Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles (original) (raw)
Paraspeckles are mammalian subnuclear bodies built on a long noncoding RNA and are enriched in RNA binding proteins with prion-like domains; two of these proteins, RBM14 and FUS, use these domains to hold paraspeckles together.
Abstract
Prion-like domains (PLDs) are low complexity sequences found in RNA binding proteins associated with the neurodegenerative disorder amyotrophic lateral sclerosis. Recently, PLDs have been implicated in mediating gene regulation via liquid-phase transitions that drive ribonucleoprotein granule assembly. In this paper, we report many PLDs in proteins associated with paraspeckles, subnuclear bodies that form around long noncoding RNA. We mapped the interactome network of paraspeckle proteins, finding enrichment of PLDs. We show that one protein, RBM14, connects key paraspeckle subcomplexes via interactions mediated by its PLD. We further show that the RBM14 PLD, as well as the PLD of another essential paraspeckle protein, FUS, is required to rescue paraspeckle formation in cells in which their endogenous counterpart has been knocked down. Similar to FUS, the RBM14 PLD also forms hydrogels with amyloid-like properties. These results suggest a role for PLD-mediated liquid-phase transitions in paraspeckle formation, highlighting this nuclear body as an excellent model system for understanding the perturbation of such processes in neurodegeneration.
Introduction
Low complexity regions containing biased amino acid composition are abundant in the proteome, particularly in regulatory molecules, yet the utility and molecular mechanism of these largely unstructured regions has been elusive. PLDs are a subset of low complexity regions, enriched in uncharged polar amino acids and glycines, with similarities to the yeast prion protein (Couthouis et al., 2011) that can be defined using a hidden Markov algorithm (Lancaster et al., 2014). PLDs are often found in RNA binding proteins that drive protein aggregation in neurodegenerative disorders such as amyotrophic lateral sclerosis (ALS; King et al., 2012).
To fully appreciate their pathophysiology and guide therapeutic strategies, we need to understand the normal physiological role of PLDs. An emerging concept is that PLDs allow proteins to “functionally aggregate,” forming higher-order assemblies and microscopically visible RNP granules (Toretsky and Wright, 2014). Concentrating proteins and RNA in a constrained space is believed to result in more efficient gene regulatory processes. The biophysical properties of PLD-mediated interactions may explain the liquid-like properties of RNP granules (Brangwynne et al., 2009) as PLDs expressed in vitro form mesh-like networks manifesting as hydrogels (Han et al., 2012; Kato et al., 2012). These hydrogels are reminiscent of, but different to, amyloid material and may represent a functional amyloid. Central to these discoveries has been research into the RNA binding protein FUS (Fused in sarcoma), with a PLD containing many repeats of [G/S]Y[G/S] (Kwon et al., 2013; Schwartz et al., 2013). The central tyrosine in the FUS repeats is essential for hydrogel formation and stress granule targeting in the cytoplasm (Kato et al., 2012).
Like cytoplasmic RNP granules, nuclear bodies also assemble without a membrane and are composed of proteins and RNA molecules selectively recruited based on protein–protein and protein–RNA interactions (Shevtsov and Dundr, 2011), although the molecular basis of these interactions is unclear. Paraspeckles are a nuclear body that are seeded by, and built on, the long noncoding RNA NEAT1 (Nuclear Paraspeckle Assembly Transcript 1/MENε/β; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009). There are ∼40 known paraspeckle proteins—mostly abundant nuclear RNA binding proteins enriched in RNA recognition motifs (RRMs), zinc finger, and K homology domains (Naganuma et al., 2012; Fong et al., 2013). Paraspeckle formation is triggered by transcription of NEAT1 followed by the recruitment of different proteins that coordinately build up the paraspeckle structure (Mao et al., 2011). Intriguingly, FUS, TDP-43, SS18L1, HNRNPA1, TAF15, and EWSR1 are all genes encoding paraspeckle proteins (Naganuma et al., 2012; Nishimoto et al., 2013) that contain PLDs and have known ALS-causing mutations (Vance et al., 2009; Couthouis et al., 2011, 2012; Chesi et al., 2013; Kim et al., 2013). Paraspeckles are also apparent in ALS motor neurons, and NEAT1 is up-regulated in the related condition frontotemporal lobar degeneration (Tollervey et al., 2011; Nishimoto et al., 2013). Furthermore, paraspeckles are stress-responsive structures induced by viral infection, proteasome inhibition, and differentiation (Sunwoo et al., 2009; Hirose et al., 2014; Imamura et al., 2014). Paraspeckles influence gene expression by nuclear retention of RNA with inverted repeats (Prasanth et al., 2005; Chen and Carmichael, 2009) and by sequestration of specific transcription factors (Hirose et al., 2014; Imamura et al., 2014).
Given many RNA binding proteins with PLDs contain nuclear localization signals (King et al., 2012), we speculated that PLDs play a role in nuclear body formation. To identify protein–protein interactions likely mediated by PLDs, we mapped the interactome network of paraspeckle proteins. We found that RBM14 (RNA binding protein 14), an essential paraspeckle component, mediates a key interaction linking several other essential proteins into the network. This interaction is driven via the PLD of RBM14, and we showed that this region possesses the same liquid-phase transition potential as FUS, forming hydrogels with amyloid-like properties in vitro. We also confirmed that the PLDs of both RBM14 and FUS are responsible for targeting these proteins to paraspeckles and are essential for the formation of paraspeckles.
Results and discussion
The paraspeckle protein–protein interactome is rich in PLD-containing proteins
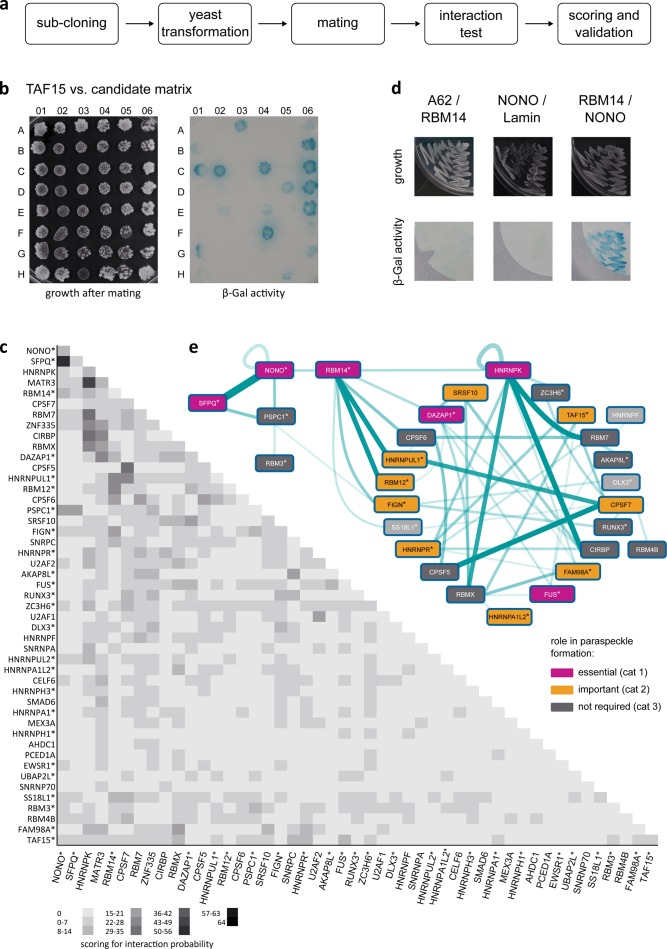
To identify the direct protein–protein interactions underpinning paraspeckles, we performed a comprehensive yeast two-hybrid screen on the known, and some putative, paraspeckle proteins (Fig. 1 c and Table S1). MATa yeast containing each candidate fused to the LexA DNA-binding domain were mated with MATα yeast containing DNA–activation domain fusions (Fig. 1, a and b). The LacZ reporter was used to screen for interactions, and interaction probability scores were assigned, with higher numbers reflecting the most stringent interactions in numerous biological replicates (Fig. 1, b–d). Interactions between key proteins were validated by cotransformation of both plasmids (Fig. 1 d) and, in some cases, by coimmunoprecipitation in HeLa cells (Fig. S1; Naganuma et al., 2012; Passon et al., 2012; Kawaguchi et al., 2015). 53 interactions between 29 paraspeckle proteins were identified that met the most stringent scoring criteria for reproducibility and strength, and these are shown in a network (Fig. 1 e). Paraspeckle proteins are unusually enriched in PLDs, as defined by the prion-like amino acid composition tool (Fig. S2; Lancaster et al., 2014). There is an overrepresentation of proteins with PLDs in the interactome network: 66% of network proteins have PLDs (19/29), whereas 55% of the starting proteins had PLDs (26/47; Fig. S2).
