The bacterial 5S rRNA loop E is recognized by the bacteria-specific L25 protein and the interaction also involves the conserved purine stack. View is rotated clockwise by 90° compared to A. (E) Superposition of Rpf2-N/5S rRNA with the ABD of histidine aminoacyl-tRNA synthetase (His-RS-ABD) in complex with tRNAHis (44). Specific RNA recognition of the respective ABD fold involves different non-overlapping surfaces. View is rotated approx. 45° compared to A and optimized to avoid overlaps. (F) Protein–protein interactions of Rpf2–Rrs1 with RpL5 and Rsa4 in the pre-60S particle. A continuous surface is formed between Rsa4 blade β7, RpL5 loop 2, Rrs1 β3s- β4s hairpin, Rpf2-C strand β7, RpL5 loop 1 and an N-terminal β-hairpin of RpL5. Loop 1 of RpL5 inserts like a nose into the Rpf2-N/Rpf2-C interface with a π-cation stacking of Tyr44 (RpL5) with Arg143 (Rpf2). The interface is completed towards 5S rRNA by an N-terminal β-hairpin of RpL5 contacting Rpf2-N at Arg133.
Rpf2 interacts with the 5S rRNA
While Rrs1 does not form any contacts with the 5S rRNA, Rpf2 is involved in a multitude of interactions (Figure 4B). The N-terminal half of the Brix domain (Rpf2-N) inserts the β3–β4 loop (KKR loop, residues 95 to 97) into the major groove of loop E of the 5S rRNA, and interactions are formed between helix α1 and the connecting region of Rpf2-N and Rpf2-C with the 5S rRNA three-way junction at loop A. Rigid body binding to the three-way junction confers stability to 5S rRNA and the 5S RNP. Interactions to loop A are non-specific to the phosphoribose backbone of C9 to U12 and C69 to G71. Residues involved are Lys37, His43 and His50 from helix α1, His61 and Lys63 from the β1-α1 loop, and Arg133 in the linker region to Rpf2-C.
Loop E of the 5S rRNA consists of several non-Watson-Crick base pairs, which create a widened and distorted major groove available for specific read-out by Rpf2 (Figure 4C). Especially G77 and G101, which form a cross-strand purine stack, are opening towards the major groove and seem to form specific interactions with the KKR loop in a similar mode as the bacteria-specific protein L25 recognizes the respective loop E in the major groove of 5S rRNA in bacterial ribosomes (41) (Figure 4D). The KKR loop is part of a stabilized type I β-turn, which is N-terminally capped by hydrogen bonding of the amide group of Asn94 with the main chain nitrogen of Arg97. The binding mode is reminiscent of arginine rich motifs (ARMs) that insert an α-helix into distorted RNA major grooves (42,43). Further rigidity to the KKR loop is provided by the subsequent proline Pro98 and two further hydrogen bonds. The interaction of Rpf2 with loop E is completed by the conserved (68-H/R)PFE motif within the β2-α2 loop of Rpf2 (Figure 4B). Phe70 is involved in a π-cation stacking interaction with Arg238 located in strand β11, which completes binding to the loop E at C100. These interactions correlate with point mutations described by Asano et al. as being involved in 5S rRNA binding (34).
The Rpf2-N/loop E interaction represents the first model for a Brix domain/RNA interaction in general. While several models have been suggested before (36), the Rpf2-N/loop E interaction was not predicted and the comparison with the ABD/tRNA interaction (44) indicates, that RNA-binding modes are completely different and concern different surfaces on the ABDs and the RNA (Figure 4E).
The Rpf2–Rrs1 complex interacts with RpL5 and Rsa4
Positioning of the Rpf2–Rrs1 complex in the pre-60S particle also reveals direct contacts with protein RpL5 of the 5S RNP and Rsa4 (Figure 4F). Rsa4 is implicated in the energy driven restructuring of the pre-60S particle by the Rea1 ATPase (19). While Rsa4 interacts with RpL5 mostly via two eukaryote specific loop insertions (loops 2 and 3) (19), the extended β3s–β4s hairpin of Rrs1 creates a continuous interaction surface between the four proteins including Rsa4 blade β7, RpL5 loop 2, Rrs1 β3s- β4s hairpin, Rpf2-C strand β7, RpL5 loop 1 and an N-terminal β-hairpin of RpL5. Although speculative, we favor the interpretation that a continuous β-sheet is formed across all four proteins. Besides the 5S rRNA, RpL5 loop 1 (residues 39 to 47) presents the second major docking site of the 5S RNP on Rpf2 and inserts in the concave interface between Rpf2-N and Rpf2-C (Figure 4F). Apparently, the conserved Tyr44 at the tip of RpL5 loop 1, which in the mature 60S subunit contacts ES12 of 25S rRNA, is here bound by π-cation stacking to Rpf2-N Arg143 and Rpf2-C Thr176. These interactions link the Rpf2–Rrs1 complex to Rsa4 and therefore might play a role in the previously suggested restructuring of the pre-60S particle (19).
