Mutations in KIAA0586 Cause Lethal Ciliopathies Ranging from a Hydrolethalus Phenotype to Short-Rib Polydactyly Syndrome (original) (raw)
Abstract
KIAA0586, the human ortholog of chicken TALPID3, is a centrosomal protein that is essential for primary ciliogenesis. Its disruption in animal models causes defects attributed to abnormal hedgehog signaling; these defects include polydactyly and abnormal dorsoventral patterning of the neural tube. Here, we report homozygous mutations of KIAA0586 in four families affected by lethal ciliopathies ranging from a hydrolethalus phenotype to short-rib polydactyly. We show defective ciliogenesis, as well as abnormal response to SHH-signaling activation in cells derived from affected individuals, consistent with a role of KIAA0586 in primary cilia biogenesis. Whereas centriolar maturation seemed unaffected in mutant cells, we observed an abnormal extended pattern of CEP290, a centriolar satellite protein previously associated with ciliopathies. Our data show the crucial role of KIAA0586 in human primary ciliogenesis and subsequent abnormal hedgehog signaling through abnormal GLI3 processing. Our results thus establish that KIAA0586 mutations cause lethal ciliopathies.
Main Text
Ciliopathies are a continuum of genetically highly heterogeneous disorders with varying severity and organ involvement and are all caused by genes involved in ciliary function or biogenesis. Among the most severe ciliopathies are the short-rib polydactyly (SRP) group of lethal skeletal dysplasia (SRPI [MIM: 613091], SRPII [MIM: 263520], SRPIII [MIM: 613091], and SRPIV [MIM: 269860]), hydrolethalus syndrome (HLS [MIM: 236680]), and Meckel syndrome (MKS [MIM: 249000]). All three represent the extreme phenotype of viable ciliopathies, namely Jeune asphyxiating thoracic dystrophy (MIM: 208500) and Ellis-van Creveld syndrome (MIM: 225500),1 acrocallosal syndrome (MIM: 200990),2 and Joubert syndrome (MIM: 213300),3–5 respectively.
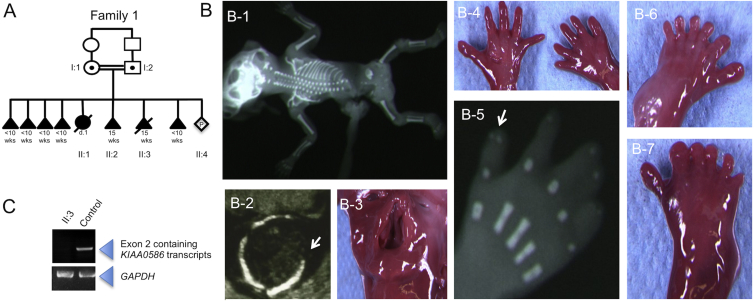
Here, we examined a consanguineous Lebanese family (family 1; Figure 1A) with two 15 gestational week (gw)-old fetuses presenting with severe hydrocephaly, polydactyly of the hands and feet, a cleft palate, and skeletal abnormalities (Figure 1B, Table 1, and supplemental case reports). We considered this phenotype to be similar to that of HLS and therefore sequenced HYLS1 (MIM: 610693) and KIF7 (MIM: 611254) but found no mutation. Because HLS is now known as a ciliopathy, we then combined a targeted capture strategy for candidate ciliary genes with next-generation sequencing, as described previously, by using DNA from fetus II:3.6,7 In brief, ciliary exome-targeted sequencing and bioinformatics filtering were conducted with a Custom SureSelect Capture Kit (Agilent Technologies) targeting 4.5 Mb of 20,168 exons (1,221 ciliary candidate genes). Agilent SureSelect libraries were prepared from 3 μg of genomic DNA sheared with a Covaris S2 Ultrasonicator according to manufacturer’s instructions. The Ovation Ultralow System (NuGEN Technologies) was used to prepare HiSeq 2500 pre-capture barcoded libraries. The ciliome capture by hybridization was performed on a pool of 10–16 barcoded pre-capture libraries. Sequencing performed on a HiSeq 2500 (Illumina) was done on pools of barcoded ciliome libraries (16 ciliome libraries per lane of HiSeq FlowCell). Paired-end reads (100 100-bp reads) were generated and mapped on a human genome reference (NCBI Genome browser build 37) with the Burrows-Wheeler Aligner (Illumina). Downstream processing was carried out with the Genome Analysis Toolkit, SAMtools, and Picard Tools according to documented best practices from the Broad Institute. All variants were annotated with a software system developed by the Paris Descartes University bioinformatics platform. Informed consent was obtained for all participating families, and the study was approved by the ethical committee of Paris Ile de France II. Genomic DNA was extracted from frozen tissue or amniocyte cultured cells for fetal subjects and from peripheral-blood samples for parents. Finally, a total of 6,263 variants were identified, and after we filtered data by removing known SNPs and synonymous coding sequence variations and by using a recessive model of inheritance, a unique nonsense homozygous mutation remained (Table S1) in KIAA0586 (MIM: 610178). KIAA0586, the ortholog of chicken KIAA0586, was considered an excellent candidate gene given that its disruption in animal models causes defects, including polydactyly and abnormal dorsoventral patterning of the neural tube, attributed to abnormal hedgehog signaling.8–11 The c.230C>G (p.Ser77∗) variant (GenBank: NM_001244189.1) segregated with the expected patterns of autosomal-recessive inheritance in all available family members and is absent from dbSNP, the NHLBI Exome Sequencing Project (ESP) Exome Variant Server (EVS), the Exome Aggregation Consortium (ExAC) Browser, and 300 Lebanese control chromosomes. This homozygous nonsense variation is located in KIAA0586 exon 2, and mRNAs produced are predicted to be targeted for nonsense-mediated decay (NMD). To test this, we extracted mRNA from the tissue of an affected individual (subject II:3, family 1) and age-matched control individuals and subsequently performed reverse transcription. Compared to control mRNA, mRNA extracted from the affected subject, showed a total absence of transcript containing KIAA0586 exon 2. Importantly, GAPDH amplification was similar in both samples (Figure 1C). We next tested KIAA0586 expression in different tissues at different human developmental stages and found those specific transcripts containing KIAA0586 exon 2 were expressed as early as 6 gw (Carnegie stage 16) in humans. They appeared ubiquitously expressed during fetal development and persisted postnatally in all human adult tissues tested (Figure S1).
Figure 1.
Pedigree and Phenotype of Family 1, Subject II:3 with a KIAA0586 Nonsense Variation
(A) Family 1 pedigree. The c.230C>G (p.Ser77∗) variant segregated with the expected patterns of autosomal-recessive inheritance in all available family members.
(B) Phenotype of affected subject II:3. A fetal X-ray shows a frontal view (B-1). Ultrasound imaging shows exencephaly (axial view) with an occipital defect (arrow; B-2). Photographs show cleft lip and palate (B-3), polysyndactyly (B-4) with duplication of the second phalange of thumbs on X-ray of the right hand (arrow; B-5), and preaxial polydactyly of the feet (B-6 and B-7).
(C) RT-PCR amplification on mRNA extracted from affected and age-matched-control tissue; amplification shows complete degradation of transcripts containing KIAA0586 exon 2 in mutant tissue. GAPDH was used as cDNA quality and quantity control.
Table 1.
