A general approach for chemical labeling and rapid, spatially controlled protein inactivation (original) (raw)
Abstract
Chemical labeling of proteins inside of living cells can enable studies of the location, movement, and function of proteins in vivo. Here we demonstrate an approach for chemical labeling of proteins that uses the high-affinity interaction between an FKBP12 mutant (F36V) and a synthetic, engineered ligand (SLF′). A fluorescein conjugate to the engineered ligand (FL-SLF′) retained binding to FKBP12(F36V) and possessed similar fluorescence properties as parental fluorescein. FL-SLF′ labeled FKBP12(F36V) fusion proteins in live mammalian cells, and was used to monitor the subcellular localization of a membrane targeted FKBP12(F36V) construct. Chemical labeling of FKBP12(F36V) fusion proteins with FL-SLF′ was readily detectable at low expression levels of the FKBP12(F36V) fusion, and the level of fluorescent staining with FL-SLF′ was proportional to the FKBP12(F36V) expression level. This FL-SLF′-FKBP12(F36V) labeling technique was tested in fluorophore assisted laser inactivation (FALI), a light-mediated technique to rapidly inactivate fluorophore-labeled target proteins. FL-SLF′ mediated FALI of a β-galactosidase-FKBP12(F36V) fusion protein, causing rapid inactivation of >90% of enzyme activity upon irradiation in vitro. FL-SLF′ also mediated FALI of a β-galactosidase fusion expressed in living NIH 3T3 cells, where β-galactosidase activity was reduced in 15 s. Thus, FL-SLF′ can be used to monitor proteins in vivo and to target rapid, spatially and temporally defined inactivation of target proteins in living cells in a process that we call FK-FALI.
Keywords: fluorescent, FKBP12, fluorophore-assisted laser inactivation
Understanding the roles of specific proteins in cellular processes is a fundamental goal of molecular biology. Methods to label proteins inside of cells are broadly useful to study the localization, movement, interactions, and microenvironments of proteins in vivo. Typically, fusion proteins are created with the target protein and a fluorescent protein from the GFP family (1). Although GFP fusions have been widely useful, there are several limitations that underscore the need for other approaches to label proteins in vivo. Fusions to GFP can alter the localization of the target protein, and GFP is large enough (27 kDa) that it can potentially affect the function of the fusion protein (2). An alternative labeling approach may succeed in those cases where GFP fails. In addition, an ideal system would allow the use of a wide range of labels, including fluorescent dyes of a variety of colors, luminescent or magnetic labels, and sensors for second messengers and other important cellular characteristics.
Chemical labeling of target proteins is an intriguing solution to these problems, because a broad range of chemicals with different properties might all be targeted to the same labeling motif. Several efforts have been made to chemically label target proteins in live cells, but these approaches have some limitations. These techniques include labeling cysteine-containing peptides with biarsenical compounds, binding of fluorescein to an antifluorescein single chain antibody (scFv), and labeling an avidin fusion protein with a biotin conjugate (3–5). Background labeling, toxicity, and suitability for only a subset of target proteins and cell types may limit the broad use of these systems (6, 7). Despite the difficulties faced by these techniques, chemical labeling is of great interest because it permits novel types of experiments by targeting chemicals with a wider range of functionality than that of fluorescent proteins. For example, intracellular targeting of a photosensitizing compound to a protein of interest can be used to rapidly inactivate that protein upon irradiation (8). GFP is not an efficient photosensitizer, presumably because the protein shell of GFP prevents the generation or the efficacy of reactive oxygen species (ROS) that mediate the inactivation (9).
Methods to inactivate or remove specific proteins from cells are essential techniques that permit direct testing of the role of a protein in a given biological process. Genetic knockout of target proteins has been an invaluable technique, but interpretation of knockout experiments is often complicated by developmental lethality of the knockout organism or concern that other proteins compensate for the defect. RNA interference (RNAi) is broadly useful, but recent concerns have been raised about the specificity of RNAi (10). RNAi depends on the gradual, natural degradation of the target protein, and can thus take hours or days to deplete the protein from cells (11). In addition, genetic knockouts and RNAi lack spatial control of the inactivation of the target protein. This deficit is significant, because many cellular signaling pathways have important spatial components, including inducible translocation, colocalization of enzymes and substrates, and local generation of second messengers. Subcellular actions of proteins remain an important source of investigation; methods to inactivate proteins in a spatially controlled manner are thus an important goal.
Fluorophore-assisted laser inactivation (FALI, also known as chromophore-assisted laser inactivation, or CALI) is a technique with great potential to inactivate proteins with spatial and temporal precision (12). In FALI, an antibody that is labeled with a suitable fluorophore is introduced into cells, which are then irradiated. Irradiation stimulates local generation of oxygen radicals by the fluorophore. These ROS chemically react with the nearby antigen and inactivate it. Because inactivation is mediated by light, FALI can be controlled spatially and temporally. Although it is a powerful technique, FALI is limited by the need to deliver a labeled antibody into cells. To circumvent this, Marek and Davis (13) recently used a fluorescein-biarsenical compound to inactivate Drosophila Synaptotagmin I that was engineered to contain a particular biarsenical-binding cysteine peptide. Although this approach has been used in Drosophila and mammalian systems (14), several reports observe toxicity or background staining (6, 7).