Positioning of the Rpf2 and Rrs1 tails in the pre-60S subunit
The crystal structure of the Rpf2–Rrs1 complex lacks the tails of both proteins, which like for most ribosomal proteins are extending from the protein core and are partially unstructured. However, positioning the Rpf2–Rrs1 complex in the pre-60S particle gives a clear indication for their location. We can attribute part of the C-terminal tails of both Rpf2 (Rpf2-Ct, residues 254 to 331) and Rrs1 (Rrs1-Ct, residues 103 to 218) and also the N-terminus of Rrs1 (Rrs1-Nt, residues 1 to 17) to previously unassigned density (Figure 5A). Binding partner for Rrs1-Nt is the RpL5 loop 2 including the C-terminal β-hairpin (residues 134 to 141), and for Rrs1-Nt and Rpf2-Ct the ubiquitin-like (Ubl) domain of Rsa4. Rpf2-Ct establishes further contacts with 25S rRNA (helices H69 and H86) at the base of the central protuberance and thus creates the docking platform for the Rpf2–Rrs1 complex on the ribosomal surface.
Figure 5.
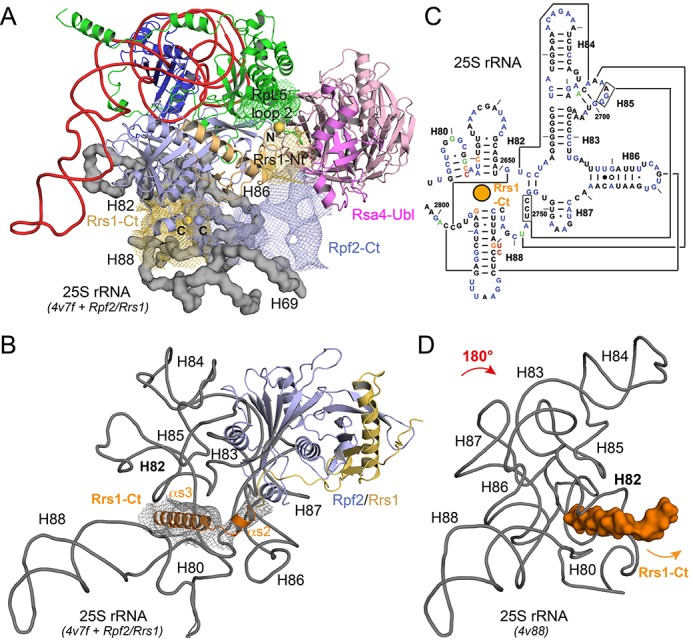
Interaction of the Rpf2–Rrs1 tails with the pre-60S particle. (A) Electron density (1 σ, coloring according proteins) of the pre-60S cryo-EM reconstruction can be assigned to the missing tails of Rpf2–Rrs1 complex and to the rearranged loop 2 of RpL5. RpL5 binds to the combined positive patch as shown in Figure 3A. Rpf2-Ct and Rrs1-Nt approach the Ubl-domain of Rsa4 and Rpf2-Ct forms the binding platform of the Rpf2–Rrs1 core on the 25S rRNA (gray surface, helices are indicated). The view is rotated by 45° to the back compared to Figure 4A to highlight the tails. (B) Rrs1-Ct includes two α-helices (α3s and α4s, orange) that wedge into the three-way junction of helices H80, H82 and H88 of 25S rRNA. The wedge fixes the rotated conformation of the central protuberance in pre-60S rRNA. (C) Secondary structure of the respective rRNA from bakers yeast with the wedge indicated as orange sphere. Figure is adapted from (48). RNA–RNA tertiary interactions of mature rRNA are given by connected lines. (D) 25S rRNA conformation of the central protuberance in the mature 60S subunit. The RNA is rotated by 180°. The wedge (orange surface) would clash with helix H82 that is now collapsed into the junction.