Figure 1.
An interactome of paraspeckle proteins. (a) Schematic of the yeast two-hybrid mating strategy. (b) Example of a growth plate (left) with 48 mated yeast spots, each containing bait protein, with different candidate fusion proteins. The code of the grid position for each candidate is in Table S1. At right is the filter lift for the plate, color is β-galactosidase (β-Gal) activity. (c) Interactions between paraspeckle proteins, with numerical values binned into grayscale, as indicated. The values reflect both the strength of the interaction as well as the number of times it occurred in the two replicate experiments, see Materials and methods. (d) Example of a cotransformation with candidate proteins and negative controls. (e) Network diagram of the interactome, excluding putative paraspeckle proteins, with a cutoff of 8 for interaction (see Materials and methods), line thickness increasing with interaction score. Color coding is relevance to paraspeckle formation, determined by siRNA knockdown (Naganuma et al., 2012). Asterisks indicate proteins with PLDs (Fig. S2).
We have previously performed siRNA knockdowns of the majority of the paraspeckle proteins to determine their importance to paraspeckle formation (Naganuma et al., 2012; summarized in Table S1). Proteins that completely, or partially, ablate paraspeckles when knocked down are colored on Fig. 1 e and comprise over half of the network: 52% of network proteins are required for paraspeckle formation (15/29), whereas only 36% of the starting protein pool are in this category (17/47; Table S1), illustrating the potential importance of protein–protein interactions in paraspeckle formation and maintenance. Furthermore, 12 of the 15 proteins required for paraspeckle formation that are in the interactome network also contain PLDs, indicating that these domains may play a key role. PLDs are emerging as important modules in gene regulation that act through a process of functional aggregation and phase transition, thus explaining their presence in numerous RNA binding proteins, transcriptional regulators, and their fusions to DNA-binding domains in chromosomal translocations in cancer. To investigate a potential for PLD-mediated functional aggregation in the context of paraspeckle formation, we focused on the PLD-containing protein RBM14, identified as directly connecting two other essential proteins, NONO (Non-POU domain-containing octamer-binding protein) and SFPQ (Splicing factor, proline- and glutamine-rich), to the rest of the network (Fig. 1 e).
The RBM14 PLD mediates protein–protein interactions and paraspeckle targeting
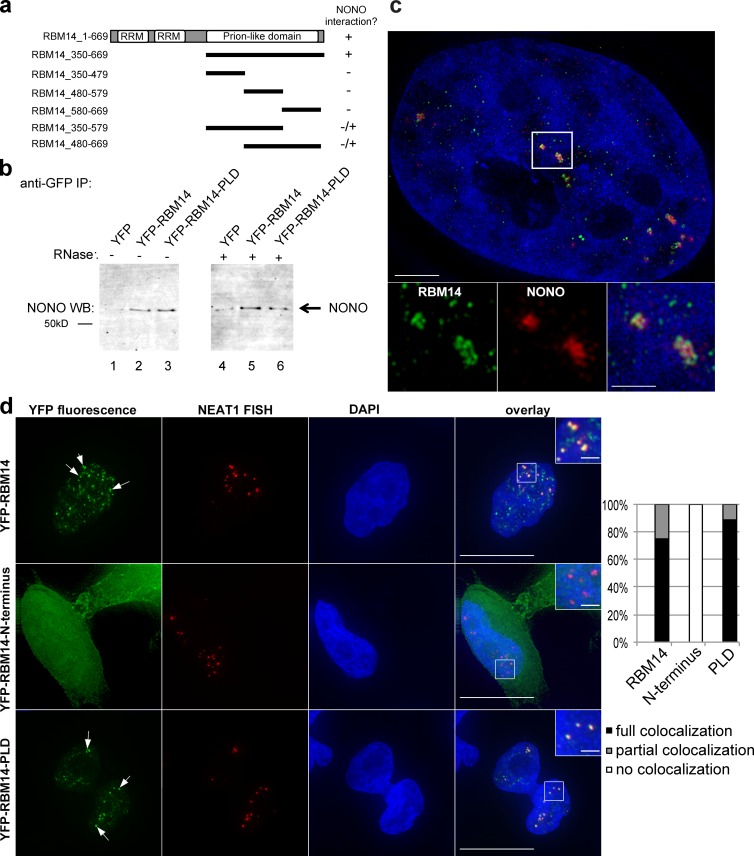
RBM14 contains two amino-terminal RRMs and a long PLD in the carboxy-terminal half of the protein, which we postulated was mediating protein–protein interactions. Indeed, yeast two-hybrid and coimmunoprecipitation experiments confirmed that the RBM14 PLD is required for its robust interaction with NONO (Fig. 2 a) and that this interaction is not dependent on RNA (Fig. 2 b). We used superresolution microscopy to confirm enrichment of NONO and RBM14 in paraspeckles (Fig. 2 c), albeit with distinct subparaspeckle patterns that may reflect unique interactions and/or functional roles. Expression of YFP-tagged fragments of RBM14, combined with FISH to detect endogenous NEAT1, showed that the PLD domain is sufficient for paraspeckle targeting (Fig. 2 d). Although the PLD and full-length RBM14 both colocalize with NEAT1 at paraspeckles, the amino-terminal fragment that contains only the RRMs (residues 1–176) does not (Fig. 2 d), suggesting that RBM14 is most probably recruited via protein–protein interactions.
Figure 2.
The RBM14 PLD mediates interaction with NONO and paraspeckle targeting. (a) Various truncations of RBM14 were tested for their ability to interact with NONO in the yeast two-hybrid assay, and the RBM14 PLD (residues 350–669) was the only fragment to recapitulate the interaction seen with full-length RBM14. (b) Western blotting for NONO showing that it is coimmunoprecipitated on GFP-trap resin from lysates containing YFP-RBM14 (lanes 2 and 5), or YFP-RBM14-PLD (lanes 3 and 6), but not YFP (lanes 1 and 4). Lysates in lanes 4–6 were treated with RNase A, which was effective at degrading RNA ∼30-fold over untreated samples, see Materials and methods. IP, immunoprecipitation; WB, Western blot. (c) Superresolution fluorescence micrograph of a HeLa nucleus: green, RBM14; red, NONO; blue, DAPI. Boxes are higher magnifications. Bars: (main image) 3 µm; (insets) 1 µm. (d) Fluorescence micrographs of representative HeLa cells transiently expressing YFP-RBM14 (top), YFP-RBM14-1-176 (middle), or YFP-RBM14-PLD (bottom). NEAT1 detected with FISH (red) to label paraspeckles. Green, YFP fluorescence; blue, DAPI. Both YFP-RBM14 and YFP-RBM14-PLD colocalize with paraspeckles, as indicated by yellow foci on overlays and arrows. Graph shows quantification of localization (see Materials and methods). Bars: (main images) 10 µm; (insets) 2 µm.