In this work, an alternative labeling platform was developed and tested for protein labeling and FALI in vitro and in vivo. This approach overcomes the limitations mentioned above because it utilizes a high-affinity, high-specificity interaction to target the fluorophore and consists of a nonendogenous protein–ligand pair for which toxicity and background binding in mammalian cells have not been observed. FKBP12 and its ligand FK506 have been widely studied for their tight protein–ligand interaction (15). This interaction has been improved by the creation of a totally synthetic FKBP12 ligand (synthetic ligand for FKBP12, SLF) that lacks the immunosuppressive effects of FK506. In an elegant structure-aided design, Clackson et al. (16) developed a protein–ligand pair, where a mutant FKBP12 (F36V) binds with a 0.094 nM _K_d to an SLF analog (SLF′) that does not bind to wild-type FKBP12. We sought to test whether this nonnative protein–ligand pair could be used to label proteins in vitro and in vivo, and whether the interaction was suitably specific to permit FALI of target proteins.
Materials and Methods
1_R_-{3-[3-carboxy-4-(6-hydroxy-3-oxo-3_H_-xanthen-9-yl)benzoylamino]-phenyl}-3-(3,4-dimethoxyphenyl)propyl _N_-[2_S_-(3,4,5-trimethoxyphenyl)-butyrl]piperidine-2S-carboxylate (FL-SLF′). The aniline analog of SLF′ (16) (7 mg, 11 μmol) was dissolved in 500 μl dimethylformamide (DMF) and added to the succinimidyl ester of 5-carboxyfluorescein (10 mg, 21 μmol, Molecular Probes) in a brown-glass vial. Triethylamine (15 μl, 108 μmol) and 1-hydroxy-7-azabenzotriazole (HOAt) (32 mg, 240 μmol) were added, and the reaction was stirred for 24 h at room temperature. DMF was removed under high vacuum, and the reaction was purified first by flash chromatography over silica (5:1 CHCl3/MeOH) and then by preparative HPLC (55% acetonitrile with 0.05% TFA in double-distilled H2O with 0.05% TFA) on a Beckman System Gold with a Rainin Dynamax-100 Å C18 5-μm column (21.5 × 250 mm). Fractions (125 μl) were collected and examined by analytical HPLC with a Rainin Dynamax 100 Å C18 5-μm column (4.6 × 250 mm). Fractions containing only the desired compound were pooled, rotovapped to remove acetonitrile, and lyophilized to yield the desired FL-SLF′ (6.8 mg, 62%). High-resolution MS (C57H56N2O14 + Na+): calculated m/z 1015.3642, found m/z 1015.3629.
Analysis of Spectral Properties of FL-SLF′. Excitation and emission spectra for fluorescein and FL-SLF′ were collected for solutions of 100 nM dye in PBS. A Gemini XS fluorescence plate reader (Molecular Devices) was used for all fluorescence measurements, and samples were measured in triplicate in 96-well plate format.
In Vitro Fluorescein-Assisted Laser Inactivation. FKBP12 and FKBP12(F36V) were cloned as N-terminal fusions to β-galactosidase (β-gal) by PCR, and the fusion was cloned into the pDEST (14) bacterial expression vector (Invitrogen) by recombination. Bacterial lysates expressing β-gal fusions were prepared by sonication and centrifugation. For FALI experiments, 1 μM fluorescein or FL-SLF′ was incubated with bacterial lysate for 30 min at 4°C before irradiation. Samples were irradiated in 20-μl droplets on a coverslip positioned on the stage of a Zeiss Axioplan microscope with the objective removed. Irradiation light was from a HbO 50 Watt mercury vapor lamp and was filtered by using a standard GFP/fluorescein filter set with a 585/20-nm excitation filter. Samples were irradiated in triplicate for each data point, and β-gal activity was measured in 96-well format by using the fluorogenic β-gal substrate 4-methylumbelliferyl-d-glucuronide (MUG) (excitation at 360 nm, emissions at 440 nm).
In Vivo Protein Labeling with FL-SLF′. Wild-type and F36V FKBP12 were cloned as C-terminal fusions to a 10-aa membrane-targeting sequence from Lyn in the retroviral construct pMG-ires.Blasticidin. NIH 3T3 were infected with retroviral particles containing this construct and selected for Blasticidin resistance. Cells were treated for 16 h with 1 μM fluorescein or FL-SLF′ for flow cytometry and microscopy experiments. Staining was also observed when shorted incubation times or lower dye concentrations were used. For flow cytometry, a FACSCalibur (Becton Dickinson) was used to analyze the fluorescence of cells that were washed with PBS with 2% FBS and treated with propidium iodide to stain dead cells. Confocal microscopy was performed by using a Zeiss LSM510 confocal microscope, and identical settings were used for each image. In confocal microscope experiments, cells were washed with Hanks' balanced salt solution (HBSS) with 2% FBS, and Hoechst dye 33342 was used as a nuclear costain to identify healthy cells. One-micrometer-thick confocal sections were taken through the middle of the cell(s) to best reveal membrane staining.