The most striking interaction is however formed by Rrs1-Ct, which directly follows the long proline-rich loop that traverses the whole Rpf2 Brix domain and is clamped by the Rpf2 termini. This region is enriched in positively charged residues and is predicted to form two α-helices (α2s and α3s; Supplementary Figure S2B), which can be assigned to distinct electron density of the cryo-EM study underneath the Rpf2–Rrs1 complex (Figure 5B). These two helices insert like a wedge into the three-way junction connecting helices H80, H82 and H88 of 25S rRNA (Figure 5B and C). The interactions involve the H87-H88 linker around C2760 and the major grooves of H82 and H88 next to the three-way junction. The two α-helices are thus placed into the fulcrum of the central protuberance of the pre-60S particle stabilising the conformation of this part of the rRNA as recently described and inhibiting the rotation of the 5S RNP (Figure 5D) (13). Rrs1-Ct further crawls along the ribosomal surface reaching up to protein RpL21, which in the pre-60S particle also contacts H82 (not shown).
In summary, the structure of the Rpf2–Rrs1 complex, its position on the pre-60S ribosomal subunit and the localization of its extended protein tails allows to correlate previous biochemical and structural data. The tails establish interactions with the N-terminal ubiquitin-like (Ubl) domain of Rsa4 (Rsa4-Ubl) and fix the 25S rRNA in an assembly competent conformation. All these interactions correlate the recruitment of the 5S RNP with the action of Rsa4.
DISCUSSION
During ribosome biogenesis, parallel pathways converge as a quality control mechanism before irreversible steps in assembly occur (4). The hierarchical assembly of the ribosome couples the association of biogenesis factors with the binding of r-proteins in a cooperative manner. During assembly pre-ribosomes gradually become more stable, with the final maturation steps centering around regions important for ribosome function, such as the central protuberance (3). In the mature 80S ribosome the 5S rRNA is involved in numerous RNA–RNA and RNA–protein interactions within the central protuberance (5,18). Our study adds to the understanding of the cascade of assembly and remodeling steps leading to the formation of this region and allows for a more detailed structural model.
The Rpf2–Rrs1 Brix domain-ligand complex
During ribosome biogenesis, RNA binding proteins have an important role in rRNA processing and preventing the formation of misfolded rRNA. Among those, the Imp4 superfamily including Rpf2 shows the characteristic Brix domain fold (40). In this study, we present the X-ray structure of the Rpf2–Rrs1 complex, showing that the Brix domain of Rpf2 is complemented by the interaction with its ligand Rrs1 in a way that was not previously anticipated. The Imp4 superfamily is divided into six families with one member in Archaea and five in Eukaryotes. Whereas a single protein seems to be sufficient for Archaea, ribosome biogenesis in Eukaryotes has evolved into a more elaborate process apparently requiring protein diversification (6,8). The topology of the Brix domain presents a duplication (although incomplete) of the anticodon-binding domain (ABD) of class IIa aminoacyl-tRNA synthetases, consisting of a central five-stranded β-sheet surrounded by three α-helices. The two ABDs of Rpf2 (Rpf2-N and Rpf2-C) are related by pseudo-twofold symmetry. Rpf2-N misses helix α3 of the ABD fold that dissolves in order to allow a tight back-to-back packing of the ABDs within the Brix domain and Rpf2-C needs Rrs1 to form a second ABD. In previous studies, the connection between the two ABDs (the β10–β11 linker) has been assigned as σ70-like motif of bacterial transcription factors and has been described as a specific insertion in the Imp4 superfamily required for ribosome biogenesis (40). It was described as putative RNA-binding motif in the context of the Brix domain in Eukaryotes (40). However, the structure of the Rpf2–Rrs1 complex reveals that this region is buried in the ABD interface and adopts a conformation completely different to σ70, which has already been discussed for the Mil Brix domain (36). According to all Brix domain structures, the conserved glycine-proline sequence central to the motif complements the hydrophobic core at the Rpf2-N/Rpf2-C interface and is therefore not available for RNA binding as proposed.