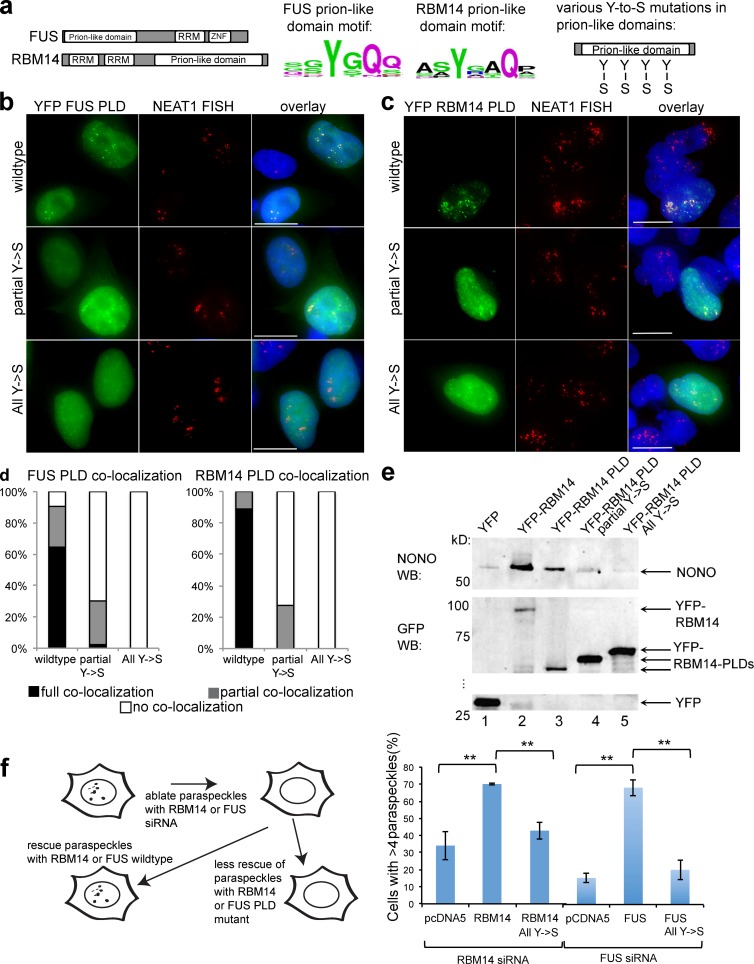
The integrity of the RBM14 and FUS PLDs is critical for paraspeckle targeting and formation
Similar to RBM14, the PLD of FUS is required for targeting FUS to paraspeckles (Shelkovnikova et al., 2014). We thus tested the importance of PLD integrity for paraspeckle targeting by these proteins, using mutants in which tyrosines in the PLD repeat motifs were mutated to serine (Fig. 3 a). FUS contains 27 repeats of Y[G/S]QQ or [S/G]Y[S/G], and RBM14 has 21 Y[G/N/A/S]AQ or [S/G]YG motifs (Table S1). The mutants we tested were either partial Y→S (half of the tyrosines mutated) or all Y→S. Fig. 3 (b and d) shows that tyrosine mutations in FUS PLD abolish paraspeckle targeting, similar to cytoplasmic stress granule targeting by FUS (Kato et al., 2012). Thus, there is likely a common PLD-dependent mechanism mediating FUS recruitment to RNPs in both subcellular compartments. RBM14 PLD mutants showed a similar trend to FUS, with YFP-RBM14-PLD partial Y→S localization showing significantly diminished paraspeckle targeting and YFP-RBM14-PLD all Y→S failing to target to paraspeckles (Fig. 3, c and d). The overexpressed PLD mutants did not disrupt paraspeckles, suggesting their expression cannot dislodge the endogenous RBM14 protein from its normal role (Fig. 3 c and Fig. S3 a). The RBM14 PLD can also promote aggregation independent of paraspeckles, as seen with numerous, additional, non-NEAT1–containing foci in cells overexpressing YFP-RBM14 PLD (Fig. S3 b). Consistent with the notion that NONO binding may be important for RBM14 paraspeckle targeting, we observed that neither RBM14 PLD tyrosine mutant could coprecipitate NONO from cell lysates (Fig. 3 e).
Figure 3.
The RBM14 PLD is essential for paraspeckle formation. (a) Schematics of secondary structure, consensus repeat motifs and mutational strategy for FUS and RBM14. ZNF, Zinc finger. (b and c) Fluorescence micrographs of representative HeLa cells transiently expressing YFP-NLS-FUS-PLD or YFP-RBM14-PLD (top), YFP-NLS-FUS-PLD partial Y→S mutant or YFP-RBM14-PLD partial Y→S mutant (middle), and YFP-NLS-FUS-PLD all Y→S mutant or YFP-RBM14-PLD All Y→S mutant (bottom). NEAT1 RNA detected with FISH (red) to label paraspeckles. Green, YFP fluorescence; blue, DAPI. The paraspeckle targeting by FUS or RBM14 PLD is lost when tyrosines are mutated. Bars, 10 µm (d) Quantification of colocalization for experiments in b and c, see Materials and methods. (e) Western blot for NONO showing it is coimmunoprecipitated on GFP-trap resin from lysates expressing YFP-RBM14 (lane 2) or YFP-RBM14-PLD (lane 3) but not YFP protein (lane 1) or either of the RBM14 PLD mutants (lanes 4 and 5). Bottom panels show anti-GFP Western of the same blot. WB, Western blot. (f) Schematic of paraspeckle rescue experiment and graph showing that transient, overexpressed, wild-type FUS or RBM14 can rescue paraspeckles after knockdown of endogenous FUS or RBM14, whereas the vector control or the Y→S mutant cannot. **, P < 0.02; means ± SD.
To assess whether the FUS and RBM14 PLD is required for paraspeckle formation, we tested the ability of wild-type and mutant constructs to rescue the ablation of these bodies by siRNA knockdown of endogenous FUS, or RBM14. We first demonstrated that knockdown of the endogenous proteins results in significantly diminished paraspeckle numbers in HeLa cells, as determined by counting NEAT1 foci. We then showed that wild-type FUS, or RBM14, but not the vector control, could rescue paraspeckle formation (Fig. 3 f). In contrast, the FUS or RBM14 PLD mutants could not rescue paraspeckle formation, giving results more similar to the vector control than wild-type FUS or RBM14 (Fig. 3 f), confirming that the PLDs of FUS and RBM14 play a critical role in paraspeckle formation. Given potential nuclear enrichment for many proteins with PLDs, nuclear body formation may represent one of the most important functions of PLDs.