In Vivo FALI with FL-SLF′. N-terminal fusions to β-gal were made for both wild-type and F36V FKBP12 in the pMG-ires.Blasticidin retroviral vector. Stable NIH 3T3 cell lines were generated, and β-gal activity was confirmed by using the fluorogenic substrates 2-[18F]-fluoro-2-deoxy-d-glucose (FDG) (flow cytometry) and 4-methylumbelliferyl-d-glucuronide (MUG) (lysates). For experiments in 96-well plates, cells were treated for 2 h with 1 μM fluorescein or FL-SLF′ and irradiated as above. Cells were then lysed and assayed for β-gal activity using MUG. For confocal microscope experiments with FALI, cells were grown on chambered coverslips and treated as above. A field of cells was located by using the ×40 objective, and the cells were irradiated by exposure to the excitation light of the arc lamp for 15 s. The arc lamp light was filtered with the GFP/fluorescein filter set described above. Then, the stage was moved so that the field included both irradiated and nonirradiated cells. The β-gal substrate 7-hydroxy-9_H_-(1,3-dichloro-9,9-dimethylacridin-2-one) β-d-galactopyranoside (DDAO)–GAL was added to 1 μg/ml, and cells were imaged for DDAO after 10 min.
Results
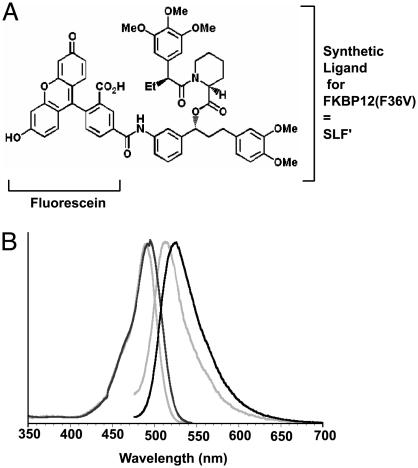
A Fluorescent Conjugate of the FKBP12(F36V) Ligand SLF′. A fluorescein conjugate of the synthetic ligand for FKBP12(F36V) (FL-SLF′) was created to test the potential to label proteins with small molecules in vivo (Fig. 1_A_). FL-SLF′ was synthesized by coupling the aniline analog of SLF′ to commercially available 5-carboxyfluorescein. The product was confirmed by HPLC and mass spectroscopy, and purified by preparative HPLC (62% yield). After purification, excitation and emission spectra were collected for fluorescein and FL-SLF′ to test whether conjugation to SLF′ altered the fluorescence of the molecule (Fig. 1_B_, normalized data). FL-SLF′ had a 5-nm excitation shift and a 10-nm emission shift compared to fluorescein, and therefore could be readily detected with standard optical filters for fluorescein in further experiments. The interaction between FL-SLF′ and recombinant FKBP12(F36V) was verified through saturation binding experiments using fluorescence polarization, and the resulting dissociation constant of FL-SLF′ for FKBP12(F36V) was similar to the published value of 0.094 nM kDa (16). Thus, the FL-SLF′ conjugate retains high-affinity binding to FKBP12(F36V) as well as the intense green fluorescence of the parent fluorescein.
Fig. 1.
Fluorescein conjugate of a synthetic ligand for FKBP12(F36V) (SLF′). (A) Structure of the FL-SLF′ conjugate. (B) Normalized excitation and emission spectra for fluorescein (gray lines) and FL-SLF′ (black lines).
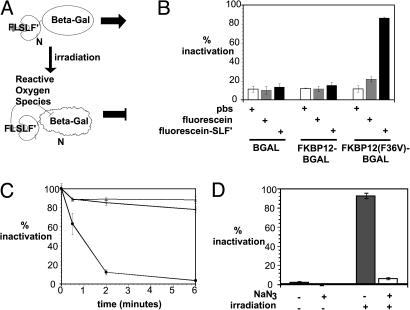
FALI in Vitro with FL-SLF′. Because fluorescein can be used as a substrate for FALI when targeted to a protein via an antibody, we tested whether FL-SLF′ could mediate FALI of an enzyme fused to FKBP12(F36V) (Fig. 2_A_). Wild-type β-gal and various FKBP12-β-gal fusions were expressed in bacteria, and the corresponding bacterial lysates were incubated with FL-SLF′, fluorescein, or a PBS control, and then irradiated by using a standard fluorescence microscope (Fig. 2_B_). FL-SLF′ mediates strong inactivation of FKBP12(F36V)-β-gal, with no significant inactivation of wild-type β-gal or FKBP12-β-gal.
Fig. 2.
FL-SLF′ mediates FALI in vitro when targeted to an FKBP12(F36V) fusion protein. (A) Mechanism of FALI with FL-SLF′. Irradiation of fluorescein drives formation of chemically active ROS, which tend to react with and inactivate the β-gal fusion partner because of proximity. (B) β-gal fusions were incubated with compound and then irradiated for 6 min. (C) FKBP12(F36V)-β-gal was incubated for various times with PBS (open triangles), fluorescein (open circles), or FL-SLF′ (filled circles). (D) FKBP12(F36V)-β-gal was incubated with FL-SLF′, treated with sodium azide (NaN3), and irradiated as indicated.
One potential advantage of FALI is that it can rapidly inactivate target proteins. To examine the time requirements for FALI, the inactivation of FKBP12(F36V)-β-gal was measured for several different irradiation times (Fig. 2_C_). When the fluorescence microscope was used, the τ1/2 for inactivation of FKBP12(F36V)β-gal with FL-SLF′ was 45 s. A τ1/2 for inactivation of 49 s was observed when the same experimental setup was used to test FALI of this fusion protein mediated by a fluorescein conjugated polyclonal anti-β-gal antibody (data not shown). This indicates that FL-SLF′-mediated FALI is at least as efficient as FALI mediated by an antibody.