Rpf2–Rrs1 in ribosome biogenesis
It has been previously shown that Rpf2 and Rrs1 are necessary for recruitment of the 5S RNP into 90S pre-ribosomes (5). In this study, we provide a detailed analysis of the direct interaction between Rpf2 and the 5S rRNA. We have also characterized the interactions between both assembly factors and the protein components of the 5S RNP by in vitro pull-down assays. RpL5 is able to bind Rpf2 and Rrs1, while RpL11 shows no interaction with these two assembly factors in contrast to the model launched by Asano et al. (34). In previous studies, we have described Syo1 as a nuclear import adaptor and chaperone for ribosome biogenesis. Syo1 shields the major docking site for the 25S rRNA on RpL11 for recruitment to the pre-60S subunit (15). Here we show that Rpf2 can bind to this 5S RNP assembled in vitro in the presence of Syo1 independent of the ribosome. Syo1 may be involved in handing over the 5S RNP to the pre-60S subunit, where Rpf2 and Rrs1 are already positioned to receive it. While this is based on the detailed analysis of the interactions between the Rpf2–Rrs1 complex and the pre-60S particle, it seems to contradict previous studies which described a Rpf2 subcomplex, independent of the ribosome (5). The Rpf2 subcomplex was identified in yeast using a mutant defective in ribosome biogenesis to identify intermediates that dissociate from the pre-ribosome (5,45–47). However, subcomplexes derived from a destabilized ribosome do not necessarily indicate whether they are bona-fide building blocks able to incorporate into nascent ribosomes. They might form only upon association with the pre-ribosome. The 5S RNP is not formed in the absence of RpL11 and therefore, it seems not surprising that the Rpf2 subcomplex was also depleted in those studies. However, further experiments are required to investigate the final steps of 5S RNP incorporation into the emerging pre-60S subunit in more detail.
Structure based model for Rpf2–Rrs1 in 5S RNP recruitment
Structural information is available for the pre-60S subunit with 5S RNP already present, but in an outward rotated orientation (13). Comparison with the mature 60S subunit (14) showed that 5S RNP is identical in both structures. The 25S rRNA helix 84 interacts with RpL11 in both cases, but the conformation of the 25S rRNA helices 82 to 89 is remarkably different. This might create binding sites for biogenesis factors (13) or the biogenesis factors might maintain these different conformations to provide a time frame for certain maturation steps. At some point, energy provided by one of these factors will be necessary to trigger the relocation of rRNA elements and move the 5S RNP to its final position. Here, the force generating ATPase Rea1 has been implicated in transmitting remodeling energy to Rsa4 (19).
Integrating the current study into this general picture allows deriving a structure-based model for 5S RNP recruitment and relocation (Figure 6). The Rpf2–Rrs1 complex is recruited to the pre-ribosome early in 60S biogenesis, maybe already to the 90S pre-ribosome (5). Structural information on early intermediates is still missing until the pre-60S particle is formed (13). The interaction of the Rrs1 C-terminal tail with the 25S rRNA suggests that the Rpf2–Rrs1 complex binds before the 5S RNP arrives to block the 25S rRNA from rearrangements. We envisage the C-terminal helices of Rrs1 to serve as a wedge that stabilizes a ‘tensed’ conformation of the rRNA like preparing a mousetrap. Upon delivery of the 5S RNP probably by Syo1, RpL11 docks to H84 and Rpf2 readily interacts with the 5S rRNA at the three-way junction. The latter interaction involves conserved positively charged regions (e.g. the KKR loop) that are stabilized in a pre-formed binding conformation by the Rpf2–Rrs1 Brix domain. This network of interactions fixes the 5S RNP in its peculiar orientation supported not only by the formation of specific contacts between Rpf2 and the 5S rRNA but also by a continuous interaction surface between RpL5, the Rpf2–Rrs1 complex and the β-propeller of Rsa4. Finally, additional contacts with the Rsa4 Ubl domain and the Rpf2–Rrs1 extensions complete the interaction network. These interactions suggest how removal of Rsa4 by the Rea1 ATPase as recently proposed would trigger rearrangement of H89 (19) and starts a series of changes. Our data indicate how the concerted removal of the Rpf2–Rrs1 complex pulls the wedge necessary (but maybe not sufficient) to allow the 25S rRNA to relax into its energetically favored final structure. The 5S RNP would no longer be fixed by Rpf2–Rrs1 and could follow the rRNA rearrangements by holding on to H84 in the next steps of pre-60S maturation.