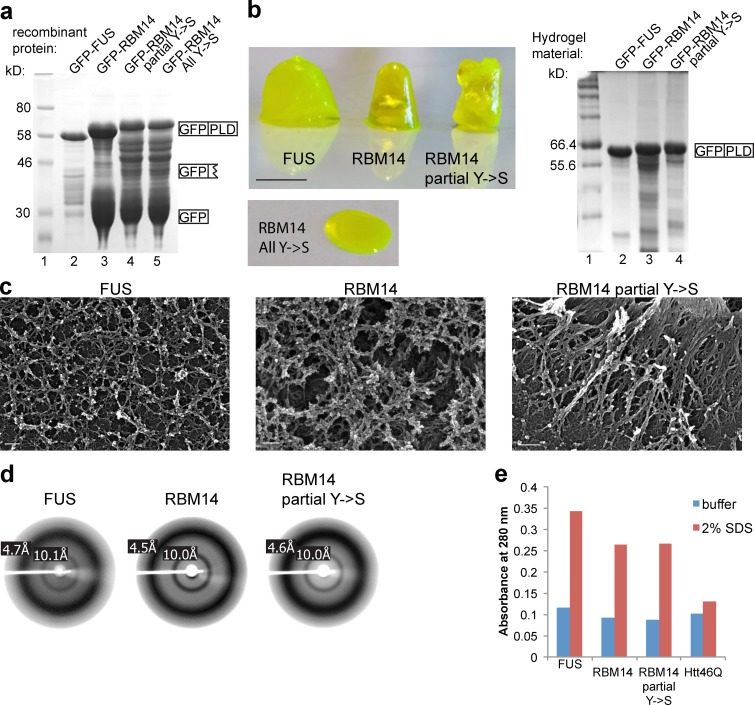
The RBM14 PLD forms a hydrogel with amyloid-like properties in vitro
Given the FUS PLD can form hydrogel in vitro, we next tested whether the RBM14 PLD shares this property. GFP-tagged RBM14-PLD and PLD mutants were expressed and purified, with GFP-FUS-PLD as a positive control (Fig. 4 a). When the samples were concentrated and cooled, all proteins, with the exception of GFP-RBM14-PLD all Y→S, formed hydrogels, composed of full-length GFP-PLDs, with a strong bias against presence of any degradation products (Fig. 4 b). Examination of their biophysical attributes confirmed similarities of RBM14 to FUS hydrogels, with scanning EM (SEM), revealing the fibril mesh networks characteristic of amyloids (Fig. 4 c) and x-ray diffraction showing the typical amyloid signature of diffraction rings at ∼4.6 and 10 Å (Fig. 4 d). Unlike pathological amyloids, however, hydrogels are relatively soluble in SDS. Fig. 4 e shows that the RBM14 and FUS hydrogels were all relatively soluble in 2% SDS, whereas aggregated huntingtin protein, typical of a pathological aggregate, was not. Collectively, these results confirm that the RBM14 PLD can form hydrogels in vitro in a similar manner to FUS PLD and that this attribute is dependent on a structurally intact PLD. Together, these findings provide new evidence for the importance of functional “tamed amyloids” in cell biology.
Figure 4.
The RBM14 PLD forms a hydrogel with amyloid-like properties. (a) Coomassie blue staining of SDS-PAGE of purified recombinant proteins with evidence of some degradation for RBM14. GFP-FUS-PLD (lane 2), GFP-RBM14-PLD (lane 3), GFP-RBM14-PLD partial Y→S (lane 4), and GFP-RBM14-PLD All Y→S (lane 5) are shown. Size markers are shown in lane 1. (b) Photos of hydrogels formed by cooled, concentrated preparations of soluble GFP-FUS-PLD (left), GFP-RBM14-PLD (middle), and GFP-RBM14-PLD partial Y→S (right). The GFP-RBM14-PLD All Y→S was incapable of forming hydrogels (bottom). Bar, 2 mm. (right) Coomassie blue staining of hydrogel material, denatured and subject to SDS-PAGE, showing that hydrogels are enriched in full-length proteins. (c) Representative SEM images showing the fibrillar nature of hydrogels made with GFP-FUS-PLD (left), GFP-RBM14-PLD (middle), and GFP-RBM14-PLD partial Y→S (right). Bars, 200 nm. (d) X-ray diffraction of hydrogels made with GFP-FUS-PLD (left), GFP-RBM14-PLD (middle), and GFP-RBM14-PLD partial Y→S (right), showing the typical amyloid rings at 4.6 and 10 Å. (e) SDS solubility assay showing GFP-FUS-PLD, GFP-RBM14-PLD, or GFP-RBM14-PLD partial Y→S hydrogels are soluble in 2% SDS, whereas the pathological form (Htt46Q) of Huntingtin protein is not.
Nuclear body and RNP granule formation are highly dynamic processes, but the molecular basis for this remains unclear. Given the abundance of tyrosine and serine residues in PLDs, phosphorylation could play a role in regulating PLD-mediated interactions underpinning bodies. Suggestive of this, we observed different SDS-PAGE mobility for RBM14 PLD mutants expressed in HeLa cells (Fig. 3 e) and found evidence that RBM14 PLD is phosphorylated in cells (Fig. S3 c). Furthermore, the FUS PLD can be phosphorylated (Gardiner et al., 2008), and phospho-FUS hydrogels have weaker interactions (Han et al., 2012).
Beyond paraspeckle formation, RBM14 is important in coregulation of transcription and splicing, centriole formation, and DNA repair (Iwasaki et al., 2001; Auboeuf et al., 2004; Yuan et al., 2014; Shiratsuchi et al., 2015). Re-examining these papers, we find the RBM14 PLD required for three out of four of these functions. Although the role of the RBM14 PLD in DNA repair is yet to be assessed, it is an attractive hypothesis that functional aggregation is involved in the dynamic formation of localized DNA repair assemblies. Notably, another essential paraspeckle protein, SFPQ, also implicated in DNA repair, requires functional aggregation via a coiled-coil motif for optimal DNA binding (Lee et al., 2015).
The regulation of RNP granule formation is emerging as a potential therapeutic application for cancer and neurodegenerative disorders. Of particular relevance is ALS, associated with cytoplasmic aggregates of normally nuclear PLD-containing proteins and caused in many cases by mutations within PLD-containing proteins. Also important is the recent discovery of the most common genetic cause of ALS: a repeat expansion in C9ORF72, resulting in formation of toxic nuclear RNA granules that sequester RNA binding proteins and production of mutant proteins that bind hydrogels (Donnelly et al., 2013; Kwon et al., 2014). Paraspeckles now join stress granules and C9ORF72 foci as RNP granules implicated in ALS pathobiology (Tollervey et al., 2011; Nishimoto et al., 2013). Six paraspeckle proteins are known to cause ALS when mutated. It is thus possible that other paraspeckle proteins are putative candidates for novel ALS genes. This growing appreciation of the importance of functional aggregation mediated by PLDs in RNA binding proteins in ALS is opening the door for future studies into how these processes are perturbed in disease.
Until now, the common feature for paraspeckle proteins was the presence of RNA binding domains, whereas the long noncoding RNA NEAT1 was thought to be the key structural determinant of paraspeckle formation. Our data do not diminish the importance of NEAT1, rather they suggest that in addition to protein–RNA interactions, there are also PLD-mediated protein–protein interactions, and together, these give rise to a dynamically regulated structure. Indeed, many paraspeckle proteins associate within cells in an RNA-dependent manner (Chesi et al., 2013; Shelkovnikova et al., 2014). Questions remain about the repeat motifs within different PLDs and how differences in these relate to functional aggregation into RNPs and other assemblies. Paraspeckle targeting could join yeast aggregation and toxicity as a useful assay to address the relative contributions of different residues within PLDs to functional aggregation (Table S1; Couthouis et al., 2011).