Clinical Data of Affected Individuals in the Four Families Studied
| Family (ID) | Subject | Age | Sex | Origin | PD | CK | BDP | CP | MTS | Brain Anomalies | Skeletal Anomalies | Other Anomalies | Variation | Exon | Inheritance |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 (HE) | II:2 | 15 gw | ND | Lebanon | + | ND | ND | + | major hydrocephaly | no clavicle | diaphragmatic hernia | no DNA | ND | ND | |
| II:3 | 15 gw | female | Lebanon | + | − | ND | + | major hydrocephaly, occipital defect | flat and wide iliac wings | fetal hydrops | c.230C>G (p.Ser77∗) | 2 | homozygous | ||
| 2 (CI) | II:2 | 29 gw | male | Romania | + | − | − | + | + | VH, polymicrogyria, absent olfactory bulb, ventriculomegaly | short ribs | tongue hamartomas, multiple frenulae | c.1815G>A (splice) | 14 | homozygous |
| II:3 | 39 gw (died 1 hr after birth) | ND | Romania | + | − | ND | ND | + | VH, abnormal gyration, mega cisterna magna | short ribs, short limbs | ND | c.1815G>A (splice) | 14 | homozygous | |
| 3 (FA) | II:1 | spontaneous fetal death (<10 weeks) | ND | Hungary | ND | ND | ND | ND | ND | anencephaly | ND | ND | no DNA | ND | ND |
| II:2 | died at 13 months | female | Hungary | + | − | − | − | + | occipital meningocele, VH, hypoplasia of the hemispheres and corpus callosum, abnormal basal ganglia, pontocerebellar hypoplasia | short ribs, short limbs, PD of hands and feet | dysplastic and low set ears, depressed nasal bridge, short neck, multiple frenulae | c.1815G>A (splice) | 14 | homozygous | |
| 4 (ME) | II:4 | died at 1 day of life | ND | Kosovo | + | − | − | + | + | described as identical to his sibling | described as identical to his sibling | described as identical to his sibling | no DNA | ND | ND |
| II:5 | 26 gw | male | Kosovo | + | − | − | + | + | Occipital meningocele (key hole), hypoplastic brain stem, VH, CC and septal agenesis, temporal polymicrogyria | Short ribs, short limbs (−6 to −8 SD), superior limb incurvation, bilateral postaxial PD of hands and feet | Retinal dysplasia with retinal coloboma, brachyphalangism, facial dysmorphism, micropenis, frenulae nodules | c.1815G>A (splice) | 14 | homozygous |
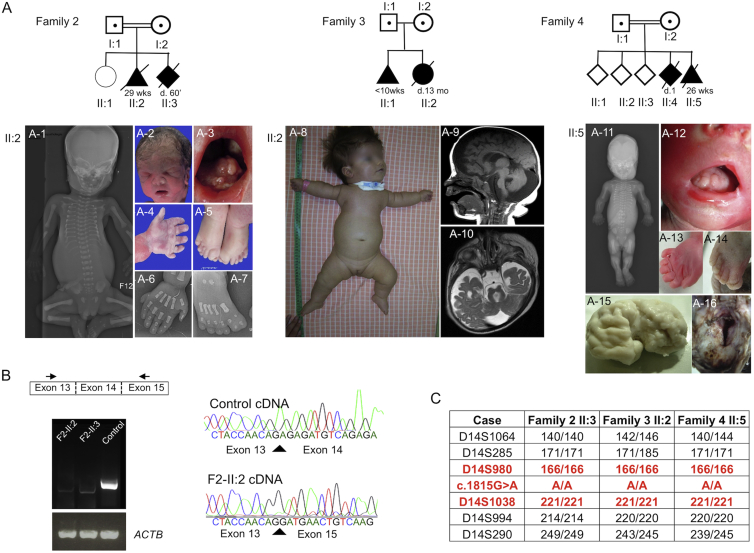
We performed additional next-generation sequencing of ciliary genes for 150 subjects presenting with lethal ciliopathies with various combinations of brain and skeletal abnormalities. This screen led us to identify a c.1815G>A (p.=) homozygous silent variant (GenBank: NM_001244189.1) in three subjects from three unrelated families (II:2 in family 2, II:2 in family 3, and II:5 in family 4) originating from Romania (family 2), Hungary (family 3), and Kosovo (family 4). The variant segregates with the expected patterns of autosomal-recessive inheritance in all available family members. All three subjects had a SRP syndrome with similar cerebral anomalies, preaxial polydactyly of the feet and postaxial polydactyly of the hands, and long-bone shortening, including short ribs. Upon neuropathological examination, all three affected subjects displayed vermis agenesis and brainstem anomalies evocative of a molar tooth sign (Figure 2A, Table 1, and supplemental case reports). The variation involves the last base of KIAA0586 exon 14, is absent from dbSNP, the EVS, and the ExAC Browser, and is predicted in silico to abolish the intron 14 donor splice site (MaxEntScan, splice site prediction by Neural Network [NNSPLICE], and Human Splicing Finder). To confirm this hypothesis, we performed RT-PCR and subsequent cDNA sequencing on total mRNA extracted from blood samples from subjects II:2 and II:3 from family 2 and from control individuals. A unique transcript lacking KIAA0586 exon 14 was observed in both affected subjects (Figure 2B) and is predicted to cause a shift in the reading frame with a premature stop codon.