In antibody mediated FALI, ROS (and singlet oxygen in particular) are required molecular mediators of the inactivation of target proteins (17). We sought to confirm that generation of singlet oxygen was in fact the mechanism of light-mediated inactivation of FKBP12(F36V)β-gal by FL-SLF′. Incubation with the singlet oxygen quencher NaN3 inhibited FL-SLF′ mediated inactivation of FKBP12(F36V)-β-gal (Fig. 2_D_). Thus, singlet oxygen is strongly implicated as a mediator of FL-SLF′ FALI. Collectively, these experiments demonstrate that FL-SLF′ can mediate extremely rapid, light-dependent inactivation of target proteins in vitro, and that ROS mediate FL-SLF′ FALI.
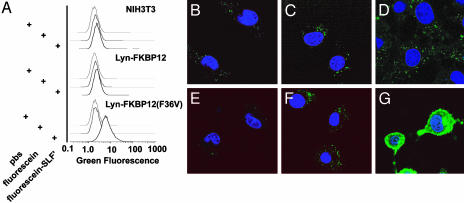
Protein Labeling and FALI in Vivo with FL-SLF′. Because SLF′ is selective for FKBP12(F36V) protein over wild type, and because SLF′ and related molecules have been successfully used as chemical inducers of dimerization (18), we sought to test with FL-SLF′ could be used to chemically label intracellular FKBP12(F36V) fusion proteins in live cells. Membrane targeted FKBP12 or FKBP12(F36V) proteins were expressed in stable NIH 3T3 cell lines. Targeting was achieved by using a 10-aa sequence from the src-like kinase Lyn, which undergoes posttranslational acylation and subsequent targeting to the cytoplasmic face of the plasma membrane (19). These cells were incubated with FL-SLF′, fluorescein, or a PBS control, washed, and analyzed by flow cytometry (Fig. 3_A_). Background staining was not detected in cells lacking an FKBP(F36V) construct. In contrast, cells expressing Lyn-FKBP12(F36V) exhibited a clear increase in green fluorescence upon treatment with FL-SLF′, but not upon treatment with fluorescein. This demonstrates that FL-SLF′ can be used to label cells that express an intracellular FKBP12(F36V) fusion protein. Flow cytometry can also be used to coherently measure cell viability (with propidium iodide uptake) and cell health (with forward and side scatter) (20, 21). FL-SLF′ was incubated in cells at a concentration as high as 10 μM, and for time periods as long as 24 h, with no observable effects on cell viability (determined by uptake of the dye propidium iodide), and no changes in the light scatter properties of the cells (data not shown). FL-SLF′ can label intracellular an FKBP12(F36V) fusion protein without overt toxicity to the cells.
Fig. 3.
FL-SLF′ specifically labels FKBP12(F36V) fusion proteins in vivo. (A) NIH 3T3 cells expressing membrane-targeted FKBP12 or FKBP12(F36V) were treated with the indicated fluorescent compounds and analyzed by flow cytometry. (B_–_G) Control NIH3T3s (B_–_D) and NIH 3T3 cells expressing a membrane-targeted FKBP12(F36V) (E_–_G) were treated with PBS (B and E), fluorescein (C and F), or FL-SLF′ (D and G) and imaged by confocal microscope.
After demonstrating that FKBP12(F36V)-expressing cells selectively stain with FL-SLF′, we sought to assess whether FL-SLF′ labeling can be used to determine the intracellular localization of the FKBP12(F36V) fusion protein. Confocal microscopy was performed on cells treated with fluorescein or FL-SLF′ (Fig. 3 B_–_G). Lyn-FKBP12(F36V)-expressing cells possessed strong plasma membrane staining when treated with FL-SLF′; a small amount of cytoplasmic staining was also observed. Lyn is synthesized in the cytoplasm and then targeted to the plasma membrane by posttranslational modification. As such, a cytoplasmic pool of newly made protein might exist that has not yet been modified and targeted to the membrane. Lyn–GFP cells have an identical cytoplasmic background staining, indicating that cytoplasmic staining is a feature of the Lyn-targeting motif and is not caused by background binding of FL-SLF′ to cellular components (data not shown).