Materials and methods
Yeast two-hybrid interaction screen
The combinatorial yeast two-hybrid interaction screen was performed adopting a method similar to Golemis et al. (2011) and Vojtek et al. (1993). In brief, yeast strains L40 (genotype: MATa, his3-Δ200, trp1-901, leu2-3, 112, ade2-101, LYS2::(lexAop)4-HIS3, URA3::(lexAop)8-lacZ, GAL4) and AMR70 (genotype: MATα, his3-Δ200, ade2-101, trp1-901, leu2-3, 112, gal4Δ, met-, gal80Δ, MEL1, URA3::GAL1UAS -GAL1TATA-lacZ) were individually transformed with all of the candidates in each of the yeast two-hybrid vectors and grown on synthetic dropout (SD) selection agar plates. The resulting 48 AMR70 transformants were grown overnight in 2 ml SD-W liquid media in deep well blocks (5 ml). One of the L40 transformants was grown in SD-W liquid media (15 ml tube). For the mating process, an SD-L-W agar plate was prewarmed at 30°C. The L40 interactor was diluted to OD600 = 1, and 2-µl drops were applied in a 6 × 8 grid on the warm agar plate. Although the drops were drying, the L40 interactors were diluted to OD600 = 1, and subsequently, 2 µl of the mixture was applied on top of the AMR70 yeast on the plate. Plates were incubated at 30°C for ∼4 d. Mating of the two strains resulted in diploid yeast growing on the selection media carrying both yeast two-hybrid plasmids. These were then tested for β-galactosidase activity. The process was repeated until all L40 transformants were tested against the grid of AMR70 candidates. The nuclear laminar protein Lamin was among the candidates and thereby served as negative control throughout the whole screen. This entire analysis was performed as a biological duplicate. Filter lift assays were performed as described in Golemis et al. (2011). β-Galactosidase activity was observed after 2 h and overnight incubation at 30°C in humidified chambers.
Yeast two-hybrid interaction screen analysis
Analysis was performed on pictures of the colored X-Gal filter lifts as well as the yeast growth on agar plates before the filter lift assay. Candidates that gave a strong blue color for all possible interactions, including the Lamin negative control, were deemed to be auto-activating the β-galactosidase reporter gene and therefore discarded from further screen analyses.
All other interactions were scored on an intensity scale from 1 to 4. As each interactor was screened as bait against all others and also present as prey within the grid, reciprocal interactions could be observed in most cases. Numerical values were assigned to each potential interaction using several rules. First, the scores based on the intensity of β-galactosidase assays were added together (in this case, the maximum was 4 + 4 + 4 + 4, in which both replicates showed maximal interaction, with bait and prey in both orientations). Second, to give greater value to recurring interactions, we multiplied the summed scores with the number of times an interaction was observed (either during the screen repetition or as duplicate within the reciprocal screen design). In this way, the example above would score (4 + 4 + 4 + 4) × 4 = 64. The NONO–SFPQ interaction was the only one to reach this value of 64 (Fig. 1 e). Thus, an overall higher score indicated a higher probability for interaction, as false positives should not reoccur during experimental repetition (either in the biological duplicate or within the reciprocal screen arrangement) and will occur just once in the intensity scoring. Fig. 1 e was made only with interactions of a minimum score of 8. One-off false positive interactions cannot reach our threshold of 8 because a one-off maximum intensity score of 4 will only ever score 4 (4 × 1 = 4), whereas interactions with low intensity on the reporter assay, yet occurring in more than one replicate or bait–prey orientation could easily reach the threshold to occur in the network shown in Fig. 1 e (for example, (1 + 1 + 1) × 3 = 9, (2 + 1 + 1) × 3 = 12, (2 + 2) × 2 = 8, (3 + 1) × 2 = 8, (4 + 1) × 2 = 10).
The resulting interactome heatmap was created using Excel and manual binning into eleven greyscales with increasing scores of 8. Network arrangement was performed from interactions scoring higher than 8 using Cytoscape 3.1.0 (National Resource of Network Biology). Line thickness and transparency corresponds to the scoring value starting from 8. Graphical adjustments (e.g., font type, layout, coloring) were made using Inkscape (v0.48).
Yeast two-hybrid interaction validation
To validate selected interactions, combinations of candidate genes (1 µg plasmid each) were cotransformed into the L40 strain and grown on SD-L-W agar plates. Lamin and A62 both served as negative controls in the yeast bait and prey plasmids (Ingley et al., 1999). Subsequently, all growing transformants were tested for β-galactosidase activity. Blue color compared with negative controls indicated protein–protein interaction.
Plasmid construction
All candidate genes are human sequences. Some candidate gene cDNAs for yeast two-hybrid experiments were amplified from human cDNA or from plasmid sources and cloned into the pDONR entry plasmid (Life Technologies). Other candidate genes were provided as gateway entry clones in pENTR (Zhu et al., 2009; Naganuma et al., 2012). These gene sequences are published on the Human Gene and Protein Database website (HGPD; http://www.HGPD.jp). Sequence variations or isoforms are indicated in Table S1. Subcloning of all candidate genes into pBTM-GW and pVP-GW yeast two-hybrid gateway destination vectors was performed by the LR-Clonase strategy according to the manufacturer’s instructions (Gateway system; Life Technologies). pBTM-GW and pVP-GW destination vectors were provided by Zhu et al. (2009). Truncations of RBM14 were amplified by PCR using primers with gateway compatible overhangs followed by BP recombination into the pENTR vector. After sequence verification, LR reactions were performed into yeast expression vectors and a mammalian YFP expression vector.
Bacterial expression plasmids encoding the FUS PLD mutants were a gift of S. McKnight (University of Texas Southwestern, Dallas, TX; Kato et al., 2012). These were used as templates for PCR amplification to clone mutated FUS 1–215 into the YFP-C1-NLS plasmid, or to replace the endogenous FUS 1–215 sequence in pcDNA5-Flag-FUS, performed with Gibson cloning (Naganuma et al., 2012). RBM14 PLD mutants (residues 350–669) were generated by gene synthesis (GenScript). RBM14 Partial Y→S has the following changes: Y365S, Y384S, Y402S, Y417S, Y432S, Y449S, Y474S, Y498S, and Y528S. RBM14 Full Y→S has the following changes: Y356S, Y365S, Y377S, Y384S, Y395S, Y402S, Y410S, Y417S, Y425S, Y432S, Y442S, Y449S, Y460S, Y474S, Y486S, Y498S, Y504S, Y528S, and Y540S. After synthesis, the mutant RBM14 PLD cDNAs were then PCR amplified and cloned into peYFP-C1-NLS using the Gibson cloning reaction (New England Biolabs, Inc.). To make full-length RBM14 mutants that were siRNA resistant, first, a Flag-tagged RBM14 construct was made with silent mutations (aAtcAgcTgcGtcGtcTctTgcAta) across the siRNA target sequence (5′-AGTCTGCAGCCTCCTCACTAGCTTA-3′). Subsequently, the Gibson method was used to replace the wild-type 350–669 residues in RBM14 with the PCR amplified partial Y→S and full Y→S 350–669 regions. To make plasmids for expressing recombinant RBM14 PLD (residues 350–669), as well as mutants, the relevant regions were PCR amplified and cloned into pHis-GFP FUS vector (gift of S. McKnight) digested with KpnI and XhoI using the Gibson method (New England Biolabs, Inc.).