Figure 2.
Pedigree, Phenotype, and Haplotype Analyses in Families 2–4 Lead to the Identification of a KIAA0586 Homozygous Splice-Site Mutation with a Founder Effect
(A) Pedigrees. In subject II:2 of family 2, an X-ray (frontal view) shows shortening of ribs and micromelia with round metaphysal ends (A-1), and pictures and X-rays show dysmorphism (A-2), lingual hamartomas (A-3), postaxial polydactyly of the hand (A-4 and A-6), and preaxial polysyndactyly of the feet (A-5 and A-7). For subject II:2 in family 3, a picture shows short thorax and micromelia (A-8), and sagittal (A-9) and axial (A-10) views of brain MRI show a micro-brain with large ventricles and large subarachnoid spaces, corpus-callosum and ponto-cerebellar hypoplasia with a large fourth ventricule and cisterna magna, and a molar tooth aspect. In subject II:5 from family 4, an X-ray (frontal view) shows shortening of ribs and micromelia with round metaphysal ends (A-11), and pictures show lingual hamartomas (A-12), postaxial polydactyly of the right hand (A-13), preaxial polydactyly of the right foot (A-14), temporal polymicrogyria (A-15), and an occipital keyhole defect (A-16).
(B) The c.1815G>A KIAA0586 variant affects the last base of exon 14 and is responsible for aberrant transcript lacking exon 14, as shown by RT-PCR and sequencing of KIAA0586 mRNA from control and affected subjects (family 2, subjects II:2 and II:3). Total RNAs were extracted from frozen blood with the Nucleospin RNA Blood Kit and on-column DNase digestion (Macherey Nagel). ACTB was used as cDNA quality and quantity control.
(C) Haplotype at KIAA0586 of affected subjects from families 2–4.
Given the Eastern European origin of the three families, a founder effect was highly suspected. The distance to the common ancestor was estimated by a likelihood-based method.12 We selected the polymorphic markers encompassing KIAA0586 and found a similar haplotype in those three families (Figure 2C). The allele frequencies of the microsatellites used were found on the CEPH genotype database, and the mutation rate was chosen as 10−3. Because the genetic distances available for closely linked markers are generally not very accurate, we computed the rates of recombination between markers by using both the overall genetic length of the haplotype and the physical distances between markers.13 The closest markers at which affected individuals do not share any alleles were determined on both sides of KIAA0586. The distance to the common ancestor was estimated to be approximately 480 years n = 16 generations ago (ninf [the lower bound of the 95% confidence interval (CI)] = 5; nsup [the upper bound of the 95% CI] = 63; nend [the total number of iterations performed] = 232; likelihood = −3.776324).