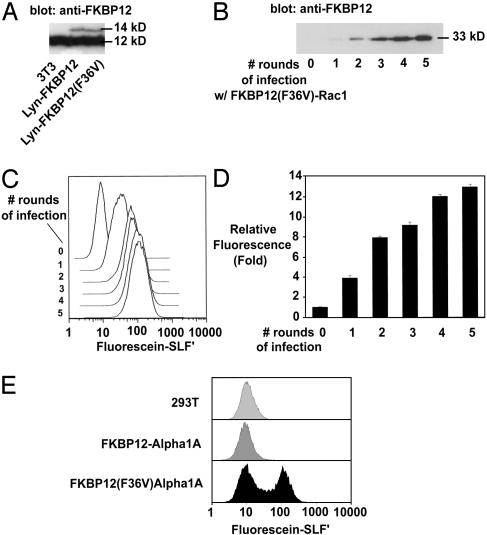
To explore possible limits on the staining, Western blots were performed on the LynFKBP12 cell lines (Fig. 4_A_). The 14-kDa Lyn-FKBP12 and Lyn-FKBP12(F36V) proteins were expressed at <10% of the level of endogenous FKBP12 (Fig. 4_A_). Although FKBP12 has been shown to be an abundant protein in NIH 3T3, this finding indicates that the LynFKBP12(F36V) fusion is expressed at a fairly low level in these cell lines (22). Thus, FL-SLF′ and FKBP12(F36V) can be used for proteins that are expressed at low levels, and higher expression levels may brighter staining. To test this, a panel of cell lines with increasing levels of FKBP12(F36V)-Rac1 protein were generated (Fig. 4_B_). Rac1 is a 21-kDa cytoplasmic GTPase implicated in cell motility and actin reorganization (23). The FKBP12(F36V)-Rac1 cell lines were treated with FL-SLF′ as above, and cellular fluorescence was determined by using flow cytometry (Fig. 4_C_). The level of FL-SLF′ staining was proportional to the level of FKBP12(F36V) fusion protein expressed, demonstrating that the FL-SLF′ can provide quantitative labeling of FKBP12(F36V) protein in vivo. After five rounds of infection, FL-SLF′ cells were on average 13.5-fold brighter than treated control cells, and the brightest 5% of cells were 50 times brighter than treated control cells lacking an FKBP12(F36V) construct (Fig. 4 C and D). Therefore, expression of FKBP12(F36V) leads to increased accumulation of FL-SLF′ in cells. Upon transfection of FKBP12(F36V) constructs, staining levels 50–100 times brighter than background are typically observed (data not shown).
Fig. 4.
FL-SLF′ specifically labels proteins expressed at varied levels in multiple cell lines. (A) Western blot with anti-FKBP12 antibody reveals low expression of Lyn-FKBP12 constructs relative to endogenous FKBP12. (B) Cell lines expressing increasing amounts of a Rac1-FKBP12(F36V) fusion. (C and D) Flow cytometry analysis of FL-SLF′-stained cells expressing increasing amounts of Rac1-FKBP12(F36V) fusion. (E) 293T cells transfected with an α1S-calcium channel fused to wild-type or F36V FKBP12 were treated with FL-SLF′ and analyzed by flow cytometry.
Some protein-labeling systems have proven to work efficiently in only a subset of cell types (6, 7). Fig. 4_E_ shows efficient labeling of an FKBP12(F36V) fusion after transient transfection in an alternate cell type, 293T cells (Fig. 4_E_). In this case, FKBP12(F36V) was fused to the N terminus of the calcium channel α1S. Control transfections with GFP revealed that ≈50% of the cells were transfected (data not shown). FL-SLF′ staining can be used in a wide variety of cell types, as FL-SLF′ has been used to successfully label FKBP12(F36V) proteins in COS-7, Jurkat, HeLa, and C2C12 myoblasts (data not shown). Additionally, FKBP12(F36V) has successfully been fused to a broad range of proteins, including a 10-aa membrane-targeting domain, a nuclear localization sequence, a transmembrane protein, and cytoplasmic proteins including 21-kDa Rac1, 116-kDa β-gal, luciferase, and β-lactamase (Figs. 3 and 4 and data not shown).
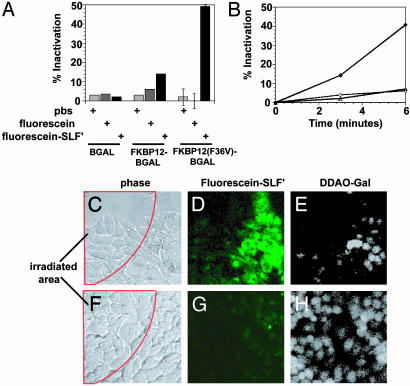
Because FL-SLF′ can be used to rapidly inactivate FKBP12(F36V) fusion proteins in vitro, and FL-SLF′ is efficiently targeted to FKBP12(F36V) in vivo, we hypothesized that FL-SLF′ could be used to target FALI in vivo. To test FALI in vivo, these NIH 3T3 cell lines stably expressing wild-type and FKBP12(F36V) fusions to β-gal were treated with FL-SLF′ and irradiated (Fig. 5_A_). Although irradiation in the absence of FL-SLF′ did not inactivate the protein in any of the cell lines, FKBP12(F36V)-β-gal-expressing cells that were treated with FL-SLF′ had a 50% reduction in β-gal activity upon irradiation (Fig. 5_A_). Because a strong time-dependence was observed with FALI in vitro, the effect of increasing irradiation times was tested in this in vivo assay (Fig. 5_B_). There was a clear, time-dependent increase in the extent of FALI inactivation, with >50% inactivation when the cells were irradiated for 6 min.
Fig. 5.
FL-SLF′ mediates light-driven inactivation of target proteins in living cells. (A) NIH 3T3 cell lines expressing β-gal fusion proteins were treated with fluorescein or FL-SLF′, irradiated for 5 min, then lysed and assayed for β-gal activity. (B) FKBP12(F36V)-β-gal-expressing cells were treated with PBS (open triangles), fluorescein (open circles), or FL-SLF′ (filled circles). (C_–_H) FKBP12(F36V)-β-gal-expressing cells (C_–_E) and β-gal expressing cells (F_–_H) were treated with FL-SLF′, then a portion of the field of view was irradiated for 15 s (C and E). Irradiation area is indicated by the red outline. Irradiation was confirmed by imaging FL-SLF′ (D and G), and then β-gal activity was assayed by addition of cell permeable dimethylacridinone (DDAO)–GAL substrate and imaging accumulation of cleaved DDAO (E and H).