Cell culture and transfection
HeLa cells were grown in DMEM supplemented with 10% fetal calf serum and 100 U/ml penicillin and streptomycin (Life Technologies). Cells were transfected with plasmids using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. For imaging experiments, cells were transfected with plasmids followed by fixation the next day. For RBM14 siRNA knockdown experiments, cells were grown on coverslips in 8-well plates and then transfected with siRNA against RBM14 (Life Technologies) using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer’s instructions. 48 h after siRNA transfection, cells were then transfected with FLAG-tagged RBM14, RBM14 partial Y→S, and RBM14 full Y→S, and pcDNA5-Flag-EGFP as a vector control followed by fixation and processing for NEAT1 FISH 16 h later. For FUS siRNA knockdown experiments, cells were grown on coverslips in 8-well plates and then transfected with siRNA against FUS (sense, 5′-UAGGAUUUCCGGAGAAUUCUUUACC-3′ and antisense, 5′-GGUAAAGAAUUCUCCGGAAAUCCUA-3′; Life Technologies) using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer’s instructions. 48 h after siRNA transfection, another round of siRNA transfection was performed. 48 h later, cells were transfected with Flag-tagged FUS, FUS all Y→S, and pcDNA5-Flag-EGFP as a vector control followed by fixation and processing for NEAT1 FISH 24 h later.
Microscopy, FISH, and immunofluorescence
For standard microscopy, images were captured using a microscope (DeltaVision Elite; GE Healthcare) using a 60×/1.42 NA objective lens, camera (CoolSNAP HQ2; Photometrics), and the softWoRx 6.0 software (GE Healthcare). Images shown are deconvolved maximum intensity projections of z stacks through each nucleus, captured at RT.
Immunofluorescent microscopy on HeLa cells was performed by first fixing cells, grown on coverslips, in 4% paraformaldehyde (in PBS). This was followed by permeabilizing in 1% Triton X-100 (in PBS) and then incubation with mouse anti-NONO (Souquere et al., 2010) and rabbit anti-RBM14 (BETHA300-331; Bethyl Laboratories, Inc.) diluted in PBS/0.05% Tween 20 for 1 h at RT. After washing, coverslips were incubated with anti–mouse Alexa Fluor 643–conjugated antibody, and anti–rabbit Alexa Fluor 488 (both Life Technologies), for 1 h, and then counterstained with DAPI and mounted with Vectorshield medium (Vector Laboratories). Superresolution microscopy was performed using the DeltaVision OMX V4 Blaze-SIM (Structured Illumination Microscopy) system (GE Healthcare). Images were taken using the 60× objective lens. Before every experiment, a reference image using TetraSpeck fluorescent microspheres (Life Technologies) was taken and used to calibrate the system, correcting for position shift. Immersion oil with a refractive index between 1.512 and 1.516 was used to match the calibrated system. Z-series images were taken at 4.5-µm optical thickness with 0.25-µm intervals. Images were taken with varying exposure times using the DAPI, FITC, and Cy5 filters. Deconvolution, followed by a quick projection, was applied on all images using the softWoRx software (GE Healthcare).
NEAT1 FISH was performed with two different methods. For all experiments with the exception of that shown in Fig. 3 e, FISH with labeled probes (Stellaris Quasar-570–conjugated NEAT1 probe; Biosearch Technologies) was performed on HeLa cells according to the manufacturer’s instructions. In brief, HeLa cells on coverslips were washed with PBS and fixed with 4% paraformaldehyde (in PBS) at RT for 15 min. The cells were then rinsed in PBS and permeabilized in 70% ethanol overnight at 4°C. Hybridization of probes at the manufacturer’s recommended dilution was performed overnight at 37°C in 100 mg/ml dextran sulfate and 10% formamide in 2× SSC. After washing, cell nuclei were counterstained with DAPI (Sigma-Aldrich).
For Fig. 3 e, NEAT1 FISH and subsequent immunofluorescence was performed as described in Naganuma et al. (2012). In brief, NEAT1 probes were made by NEAT1 cDNA fragment (Naganuma et al., 2012) using Fluorescein RNA Labeling Mix (Roche) according to the manufacturer’s instructions. Hybridization of NEAT1 FITC-labeled probes were performed for 16 h at 55°C in 2× SSC/50% formamide with Denhardt’s, 10 mM EDTA, 100 µg/ml yeast tRNA, 0.01% Tween 20, and 5% Dextran. Slides were washed two times in 2× SSC/50% formamide for 30 min at 55°C and one time in NTET buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 500 mM NaCl, and 0.1% Tween 20) for 15 min at 37°C before incubating with 10 µg/ml RNase in NTET buffer for 1 h at 37°C. After washing once in 2× SSC/0.01% Tween 20 for 30 min at 55°C and twice in 0.01% SSC/0.01% Tween 20 for 30 min at 55°C, slides were incubated with mouse anti-FLAG (1:1,000 dilution; Sigma-Aldrich) and rabbit anti-Fluorescein (1:1,000 dilution; Abcam) overnight at 4°C. After washing in TBST for 5 min at RT, slides were incubated with anti–mouse Alexa Fluor 488 goat anti–rabbit antibody and Alexa Fluor 568 goat anti–mouse antibody (1:1,000 dilution; both obtained from Life Technologies) for 1 h at RT in shielding light. Slides were washed three times with TBST in shielding light before mounting VECTASHIELD with DAPI. Images were taken using a microscope (FV1000D-IX81) and FV10-ASW software (both obtained from Olympus). Numbers of NEAT1 foci were counted in FLAG-positive cells in about one hundred cell nuclei. The cells with more than four NEAT1 foci in a nucleus were counted as cells with normally formed paraspeckles.
For quantification of fluorescence colocalization, linescan analysis was performed for red (NEAT1 FISH) and green fluorescence (YFP proteins) channels. For each YFP fluorescent protein, 40 transfected cells were chosen in an unbiased manner. Colocalization was determined quantitatively by analyzing coincidence of fluorescence peaks in linescans. Peaks were identified where the first derivative of the fluorescence signal passes through 0 and thresholded using 1 SD of the cumulative first derivative as a cutoff. Cells were scored “full colocalization” if, for every red (NEAT1) peak, there was a coincident green fluorescent (YFP protein) peak; scored “partial colocalization” if only some red peaks had a corresponding green peak, even if only just above background fluorescence; and “no colocalization” if there was no green peak for any red peak.
Coimmunoprecipitation experiments
HeLa cells were transfected with YFP-fusion plasmids in 6-well plates. The next day, cells were washed with PBS and lysed with radioimmunoprecipitation assay buffer (100 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.5% SDS, 0.5% sodium deoxycholate, 1% NP-40, and protease inhibitor cocktail tablet [Roche]) on ice for 15 min. Lysates were passed through a QIAshredder (QIAGEN). The lysates of YFP, YFP-RBM14 partial Y→S and YFP-RBM14 full Y→S were diluted with mock transfected lysate to adjust for the amount of extracted YFP fusion proteins. In some cases, the lysates were incubated with 50 µg/ml RNase A for 15 min at 30°C before immunoprecipitation. RNase A was demonstrated to be efficient at degrading RNA by spectrophotometric analysis of total RNA levels as a result of purifying RNA in control and treated samples using a RNA column (QIAGEN) according to the manufacturer’s instructions. Lysates containing equal amounts of YFP fusions were incubated with 20 µl magnetic GFP-Trap beads (ChromoTek) at 4°C for 1 h under constant mixing. The beads were washed three times with radioimmunoprecipitation assay buffer and two times with wash buffer (10 mM Tris-HCl, pH 7.5, 300 mM NaCl, 0.5 mM EDTA, and 0.05% NP-40). For phosphatase treatment, the beads in each immunoprecipitation were split, one half was resuspended in CutSmart phosphatase buffer, and the other half was resuspended in CutSmart buffer plus calf intestinal alkaline phosphatase (New England Biolabs, Inc.); both samples were incubated at 37°C for 1 h. Bound proteins were eluted by directly adding SDS loading buffer to the beads. The resultant protein mixtures were separated by 1D SDS-PAGE and transferred onto membranes for Western blotting. Coimmunoprecipitated proteins were detected with mouse anti-NONO antibody (Souquere et al., 2010), and immune-precipitated YFP fusions were detected with anti-GFP (Roche) and anti–mouse antibody (800 nm; LI-COR Biosciences). The signal was visualized with the Odyssey imager (LI-COR Biosciences).