In animal and cell models, TALPID3 has been involved in very early stages of ciliogenesis, before the formation and docking of a primary ciliary vesicle at the distal appendage.14 TALPID3 interacts with CP110 and co-localizes to the distal ends of centrioles. CP110 and its protein-interaction network (including CEP97, CEP290, and KIF24) have been found to modulate cilium assembly (reviewed in Tsang et al.15). In addition, TALPID3 localizes near the distal appendages,14 which localize to the site of centriole-to-membrane docking and are required for the centriole to be converted to a basal body and thus for ciliogenesis.16
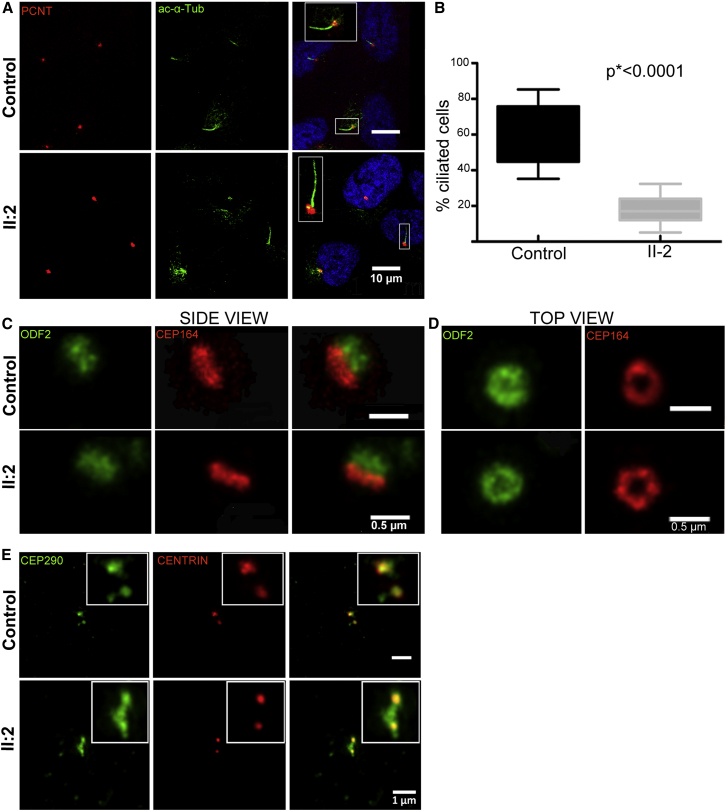
In order to examine the effect of KIAA0586 loss on cilia assembly in humans, we induced ciliogenesis by using serum-starvation-mediated cell-cycle arrest in confluent fibroblasts from both affected (family 2, subject II:2) and control subjects and visualized cilia by co-immunostaining with antibodies to ARL13B (a cilia marker), glutamylated tubulin (GT335) or acetylated α-tubulin, and pericentrin (Figure 3A and data not shown). In control fibroblasts with wild-type KIAA0586, cilia were evident in 60% of the total stained cells by 48 hr of serum starvation, and nearly all were co-stained with all markers. In mutant fibroblasts, only 20% of cells presented with cilia (Figures 3A and 3B). This result indicates that KIAA0586 is necessary for cilia biogenesis in humans, in accordance with its function in animal and cell models.
Figure 3.
Analysis of Ciliogenesis in KIAA0586 Mutant Cells
Cells from affected individuals were obtained from subject II:2 in family 2.
(A and B) In control cells, primary cilia co-stained with acetylated alpha tubulin (ac-α-Tub, clone 6-11B-1, Sigma) and pericentrin (PCNT, Abcam Ab4448) protrude from 60% of cells (n = 500, three independent experiments), whereas only 20% of mutant cells (n = 500) bear a primary cilia. The graph in (B) is a boxplot representing the distribution of the data. The boxes contain 50% of the values and the whiskers indicate the minimum and the maximum of the distribution. The p values were calculated with the Mann-Whitney two-tailed test. Confocal images were taken with a Leica SP5 confocal microscope.
(C and D) High-resolution imaging of control and mutant cell centrioles co-stained with ODF2 (centriolar subdistal appendage protein; Abnova, H00004957-M01) and CEP164 (a centriolar distal appendage protein; Sigma, HPA037606) showed no defect in mutant cells, neither on side view (C) nor on top view (D). Super-resolution microscopy was performed with a Leica TCS SP8 STED (Stimulated Emission Depletion).
(E) Immunostaining with CEP290 (Novus Biologicals, NB100-86991) and centrin (Clone 20H5, Millipore) antibodies, showing abnormal extended pattern of CEP290.
We then checked centriolar morphogenesis and targeting of crucial proteins and found no differences in the staining patterns of CEP164 (distal appendages of mother centrioles) and ODF2 (subdistal appendages), indicating that assembly of these proteins to centrioles is not affected by KIAA0586 loss (Figures 3C and 3D). Finally, as suggested by TALPID3 siRNA depletion studies in which TALPID3 is required for centriolar satellite dispersal preceding the formation of mature ciliary vesicles,14 we compared the distribution of the CP110-interacting protein CEP290 in affected cells to the distribution in control cells and found an abnormal extended pattern of this satellite protein in asynchronously growing conditions (Figure 3E).