FALI depends on the length of irradiation time and the strength of the light source that is used for irradiation (12, 17). In the irradiation experiments above, the microscope objective was removed so that the irradiation light was diffuse and could irradiate relatively large volumes of solution or large areas of cells. For example, after removing the objective from the microscope, the microscope was used to irradiate an entire well of a 96-well plate (0.25 cm2). Because the generation of singlet oxygen is proportional to the total amount of energy delivered to the sample, irradiation sources that focus the same intensity of light on a much smaller area achieve better inactivation with even shorter irradiation times (24). To test this, we focused the irradiation light much more finely by using the ×40 objective lens to irradiate a small region (≈0.0016 cm2), containing 10–20 cells (Fig. 5 C and F). In this case, it was only necessary to irradiate cells for 15 s to achieve strong photobleaching of FL-SLF′ (Fig. 5 D and G). Because photobleaching and ROS generation are parallel biophysical processes, this rapid photobleaching indicates that ROS are generated very quickly in this setting (25). This 15-s irradiation time was sufficient to inactivate FKBP12(F36V)-β-gal (Fig. 5 E and H). Thus, FL-SLF′ can be used to selectively label FKBP12(F36V) fusion proteins, and the fluorescein moiety of FL-SLF′ can be used to target FALI to FKBP12(F36V) fusion proteins. We have named this technique FK-FALI, for FKBP-fluorophore assisted laser inactivation.
Discussion
Protein Labeling with FL-SLF′. A method to label proteins with small molecules was created in this work. This labeling technique is effective in vitro and in vivo, and should have broad uses in studying intracellular signaling. The approach takes advantage of the high-affinity interaction of an FKBP12 mutant (F36V) and a synthetic ligand based on FK506. This interaction has a subnanomolar _K_d and >1,000-fold selectivity for the F36V mutant of FKBP12 over wild type (16). The high affinity and specificity of this protein–ligand interaction suggested that it might be used to label proteins with chemical conjugates to SLF′. Because dimeric forms of synthetic ligands for FKBPs retain high affinities, we reasoned that conjugation of fluorescent dyes or other small molecules to the same site would not affect binding (16, 26, 27).
In addition to their high-affinity, high-specificity interaction with FKBP12(F36V), SLF′-based molecules have been shown to be readily cell-permeable and nontoxic (28). After confirming that the FL-SLF′ retains a high-affinity interaction with FKBP12(F36V) in vitro, it was used to label FKBP12(F36V) fusion proteins in cells.
FL-SLF′ very effectively labeled FKBP(F36V) fusion proteins in vivo. Flow cytometry showed that there was very little background staining of control cells, and in several cases a >10-fold increase in f luorescence in cells expressing FKBP12(F36V) (Figs. 3_A_ and 4). The absence of background staining in control cells permits useful and accurate staining of proteins expressed at low levels, such as the Lyn-FKBP12(F36V) fusion (Figs. 3 and 4_A_), whereas other labeling systems have noted problems with proteins which are expressed at low levels (6, 7). The ability to label proteins that are expressed at or below physiologic levels is significant because overexpression can alter biological phenotypes (1). FL-SLF′ staining is directly proportional to the expression level of the FKBP12(F36V) fusion (Fig. 4 B and C), and staining levels >100-fold higher than control cells are observed in high expressers. Thus, the FL-SLF′ labeling approach is well suited for proteins across a broad range of expression levels.
The FL-SLF′ labeling system has several advantages over other methods for fluorescent labeling of proteins in vivo. Direct chemical modification and microinjection of proteins into cells has been performed, but with great concern about the invasiveness of this procedure. The biarsenical compound approach developed by Roger Tsien and colleagues (3) is an elegant solution that uses a small peptide epitope for labeling. However, background staining and potential toxicity may limit its use to a subset of cell types (3, 6, 7). Recently, a novel method was presented in which a human alkyltransferase covalently modifies its fusion partner with a fluorescent substrate (29). This work presents an interesting approach, but currently has a low efficiency and can only be used in cells that have been modified to remove the endogenous alkyltransferase (29).
In Vitro and in Vivo FALI with FL-SLF′. FL-SLF′ effectively targeted destruction of an FKBP12(F36V)–β-gal fusion in vitro, without inactivation of a wild-type FKBP12–β-gal fusion. This demonstrates the specificity of the interaction between FL-SLF′ and FKBP12(F36V), and indicates that the fluorescein moiety must be brought very close to β-gal to cause inactivation. Beck et al. (17) determined that the half maximal radius of FALI with fluorescein was 40 Å, which is less than the typical expected distance between proteins in vivo.
The distance requirement for FALI highlights an advantage of small-molecule FALI over antibody-mediated schemes. FKBP12(F36V)-FL-SLF′ is expected to bring the fluorescein moiety closer to the active regions of the target protein than will an antibody, because antibodies are comparatively large. The rates of inactivation of FKBP12(F36V)β-gal by FL-SLF′ and by a fluorescein labeled antibody were almost identical (45 and 49 s, respectively). These data suggest that FL-SLF′-mediated FALI is in fact more efficient per fluorescein molecule than antibody-mediated FALI. Antibodies can be labeled with multiple fluorophore molecules per antibody molecule, and the fluorescein-labeled anti-β-gal antibody used in this study had 3.3 molecules of fluorescein per antibody molecule (per manufacturer). In addition, polyclonal antibodies are typically used for antibody-mediated FALI, and multiple antibody molecules might bind to a single target molecule. Therefore, FL-SLF′-mediated FALI is likely more efficient per fluorescein molecule than antibody-mediated FALI. This suggests that interaction with FKBP12(F36V) positions the fluorescein molecule closer to β-gal than is possible with antibodies. As such, it should also have lower levels of inactivation of bystander molecules, because the singlet oxygen molecules are more closely targeted to the protein of interest. If required, it is possible to alter the radius of damage caused by singlet oxygen, by subsitution of H2O with D2O to increase the effective range of singlet oxygen, or by incubation of cell permeable singlet oxygen quenchers to decrease the range (14, 17).