Figure 6.
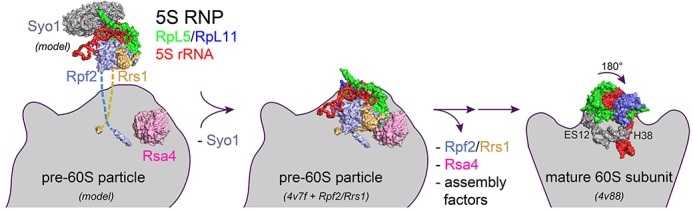
Scheme for the role of Rpf2–Rrs1 in ribosome biogenesis. (Left) An assembled 5S RNP is delivered by the symportin Syo1, which chaperones the RpL11 binding site for H84 of 25S rRNA (15). The 5S RNP is recruited to the pre-60S particle (only Rsa4 is highlighted) by the interaction with the Rpf2–Rrs1 core complex that is tethered to the pre-particle by the extended protein tails (model). Syo1 is released. (Middle) The pre-60S particle as observed by cryo-EM studies (13) with the now modeled Rpf2–Rrs1 complex. View is approximately as in Figure 5A. (Right) The central protuberance of pre-60S including the 5S RNP rotates by 180° upon retrieval of Rpf2–Rrs1 and Rsa4 and further maturation to the 60S subunit (14). All panels correspond to the crown view.
Our study provides first insights into the scaffolding function and RNA binding properties of Brix domains and their role in ribosome biogenesis. The structural and functional interplay of Rpf2 and Rrs1 may serve as a paradigm for other ribosome biogenesis factors comprising Brix domains. However, their binding partners and RNA binding modes need to be identified.
Note: While this study was written up for publication, a similar study was published by Asano et al. in Nucleic Acids Res. (34) which also reported the structure of the Rpf2–Rrs1 complex.
ACCESSION CODES
Coordinates and structure factors for the Rpf2-Rrs1 complex have been deposited in the PDB under accession number 5BY8.
Supplementary Material
SUPPLEMENTARY DATA
Acknowledgments
We gratefully acknowledge support of J. Kopp, C. Siegmann and G. Müller from the BZH/Cluster of Excellence: CellNetworks crystallization platform and B. Segnitz for excellent technical assistance. We thank R. Beckmann and E. Hurt for stimulating discussions, M. Thoms and G. Bange for their contributions in the early stages of the project, and M. McDowell and Y. L. Ahmed for helpful comments on the manuscript. Data collection was performed at the MASSIF-1 beamline at the European Synchrotron Radiation Facility (ESRF), Grenoble/France. I.S. is an investigator of the Cluster of Excellence: CellNetworks and acknowledges support through Ectop1 and by a grant from the Deutsche Forschungsgemeinschaft (SFB 638).
Author contributions: S.K. and F.R.C. performed experiments, F.R.C. and I.S. designed experiments, all authors analyzed data and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Deutsche Forschungsgemeinschaft [SFB 638].
Conflict of interest statement. None declared.