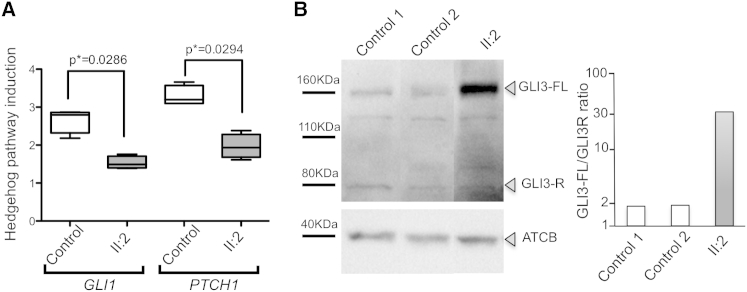
We next tested the transduction of the SHH pathway in mutant fibroblasts by assaying the expression of PTCH1 and GLI1, two transcriptional targets of SHH signaling, after pathway activation by the addition of smoothened agonist. We found that both PTCH1 and GLI1 were less induced in mutant fibroblasts than in controls (Figure 4A). We also analyzed GLI3 processing by western blot and found that KIAA0586 mutant fibroblasts exhibited increased amounts of full-length, unprocessed GLI3 (GLI3-FL) and an abnormal GLI3-FL/GLI3-R ratio (Figure 4B and Figure S3). Thus, KIAA0586 mutant cells exhibit abnormal SHH responsiveness, suggesting that at least some of the defects in affected individuals with KIAA0586 variations might be secondary to abnormal SHH signaling, as observed in animal models.8–11,17
Figure 4.
SHH Signaling in KIAA0586 Mutant Cells
(A) KIAA0586 mutant fibroblasts from subject II:2 in family 2 showed an altered response to smoothened agonist (SAG; sc-202814, Santa Cruz; 5 μM for 18 hr), given that they induced less GLI1 and PTCH1 expression than did control cells. Primers used for real-time qRT-PCR are listed in Table S2. Values from five independent experiments were normalized to GAPDH and are presented as relative expression levels. The boxes contain 50% of the values and the whiskers indicate the minimum and the maximum of the distribution. The p values were calculated with the Mann-Whitney two-tailed test.
(B) Western blot analysis with a GLI3 antibody (AF3690, R&D Systems) showed that the amount of processing of GLI3-FL into its repressor form, GLI3-R, was much lower in KIAA0586 mutant fibroblasts than in control cells. The samples were not run in contiguous lanes, and in the figure presented here, lanes were spliced together (the entire photograph of the immunoblot is presented in Figure S3). A graphical evaluation of the GLI3-FL/GLI3-R ratio, with actin as a loading control and ImageJ software for densitometry, is presented.
In this study, using a targeted high-throughput sequencing strategy for candidate ciliary genes, we identified homozygous nonsense and splicing KIAA0586 mutations as responsible for lethal ciliopathies. This candidate-gene strategy previously succeeded in identifying ciliopathy genes, including TCTN3 (MIM: 613847), which is associated with Mohr-Majewski syndrome (OFD4 [MIM: 258860]).6
The phenotype of _KIAA0586_-affected subjects ranges from a hydrolethalus phenotype to SRP syndrome in four unrelated families. In particular, cerebral phenotypes range from anencephaly or large occipital meningocele to vermian agenesis associated with brainstem anomalies; such anomalies are classically suggestive of ciliopathies. Importantly, the only living SRP-affected individual reported in this study also presented with a molar tooth sign on brain MRI, consistent with the conclusions of neuropathological examination on fetal subjects. All affected subjects display skeletal anomalies, including a consistent polydactyly, short ribs, and micromelia with round femoral ends in families 2– 4. In addition, most of the affected subjects have a large median cleft palate. These features are reminiscent of the spectrum of SRPII and SRPIV.1,18,19
Thus, fetuses and individuals reported in this study all display lethal phenotypes consistent with a hedgehog-signaling defect and similar to the phenotype described in KIAA0586 animal models.8–11,17 In particular, the KIAA0586 chicken with a frameshift mutation in exon 7 (corresponding to human exon 9) closely models the human SRP ciliopathy phenotype, including a small rib cage and polydactyly, as well as lung anomalies.17 Interestingly, the c.1815G>A variation identified in families 2– 4 leads to the deletion of human exon 14, corresponding to mouse exon 12 (Figure S2), which has been shown to be essential for the function of the protein.9,10 Indeed, Talpid3 −/− mice in which exons 11 and 12 are constitutively deleted show abnormal Shh signaling and embryonic lethality as early as embryonic day 10.5. The c.230C>G (p.Ser77∗) nonsense variation of family 1 leads to NMD of exon-2-containing transcripts shown to be expressed early in human embryos and throughout fetal development.