The rapid inactivation mediated by FK-FALI is in contrast to RNAi and other methods to remove or inactivate a target protein, which can take hours or days to reduce active protein levels (11). Rapid inactivation also avoids the common problems with genetic knockouts, i.e., compensation and embryonic lethality when the protein of interest is required for viability. FK-FALI may be particularly powerful when combined with small interfering RNA or genetic knockouts, as the latter could be used to remove the endogenous protein, permitting loss-of-function studies with exceptionally high spatial and temporal resolution. FK-FALI permits inactivation of proteins on the second time scale, and subsecond time scale inactivation might be possible with more efficient light sources. In this work, a standard epifluorescence microscope was used because this tool is widely available and can be used by other investigators with minimal difficulty. Laser-mediated irradiation is very common in FALI, and light sources including the 488-nm laser line typically used in confocal microscopy might further reduce the time required for FALI irradiation (9).
FK-FALI can also be spatially defined. Inactivation of the protein from a subset of the cells in a field of view demonstrates this principle. In fact, when a laser is used to irradiate a sample, very fine spatial control should be possible. In related work, we demonstrated this possibility by using FK-FALI to inactivate the small GTPase Rac1 with subcellular resolution. This work demonstrated the spatial and temporal requirements of Rac1 activity for membrane ruffling and cell motility (K.M.M. and G.P.N., unpublished data). It is increasingly clear that cell biology is spatially and temporally controlled, and many proteins possess activities that are spatially and temporally restricted (30). FK-FALI can permit experiments to directly test the spatiotemporal requirements of target proteins by rapidly inactivating the protein in defined regions of the cell.
In addition to future applications based on the FL-SLF′ molecule itself, it should be possible to conjugate a wide variety of fluorescent dyes or other small molecules to SLF′ to create a broad family of chemicals that could all be targeted to proteins via the same FKBP12(F36V) fusion. Possibilities include the targeting of fluorescent dyes with different excitation and emissions spectra, fluorescent sensors like calcium dyes, crosslinkers or caged compounds, or affinity reagents such as biotin. FKBP12(F36V)-targeted fluorescent dyes might be useful in fluorescence resonance energy transfer (FRET), because FRET requires close proximity between the pair of fluorescent molecules, and FKBP12(F36V)-targeting likely permits closer association of the fluorescent molecules than can be achieved with pairs of fluorescent proteins (31). For multicolor imaging, FL-SLF′-FKBP12(F36V) could be combined with fluorescent proteins or other chemical labeling techniques, such as the biarsenical technique or short, constrained peptides that bind with high affinity to Texas red (32). Additionally, it will be interesting to test whether multimerization of FKBP12(F36V) increases the efficiency of labeling by avidity and by permitting multiple fluorescent molecules per fusion protein. This might increase the strength of the signal and may increase the potency of FK-FALI.
In summary, we have created a protein labeling system that can be used to monitor protein localization and permits rapid, spatially and temporally defined knockout of target proteins. The modular synthetic approach that was taken should permit creation of a broad set of chemicals and fluorescent dyes for protein labeling in vitro and in vivo.
Acknowledgments
We thank Christopher Amos (Stanford University) for providing intermediates used in the synthesis of FL-SLF′. This work was supported by National Heart, Lung, and Blood Center Contract NO1-HV-281831, National Institutes of Health Grant A135304, and a Stanford Graduate Fellowship (to K.M.M.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RNAi, RNA interference; FALI, fluorophore-assisted laser inactivation; β-gal, β-galactosidase; ROS, reactive oxygen species.