For Fig. S1, HeLa cells (107 cells) were lysed with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, Complete EDTA-free [Roche], and PhoSTOP [Roche]) and placed on ice for 30 min, and the supernatant was recovered by centrifugation at 10,000 g for 10 min. Antibodies were mixed with Dynabeads protein G (Invitrogen) for 1 h followed by washing twice in lysis buffer. The remaining supernatants were mixed with antibody–beads conjugates and rotated overnight at 4°C, and then the beads were washed five times with lysis buffer. Immunoprecipitations were performed with rabbit polyclonal antibody to RBM14 (Bethyl Laboratories) and mouse monoclonal to HNRNPK (Abcam). Western blotting was carried out with rabbit polyclonal to HNRNPK (Bethyl Laboratories), rabbit polyclonal to HNRNPUL1 (Abcam) and rabbit polyclonal to RBMX (Abcam).
Protein expression and purification
GFP-FUS PLD, GFP-RBM14 PLD, and GFP-RBM14 PLD mutant proteins were overexpressed in Escherichia coli BL21(DE3) cells with 0.5 mM IPTG at 16°C overnight. Harvested cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 20 mM imidazole, 100 mM DTT, 0.1 mM PMSF, and protease inhibitor cocktail, EDTA-free [Roche]) and incubated at 60°C for 10 min with agitation at 2-min intervals, to heat denature proteases. Cells were lysed using a high-pressure homogenizer (EmulsiFlex-C5; Avestin) at 200 kPa. The lysates were spun at 24,000 g at 4°C for 30 min. The soluble fraction was mixed with preequilibrated Profinity IMAC Ni2+-charged resin (Bio-Rad Laboratories, Inc.) at 4°C for 30 min. The resin mixture was poured into a Econo-Pac gravity-flow column, washed with 10× resin volume of lysis buffer, and the bound proteins were eluted with elution buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 500 mM imidazole, 100 mM DTT, 0.1 mM PMSF, protease inhibitor cocktail, and EDTA-free [Roche]). EDTA was added to a final concentration of 0.5 mM. Samples were subject to 1D SDS-PAGE and stained with Coomassie blue.
Formation of FUS and RBM14 hydrogels
The purified proteins were concentrated using Amicon Ultra Centrifugal filters (EMD Millipore) to ∼40 mg/ml. The protein solutions were then dialyzed against gelation buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 100 mM DTT, 0.5 mM EDTA, and 0.1 mM PMSF) at 4°C for 2 h. The dialyzed protein solutions were left in 0.2-ml PCR tubes at 4°C for ≥48 h to gelate.
X-ray diffraction
GFP-FUS, GFP-RBM14, and GFP-RBM14 partial Y→S mutant hydrogels were dialyzed against distilled water overnight. The hydrogels were then lyophilized, and the solid materials were mounted on a nylon CryoLoop. X-ray diffraction images were collected at RT using a Bruker MICROSTAR generator (wavelength = 1.54 Å) equipped with a Mar345dtb detector. The sample to detector distance was 350 mm. Each of the samples was oscillated 1° during a 20-min exposure.
SEM
GFP-FUS, GFP-RBM14, and GFP-RBM14 partial Y→S mutant hydrogels were mounted onto 10-mm coverslips that were coated with poly-l-lysine. The hydrogels were then fixed with 2.5% glutaraldehyde at 4°C for 2 h before they were dehydrated in a series of ethanol washes (50%, 70%, 90% and twice in absolute “dry” ethanol). The samples were critical point dried, mounted on SEM aluminum stubs with double-sided carbon tape, and coated with gold. Images were collected using a variable-pressure field-emission scanning electron microscope (1555; Carl Zeiss; Centre for Microscopy, Characterisation and Analysis, University of Western Australia) at 10 kV.
Htt46Q-CFP aggregation and SDS solubility assay
6 µmol of purified Htt46Q-CFP protein was incubated at 37°C for 72 h with 0.1% sodium azide to induce aggregation. For the SDS solubility assay, Htt46Q aggregate material and hydrogel fragments of GFP-FUS, GFP-RBM14 wild type, and GFP-RBM14 partial mutant were incubated with and without 2% SDS at 37°C for 10 min. The samples were then passed through a 0.2-μm spin filter to remove solid material, and the UV absorbance of the flow through was measured to monitor the amount of monomeric protein that passed through the filter.
Online supplemental material
Fig. S1 shows selected coimmunoprecipitations for validation of the yeast two-hybrid screen. Fig. S2 shows the PLD prediction algorithm output for each of the proteins used in this study. Fig. S3 shows that RBM14 PLD mutants do not act as dominant negatives to disrupt endogenous paraspeckles, but that wild-type RBM14 PLDs are capable of aggregating in the absence of NEAT1, and that RBM14 PLD is phosphorylated in vivo. Table S1 is a list of proteins and their attributes used in this study and is provided online as an Excel file. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201504117/DC1.
Supplementary Material
Supplemental Material
Acknowledgments
We acknowledge the University of Western Australia Centre for Microscopy Characterisation and Analysis and the University of Melbourne Bio21 imaging facility. We gratefully acknowledge Steve McKnight for plasmids.