Finally, ciliogenesis was severely impaired in KIAA0586 mutant cells, and the SHH signaling pathway was abnormal, characteristic of other severe human ciliopathies, such as Morh-Majewski or hydrolethalus syndromes, associated with TCTN3 and KIF7 variations,2,6 respectively, or Meckel syndrome.
Overall, these results highlight the conserved function of KIAA0586 among species and its involvement in human ciliopathies. In view of the wide phenotypic spectrum of ciliopathy genes, the lethal phenotype reported here might represent the severe end of the phenotypic spectrum associated with KIAA0586 variations.
Acknowledgments
We thank the families for their participation. We also thank Leila Hakkakian, Judite De Oliveira, and Nadège Gigot for technical assistance and Prof. Andrew Green for referral of affected subjects, and we are grateful to the French Society of Fetal Pathology for participating in the study. This work was supported by grants from the Agence Nationale de la Recherche (ANR; 2010 fetal ciliopathies grant BLAN-1122-01 and 2013 cilia, axonal guidance, and corpus callosum grant ANR-13-BSV1-0027 to T.A.B.). C.A. is funded by the Fondation pour la Recherche Médicale and K.P. by grants from ANR. The Institut Imagine is supported by an ANR grant (ANR-A0-IAHU-01).
Published: July 9, 2015
Footnotes
Web Resources
The URLs for data presented herein are as follows:
- Broad Institute GATK Best Practices, https://www.broadinstitute.org/gatk/guide/best-practices
- ClustalW2, http://www.ebi.ac.uk/Tools/msa/clustalw2/
- Human Splicing Finder, http://www.umd.be/HSF/
- MaxEntScan, http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq_acc.html
- MultAlin, http://multalin.toulouse.inra.fr/multalin/
- OMIM, http://www.omim.org/
- PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
- Primer3Plus, http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/
- SIFT, http://sift.bii.a-star.edu.sg/
- Splice Site Prediction by Neural Network (NNSPLICE), http://www.fruitfly.org/seq_tools/splice.html
- UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
Document S1. Case reports, Figures S1–S3, and Tables S1 and S2
Document S2. Article plus Supplemental Data
References
- 1.Huber C., Cormier-Daire V. Ciliary disorder of the skeleton. Am. J. Med. Genet. C. Semin. Med. Genet. 2012;160C:165–174. doi: 10.1002/ajmg.c.31336. [DOI] [PubMed] [Google Scholar]
- 2.Putoux A., Thomas S., Coene K.L.M., Davis E.E., Alanay Y., Ogur G., Uz E., Buzas D., Gomes C., Patrier S. KIF7 mutations cause fetal hydrolethalus and acrocallosal syndromes. Nat. Genet. 2011;43:601–606. doi: 10.1038/ng.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baala L., Romano S., Khaddour R., Saunier S., Smith U.M., Audollent S., Ozilou C., Faivre L., Laurent N., Foliguet B. The Meckel-Gruber syndrome gene, MKS3, is mutated in Joubert syndrome. Am. J. Hum. Genet. 2007;80:186–194. doi: 10.1086/510499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delous M., Baala L., Salomon R., Laclef C., Vierkotten J., Tory K., Golzio C., Lacoste T., Besse L., Ozilou C. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat. Genet. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- 5.Valente E.M., Logan C.V., Mougou-Zerelli S., Lee J.H., Silhavy J.L., Brancati F., Iannicelli M., Travaglini L., Romani S., Illi B. Mutations in TMEM216 perturb ciliogenesis and cause Joubert, Meckel and related syndromes. Nat. Genet. 2010;42:619–625. doi: 10.1038/ng.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas S., Legendre M., Saunier S., Bessières B., Alby C., Bonnière M., Toutain A., Loeuillet L., Szymanska K., Jossic F. TCTN3 mutations cause Mohr-Majewski syndrome. Am. J. Hum. Genet. 2012;91:372–378. doi: 10.1016/j.ajhg.