References
- 1.Tsien, R. Y. (1998) Annu. Rev. Biochem. 67**,** 509-544. [DOI] [PubMed] [Google Scholar]
- 2.Zacharias, D. A., Violin, J. D., Newton, A. C. & Tsien, R. Y. (2002) Science 296**,** 913-916. [DOI] [PubMed] [Google Scholar]
- 3.Griffin, B. A., Adams, S. R. & Tsien, R. Y. (1998) Science 281**,** 269-272. [DOI] [PubMed] [Google Scholar]
- 4.Farinas, J. & Verkman, A. S. (1999) J. Biol. Chem. 274**,** 7603-7606. [DOI] [PubMed] [Google Scholar]
- 5.Wu, M. M., Llopis, J., Adams, S., McCaffery, J. M., Kulomaa, M. S., Machen, T. E., Moore, H. P. & Tsien, R. Y. (2000) Chem. Biol. 7**,** 197-209. [DOI] [PubMed] [Google Scholar]
- 6.Griffin, B. A., Adams, S. R., Jones, J. & Tsien, R. Y. (2000) Methods Enzymol. 327**,** 565-578. [DOI] [PubMed] [Google Scholar]
- 7.Stroffekova, K., Proenza, C. & Beam, K. G. (2001) Pflügers Arch. 442**,** 859-866. [DOI] [PubMed] [Google Scholar]
- 8.Jay, D. G. & Keshishian, H. (1990) Nature 348**,** 548-550. [DOI] [PubMed] [Google Scholar]
- 9.Surrey, T., Elowitz, M. B., Wolf, P. E., Yang, F., Nedelec, F., Shokat, K. & Leibler, S. (1998) Proc. Natl. Acad. Sci. USA 95**,** 4293-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson, A. L., Bartz, S. R., Schelter, J., Kobayashi, S. V., Burchard, J., Mao, M., Li, B., Cavet, G. & Linsley, P. S. (2003) Nat. Biotechnol. 21**,** 635-637. [DOI] [PubMed] [Google Scholar]
- 11.Wilson, J. A. & Richardson, C. D. (2003) Curr. Opin. Mol. Ther. 5**,** 389-396. [PubMed] [Google Scholar]
- 12.Jay, D. G. (1988) Proc. Natl. Acad. Sci. USA 85**,** 5454-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marek, K. W. & Davis, G. W. (2002) Neuron 36**,** 805-813. [DOI] [PubMed] [Google Scholar]
- 14.Tour, O., Meijer, R. M., Zacharias, D. A., Adams, S. R. & Tsien, R. Y. (2003) Nat. Biotechnol. 21**,** 1505-1508. [DOI] [PubMed] [Google Scholar]
- 15.Dumont, F. J. (2000) Curr. Med. Chem. 7**,** 731-748. [DOI] [PubMed] [Google Scholar]
- 16.Clackson, T., Yang, W., Rozamus, L. W., Hatada, M., Amara, J. F., Rollins, C. T., Stevenson, L. F., Magari, S. R., Wood, S. A., Courage, N. L., et al. (1998) Proc. Natl. Acad. Sci. USA 95**,** 10437-10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck, S., Sakurai, T., Eustace, B. K., Beste, G., Schier, R., Rudert, F. & Jay, D. G. (2002) Proteomics 2**,** 247-255. [DOI] [PubMed] [Google Scholar]
- 18.Amara, J. F., Clackson, T., Rivera, V. M., Guo, T., Keenan, T., Natesan, S., Pollock, R., Yang, W., Courage, N. L., Holt, D. A. & Gilman, M. (1997) Proc. Natl. Acad. Sci. USA 94**,** 10618-10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovarova, M., Tolar, P., Arudchandran, R., Draberova, L., Rivera, J. & Draber, P. (2001) Mol. Cell. Biol. 21**,** 8318-8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeh, C. J., Hsi, B. L. & Faulk, W. P. (1981) J. Immunol. Methods 43**,** 269-275. [DOI] [PubMed] [Google Scholar]
- 21.Loken, M. R., Sweet, R. G. & Herzenberg, L. A. (1976) J. Histochem. Cytochem. 24**,** 284-291. [DOI] [PubMed] [Google Scholar]
- 22.Siekierka, J. J., Wiederrecht, G., Greulich, H., Boulton, D., Hung, S. H., Cryan, J., Hodges, P. J. & Sigal, N. H. (1990) J. Biol. Chem. 265**,** 21011-21015. [PubMed] [Google Scholar]
- 23.Ridley, A. J. (2001) J. Cell Sci. 114**,** 2713-2722. [DOI] [PubMed] [Google Scholar]
- 24.Inoue, T., Kikuchi, K., Hirose, K., Iino, M. & Nagano, T. (2003) Chem. Biol. 10**,** 503-509. [DOI] [PubMed] [Google Scholar]
- 25.Greenbaum, L., Rothmann, C., Lavie, R. & Malik, Z. (2000) Biol. Chem. 381**,** 1251-1258. [DOI] [PubMed] [Google Scholar]
- 26.Briesewitz, R., Ray, G. T., Wandless, T. J. & Crabtree, G. R. (1999) Proc. Natl. Acad. Sci. USA 96**,** 1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braun, P. D., Barglow, K. T., Lin, Y. M., Akompong, T., Briesewitz, R., Ray, G. T., Haldar, K. & Wandless, T. J. (2003) J. Am. Chem. Soc. 125**,** 7575-7580. [DOI] [PubMed] [Google Scholar]
- 28.Iuliucci, J. D., Oliver, S. D., Morley, S., Ward, C., Ward, J., Dalgarno, D., Clackson, T. & Berger, H. J. (2001) J. Clin. Pharmacol. 41**,** 870-879. [DOI] [PubMed] [Google Scholar]
- 29.Keppler, A., Gendreizig, S., Gronemeyer, T., Pick, H., Vogel, H. & Johnsson, K. (2003) Nat. Biotechnol. 21**,** 86-89. [DOI] [PubMed] [Google Scholar]
- 30.Weijer, C. J. (2003) Science 300**,** 96-100. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, J., Campbell, R. E., Ting, A. Y. & Tsien, R. Y. (2002) Nat. Rev. Mol. Cell Biol. 3**,** 906-918. [DOI] [PubMed] [Google Scholar]
- 32.Marks, K. M., Rosinov, M. & Nolan, G.P. (2004) Chem. Biol. 11**,** 347-356. [DOI] [PubMed] [Google Scholar]