REFERENCES
- 1.Tschochner H., Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 2.Henras A.K., Soudet J., Gérus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gamalinda M., Ohmayer U., Jakovljevic J., Kumcuoglu B., Woolford J., Mbom B., Lin L., Woolford J.L. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev. 2014;28:198–210. doi: 10.1101/gad.228825.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talkish J., Zhang J., Jakovljevic J., Horsey E.W., Woolford J.L. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:8646–8661. doi: 10.1093/nar/gks609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J., Harnpicharnchai P., Jakovljevic J., Tang L., Guo Y., Oeffinger M., Rout M.P., Hiley S.L., Hughes T., Woolford J.L. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–2592. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenhaber F., Wechselberger C., Kreil G. The Brix domain protein family—a key to the ribosomal biogenesis pathway. Trends Biochem. Sci. 2001;26:345–347. doi: 10.1016/s0968-0004(01)01851-5. [DOI] [PubMed] [Google Scholar]
- 7.Koonin E. V, Wolf Y.I., Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001;11:240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayer C., Suck D., Poch O. The archaeal homolog of the Imp4 protein, a eukaryotic U3 snoRNP component. Trends Biochem. Sci. 2001;26:143–144. doi: 10.1016/s0968-0004(00)01779-5. [DOI] [PubMed] [Google Scholar]
- 9.Tsuno A., Miyoshi K., Tsujii R., Miyakawa T., Mizuta K. RRS1, a conserved essential gene, encodes a novel regulatory protein required for ribosome biogenesis in saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:2066–2074. doi: 10.1128/mcb.20.6.2066-2074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nariai M., Tanaka T., Okada T., Shirai C., Horigome C., Mizuta K. Synergistic defect in 60S ribosomal subunit assembly caused by a mutation of Rrs1p, a ribosomal protein L11-binding protein, and 3′-extension of 5S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2005;33:4553–4562. doi: 10.1093/nar/gki772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyoshi K., Shirai C., Horigome C., Takenami K., Kawasaki J., Mizuta K. Rrs1p, a ribosomal protein L11-binding protein, is required for nuclear export of the 60S pre-ribosomal subunit in Saccharomyces cerevisiae. FEBS Lett. 2004;565:106–110. doi: 10.1016/j.febslet.2004.03.087. [DOI] [PubMed] [Google Scholar]
- 12.Bradatsch B., Leidig C., Granneman S., Gnädig M., Tollervey D., Böttcher B., Beckmann R., Hurt E. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat. Struct. Mol. Biol. 2012;19:1234–1241. doi: 10.1038/nsmb.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leidig C., Thoms M., Holdermann I., Bradatsch B., Berninghausen O., Bange G., Sinning I., Hurt E., Beckmann R. 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat. Commun. 2014;5:3491. doi: 10.1038/ncomms4491. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 15.Calviño F.R., Kharde S., Ori A., Hendricks A., Wild K., Kressler D., Bange G., Hurt E., Beck M., Sinning I. Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA-binding site. Nat. Commun. 2015;6:6510. doi: 10.1038/ncomms7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kressler D., Bange G., Ogawa Y., Stjepanovic G., Bradatsch B., Pratte D., Amlacher S., Strauss D., Yoneda Y., Katahira J., et al. Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science. 2012;338:666–671. doi: 10.1126/science.1226960. [DOI] [PubMed] [Google Scholar]
- 17.Bange G., Murat G., Sinning I., Hurt E., Kressler D. New twist to nuclear import: When two travel together. Commun. Integr. Biol. 2013;6:e24792. doi: 10.4161/cib.24792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohmayer U., Gamalinda M., Sauert M., Ossowski J., Pöll G., Linnemann J., Hierlmeier T., Perez-Fernandez J., Kumcuoglu B., Leger-Silvestre I., et al. Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae. PLoS One. 2013;8:e68412. doi: 10.1371/journal.pone.0068412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bassler J., Paternoga H., Holdermann I., Thoms M., Granneman S., Barrio-Garcia C., Nyarko A., Stier G., Clark S.A., Schraivogel D., et al. A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation. J. Cell Biol. 2014;207:481–498. doi: 10.1083/jcb.201408111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bogomolovas J., Simon B., Sattler M., Stier G. Screening of fusion partners for high yield expression and purification of bioactive viscotoxins. Protein Expr. Purif. 2009;64:16–23. doi: 10.1016/j.pep.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Van Duyne G.D., Standaert R.F., Karplus P.A., Schreiber S.L., Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 23.Wild K., Bange G., Bozkurt G., Segnitz B., Hendricks A., Sinning I. Structural insights into the assembly of the human and archaeal signal recognition particles. Acta Crystallogr. D Biol. Crystallogr. 2010;66:295–303. doi: 10.1107/S0907444910000879. [DOI] [PubMed] [Google Scholar]