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Failler M., Gee H.Y., Krug P., Joo K., Halbritter J., Belkacem L., Filhol E., Porath J.D., Braun D.A., Schueler M. Mutations of CEP83 cause infantile nephronophthisis and intellectual disability. Am. J. Hum. Genet. 2014;94:905–914. doi: 10.1016/j.ajhg.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davey M.G., Paton I.R., Yin Y., Schmidt M., Bangs F.K., Morrice D.R., Smith T.G., Buxton P., Stamataki D., Tanaka M. The chicken talpid3 gene encodes a novel protein essential for Hedgehog signaling. Genes Dev. 2006;20:1365–1377. doi: 10.1101/gad.369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin Y., Bangs F., Paton I.R., Prescott A., James J., Davey M.G., Whitley P., Genikhovich G., Technau U., Burt D.W., Tickle C. The Talpid3 gene (KIAA0586) encodes a centrosomal protein that is essential for primary cilia formation. Development. 2009;136:655–664. doi: 10.1242/dev.028464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bangs F., Antonio N., Thongnuek P., Welten M., Davey M.G., Briscoe J., Tickle C. Generation of mice with functional inactivation of talpid3, a gene first identified in chicken. Development. 2011;138:3261–3272. doi: 10.1242/dev.063602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben J., Elworthy S., Ng A.S.M., van Eeden F., Ingham P.W. Targeted mutation of the talpid3 gene in zebrafish reveals its conserved requirement for ciliogenesis and Hedgehog signalling across the vertebrates. Development. 2011;138:4969–4978. doi: 10.1242/dev.070862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genin E., Tullio-Pelet A., Begeot F., Lyonnet S., Abel L. Estimating the age of rare disease mutations: the example of Triple-A syndrome. J. Med. Genet. 2004;41:445–449. doi: 10.1136/jmg.2003.017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picard C., Fieschi C., Altare F., Al-Jumaah S., Al-Hajjar S., Feinberg J., Dupuis S., Soudais C., Al-Mohsen I.Z., Génin E. Inherited interleukin-12 deficiency: IL12B genotype and clinical phenotype of 13 patients from six kindreds. Am. J. Hum. Genet. 2002;70:336–348. doi: 10.1086/338625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi T., Kim S., Lin Y.-C., Inoue T., Dynlacht B.D. The CP110-interacting proteins Talpid3 and Cep290 play overlapping and distinct roles in cilia assembly. J. Cell Biol. 2014;204:215–229. doi: 10.1083/jcb.201304153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsang W.Y., Dynlacht B.D. CP110 and its network of partners coordinately regulate cilia assembly. Cilia. 2013;2:9. doi: 10.1186/2046-2530-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanos B.E., Yang H.-J., Soni R., Wang W.-J., Macaluso F.P., Asara J.M., Tsou M.-F.B. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 2013;27:163–168. doi: 10.1101/gad.207043.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davey M.G., McTeir L., Barrie A.M., Freem L.J., Stephen L.A. Loss of cilia causes embryonic lung hypoplasia, liver fibrosis, and cholestasis in the talpid3 ciliopathy mutant. Organogenesis. 2014;10:177–185. doi: 10.4161/org.28819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El Hokayem J., Huber C., Couvé A., Aziza J., Baujat G., Bouvier R., Cavalcanti D.P., Collins F.A., Cordier M.-P., Delezoide A.-L. NEK1 and DYNC2H1 are both involved in short rib polydactyly Majewski type but not in Beemer Langer cases. J. Med. Genet. 2012;49:227–233. doi: 10.1136/jmedgenet-2011-100717. [DOI] [PubMed] [Google Scholar]
- 19.Schmidts M. Clinical genetics and pathobiology of ciliary chondrodysplasias. J. Pediatr. Genet. 2014;3:46–94. doi: 10.3233/PGE-14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Case reports, Figures S1–S3, and Tables S1 and S2
Document S2. Article plus Supplemental Data