This research was supported by a Deutscher Akademischer Austauschdienst (DAAD) Fellowship to S. Hennig, Cancer Council of Western Australia Fellowship to A.H. Fox, Ministry of education, culture, sports, science and technology Grant (of Japan) 26113002 to T. Hirose, and National Health and Medical Research Council (of Australia) grants 1030695 and 1050585 to A.H. Fox and C.S. Bond.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
ALS
amyotrophic lateral sclerosis
PLD
prion-like domain
RRM
RNA recognition motif
SD
synthetic dropout
SEM
scanning EM
References
- Auboeuf D., Dowhan D.H., Li X., Larkin K., Ko L., Berget S.M., and O’Malley B.W.. 2004. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol. Cell. Biol. 24:442–453. 10.1128/MCB.24.1.442-453.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., and Hyman A.A.. 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 324:1729–1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Chen L.L., and Carmichael G.G.. 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 35:467–478. 10.1016/j.molcel.2009.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesi A., Staahl B.T., Jovičić A., Couthouis J., Fasolino M., Raphael A.R., Yamazaki T., Elias L., Polak M., Kelly C., et al. . 2013. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat. Neurosci. 16:851–855. 10.1038/nn.3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C.M., Hutchinson J.N., Sara S.A., Ensminger A.W., Fox A.H., Chess A., and Lawrence J.B.. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 33:717–726. 10.1016/j.molcel.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J., Hart M.P., Shorter J., DeJesus-Hernandez M., Erion R., Oristano R., Liu A.X., Ramos D., Jethava N., Hosangadi D., et al. . 2011. A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl. Acad. Sci. USA. 108:20881–20890. 10.1073/pnas.1109434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J., Hart M.P., Erion R., King O.D., Diaz Z., Nakaya T., Ibrahim F., Kim H.J., Mojsilovic-Petrovic J., Panossian S., et al. . 2012. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 21:2899–2911. 10.1093/hmg/dds116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly C.J., Zhang P.W., Pham J.T., Haeusler A.R., Mistry N.A., Vidensky S., Daley E.L., Poth E.M., Hoover B., Fines D.M., et al. . 2013. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 80:415–428. 10.1016/j.neuron.2013.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K.W., Li Y., Wang W., Ma W., Li K., Qi R.Z., Liu D., Songyang Z., and Chen J.. 2013. Whole-genome screening identifies proteins localized to distinct nuclear bodies. J. Cell Biol. 203:149–164. 10.1083/jcb.201303145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner M., Toth R., Vandermoere F., Morrice N.A., and Rouse J.. 2008. Identification and characterization of FUS/TLS as a new target of ATM. Biochem. J. 415:297–307. 10.1042/BJ20081135 [DOI] [PubMed] [Google Scholar]
- Golemis E.A., Serebriiskii I., Finley R.L. Jr., Kolonin M.G., Gyuris J., and Brent R.. 2011. Interaction trap/two-hybrid system to identify interacting proteins. Curr. Protoc. Cell Biol. Chapter 17:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., and McKnight S.L.. 2012. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 149:768–779. 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Hirose T., Virnicchi G., Tanigawa A., Naganuma T., Li R., Kimura H., Yokoi T., Nakagawa S., Bénard M., Fox A.H., and Pierron G.. 2014. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell. 25:169–183. 10.1091/mbc.E13-09-0558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K., Kato A., Kawaguchi Y., Sato H., Yoneda M., et al. . 2014. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 53:393–406. 10.1016/j.molcel.2014.01.009 [DOI] [PubMed] [Google Scholar]
- Ingley E., Williams J.H., Walker C.E., Tsai S., Colley S., Sayer M.S., Tilbrook P.A., Sarna M., Beaumont J.G., and Klinken S.P.. 1999. A novel ADP-ribosylation like factor (ARL-6), interacts with the protein-conducting channel SEC61beta subunit. FEBS Lett. 459:69–74. 10.1016/S0014-5793(99)01188-6 [DOI] [PubMed] [Google Scholar]
- Iwasaki T., Chin W.W., and Ko L.. 2001. Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM). J. Biol. Chem. 276:33375–33383. 10.1074/jbc.M101517200 [DOI] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. . 2012. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 149:753–767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T., Tanigawa A., Naganuma T., Ohkawa Y., Souquere S., Pierron G., and Hirose T.. 2015. SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc. Natl. Acad. Sci. USA. 112:4304–4309. 10.1073/pnas.1423819112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Kim N.C., Wang Y.D., Scarborough E.A., Moore J., Diaz Z., MacLea K.S., Freibaum B., Li S., Molliex A., et al. . 2013. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 495:467–473. 10.1038/nature11922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King O.D., Gitler A.D., and Shorter J.. 2012. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 1462:61–80. 10.1016/j.brainres.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H., Han T., Xie S., Corden J.L., and McKnight S.L.. 2013. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 155:1049–1060. 10.1016/j.cell.2013.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I., Xiang S., Kato M., Wu L., Theodoropoulos P., Wang T., Kim J., Yun J., Xie Y., and McKnight S.L.. 2014. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 345:1139–1145. 10.1126/science.1254917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster A.K., Nutter-Upham A., Lindquist S., and King O.D.. 2014. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics. 30:2501–2502. 10.1093/bioinformatics/btu310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Sadowska A., Bekere I., Ho D., Gully B.S., Lu Y., Iyer K.S., Trewhella J., Fox A.H., and Bond C.S.. 2015. The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation. Nucleic Acids Res. 43:3826–3840. 10.1093/nar/gkv156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y.S., Sunwoo H., Zhang B., and Spector D.L.. 2011. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 13:95–101. 10.1038/ncb2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganuma T., Nakagawa S., Tanigawa A., Sasaki Y.F., Goshima N., and Hirose T.. 2012. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 31:4020–4034. 10.1038/emboj.2012.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto Y., Nakagawa S., Hirose T., Okano H.J., Takao M., Shibata S., Suyama S., Kuwako K., Imai T., Murayama S., et al. . 2013. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain. 6:31 10.1186/1756-6606-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passon D.M., Lee M., Rackham O., Stanley W.A., Sadowska A., Filipovska A., Fox A.H., and Bond C.S.. 2012. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc. Natl. Acad. Sci. USA. 109:4846–4850. 10.1073/pnas.1120792109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth K.V., Prasanth S.G., Xuan Z., Hearn S., Freier S.M., Bennett C.F., Zhang M.Q., and Spector D.L.. 2005. Regulating gene expression through RNA nuclear retention. Cell. 123:249–263. 10.1016/j.cell.2005.08.033 [DOI] [PubMed] [Google Scholar]
- Sasaki Y.T., Ideue T., Sano M., Mituyama T., and Hirose T.. 2009. MENε/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA. 106:2525–2530. 10.1073/pnas.0807899106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J.C., Wang X., Podell E.R., and Cech T.R.. 2013. RNA seeds higher-order assembly of FUS protein. Cell Reports. 5:918–925. 10.1016/j.celrep.2013.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelkovnikova T.A., Robinson H.K., Troakes C., Ninkina N., and Buchman V.L.. 2014. Compromised paraspeckle formation as a pathogenic factor in FUSopathies. Hum. Mol. Genet. 23:2298–2312. 10.1093/hmg/ddt622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov S.P., and Dundr M.. 2011. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 13:167–173. 10.1038/ncb2157 [DOI] [PubMed] [Google Scholar]
- Shiratsuchi G., Takaoka K., Ashikawa T., Hamada H., and Kitagawa D.. 2015. RBM14 prevents assembly of centriolar protein complexes and maintains mitotic spindle integrity. EMBO J. 34:97–114. 10.15252/embj.201488979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquere S., Beauclair G., Harper F., Fox A., and Pierron G.. 2010. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol. Biol. Cell. 21:4020–4027. 10.1091/mbc.E10-08-0690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H., Dinger M.E., Wilusz J.E., Amaral P.P., Mattick J.S., and Spector D.L.. 2009. MEN ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 19:347–359. 10.1101/gr.087775.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., König J., Hortobágyi T., Nishimura A.L., Zupunski V., et al. . 2011. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14:452–458. 10.1038/nn.2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toretsky J.A., and Wright P.E.. 2014. Assemblages: functional units formed by cellular phase separation. J. Cell Biol. 206:579–588. 10.1083/jcb.201404124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C., Rogelj B., Hortobágyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. . 2009. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 323:1208–1211. 10.1126/science.1165942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek A.B., Hollenberg S.M., and Cooper J.A.. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 74:205–214. 10.1016/0092-8674(93)90307-C [DOI] [PubMed] [Google Scholar]
- Yuan M., Eberhart C.G., and Kai M.. 2014. RNA binding protein RBM14 promotes radio-resistance in glioblastoma by regulating DNA repair and cell differentiation. Oncotarget. 5:2820–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T., Wang W., Yang X., Wang K., and Cui Z.. 2009. Construction of two gateway vectors for gene expression in fungi. Plasmid. 62:128–133. 10.1016/j.plasmid.2009.06.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material