- 24.Kabsch W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 28.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 30.Van Noort V., Bradatsch B., Arumugam M., Amlacher S., Bange G., Creevey C., Falk S., Mende D.R., Sinning I., Hurt E., et al. Consistent mutational paths predict eukaryotic thermostability. BMC Evol. Biol. 2013;13:7. doi: 10.1186/1471-2148-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyoshi K., Tsujii R., Yoshida H., Maki Y., Wada A., Matsui Y., Toh-e A., Mizuta K. Normal assembly of 60 S ribosomal subunits is required for the signaling in response to a secretory defect in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:18334–18339. doi: 10.1074/jbc.M201667200. [DOI] [PubMed] [Google Scholar]
- 32.Miles T.D., Jakovljevic J., Horsey E.W., Harnpicharnchai P., Tang L., Woolford J.L. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell. Biol. 2005;25:10419–10432. doi: 10.1128/MCB.25.23.10419-10432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holm L., Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asano N., Kato K., Nakamura A., Komoda K., Tanaka I., Yao M. Structural and functional analysis of the Rpf2-Rrs1 complex in ribosome biogenesis. Nucleic Acids Res. 2015;43:4746–4757. doi: 10.1093/nar/gkv305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaser A., Bogengruber E., Hallegger M., Doppler E., Lepperdinger G., Jantsch M., Breitenbach M., Kreil G. Brix from Xenopus laevis and Brx1p from yeast define a new family of proteins involved in the biogenesis of large ribosomal subunits. Biol. Chem. 2001;382:1637–1647. doi: 10.1515/BC.2001.199. [DOI] [PubMed] [Google Scholar]
- 36.Ng C.L., Waterman D., Koonin E. V, Antson A.A., Ortiz-Lombardía M. Crystal structure of Mil (Mth680): internal duplication and similarity between the Imp4/Brix domain and the anticodon-binding domain of class IIa aminoacyl-tRNA synthetases. EMBO Rep. 2005;6:140–146. doi: 10.1038/sj.embor.7400328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fox N.K., Brenner S.E., Chandonia J.-M. SCOPe: Structural Classification of Proteins–extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 2014;42:D304–D309. doi: 10.1093/nar/gkt1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Logan D.T., Cura V., Touzel J.P., Kern D., Moras D. Crystallisation of the glycyl-tRNA synthetase from Thermus thermophilus and initial crystallographic data. J. Mol. Biol. 1994;241:732–735. doi: 10.1006/jmbi.1994.1547. [DOI] [PubMed] [Google Scholar]
- 39.Cusack S. Sequence, structure and evolutionary relationships between class 2 aminoacyl-tRNA synthetases: an update. Biochimie. 1993;75:1077–1081. doi: 10.1016/0300-9084(93)90006-e. [DOI] [PubMed] [Google Scholar]
- 40.Wehner K.A., Baserga S.J. The sigma(70)-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell. 2002;9:329–339. doi: 10.1016/s1097-2765(02)00438-0. [DOI] [PubMed] [Google Scholar]
- 41.Lu M., Steitz T.A. Structure of Escherichia coli ribosomal protein L25 complexed with a 5S rRNA fragment at 1.8-A resolution. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2023–2028. doi: 10.1073/pnas.97.5.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grotwinkel J.T., Wild K., Segnitz B., Sinning I. SRP RNA remodeling by SRP68 explains its role in protein translocation. Science. 2014;344:101–104. doi: 10.1126/science.1249094. [DOI] [PubMed] [Google Scholar]
- 43.Weeks K.M., Crothers D.M. Major groove accessibility of RNA. Science. 1993;261:1574–1577. doi: 10.1126/science.7690496. [DOI] [PubMed] [Google Scholar]
- 44.Tian Q., Wang C., Liu Y., Xie W. Structural basis for recognition of G-1-containing tRNA by histidyl-tRNA synthetase. Nucleic Acids Res. 2015;43:2980–2990. doi: 10.1093/nar/gkv129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harnpicharnchai P., Jakovljevic J., Horsey E., Miles T., Roman J., Rout M., Meagher D., Imai B., Guo Y., Brame C.J., et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 46.Horsey E.W., Jakovljevic J., Miles T.D., Harnpicharnchai P., Woolford J.L. Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA. 2004;10:813–827. doi: 10.1261/rna.5255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fabian G.R., Hopper a K. RRP1, a Saccharomyces cerevisiae gene affecting rRNA processing and production of mature ribosomal subunits. J. Bacteriol. 1987;169:1571–1578. doi: 10.1128/jb.169.4.1571-1578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leshin J.A., Heselpoth R., Belew A.T., Dinman J. High throughput structural analysis of yeast ribosomes using hSHAPE. RNA Biol. 2011;8:478–487. doi: 10.4161/rna.8.3.14453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY DATA